©The Author(s) 2025.
World J Hepatol. Nov 27, 2025; 17(11): 112364
Published online Nov 27, 2025. doi: 10.4254/wjh.v17.i11.112364
Published online Nov 27, 2025. doi: 10.4254/wjh.v17.i11.112364
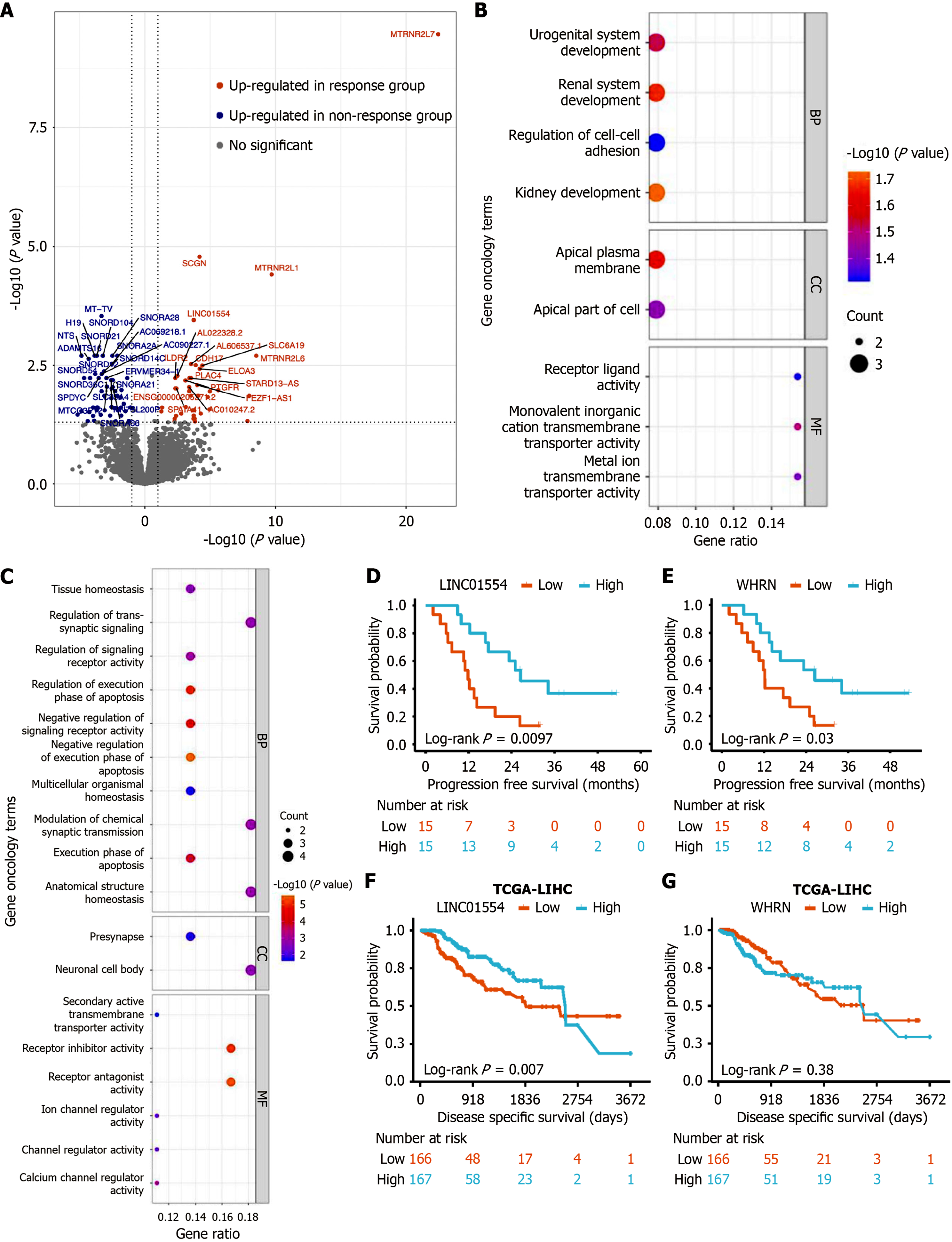
Figure 1 Differentially expressed genes.
A: Volcano plot showing differentially expressed genes between the response and non-response groups; B: Gene Ontology annotation analysis of highly expressed genes in the non-response group; C: Gene Ontology annotation analysis of highly expressed genes in the response group; D and E: Kaplan-Meier curves for long intergenic non-protein coding RNA 01554 (D) and whirlin (E), respectively, comparing high and low expression in the study population; F and G: Disease-specific survival for long intergenic non-protein coding RNA 01554 (F) and whirlin (G), based on high and low expression, as derived from The Cancer Genome Atlas database. LINC01554: Long intergenic non-protein coding RNA 01554; WHRN: Whirlin; TCGA: The Cancer Genome Atlas; LIHC: Liver hepatocellular carcinoma.
