Published online Feb 15, 2003. doi: 10.3748/wjg.v9.i2.205
Revised: August 1, 2002
Accepted: September 4, 2002
Published online: February 15, 2003
AIM: To study the expression pattern of ETS2 (erythroblastosis virus oncogene homolog 2) in human esophageal squamous cell carcinoma (ESCC).
METHODS: Reverse transcription polymerase chain reaction (RT-PCR) and Northern blot were performed to examine the expression level of ETS2 mRNA in 37 pairs of ESCC tissue samples. Western blot and immunohistochemistry were carried out to check the expression level of ETS2 protein in 30 pairs of ESCC tissue specimens.
RESULTS: RT-PCR and Northern blot analysis showed that ETS2 mRNA upregulated in 75.7% (28/37) examined ESCC tissues relative to matched normal tissues. From those 37 cases, 14 cases were randomly selected to perform Western blot and the results revealed that ETS2 protein overexpressed in 71.4% (10/14) checked ESCC tissues compared with the corresponding normal tissues. Moreover, the expression patterns of ETS2 protein in those 14 cases were identical to those of ETS2 mRNA displayed by RT-PCR or Northern Blot. Immunohistochemistry analysis showed that the expression level of ETS2 protein rose in 75% (12/16) tumor epithelial cells contrasted to the normal cells. Altogether the expression level of ETS2 protein increased in 73.3% (22/30) checked ESCC tissue samples contrary to their normal counterparts.
CONCLUSION: The results suggested that ETS2 overexpressed in paired human ESCC tissue samples at both mRNA and protein levels and may be associated with the tumorigenesis of esophagus.
- Citation: Li X, Lu JY, Zhao LQ, Wang XQ, Liu GL, Liu Z, Zhou CN, Wu M, Liu ZH. Overexpression of ETS2 in human esophageal squamous cell carcinoma. World J Gastroenterol 2003; 9(2): 205-208
- URL: https://www.wjgnet.com/1007-9327/full/v9/i2/205.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i2.205
Erythroblastosis virus oncogene homolog 2 (ETS2) is a pro-oncogene, which is located in human chromosomal region 21q22.3 and encodes a 56 kD protein that is phosphorylated by a Ca2+-dependent mitogenic signal process[1,2]. ETS2 gene expresses in various tissues, including blood, breast and prostate. ETS2 may be involved in the regulation of cellular proliferation and differentiation and may play a critical role in T-cell activation and cytokines production[3-6].
As a member of ETS oncogene family, ETS2 gene has the oncogenic potential. It is similarly transposed as a consequence of nonrandom chromosomal translocations; especially, the t (8;21) (q22;q22) translocation is the most frequently noted breakpoint involving chromosome 21 for acute myelogenous leukemia[7]. An acute non-lymphoblastic leukemia with a complex t (6;18;21) chromosomal translocation, has so far been associated with higher expression level of ETS2[8]. It had been reported that ETS2 gene was associated with the growth and invasion of breast carcinoma cells and was required to maintain the transformed state for human prostate cancer cells[9,10].
Esophageal cancer ranks among the 10 most frequent cancers in the world, with a predominant distribution in developing countries. Our previous study showed that genetic susceptibility to esophageal cancer was one of the important causes for the high prevalence and familial aggregation of this disease in some areas of northern China[11]. We observed the upregulation of ETS2 gene in human esophageal squamous cell carcinoma (ESCC) using cDNA microarray technique[12]. To our best knowledge, this study first investigated the expression patterns of ETS2 gene and ETS2 protein in ESCC by reverse transcription polymerase chain reaction (RT-PCR), Northern blot, Western blot and immunohistochemistry. Both the expression levels of ETS2 gene and ETS2 protein increased in ESCC tissues contrary to their normal counterparts. The results were consistent with the microarray results. Therefore, it indicated that ETS2 gene might be related to the formation of human ESCC and its further study may provide the insight into the mechanisms of carcinogenesis of esophagus.
Samples of ESCC and matched normal esophagus tissues were collected from 37 patients who had not receive chemotherapy or radiotherapy before surgery. The tissues were immediately stored in liquid nitrogen until analysis and each sample was confirmed by histological examination. Total RNA and total protein of the samples were extracted using Trizol solution (Life Technologies, Rochville, ML) per manufacturer's protocol. Total RNA was quantitated and assessed for purity by means of UV spectrophotometry and electrophoresis in denaturing formaldehyde gel. The standard curve of protein was prepared to determine the protein concentration of tissue samples with Bicinchoninic Acid Protein Assay Kit (SIGMA, St. Louis, MO). Sixteen pairs of ESCC paraffin slides were obtained from Department of Pathology, Cancer Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College.
