Published online Apr 15, 2003. doi: 10.3748/wjg.v9.i4.655
Revised: November 7, 2002
Accepted: November 14, 2002
Published online: April 15, 2003
AIM: To appraise the correlation of mutation and methylation of hMSH1 with microsatellite instability (MSI) in gastric cancers.
METHODS: Mutation of hMLH1 was detected by Two-dimensional electrophoresis (Two-D) and DNA sequencing; Methylation of hMLH1 promoter was measured with methylation-specific PCR; MSI was analyzed by PCR-based methods.
RESULTS: Sixty-eight cases of sporadic gastric carcinoma were studied for mutation and methylation of hMLH1 promoter and MSI. Three mutations were found, two of them were caused by a single bp substitution and one was caused by a 2 bp substitution, which displayed similar Two-D band pattern. Methylation of hMLH1 promoter was detected in 11 (16.2%) gastric cancer. By using five MSI markers, MSI in at least one locus was detected in 17/68 (25%) of the tumors analyzed. Three hMLH1 mutations were all detected in MSI-H (≥ 2 loci, n = 8), but no mutation was found in MSI-L (only one locus, n = 9) or MSS (tumor lacking MSI or stable, n = 51). Methylation frequency of hMLH1 in MSI-H (87.5%, 7/8) was significantly higher than that in MSI-L (11.1%, 1/9) or MSS (5.9%, 3/51) (P < 0.01-0.001), but no difference was found between MSI-L and MSS (P > 0.05).
CONCLUSION: Both mutation and methylation of hMLH1 are involved in the MSI pathway but not related to the LOH pathway in gastric carcinogenesis.
- Citation: Fang DC, Wang RQ, Yang SM, Yang JM, Liu HF, Peng GY, Xiao TL, Luo YH. Mutation and methylation of hMLH1 in gastric carcinomas with microsatellite instability. World J Gastroenterol 2003; 9(4): 655-659
- URL: https://www.wjgnet.com/1007-9327/full/v9/i4/655.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i4.655
Our previous studies indicated that genetic instability may play an important role in gastric carcinogenesis[1]. There are at least two distinct genetic instabilities in gastric tumorigenesis: one is the chromosomal instability (or suppressor pathway) and the other is microsatellite instability (or MSI pathway). In the former, perhaps including tumors with low-frequency MSI (MSI-L) as well as microsatellite stable (MSS), accumulation of loss of tumor suppressor genes such as p53, Rb, APC, MCC and DCC play an important role in their carcinogenesis; whereas in the latter, consisting of a small subset of gastric cancer with high-frequency MSI (MSI-H), defective repair of mismatched bases results in an increased mutation rate at the nucleotide level, and the consequent widespread MSI[2-4].
Mismatch repair is required for the cell to accurately copy its genome during cellular proliferation. Deficiencies of this system result in mutation rates 100-fold greater than those observed in normal cells[5]. MSI is a hallmark of mismatch repair gene (MMR)-deficient cancers. MSI in tumors from patients with hereditary non-polyposis colorectal cancer (HNPCC) is caused by germline mutations in MMR genes, principally hMSH2 and hMLH1[6-10]. In contrast, somatic mutations in MMR genes are relatively rare in sporadic MSI+ colon cancers[9,11]. Rather, the majority of negative mutation, MSI+ cases involve hypermethylation of the hMLH1 promoter and subsequent lack of expression of hMLH1[12-16]. The details of the mechanisms of this epigenetic gene silencing remain to be elucidated in gastric cancer. The aim of this study was to define the mutation and methylation of hMLH1 in gastric carcinomas with MSI.
Sixty-eight cancer and corresponding normal tissues were obtained from surgically resected gastric carcinoma in our hospital. Each specimen was frozen immediately and stored at -80 °C until analyzed. A 5 µm section was cut from each tissue and stained with hematoxylin/eosin in order to ascertain whether the cancer cells in the tissues were predominant or not. Genomic DNA was isolated by standard proteinase-K digestion and phenol-chloroform extraction protocols. Of the 68 patients with gastric cancer, 45 were men and 23 were women with an age range of 30-76 years (a mean of 56.2 years at diagnosis). None of the patients included in the present series had a family history suggestive of HNPCC or had received chemotherapy or radiation therapy.
