Published online Jun 15, 2003. doi: 10.3748/wjg.v9.i6.1333
Revised: December 24, 2002
Accepted: January 2, 2003
Published online: June 15, 2003
AIM: Human hepatitis B virus enhancer II B1 binding factor (hB1F) was cloned and characterized as a novel member of the Ftz-F1 (NR5A) nuclear receptor subfamily. Although progresses have recently been made, its biological function remains largely unidentified. The aim of this study was to establish an hB1F transgenic mouse model to promote the functional study of hB1F.
METHODS: Transgene fragments were microinjected into fertilized eggs of mice. The manipulated embryos were transferred into the oviducts of pseudopregnant female mice. The offsprings were identified by PCR and Southern blot analysis. Transgene expression was analyzed with RT-PCR and Western blot analysis. Transgenic founder mice were used to establish transgenic mouse lineages. The F1 and F2 mice were identified by PCR analysis.
RESULTS: Seven mice were identified as carrying copies of transgene. RT-PCR and Western blotting results showed that the transgene was expressed in heart, liver, lung, kidney and stomach in one of the transgenic mouse lineages. Genetic analysis of the transgenic mice demonstrated that the transgene was integrated into the chromosome at a single site, and was transmitted stably.
CONCLUSION: In this study we established an hB1F transgenic mouse model, which will facilitate the investigation of the biological function of hB1F in vivo.
- Citation: Wang SL, Yang H, Xie YH, Wang Y, Li JZ, Wang L, Wang ZG, Fu JL. Establishment of transgenic mice carrying the gene of human nuclear receptor NR5A2 (hB1F). World J Gastroenterol 2003; 9(6): 1333-1336
- URL: https://www.wjgnet.com/1007-9327/full/v9/i6/1333.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i6.1333
Nuclear receptor (NR) is a superfamily of eukaryotic transcription factors that are crucial for gene regulation and development. Members of this superfamily include receptors for steroid and non-steroid hormones as well as a large number of orphan receptors whose regulatory ligands have not been identified[1,2]. Fushi tarazu factor 1 (Ftz-F1) is one of the six subfamilies of the NR superfamily[3], which has been nominated as NR5[4]. Members of the Ftz-F1 subfamily all possess a particular Ftz-F1 box located at the C-terminal of the DNA-binding domain (DBD) and bind to their response elements as monomer.
Human hepatitis B virus (HBV) enhancer II B1 binding factor (hB1F) has been cloned and characterized as a novel member of the Ftz-F1 subfamily[5]. hB1F (also known as NR5A2, CPF, and hFTF) binds specifically to the B1 element in the enhancer II of HBV and activates its function, thus regulating the viral gene expression and replication. RT-PCR and 3’-RACE have previously revealed that utilization of two polyadenylation signals results in the 3.8 and 5.2 kb transcripts[6,7]. hB1F is the human homologue of the mouse transcription factor mLRH-1. It has been reported that NR5A2 may play an important role in regulating the liver-specific expression of several genes[8]. Recent findings demonstrate that NR5A2 is a critical transcription factor in the bile acid biosynthesis pathway[9,10] and may also regulate the expression of bile acid and cholesterol transporters in the liver and intestine[11,12].
To facilitate the functional studies of hB1F, we reported here the establishment of transgenic mice carrying hB1F gene. 7 transgenic mice were identified by PCR and Southern blotting and used as founders to establish transgenic mouse lineages. The results of the F1 identification with PCR showed that the transgenes could be transmitted stably. RT-PCR and Western blotting analysis demonstrated that the transgenes expressed in multiple tissues of transgenic mice.
pcDNA3-hB1F contains the full length hB1F cDNA under the control of CMV promoter. A Flag tag was placed upstream to the hB1F coding sequence to facilitate the characterization of the expressed protein.
C57 and CBA mice were maintained by Shanghai Nanfang Research Center for Model Organisms. Transgenic mice were raised, and bred in the Laboratory Animal Centre of Second Military Medical University.
