©The Author(s) 2025.
World J Hepatol. Apr 27, 2025; 17(4): 102027
Published online Apr 27, 2025. doi: 10.4254/wjh.v17.i4.102027
Published online Apr 27, 2025. doi: 10.4254/wjh.v17.i4.102027
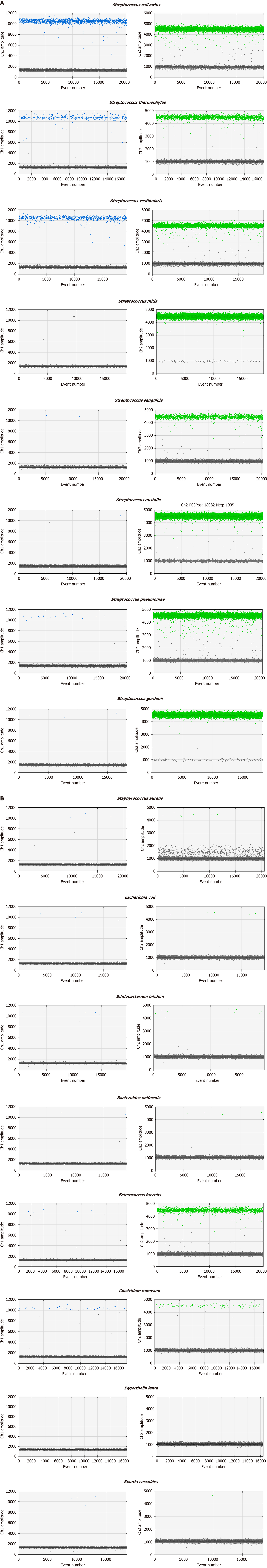
Figure 1 Specificity of primers and probes for Streptococcus salivarius.
Positive droplets of Streptococcus salivarius with channel amplitude signals (Blue signal: tnpA; Green signal: 16S rRNA). A: Digital PCR (dPCR) assay targeting both the tnpA and 16S rRNA gene to 8 kinds of Streptococcus spp.; B: dPCR assay targeting both the tnpA and 16S rRNA gene to 8 kinds of common microorganisms of the genitourinary tract.
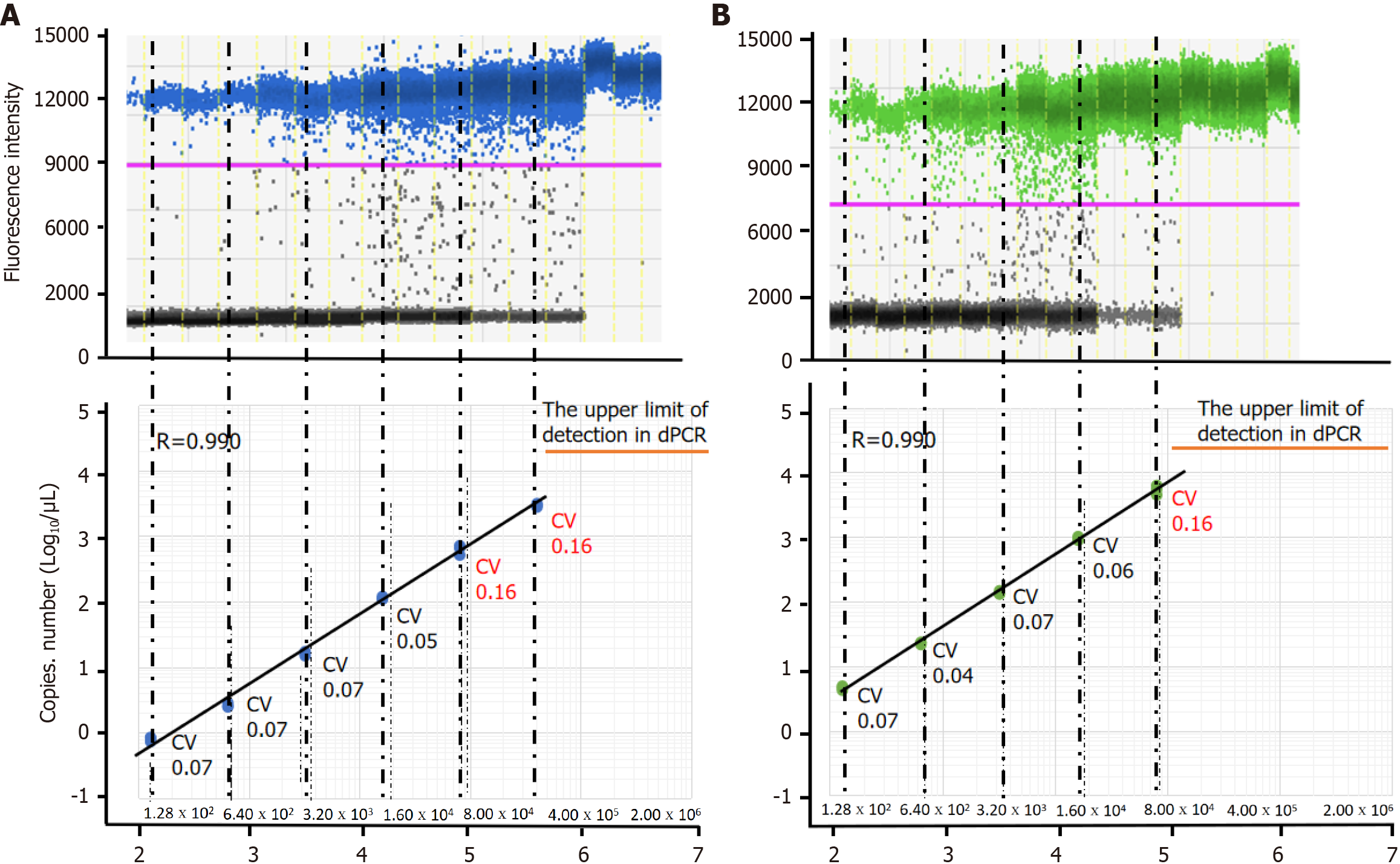
Figure 2 Comparison of bacterial load measured using digital PCR and bacterial titremeasured using plaque assay.
A: Profiles of digital PCR (dPCR) assays targeting the tnpA on bacterial suspensions of Streptococcus salivarius (S. salivarius) strain; B: Profiles of dPCR assays targeting 16S rRNA in S. salivarius bacterial suspensions. Fluorescence amplitude plot of dPCR assays on bacterial suspensions at 5-fold diluted concentrations from 2.00 × 106 to 1.28 × 102 colony forming unit/mL. The blue line represents a threshold to distinguish between positive and negative droplets based on the fluorescence signal. Droplets with a fluorescence signal above this threshold are classified as positive, indicating the presence of amplification. dPCR: Digital PCR; CV: Coefficients of variability; CFU: Colony forming unit.