PCR and heteroduplexing Primer pairs for long-chain and short-chain PCR and GC-clamped primers used were shown in Table 1[17]. PCR reactions were carried out in 50 µl reactions in thin-walled tubes in a Perkin-Elmer 2400 thermocycler. A total of 200-400 ng of genomic DNA, varying concentrations of each primer, and the LA PCR kit (TaKaRa, Otsu, Shiga, Japan) were used for long PCR. Final concentrations of each LA PCR primer pair were as follows: hMLH1-4F and hMLH1-4R, 0.16 µM each; hMLH5-10F and hMLH5-10R, 0.125 µM each; hMLH11-13F and hMLH11-13R, 0.094 µM each; and hMLH14-19F and hMLH14-19R, 0.125 µM each. The reactions were carried out according to the manufacturer’s instructions. In brief, the conditions were as follows: a hot start of 94 °C for 2 min, with the addition of Taq Pol in between, followed by eight cycles of 98 °C × 20 sec, 69 °C × 1 min (with decrements of 0.5 °C/cycle), and 68 °C × 12 min; six cycles of 96 °C × 20 sec, and 68 °C × 12 min; 16 cycles of 96 °C × 20 sec, and 68 °C × 12 min (with increments of 15 sec/cycle), and finally a chain extension of 72 °C for 10 min.
| A. Primer pairs for long-distance PCR | ||||
| Exons 1-4 | ||||
| MLH1-4F GCG.GCT.AAG.CTA.CAG.CTG.AAG.GAA.GAA.CGT.GAa | ||||
| MLH1-4R GGC.GAG.ACA.GGA.TTA.CTC.TGA.GAC.CTA.GGC.CC | ||||
| Product size-10.8 kb | ||||
| Exons 5-10 | ||||
| MLH5-10F GCG.CCC.CTT.GGG.ATT.AGT.ATC.TAT.CTC.TCT.ACT.GG | ||||
| MLH5-10R GCG.CTC.ATC.TCT.TTC.AAA.GAG.GAG.AGC.CTG | ||||
| Product size-10.5 kb | ||||
| Exons 11-13 | ||||
| MLH11-13F CGG.CTT.TTT.CTC.CCC.CTC.CCA.CTA.TCT.AAG.G | ||||
| MLH11-13R GGG.TTA.GTA.AAG.GAA.GAG.GAG.CTT.GCC.C | ||||
| Product size-8.7 kb | ||||
| Exons 14-19 | ||||
| MLH14-19F GGT.GCT.TTG.GTC.AAT.GAA.GTG.GGG.TTG.GTA.G | ||||
| MLH14-19R GCG.CGC.GTA.TGT.TGG.TAC.ACT.TTG.TAT.ATC.ACA.C | ||||
| Product size = 10.5 kb | ||||
| B. Primer pairs for short PCR | ||||
| Exon | Clampb | Product size | Tmc | Primer sequence |
| 1 | 5 | 258 | 64.13 | GCA.CTT.CCG.TTG.AGC.ATC |
| 40 | CCG.TTA.AGT.CGT.AGC.CCT | |||
| 2 | 40 | 187 | 38.14 | ATA.AAT.TAT.TTT.CTG.TTT |
| CAT.CCT.GCT.ACT.TTG.AGG | ||||
| 3 | 40 | 237 | 32.22 | GGA.AAA.TGA.GTA.ACA.TGA |
| 2 | TGT.CAT.CAC.AGG.AGG.ATA | |||
| 4 | 2 | 218 | 36.26 | ACC.CAG.CAG.TGA.GTT.TT |
| 40 | GCC.CAA.AAT.ACA.TTT.CAG | |||
| 5 | 40 | 170 | 30.19 | ATA.TTA.ATT.TGT.TAT.ATT |
| CAA.TTT.ACT.CTC.CCA.TGT | ||||
| 6 | 40 | 228 | 35.58 | TTT.CAA.GTA.CTT.CTA.TGA |
| ACT.TTG.TAG.ACA.AAT.CTC | ||||