The 6.9 kb linearized pcDNA3-hB1F was purified from agarose gel with QIAGEN gel extraction kit (Qiagen, CA, USA), adjusted to a final concentration of 1 μg/mL in TE buffer and used as the DNA solution in microinjection. The F1 female hybrids of C57 and CBA mice were hormonally superovulated and mated with F1 male hybrids. Next morning the fertilized one-cell eggs were collected from the oviduct. The eggs were microinjected with the DNA solution under a microscope. The injected fertilized eggs were transplanted into the oviduct of pseudo-pregnant F1 hybrids of C57 and CBA mice.
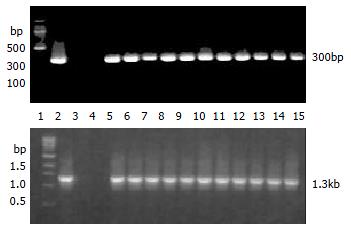
Founder (G0) mice were identified by PCR and Southern blotting analysis. For PCR, DNA was extracted from tails with Genomic DNA MINIPREPES kit (Sangon Inc. China). PCR amplification was performed with hB1F primers (P1: 5'-CCGACAAGTGGTACATGGAA-3' P2: 5'-CTGCTGCGGGTAGTTACACA-3') and pcDNA3 primers (P3: 5'-ATGCGGTGGGCT CTATG-3' P4: 5'-CGGCTTCCATCCGAGTA-3') which would produce 300 bp and 1353 bp fragments from mice carrying the transgene, respectively. For Southern blotting, genomic DNA was digested overnight with HindIII and subjected to electrophoresis on a 1.0% agarose gel. DNA was transferred onto nylon membrane (MILLIPORE Co., Ltd, London, UK). Hybridization was performed under a stringent condition with a randomly-primed (α-32P)-labeled hB1F probe.
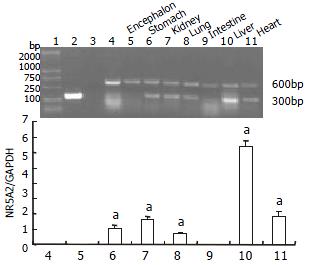
One of the transgenic mouse lineages was used to study the expression of transgene. Total RNA was isolated from tissues with the TRIzol reagent (Invitrogen, CA, USA), according to manufacturer's instructions. First strand cDNA was synthesized by reverse transcription (Promega, USA). Semiquantitative RT-PCR reactions were performed using primer pairs 5'-CCGACAAGTGGT ACATGGAA-3' and 5'-CTGCTGCGGGTAGTTACACA-3' for hB1F cDNA, and 5'-AACTTTGGCATTGTGGAAGG-3' and 5'-TGTGAGGGAGATGCTCAGT G-3' for mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA, which resulted in the generation of 300 bp and 600 bp products, respectively. PCR was performed for 30 cycles at 94 °C for 1 min, at 57 °C for 1 min, and at 72 °C for 1 min. PCR products were electrophoresed on 1.5% agarose gels. Signals were quantified by density analysis of the digital images using BioStar image software, version 2.
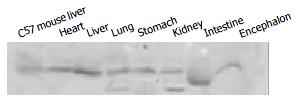
For Western blotting, protein samples from tissues were prepared according the protocols by the manufacturer (Santa Cruz Biotechnology, Inc. WA, USA). 50 μg protein of each sample was electrophoresed on 10% SDS-polyacrylamide gel and transferred to PVDF membrane. Membranes were blocked with 5% (w/v) non-fat milk in Tween-TBS (TBST) overnight at 4°Cand incubated with anti-Flag antibody (Sigma, USA) at a dilution of 1:500 in TBST for 2 h at room temperature. Membranes were washed three times with TBST and incubated with a secondary antibody (horseradish peroxidase-conjugated anti-mouse IgG) at a dilution of 1:2000 at room temperature for 1 h. Immunodetection was carried out with an enhanced chemiluminescence kit (Amersham Pharmacia Biotech, USA).
To study the transmission of transgene in mice, transgenic founder mice were mated to normal C57 mice to produce the first generation (F1) transgenic mice which were identified by PCR analysis using hB1F primers P1 and P2, and primers P5 (5'-GTTGGAGGTCGCTGAGTA-3') and P6 (5'- AGTAGGAAAGTCCCATAAGGTC-3'), yielding 300 bp and 500 bp products, respectively, from mice carrying the transgene. The F1 mice of the same founder carrying the transgene were mated between brother and sister mice to produce the second generation (F2). The F2 transgenic mice were identified by the same methods for the F1 transgenic mice.
