©The Author(s) 2018.
World J Hepatol. Jan 27, 2018; 10(1): 155-165
Published online Jan 27, 2018. doi: 10.4254/wjh.v10.i1.155
Published online Jan 27, 2018. doi: 10.4254/wjh.v10.i1.155
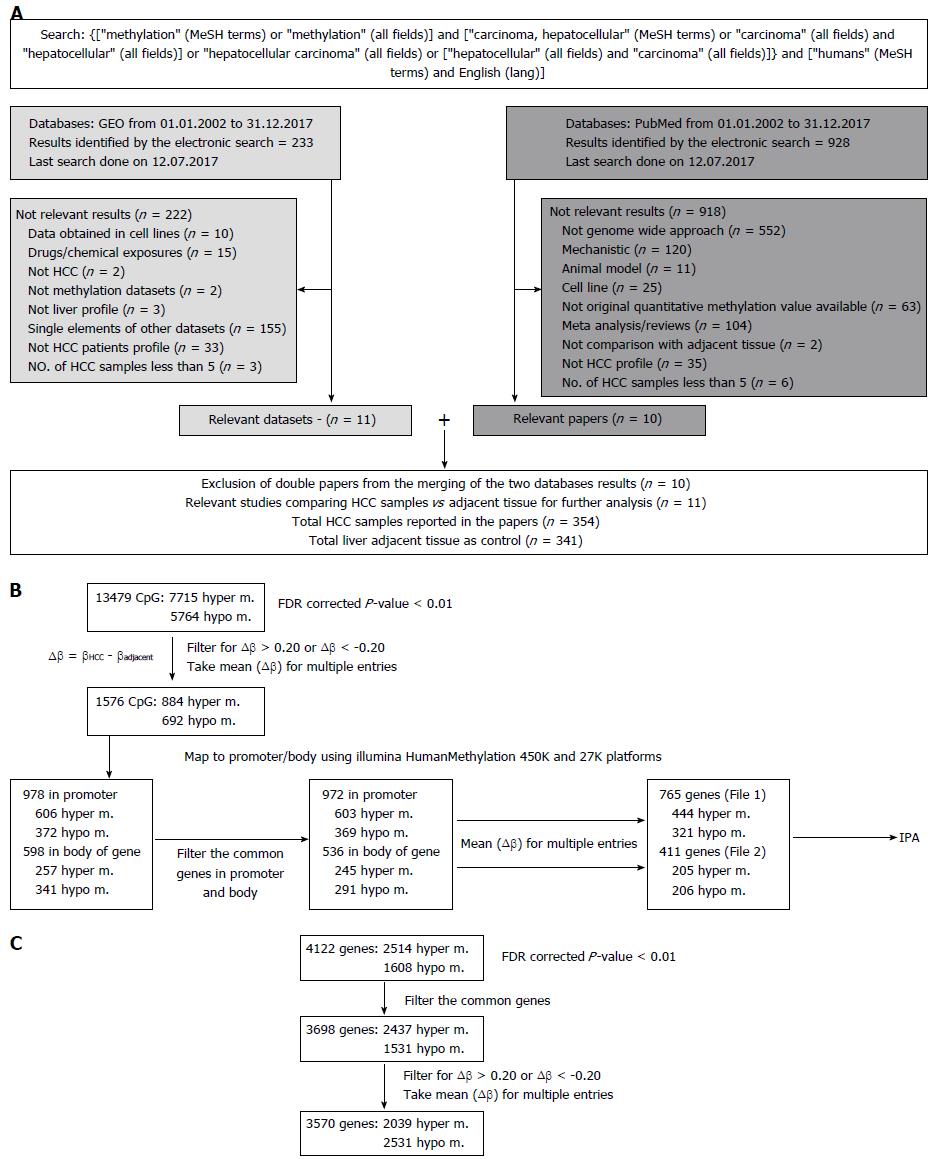
Figure 1 Flow chart.
A: Workflow of Data collection, analysis and database compiling; B: Bioinformatics flow for selecting differentially methylated CpG sites from HCC meta-analysis; C: Bioinformatics flow for selecting differentially methylated genes from HCC meta-analysis. HCC: Hepatocellular carcinoma.