| 7 | 194 | 30.88 | GAC.ATC.TAG.TGT.GTG.TTT | |
| 40 | CCC.CTT.TTT.TCT.TTT.CAT | |||
| 8 | 5 | 213 | 42.21 | GAC.AAT.AAA.TCC.TTG.TGT |
| 50 | AAG.ATT.TTT.TTA.TAT.AGG | |||
| 9 | 40 | 249 | 33.73 | TTT.GAG.TTT.TGA.GTA.TTT |
| TGG.GTG.TTT.CCT.GTG.AGT | ||||
| 10 | 50 | 240 | 41.47 | CAC.CCC.TCA.GGA.CAG.TTT |
| ACA.TCT.GTT.CCT.TGT.GAG | ||||
| 11.1 | 50 | 145 | 40.58 | AGG.TAA.TTG.TTC.TCT.CTT |
| GAA.GTG.AAC.TTC.ATG.CTT | ||||
| 11.2 | 40 | 224 | 60.81 | TCC.CAA.GAA.TGT.GGA.TGT |
| 2 | AAA.GGC.CCC.AGA.GAA.GTA | |||
| 12.1 | 40 | 184 | 44.53 | TTT.TTT.TTT.TTT.TAA.TAC.A |
| AAT.CTG.TAC.GAA.CCA.TCT | ||||
| 12.2 | 8 | 366 | 53.23 | TGG.AAG.TAG.TGA.TAA.GGT |
| 40 | TGT.ACT.TTT.CCC.AAA.AGG | |||
| 13 | 40 | 272 | 49.06 | ATC.TGC.ACT.TCC.TTT.TCT |
| AAA.ACC.TTG.GCA.GTT.GAG | ||||
| 14 | 45 | 235 | 48.94 | TAC.TTA.CCT.GTT.TTT.TGG |
| 5 | GTA.GTA.GCT.CTG.CTT.GTT | |||
| 15 | 40 | 179 | 29.97 | CAG.CTT.TTC.CTT.AAA.GTC |
| CAG.TTG.AAA.TGT.CAG.AAG | ||||
| 16 | 261 | 47.56 | CTT.GCT.CCT.TCA.TGT.TCT.TG | |
| 40 | AGA.AGT.ATA.AGA.ATG.GCT.GTC | |||
| 17 | 40 | 199 | 47.01 | ATT.ATT.TCT.TGT.TCC.CTT |
| AAT.GCT.TAG.TAT.CTG.CCT | ||||
| 18 | 45 | 215 | 46.67 | CCT.ATT.TTG.AGG.TAT.TGA.AT |
| GCC.AGT.GTG.CAT.CAC.CA | ||||
| 19 | 282 | 43.43 | TGT.TGG.GAT.GCA.AAC.AGG | |
| 40 | ATC.CCA.CAG.TGC.ATA.AAT | |||
After checking and visualizing the long PCR products on a 0.8% agarose gel, 1µl of the long PCR amplicons was used as template for subsequent extensive multiplex short PCR. The short PCR was performed in two multiplex groups of 11 and 10 amplicons, respectively. The final concentrations of each primer were shown in Table 2. The final concentrations of the PCR mix included 1 × PCR buffer, 7 mM MgCl2, and 0.25 mM each dNTP. Cycling conditions included a hot start of 3 min at 94 °C with the Taq Pol added after 2 min, followed by five cycles of 94 °C × 1 min, 52 °C × 45 sec (decrements of 1 °C/cycle), and 72 °C × 1 min; 15 cycles of 94 °C × 1 min, 48 °C × 45 sec, and 72 °C × 1 min, 30 sec (with increments of 2 sec/cycle); 15 cycles of 94 °C × 1 min, 38 °C × 45 sec, and 72 °C × 1 min 30 sec. Each PCR reaction was teminated with a round of heteroduplexing: 72 °C × 10 min, 98 °C × 10 min, 45 °C × 30 min, and finally 37 °C × 30 min. Each tube reaction was directly mixed with 1/10 volume of 10 × loading buffer, 6.5 µl of multiplex group I and 8.5 µl of multiplex group II were loaded onto a slab gel for size separation.