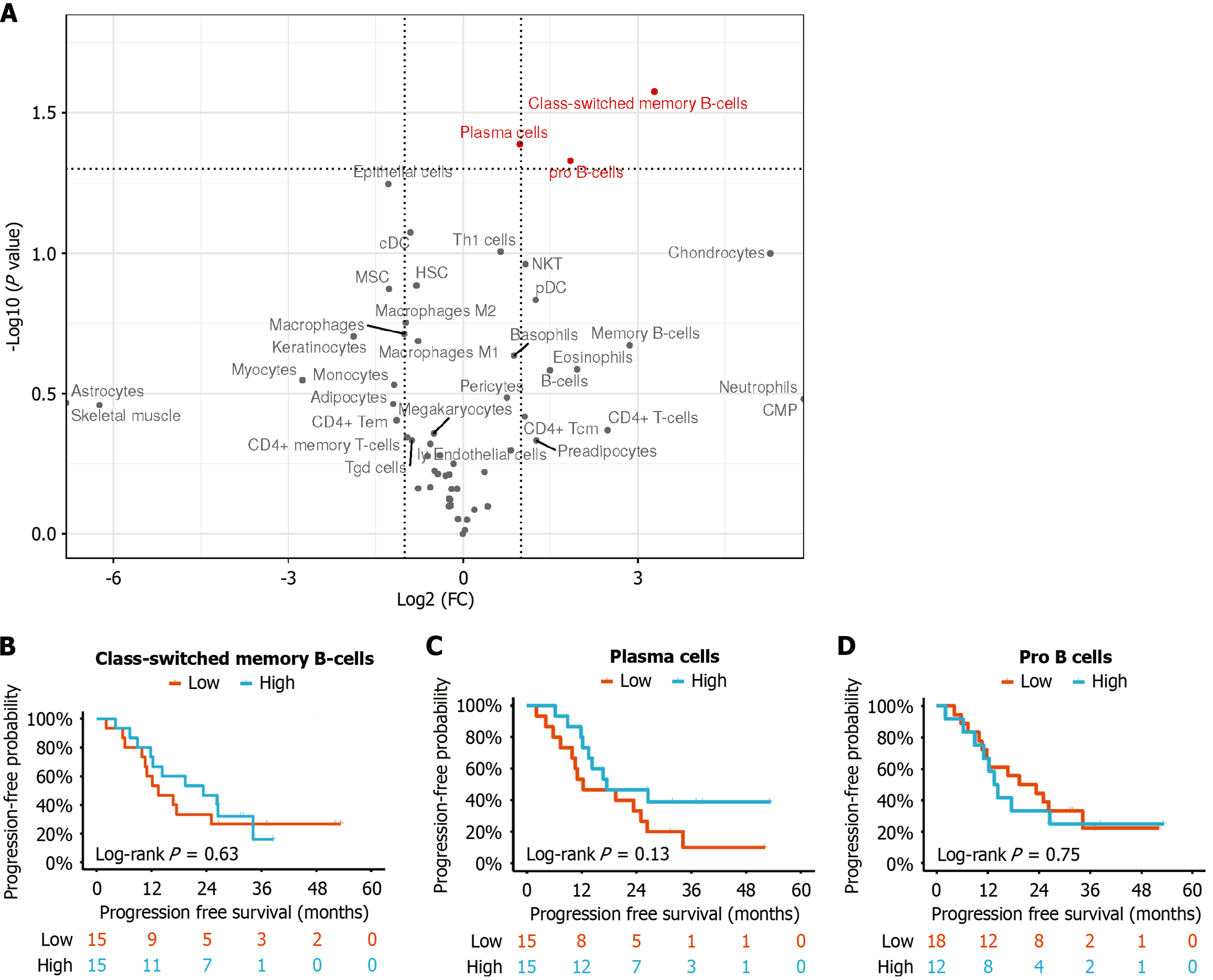
Figure 2 Immune cell infiltration scores and survival analysis.
A: Volcano plot showing the differences in immune cell infiltration scores between the response and non-response groups; B-D: Kaplan-Meier curves for class-switched memory B cells (B), plasma cells (C) and pro-B cells (D), respectively, comparing high and low expression levels in the study population. FC: Fold change.