Before reverse-transcription, 5 μg total RNA of each sample was treated with 20 units DNase I (Promega, Madison, WI), 40 units RNasin (Promega, Madison, WI) at 37 °C for 15 min to remove contaminated genomic DNA. Then first strand cDNA was synthesized with SuperScript Preamplification System For First Strand cDNA Synthesis kit (Life Technologies, Rochville, ML). Two microliters of reverse-transcription product were used as the template to amplify specific fragment of ETS2 gene. PCR conditions were as follows: initial denaturation at 94 °C for 3 min, followed by 28 cycles of 94 °C for 30 s, 58 °C for 30 s and 72 °C for 1 min. The expression of housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), was used to normalize the template input. The sequences of the PCR primer pairs of ETS2 and GAPDH were as follows: ETS2, 5'-GTGGACCTATTCAGCTGTGG-3' 5'TTCCCCGACGTCTTGTGGAT-3' GAPDH, 5'-ACCACAGTCCATGCCATCAC-3' 5'-TCCACCACCCTGTTGCTGTA-3'
Briefly, 30 μg total RNA of each sample was dissolved in loading buffer containing formamide and formaldehyde, heated at 70 °C for 10 min, electrophoresed on the 2% formaldehyde agarose gel and transferred to a positively charged nylon membrane. The membrane was prehybridized in 5 mL hybridization solution (6 × SSC, 2 × Denhart's solution, 0.1% SDS, 100 μg/mL denatured salmon sperm DNA) at 68 °C for 2 h. Then 32P-labelled probe (bases 1202-1952) was added and hybridization was performed at 68 °C for 18 h. The membrane was washed twice at room temperature in 2 × SSC, 0.1% SDS for 20 min, once at 65 °C in 0.5 × SSC, 0.1% SDS for 20 min and exposed to X-ray films at -70 °C for 72 h.
Anti-ETS2 antibody was purchased from Santa Cruz biotechnology, Inc (Santa Cruz, CA). Thirty-microgram total protein of each sample was mixed with 20 μL sample buffer (100 mM Tris·Cl, 200 mM DTT, 4% SDS, 20% glycerol and 0.2% bromphenol blue) before separation by SDS-PAGE (10%) electrophoresis system. Samples were then transferred to PVDF membrane and nonspecific binding was blocked with 5% nonfat dry milk for 1 h at room temperature. Then the filter was incubated with anti-ETS2 antibody (1:500) for 2 h at room temperature. An enhanced chemiluminescence system (Santa Cruz, Santa Cruz, CA) was used for signal detection.
Four-micron paraffin-embedded slides were dewaxed in xylene, rehydrated in ethanol and treated with H2O2 to block the endogenous peroxidase activity. Antigen retrieval was achieved by microwaving in a citrate buffer (pH6.0) for 15 min. The slides were incubated with anti-ETS2 antibody at a 1:50 dilution for 2 h at 37 °C. Biotinylated secondary antibody and peroxidase-conjugated streptavidin steps were performed using UltraSensitiveTM S-P kit (Maxim Biotech, Fujian, China) according to the manufacturer's protocol. DAB was used as the chromogen and hematoxylin as the counterstain. Negative control was performed by substituting PBS for the primary antibody.
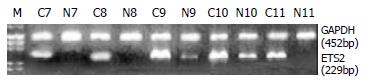
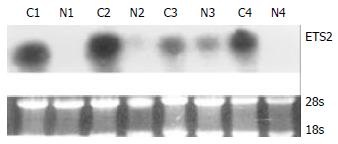
RT-PCR and Northern blot were performed to confirm the differential expression of ETS2 in ESCC at transcriptional level. RT-PCR analysis showed that ETS2 gene overexpressed in 24 pairs of tumor versus normal tissues among a total of 31 tested cases (Figure 1). Northern blot analysis showed that the expression level of ETS2 gene increased in 4 of 6 cases of tumor tissues relative to the matched normal tissues (Figure 2).
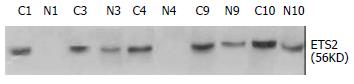
Western blot and immunohistochemistry were conducted to verify the differential expression of ETS2 in ESCC at translational level. From the above mentioned 37 cases, 14 pairs of ESCC tissue samples were randomly selected to carry out Western blot and the results showed that ETS2 protein was upregulated in 71.4% (10/14) examined ESCC tissues relative to the corresponding normal tissues (Figure 3). The expression patterns of ETS2 protein in those 14 cases were identical to those of ETS2 mRNA revealed by RT-PCR or Northern Blot.