| Multipex group I | Multipex group II | ||
| Exon | Final concentration | Exon | Final concentration |
| 11.1 | 0.375 µM | 5 | 1.5 µM |
| 15 | 0.5 µM | 2 | 1.25 µM |
| 12.1 | 0.375 µM | 7 | 1.75 µM |
| 17 | 0.5 µM | 4 | 1.625 µM |
| 8 | 0.375 µM | 11.2 | 0.5 µM |
| 18 | 1.25 µM | 6 | 1.625 µM |
| 14 | 1.25 µM | 9 | 1.5 µM |
| 3 | 0.625 µM | 1 | 1.25 µM |
| 10 | 0.625 µM | 13 | 1.625 µM |
| 16 | 1.0 µM | 12.2 | 0.325 µM |
| 19 | 1.75 µM | ||
Two-dimensional electrophoresis For two-dimensional electrophoresis, the DGGE instrument was from CBS Scientific Co. (Solana Beach, CA. Amplicons from each of the two mu ltiplex reactio ns were mixed and sub jected to electrophoresis in 0.5 × TAE (Tris-Acetate EDTA). A 10% polyacrylamide, 0.75 mm thick slab gel was used, the amplicons were fractionated at 140 V for 7.5 hr at 50 °C. The separation pattern was detected by SYBR green staining and UV-transillumination of the slab gel. The 120-to 420-bp region in the middle of each lane was quickly cut out and applied to a 1 mm thick DGGE gel.
The DGGE gel was prepared as a 1 mm thick slab gel with a 10%-6.5% reverse polyacrylamide gradient containing 25%-70% urea/formamide (UF) and 3%-9% glycerol gradients. Electrophoresis was carried out for 16 hr at 56 °C and 90-110 V. After electrophoresis, the gels were stained with SYBR-green I and II for 30 min. The DGGE band patterns were detected and documented with a gel documentation system (Oncor, Gaithersburg, MD).
Sequence analysis Amplicons were prepared by PCR such that a standard M13 primer site was incorporated at one end. These products were sequenced on an ABI 377 sequencer (Foster City, CA) with kits containing Taq FS DNA polymerase and dye primer technology, as recommended by the manufacturer.
DNA methylation patterns in the CpG islands of hMLH1 gene were determined by Methylation-specific PCR (MSP) as described[18]. The primer sequences of hMLH1 for unmethylated reaction were 5’-TTTTGATGTAGATGTTTTATTAGGGTTGT-3’ (sense) and 5’-ACCACCTCATCATAACTACCCACA-3’ (antisense), and for methylated reaction were 5’-ACGTAGACGTTTTATTAGGGTCGC-3’ (sense) and 5’-CCTCATCGTAACTACCCGCG-3’ (antisense).
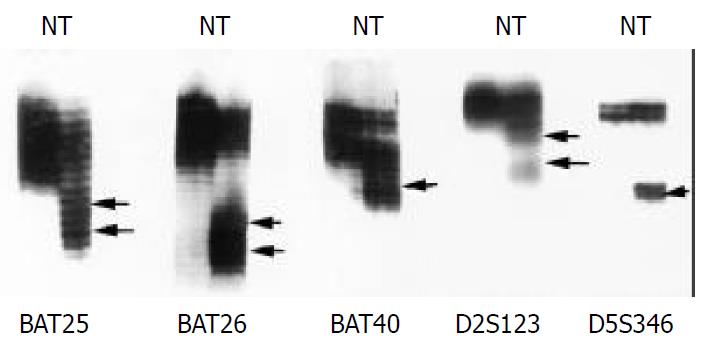
MSI analyses included five microsatellite markers: BAT25, BAT26, BAT40, D2S123, and D5S346. PCR was performed as previous described[1]. MSI was defined as the presence of band shift in the tumor DNA that was not present in the corresponding normal DNA. Based on the number of mutated MSI markers in each tumor, carcinomas were characterized as MSI-H if they manifested instability in two or more markers, MSI-L if unstable in only one marker, and MSS if they showed no instability in any marker[19,20].
Chi-square test with Yates’ correction were used. A P value < 0.05 was considered significant.
Alterations of electrophoretic patterns of PCR products of five microsatellite markers were compared between the tumor and the normal DNA in each patient (Figure 1). MSI affecting at least one locus was observed in 17 (25%) of 68 tumors, among which eight (11.8%) were MSI-H, nine (13.2%) were MSI-L, and fifty-one (75%) were MSS.