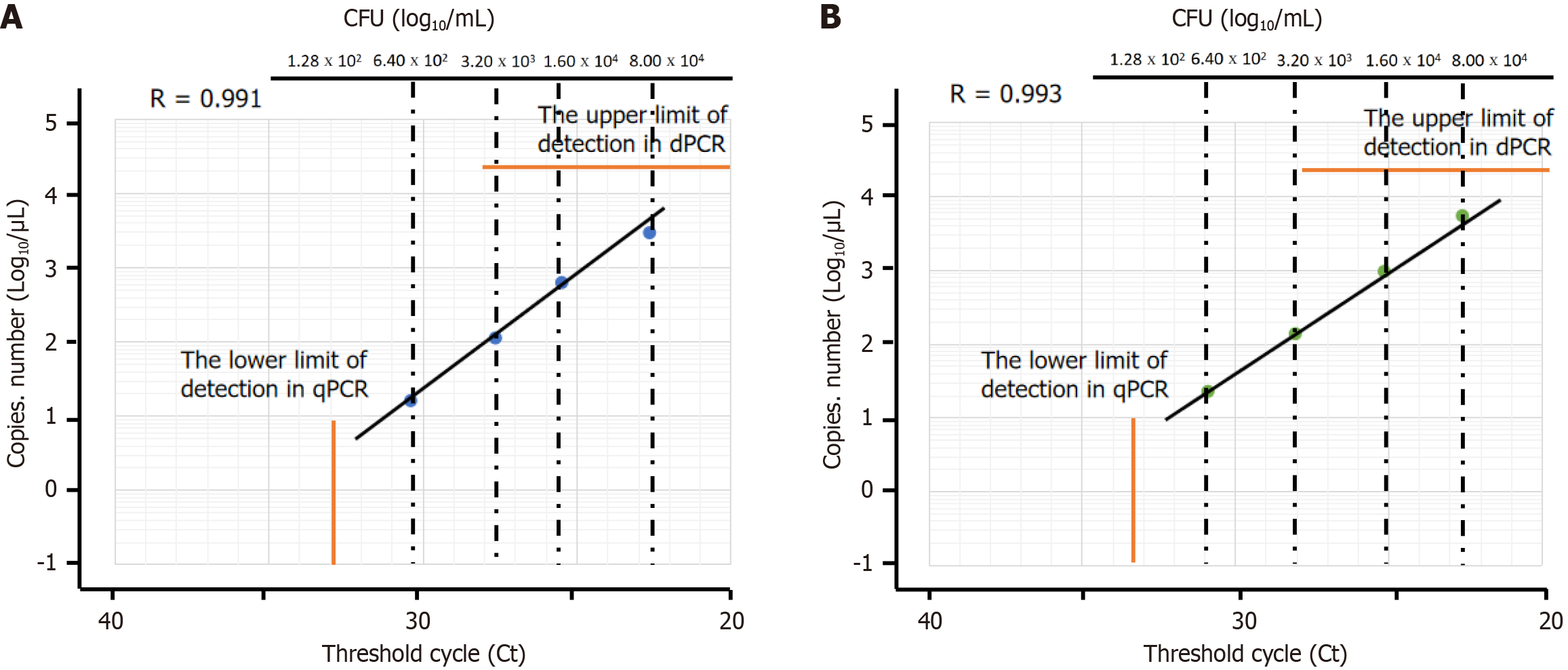
Figure 3 Comparison of bacterial load measured using digital PCR and quantitative PCR.
A: Quantitative PCR (qPCR) can reliably detect bacterial concentrations down to 6.40 × 102 colony forming unit (CFU)/mL using a 1 μL sample volume; B: Digital PCR has a higher sensitivity than qPCR, as it can detect lower concentrations of bacteria (6.40 × 102 CFU/mL) with greater accuracy and precision. dPCR: Digital PCR; qPCR: Quantitative PCR; CFU: Colony forming unit.