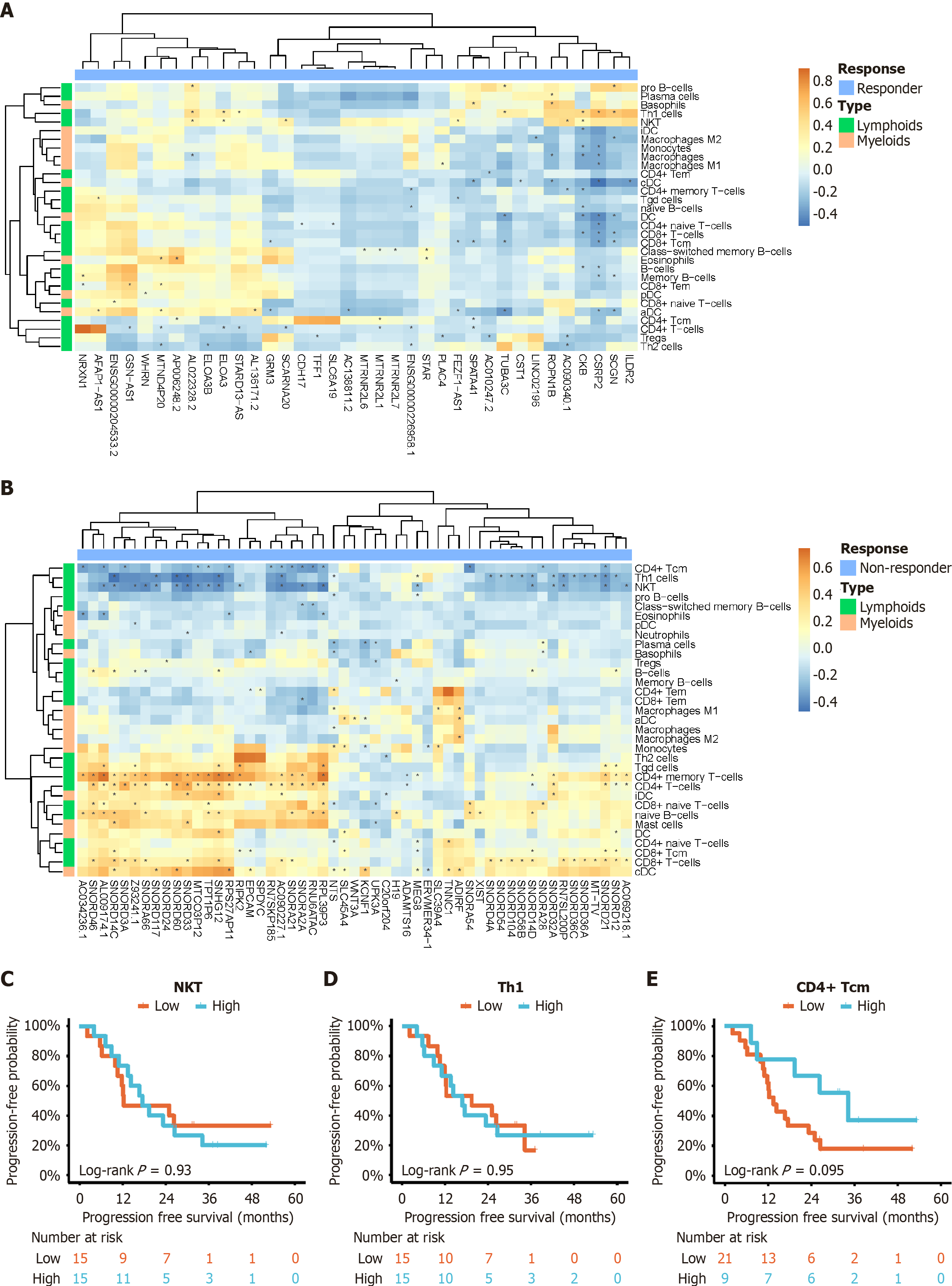
Figure 3 Correlation analysis.
A: Correlation between upregulated genes in the response group and immune cell infiltration scores; B: Correlation between upregulated genes in the non-response group and immune cell infiltration scores; C-E: Kaplan-Meier curves for natural killer T cells (C), T helper 1 cells (D), and CD4+ central memory T cells (E), respectively, comparing high and low expression levels in the study population. NKT: Natural killer T cells; Th1: T helper 1 cells; CD4+ Tcm: CD4+ central memory T cells.