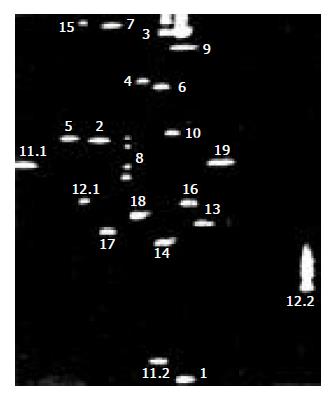
Mutations and sequence alteration in various exons manifested as the four-spot pattern denoting a heterozygous variant (Figure 2). We found three mutations in 68 (4.4%) gastric cancer by DNA sequencing. Two mutations were caused by a single bp substitution (exon 8 at codon 226, CGG→CTG, Arg→Leu; exon 9 at codon 252, TCA→TTA, Ser→Phe);. One identical change was caused by a 2 bp substitution (exon 12 at codon 409, CAG→CGT, Gln→Arg), which displayed similar DGGE band pattern. A comparison of MSI status with hMLH1 mutation was shown in Table 3. Three hMLH1 mutations were all detected in MSI-H, but no mutation was found in MSI-L or MSS.
| MSI Status | No.of cases | hMLH1 mutation |
| MSI-H | 8 | 3 |
| MSI-L | 9 | 0 |
| MSS | 51 | 0 |
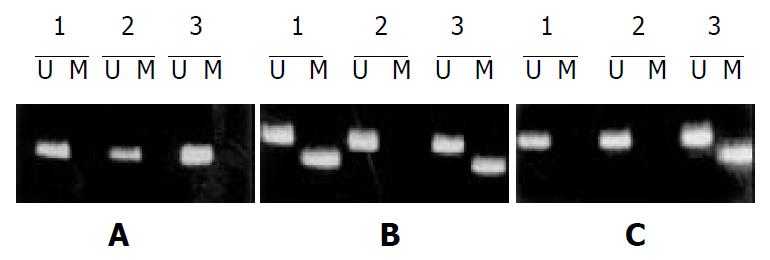
To examine methylation of promoter region of hMLH1, we adapted MSP for the 5’ CpG islands in this gene. The region chosen spaned the area of greatest CpG density immediately 5’ to the transcription starting site, in an area previously studied for methylation changes[15]. In gastric mucosal samples without cancer, only unmethylated hMLH1 genes were present. Eleven of 68 (16.2%) gastric cancers exhibited prominent methylation, which always had both methylated and nonmethylated hMLH1 genes (Figure 3). A comparison of MSI status with hMLH1 methylation status was shown in Table 4. Seven of 8 (87.5%) cancers with MSI-H exhibited prominent methylation, whereas methylated hMLH1 gene was only found in 1/9 (11.1%) gastric cancer with MSI-L and 3/51 (5.9%) with MSS, suggesting that hMLH1 methylation was more correlated with gastric cancers with MSI-H than that with MSI-L or with MSS (P < 0.01-0.001).
A significant portion of gastric cancers exhibit defective DNA mismatch repair, manifested as MSI. There is now evidence that MSI cancer comprises distinctive MSI-H and MSI-L categories[21-23]. MSI-H cancers are distingushed clinicopathologically and in their spectrum of genetic alterations from cancers showing MSI-L and MSS cancers[24-34]. Our previous studies indicated that MSI-H gastric cancers often showed lower frequency of LOH in APC, MCC and DCC genes than do MSI-L and MSS cancers[1]. In the present study, all three hMLH1 mutations were detected in MSI-H, but no mutation was found in those showing MSI-L or MSS. This result further indicates that hMLH1 is mutational target in MSI-H tumor cells and supported the notion that MIS-H tumors identified an alterative pathway of tumorigenesis that had been proposed by Vogelstein and co-workers[35].
Human cancers with MSI-H phenotype develop due to defects in DNA mismatch repair genes. Silencing of a DNA mismatch repair gene, hMLH1 gene, by promoter hypermethylation is a frequent cause of MSI-H phenotype[36-42]. In this study, 11 (16.2%) of 68 gastric cancers exhibited prominent hMLH1 methylation, which is similar to previous studies[36]. It has been reported that MSI-H is related to methylation of the hMLH1 promoter but not hMSH1 mutations in sporadic gastric carcinomas[43]. It was also found that MSI-L gastric carcinomas share the hMLH1 methylation status of MSI-H carcinomas but not their clinicopathological profile[44]. In this study, 7/8 (87.5%) cancers with MSI-H exhibited prominent methylation of the hMLH1 promoter, suggesting that methylation of the hMLH1 promoter is correlated with MSI-H gastric cancer. The frequency of methylation in MSI-H cancers is significantly higher than that in cancers with MSI-L and MSS, however no difference was found between cancers with MSI-L and MSS, indicating that MSI-H tumors identify a different methylation pathway from cancers with MSI-L and MSS, and MSI-L cancers involves the same methylation pathway as MSS tumors. This finding is in agreement with our recently published data on molecular pathway in the gastric carcinogenesis[2].