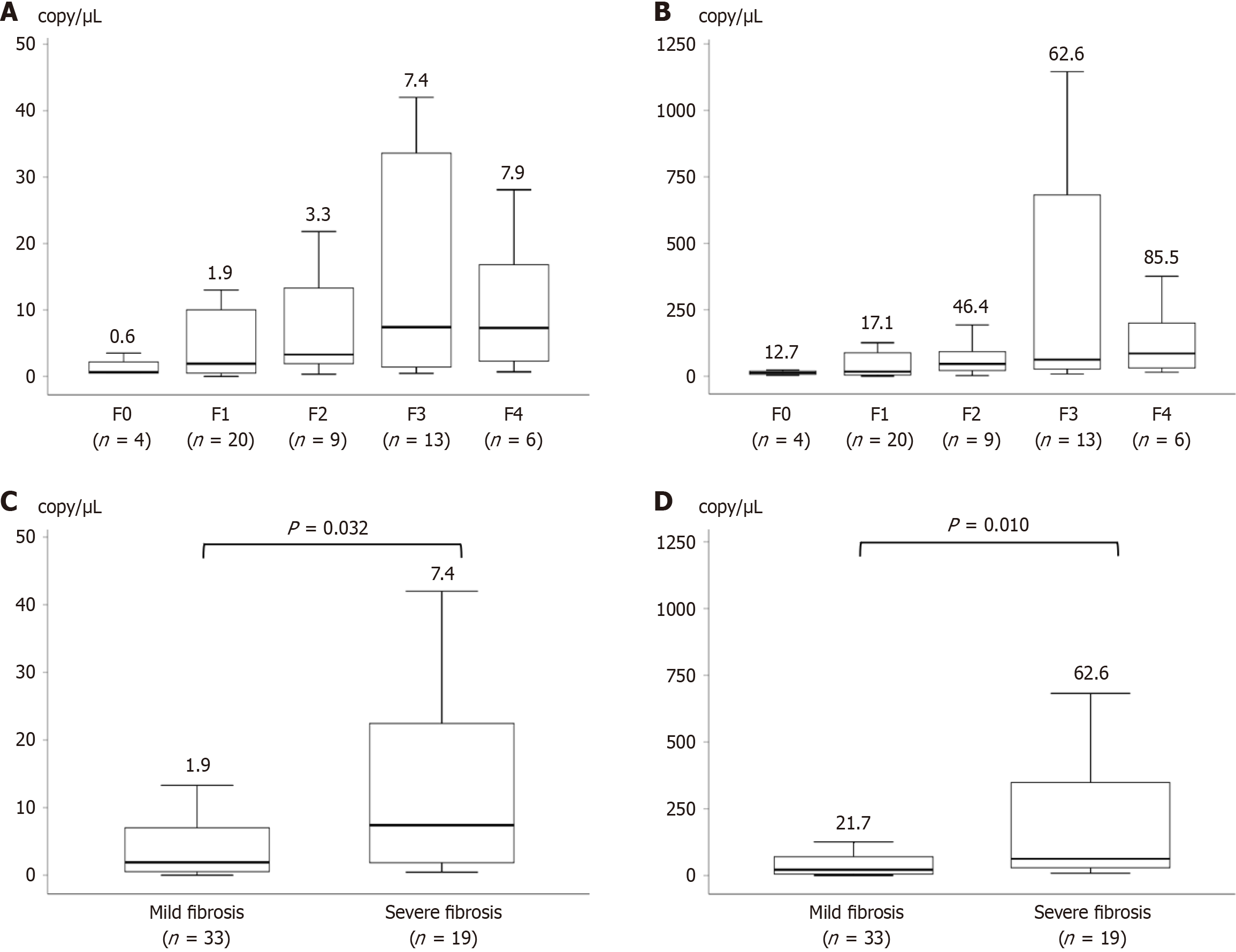
Figure 4 Correlation between the bacterial load.
A: Correlation between fibrosis stage and the bacterial load measured using digital PCR (dPCR) with tnpA; B: Correlation between fibrosis stage and the bacterial load measured using dPCR with 16S rRNA; C: Comparisons between mild and severe fibrosis in the bacterial load measured using dPCR with tnpA; D: Comparisons between mild and severe fibrosis in the bacterial load measured using dPCR with 16S rRNA.