The transgene fragment containing the full length hB1F cDNA was microinjected into the male pronucleus of 653 fertilized oocytes of F1 hybrids between C57 and CBA mice. The injected eggs were implanted into the oviducts of 24 pseudo-pregnant foster mothers, of which 13 mice became pregnant and gave birth to 97 offsprings. Eleven offspring mice were identified to carry the hB1F cDNA as demonstrated by PCR analysis (Figure 1), seven out of which were further confirmed by Southern blotting analysis (Figure 2). The ratio of transgene integration was 11.3% and 7.2%, respectively, by PCR and Southern blotting analysis.
As the transgene was driven by the CMV promoter, to study if it can express in multiple tissues of transgenic mice, we analysed the tissue expression profile of the hB1F transgene by RT-PCR and Western blot. The results (Figure 3, Figure 4) showed that the hB1F transgene was expressed in the heart, liver, lung, stomach and kidney, but not in intestine and encephalon.
To establish the transgenic mouse lineages, founder mice were mated to C57 mice to produce F1 mice. Among 128 mice of the first generation, 60 were identified as carrying hB1F cDNA transgene by PCR analysis (Figure 5). The ratio of transgene transmission was 46.9%. The F1 mice from the same founder were mated each other to produce the F2 mice. 45 out of 64 F2 mice were hB1F transgenic mice, with a ratio of transgene transmission of 70.3%. These results showed that the inheritance of hB1F transgene was in accordance with Mendel's laws, and the transgene was integrated into the chromosome in a single site and could be transmitted stably.
hB1F (NR5A2) is a novel member of the FTZ-F1 nuclear receptor subfamily. It was originally cloned based on its interaction with hepatitis B virus (HBV) enhancer II B1 element[5]. It has been shown to be a critical regulator of HBV gene expression and replication. Recent studies have revealed that hB1F is mainly involved in the regulation of cholesterol related gene expresssion in the hepatic-intestinal system. hB1F and its mouse homolog mLRH-1 are essential for the transcription of the gene encoding cholesterol 7a hydroxylase (CYP7A1), the first and rate limiting enzyme in bile acid biosynthesis[6,9,10]. Together with two other nuclear receptors, FXR and SHP, and hB1F/mLRH-1 can mediate the feedback effect of bile acids on the transcription of CYP7A1. Moreover, hB1F/mLRH-1 can also regulate the expression of sterol 12α-hydrolyase (CYP8B1)[13], multidrug resistance protein-3 (MRP-3)[11], cholesterol ester transport protein (CETP)[12] and aromatases[14]. There is evidence that mLRH-1 may also regulate the expression of several critical transcritional factors, such as HNF1, HNF3β and HNF4[15]. It is possible that hB1F/mLRH-1 may exert broad influences on many genes by regulating these important factors. Since the regulation of cholesterol and bile acid homeostats is complicated, prone to be influenced by various genetic and environmental factors, it is vital to establish a transgenic or knock-out animal model for the studies of the biological role that hB1F plays in a living organism. The strategy to knock out mLRH-1 in mice has not been successful, due to early embryo lethality (personal communication), which suggests that hB1F/mLRH-1 has yet unidentified crucial functions during embryogenesis. The attempt to generate hB1F transgenic mice has not been reported either. To our knowledge, we have generated the first transgenic mice carrying and expressing the hB1F transgene.
In vivo hB1F/mLRH-1 is mainly expressed in the liver and pancreas. However, weak expression of hB1F has also been detected in other tissues such as heart, lung, skeletal muscle and intestine[5,6]. Recent studies have also demonstrated that hB1F is expressed abundantly in human ovary, testis[16] and preadipocytes[14], as well as in human adrenal and placenta in small amount[16], thus hB1F has a broader expression tissue profile than previously thought. In this study, the hB1F transgene was expressed in multiple tissues, most of which are capable of expressing endogenous hB1F in vivo. Therefore, the hB1F transgenic mice we generated in this study is an invaluable animal model, for investigating hB1F function not only in hepatic tissues, but also in other tissues, which may ultimately reveal the yet unidentified biological functions of hB1F.