It has been found that colorectal cancer cell lines only had methylated hMLH1 genes, but had absent unmethylated hMLH1 genes[45]. In the present study, unlike the situation with the cell lines, however, the primary MSI+ gastric cancers always had both methylated and nonmethylated hMLH1 genes. It is likely that a significant fraction of the unmethylated genes is derived from the non-neoplastic cells (stromal, inflammatory, vascular, etc.), which are invariably present within the primary tumors but are not found in cultured cell lines. It has been reported that methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines[13]. whereas demethylation of hMLH1 promoter results in reexpression of hMLH1 in tumor cells tested. Not only was the protein expressed, but MMR activity was restored. DNA methylation is a fundamental feature of the genomes and the control of their functions therefore it is a candidate for pharmacological manipulation that might have important therapeutic advantage.
| 1. | Fang DC, Jass JR, Wang DX, Zhou XD, Luo YH, Young J. Infrequent loss of heterozygosity of APC/MCC and DCC genes in gastric cancer showing DNA microsatellite instability. J Clin Pathol. 1999;52:504-508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 2. | Fang DC, Yang SM, Zhou XD, Wang DX, Luo YH. Telomere erosion is independent of microsatellite instability but related to loss of heterozygosity in gastric cancer. World J Gastroenterol. 2001;7:522-526. [PubMed] |
| 3. | Martins C, Kedda MA, Kew MC. Characterization of six tumor suppressor genes and microsatellite instability in hepatocellular carcinoma in southern African blacks. World J Gastroenterol. 1999;5:470-476. [PubMed] |
| 4. | Wu BP, Zhang YL, Zhou DY, Gao CF, Lai ZS. Microsatellite instability, MMR gene expression and proliferation kinetics in colorectal cancer with famillial predisposition. World J Gastroenterol. 2000;6:902-905. [PubMed] |
| 5. | Thomas DC, Umar A, Kunkel TA. Microsatellite instability and mismatch repair defects in cancer. Mutat Res. 1996;350:201-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 6. | Peltomäki P. Microsatellite instability and hereditary non-polyposis colon cancer. J Pathol. 1995;176:329-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 7. | Cai Q, Sun MH, Lu HF, Zhang TM, Mo SJ, Xu Y, Cai SJ, Zhu XZ, Shi DR. Clinicopathological and molecular genetic analysis of 4 typical Chinese HNPCC families. World J Gastroenterol. 2001;7:805-810. [PubMed] |
| 8. | Modrich P. Mismatch repair, genetic stability, and cancer. Science. 1994;266:1959-1960. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 287] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 9. | Papadopoulos N, Lindblom A. Molecular basis of HNPCC: mutations of MMR genes. Hum Mutat. 1997;10:89-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 10. | Zhao B, Wang ZJ, Xu YF, Wan YL, Li P, Huang YT. Report of 16 kindreds and one kindred with hMLH1 germline mutation. World J Gastroenterol. 2002;8:263-266. [PubMed] |
| 11. | Aaltonen LA, Peltomäki P, Leach FS, Sistonen P, Pylkkänen L, Mecklin JP, Järvinen H, Powell SM, Jen J, Hamilton SR. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812-816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1810] [Cited by in RCA: 1817] [Article Influence: 55.1] [Reference Citation Analysis (1)] |
| 12. | Miyakura Y, Sugano K, Konishi F, Ichikawa A, Maekawa M, Shitoh K, Igarashi S, Kotake K, Koyama Y, Nagai H. Extensive methylation of hMLH1 promoter region predominates in proximal colon cancer with microsatellite instability. Gastroenterology. 2001;121:1300-1309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 147] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 13. | Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808-811. [PubMed] |
| 14. | Garinis GA, Patrinos GP, Spanakis NE, Menounos PG. DNA hypermethylation: when tumour suppressor genes go silent. Hum Genet. 2002;111:115-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 87] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 15. | Jones PA. DNA methylation and cancer. Oncogene. 2002;21:5358-5360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 211] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 16. | Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400-5413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1124] [Cited by in RCA: 1102] [Article Influence: 45.9] [Reference Citation Analysis (0)] |