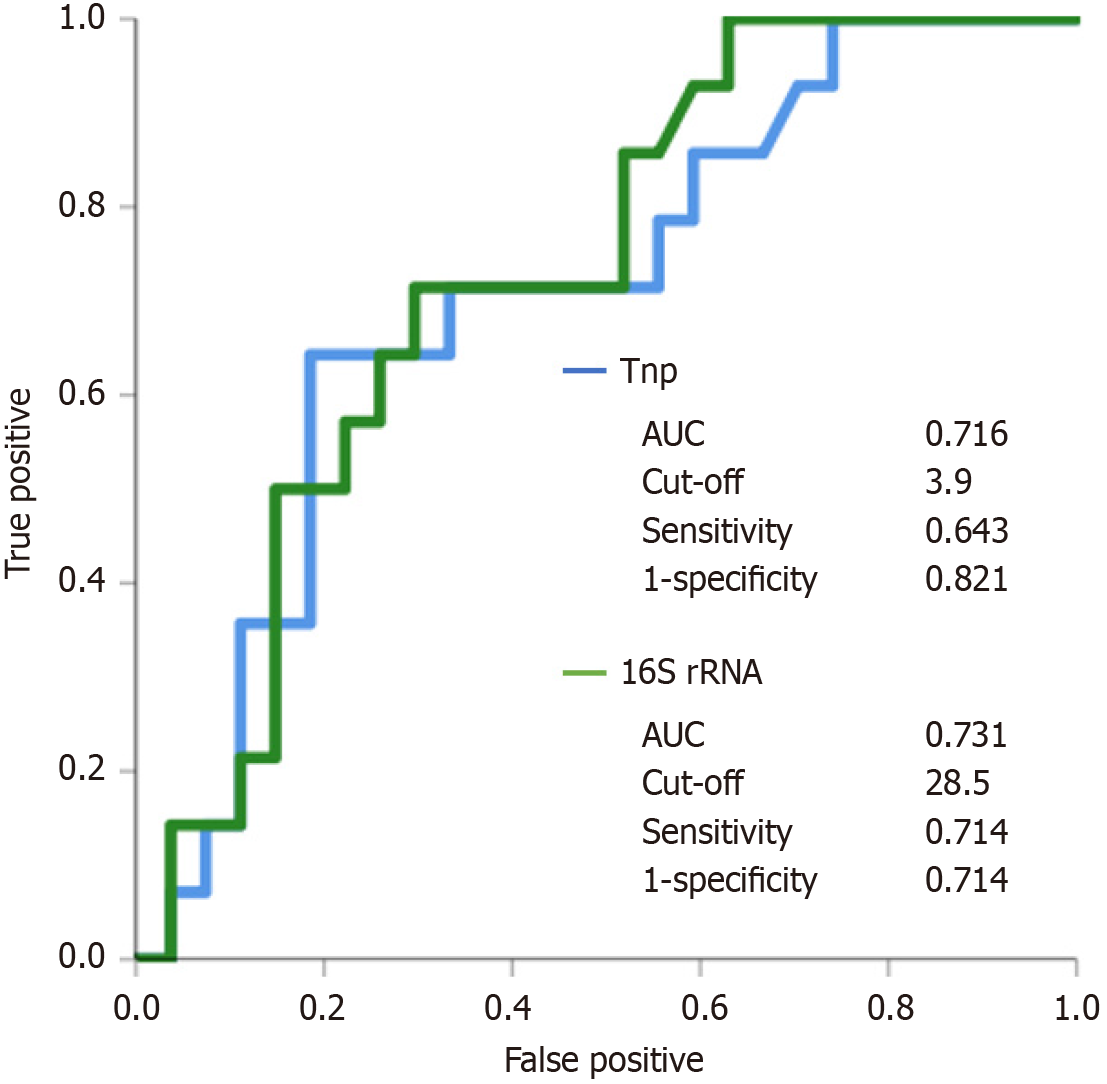
Figure 5 Severe fibrosis receiver operating characteristic curves for patients with chronic liver disease.
AUC: Area under the curve.
- Citation: Iwasaki S, Také A, Uojima H, Horio K, Sakaguchi Y, Gotoh K, Satoh T, Hidaka H, Tanaka Y, Hayashi S, Kusano C. Quantification of Streptococcus salivarius using the digital polymerase chain reaction as a liver fibrosis marker. World J Hepatol 2025; 17(4): 102027
- URL: https://www.wjgnet.com/1948-5182/full/v17/i4/102027.htm
- DOI: https://dx.doi.org/10.4254/wjh.v17.i4.102027