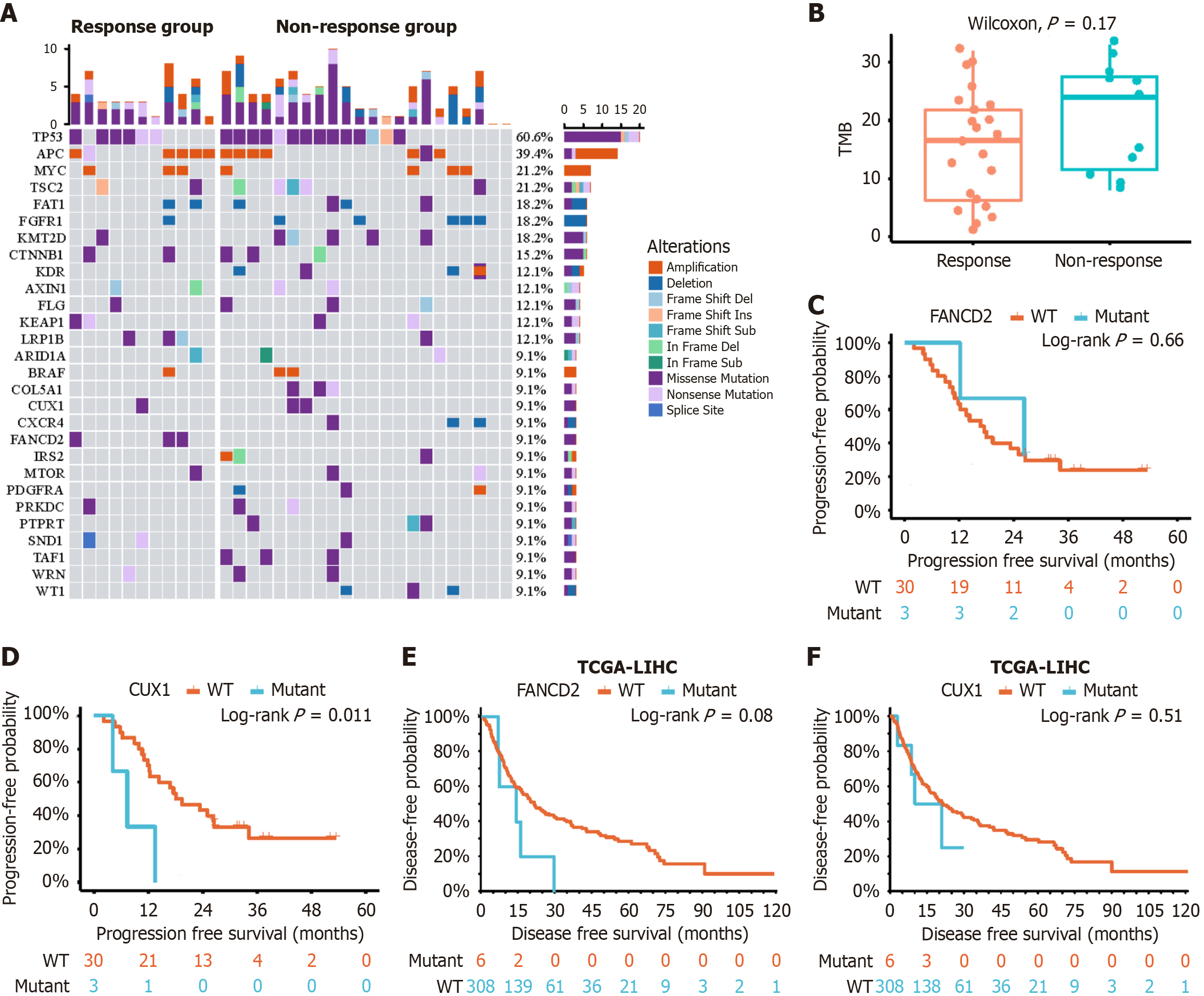
Figure 4 Mutational landscape and survival analysis.
A: Oncoplot illustrating mutations detected by whole exome sequencing; B: Boxplots depicting comparisons of tumor mutational burden between the response group and the non-response group; C and D: Kaplan-Meier curves for survival analysis based on mutations in Fanconi anemia complementation group D2 (C) and cut-like homeobox 1 (D) within the study population; E and F: Kaplan-Meier curves for survival analysis for mutations in Fanconi anemia complementation group D2 (E) and cut-like homeobox 1 (F), utilizing data derived from The Cancer Genome Atlas. FANCD2: Fanconi anemia complementation group D2; WT: Wild type; CUX1: Cut-like homeobox 1; TCGA: The Cancer Genome Atlas; LIHC: Liver hepatocellular carcinoma.
- Citation: Wang LJ, Cui Y, Huang LF, Zhang JQ, Zhao TT, Wang HW, Liu M, Jin KM, Wang K, Xing BC. Molecular biomarkers of sintilimab plus lenvatinib in hepatitis-B-virus-associated hepatocellular carcinoma. World J Hepatol 2025; 17(11): 112364
- URL: https://www.wjgnet.com/1948-5182/full/v17/i11/112364.htm
- DOI: https://dx.doi.org/10.4254/wjh.v17.i11.112364