| 17. | Wu Y, Nyström-Lahti M, Osinga J, Looman MW, Peltomäki P, Aaltonen LA, de la Chapelle A, Hofstra RM, Buys CH. MSH2 and MLH1 mutations in sporadic replication error-positive colorectal carcinoma as assessed by two-dimensional DNA electrophoresis. Genes Chromosomes Cancer. 1997;18:269-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 18. | Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821-9826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4183] [Cited by in RCA: 4262] [Article Influence: 142.1] [Reference Citation Analysis (12)] |
| 19. | Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248-5257. [PubMed] |
| 20. | Sood AK, Holmes R, Hendrix MJ, Buller RE. Application of the National Cancer Institute international criteria for determination of microsatellite instability in ovarian cancer. Cancer Res. 2001;61:4371-4374. [PubMed] |
| 21. | Jass JR, Biden KG, Cummings MC, Simms LA, Walsh M, Schoch E, Meltzer SJ, Wright C, Searle J, Young J. Characterisation of a subtype of colorectal cancer combining features of the suppressor and mild mutator pathways. J Clin Pathol. 1999;52:455-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 163] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 22. | Wahlberg SS, Schmeits J, Thomas G, Loda M, Garber J, Syngal S, Kolodner RD, Fox E. Evaluation of microsatellite instability and immunohistochemistry for the prediction of germ-line MSH2 and MLH1 mutations in hereditary nonpolyposis colon cancer families. Cancer Res. 2002;62:3485-3492. [PubMed] |
| 23. | Kambara T, Matsubara N, Nakagawa H, Notohara K, Nagasaka T, Yoshino T, Isozaki H, Sharp GB, Shimizu K, Jass J. High frequency of low-level microsatellite instability in early colorectal cancer. Cancer Res. 2001;61:7743-7746. [PubMed] |
| 24. | Miyoshi E, Haruma K, Hiyama T, Tanaka S, Yoshihara M, Shimamoto F, Chayama K. Microsatellite instability is a genetic marker for the development of multiple gastric cancers. Int J Cancer. 2001;95:350-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 25. | Yamamoto H, Min Y, Itoh F, Imsumran A, Horiuchi S, Yoshida M, Iku S, Fukushima H, Imai K. Differential involvement of the hypermethylator phenotype in hereditary and sporadic colorectal cancers with high-frequency microsatellite instability. Genes Chromosomes Cancer. 2002;33:322-325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 26. | Halford S, Sasieni P, Rowan A, Wasan H, Bodmer W, Talbot I, Hawkins N, Ward R, Tomlinson I. Low-level microsatellite instability occurs in most colorectal cancers and is a nonrandomly distributed quantitative trait. Cancer Res. 2002;62:53-57. [PubMed] |
| 27. | Young J, Simms LA, Biden KG, Wynter C, Whitehall V, Karamatic R, George J, Goldblatt J, Walpole I, Robin SA. Features of colorectal cancers with high-level microsatellite instability occurring in familial and sporadic settings: parallel pathways of tumorigenesis. Am J Pathol. 2001;159:2107-2116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 283] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 28. | Lee JH, Abraham SC, Kim HS, Nam JH, Choi C, Lee MC, Park CS, Juhng SW, Rashid A, Hamilton SR. Inverse relationship between APC gene mutation in gastric adenomas and development of adenocarcinoma. Am J Pathol. 2002;161:611-618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 112] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 29. | Duval A, Hamelin R. Genetic instability in human mismatch repair deficient cancers. Ann Genet. 2002;45:71-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 52] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 30. | Morán A, Iniesta P, de Juan C, González-Quevedo R, Sánchez-Pernaute A, Díaz-Rubio E, Ramón y Cajal S, Torres A, Balibrea JL, Benito M. Stromelysin-1 promoter mutations impair gelatinase B activation in high microsatellite instability sporadic colorectal tumors. Cancer Res. 2002;62:3855-3860. [PubMed] |
| 31. | Peiró G, Diebold J, Lohse P, Ruebsamen H, Lohse P, Baretton GB, Löhrs U. Microsatellite instability, loss of heterozygosity, and loss of hMLH1 and hMSH2 protein expression in endometrial carcinoma. Hum Pathol. 2002;33:347-354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 42] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 32. | Wu CW, Chen GD, Jiang KC, Li AF, Chi CW, Lo SS, Chen JY. A genome-wide study of microsatellite instability in advanced gastric carcinoma. Cancer. 2001;92:92-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 33. | Ogata S, Tamura G, Endoh Y, Sakata K, Ohmura K, Motoyama T. Microsatellite alterations and target gene mutations in the early stages of multiple gastric cancer. J Pathol. 2001;194:334-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 34. | Tamura G, Sato K, Akiyama S, Tsuchiya T, Endoh Y, Usuba O, Kimura W, Nishizuka S, Motoyama T. Molecular characterization of undifferentiated-type gastric carcinoma. Lab Invest. 2001;81:593-598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 57] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 35. | Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4616] [Cited by in RCA: 4493] [Article Influence: 118.2] [Reference Citation Analysis (0)] |