In conclusion, we reported here the successful generation of a transgenic mouse model expressing the orphan nuclear receptor hB1F (NR5A2). Future studies will focus on the physiological and pathological changes in this mouse model using powerful analytic methods e.g. microarray comparisons of gene expression profiles between normal and transgenic mice.
| 1. | Beato M, Herrlich P, Schütz G. Steroid hormone receptors: many actors in search of a plot. Cell. 1995;83:851-857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1335] [Cited by in RCA: 1291] [Article Influence: 41.6] [Reference Citation Analysis (0)] |
| 2. | Kastner P, Mark M, Chambon P. Nonsteroid nuclear receptors: what are genetic studies telling us about their role in real life. Cell. 1995;83:859-869. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 773] [Cited by in RCA: 758] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 3. | Laudet V. Evolution of the nuclear receptor superfamily: early diversification from an ancestral orphan receptor. J Mol Endocrinol. 1997;19:207-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 377] [Cited by in RCA: 349] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 4. | Nuclear Receptors Nomenclature Committee. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 851] [Cited by in RCA: 842] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 5. | Li M, Xie YH, Kong YY, Wu X, Zhu L, Wang Y. Cloning and characterization of a novel human hepatocyte transcription factor, hB1F, which binds and activates enhancer II of hepatitis B virus. J Biol Chem. 1998;273:29022-29031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 82] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 6. | Nitta M, Ku S, Brown C, Okamoto AY, Shan B. CPF: an orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7alpha-hydroxylase gene. Proc Natl Acad Sci U S A. 1999;96:6660-6665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 226] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 7. | Zhang CK, Lin W, Cai YN, Xu PL, Dong H, Li M, Kong YY, Fu G, Xie YH, Huang GM. Characterization of the genomic structure and tissue-specific promoter of the human nuclear receptor NR5A2 (hB1F) gene. Gene. 2001;273:239-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 29] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 8. | Galarneau L, Paré JF, Allard D, Hamel D, Levesque L, Tugwood JD, Green S, Bélanger L. The alpha1-fetoprotein locus is activated by a nuclear receptor of the Drosophila FTZ-F1 family. Mol Cell Biol. 1996;16:3853-3865. [PubMed] |
| 9. | Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6:517-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1411] [Cited by in RCA: 1549] [Article Influence: 59.6] [Reference Citation Analysis (0)] |
| 10. | Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1110] [Cited by in RCA: 1151] [Article Influence: 44.3] [Reference Citation Analysis (0)] |
| 11. | Inokuchi A, Hinoshita E, Iwamoto Y, Kohno K, Kuwano M, Uchiumi T. Enhanced expression of the human multidrug resistance protein 3 by bile salt in human enterocytes. A transcriptional control of a plausible bile acid transporter. J Biol Chem. 2001;276:46822-46829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 82] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 12. | Luo Y, Liang CP, Tall AR. The orphan nuclear receptor LRH-1 potentiates the sterol-mediated induction of the human CETP gene by liver X receptor. J Biol Chem. 2001;276:24767-24773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 109] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 13. | del Castillo-Olivares A, Gil G. Alpha 1-fetoprotein transcription factor is required for the expression of sterol 12alpha -hydroxylase, the specific enzyme for cholic acid synthesis. Potential role in the bile acid-mediated regulation of gene transcription. J Biol Chem. 2000;275:17793-17799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 105] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 14. | Clyne CD, Speed CJ, Zhou J, Simpson ER. Liver receptor homologue-1 (LRH-1) regulates expression of aromatase in preadipocytes. J Biol Chem. 2002;277:20591-20597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 180] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 15. | Pare JF, Roy S, Galarneau L, Belanger L. The mouse fetoprotein transcription factor (FTF) gene promoter is regulated by three GATA elements with tandem E box and Nkx motifs, and FTF in turn activates the Hnf3beta, Hnf4alpha, and Hnf1alpha gene promoters. J Biol Chem. 2001;276:13136-13144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 74] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 16. | Sirianni R, Seely JB, Attia G, Stocco DM, Carr BR, Pezzi V, Rainey WE. Liver receptor homologue-1 is expressed in human steroidogenic tissues and activates transcription of genes encoding steroidogenic enzymes. J Endocrinol. 2002;174:R13-R17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 111] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
Edited by Xia HHX