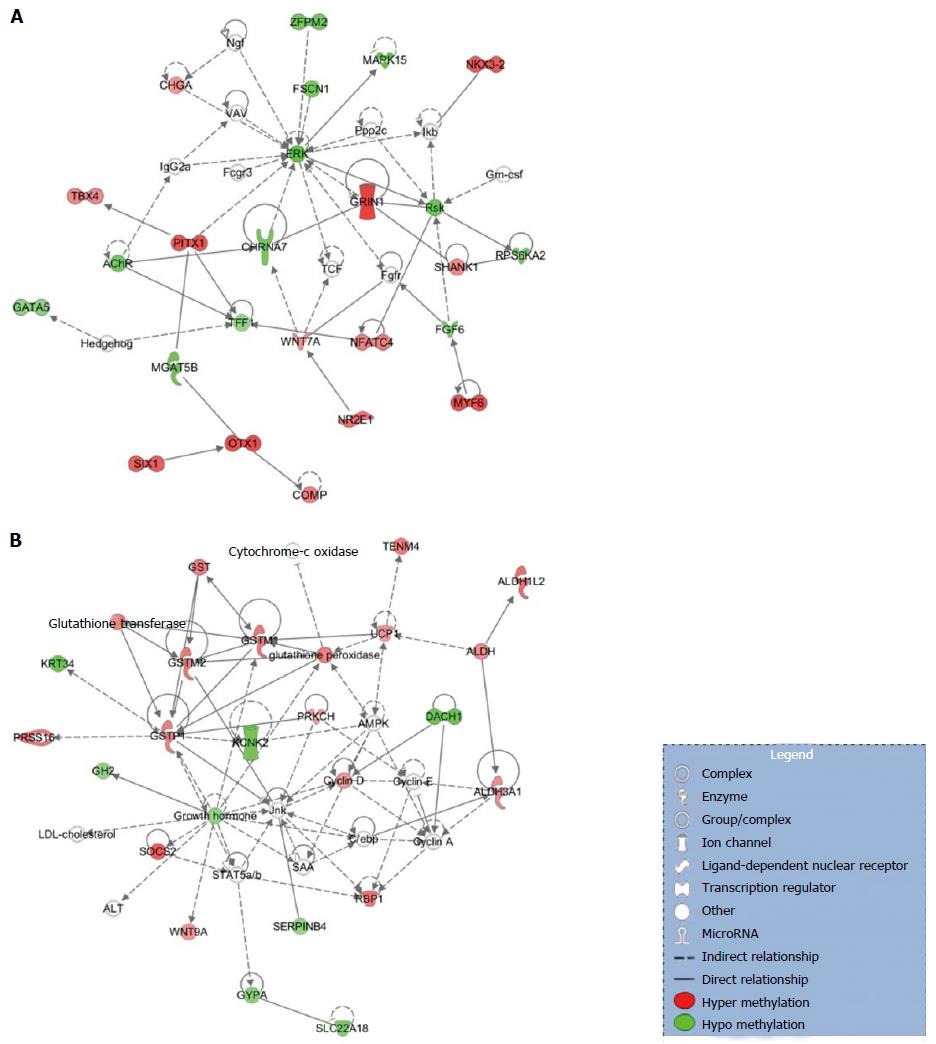
Figure 2 Networks associated with differentially methylated CpG sites in the promoter regions of genes.
A: Organismal development, organismal injury and abnormalities, cellular development; B: Lipid metabolism, small molecule biochemistry, cell death and survival; C: Cell-to-cell signaling and interaction, drug metabolism, small molecule biochemistry.
Figure 3 Networks associated with differentially methylated CpG sites in body of the gene.
A: Cellular assembly and organization, cellular function and maintenance; B: Drug metabolism, glutathione depletion in liver, small molecule biochemistry.
Figure 4 Venn diagram intersecting three lists: (1) reported differentially methylated genes in HCC from studies not providing information on the corresponding CpG sites; (2) identified differentially methylated genes in HCC corresponding to CpG sites in promoter; and (3) identified differentially methylated genes in HCC corresponding to CpG sites in body of the gene.
- Citation: Bhat V, Srinathan S, Pasini E, Angeli M, Chen E, Baciu C, Bhat M. Epigenetic basis of hepatocellular carcinoma: A network-based integrative meta-analysis. World J Hepatol 2018; 10(1): 155-165
- URL: https://www.wjgnet.com/1948-5182/full/v10/i1/155.htm
- DOI: https://dx.doi.org/10.4254/wjh.v10.i1.155