| 36. | Fleisher AS, Esteller M, Tamura G, Rashid A, Stine OC, Yin J, Zou TT, Abraham JM, Kong D, Nishizuka S. Hypermethylation of the hMLH1 gene promoter is associated with microsatellite instability in early human gastric neoplasia. Oncogene. 2001;20:329-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 96] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 37. | Murata H, Khattar NH, Kang Y, Gu L, Li GM. Genetic and epigenetic modification of mismatch repair genes hMSH2 and hMLH1 in sporadic breast cancer with microsatellite instability. Oncogene. 2002;21:5696-5703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 63] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 38. | Norrie MW, Hawkins NJ, Todd AV, Meagher AP, O'Connor TW, Ward RL. The role of hMLH1 methylation in the development of synchronous sporadic colorectal carcinomas. Dis Colon Rectum. 2002;45:674-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 39. | Sakata K, Tamura G, Endoh Y, Ohmura K, Ogata S, Motoyama T. Hypermethylation of the hMLH1 gene promoter in solitary and multiple gastric cancers with microsatellite instability. Br J Cancer. 2002;86:564-567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 40. | Baranovskaya S, Soto JL, Perucho M, Malkhosyan SR. Functional significance of concomitant inactivation of hMLH1 and hMSH6 in tumor cells of the microsatellite mutator phenotype. Proc Natl Acad Sci USA. 2001;98:15107-15112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 41. | Deng G, Peng E, Gum J, Terdiman J, Sleisenger M, Kim YS. Methylation of hMLH1 promoter correlates with the gene silencing with a region-specific manner in colorectal cancer. Br J Cancer. 2002;86:574-579. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 82] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 42. | Hawkins N, Norrie M, Cheong K, Mokany E, Ku SL, Meagher A, O'Connor T, Ward R. CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology. 2002;122:1376-1387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 283] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 43. | Bevilacqua RA, Simpson AJ. Methylation of the hMLH1 promoter but no hMLH1 mutations in sporadic gastric carcinomas with high-level microsatellite instability. Int J Cancer. 2000;87:200-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 44. | Pinto M, Oliveira C, Machado JC, Cirnes L, Tavares J, Carneiro F, Hamelin R, Hofstra R, Seruca R, Sobrinho-Simões M. MSI-L gastric carcinomas share the hMLH1 methylation status of MSI-H carcinomas but not their clinicopathological profile. Lab Invest. 2000;80:1915-1923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 34] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 45. | Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870-6875. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1343] [Cited by in RCA: 1380] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 46. | Machover D, Zittoun J, Saffroy R, Broët P, Giraudier S, Magnaldo T, Goldschmidt E, Debuire B, Orrico M, Tan Y. Treatment of cancer cells with methioninase produces DNA hypomethylation and increases DNA synthesis. Cancer Res. 2002;62:4685-4689. [PubMed] |
| 47. | Nichol K, Pearson CE. CpG methylation modifies the genetic stability of cloned repeat sequences. Genome Res. 2002;12:1246-1256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 44] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 48. | Karpf AR, Jones DA. Reactivating the expression of methylation silenced genes in human cancer. Oncogene. 2002;21:5496-5503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 203] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 49. | Christman JK. 5-Azacytidine and 5-aza-2'-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483-5495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 991] [Cited by in RCA: 1100] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
Edited by Wu XN