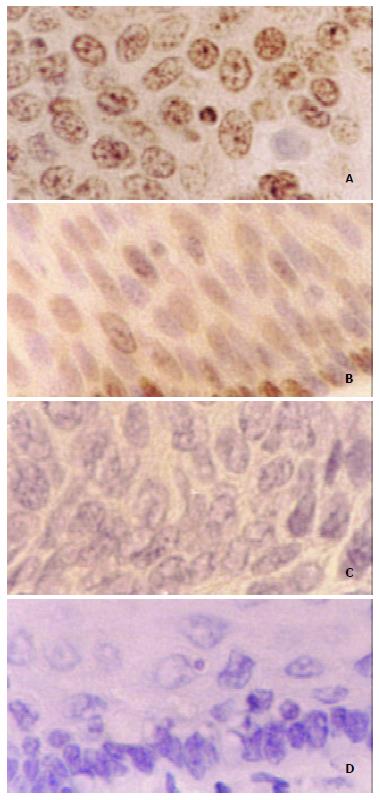
Immunohistochemistry results displayed that 75% (12/16) checked ESCC cases existed diffuse and strong staining in the nucleus of tumor cells, while sporadic and weak staining was observed in the nucleus of the matched normal esophageal epithelial cells (Figure 4).
The understanding of the molecular basis of tumor development has progressed dramatically in the last two decades. It is well known that tumor is essentially a genetic disease. So it is important to demonstrate what genes are and how they work in carcinogenesis. Identifying the genetic differences between normal and tumor cells or tissues will discover the genes that directly cause tumor or be associated with tumorigenesis and provide novel markers for early detection and appropriate therapy.
In our previous study, ETS2 gene showed upregulation in human ESCC tissue[12]. To verify this differential expression, we explored the expression pattern of ETS2 in paired ESCC tissue samples at both mRNA and protein levels. The results of RT-PCR and Northern blot revealed that ETS2 overexpressed in 75.7% (28/37) examined tumor tissues relative to the corresponding normal tissues. From those 37 cases, 14 cases were randomly selected to perform Western blot and the results showed that the expression level of ETS2 protein elevated in 71.4% (10/14) checked ESCC tissues compared with the corresponding normal tissues. Moreover, the expression patterns of ETS2 protein in those 14 cases were identical to those of ETS2 mRNA displayed by RT-PCR or Northern Blot. Immunohistochemistry analysis showed that the expression level of ETS2 protein raised in 75% (12/16) examined tumor epithelial cells contrasted to the normal counterparts. Altogether ETS2 protein overexpressed in 73.3% (22/30) tested ESCC tissues relative to the matched normal tissues. Therefore, the data suggested that ETS2 abnormally expressed not only in the transcriptional level but also in the translational level for human ESCC and the increasing transcription of ETS2 mRNA in ESCC may result in increasing translation of ETS2 protein.
As a transcription factor, ETS2 protein controls the transcription of some important genes participating in a number of biological processes including cell growth and apoptosis. Although many studies have been done, the accurate function of ETS2 in biological and pathophysiologic state is still unclear. Previous studies showed that the overexpression of ETS2 led to different results in different cells. Some promoted tumorigenesis[13,14], while others were arresting[15]. So it indicated that ETS2 could mediate multiple different signal pathways and might be involved in carcinogenesis with some unlike ways. In the present study, we found the overexpression of ETS2 in ESCC at both mRNA and protein levels. It suggested that ETS2 might be associated with the formation of ESCC. But how ETS2 acts during the neoplasia of esophagus and whether its function is tissue-specific or organ-specific are still unknown. Although a lot of work was focused on the chromosomal aberrations of human ESCC[16-18] and even in several studies frequent loss of 21q was observed[19,20], 21q had not yet been investigated in detail. Hence the variation of 21q22.3 in human ESCC, and which ETS2 gene is located in remain unclear. Further investigations will help us to make clear whether the overexpression of ETS2 is caused by the gain of 21q22.3 in human ESCC. As mentioned above, the higher expression level of ETS2 was found in an acute non-lymphoblastic leukemia with a complex t (6;18;21) chromosomal translocation[8]. Another speculation is that the upregulation of ETS2 in human ESCC may be associated with the chromosomal translocation of 21q22.3. More studies about chromosomal aberrations will be helpful to demonstrate the precise function of ETS2 and its molecular mechanisms in tumorigenesis of esophagus.
| 1. | Watson DK, McWilliams-Smith MJ, Nunn MF, Duesberg PH, O'Brien SJ, Papas TS. The ets sequence from the transforming gene of avian erythroblastosis virus, E26, has unique domains on human chromosomes 11 and 21: Both loci are transcriptionally active. Proc Natl Acad Sci USA. 1985;82:7294-7298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 123] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 2. | Fujiwara S, Fisher RJ, Seth A, Bhat NK, Showalter SD, Zweig M, Papas TS. Characterization and localization of the products of the human homologs of the v-ets oncogene. Oncogene. 1988;2:99-103. [PubMed] |
| 3. | Villena JA, Martin I, Viñas O, Cormand B, Iglesias R, Mampel T, Giralt M, Villarroya F. ETS transcription factors regulate the expression of the gene for the human mitochondrial ATP synthase beta-subunit. J Biol Chem. 1994;269:32649-32654. [PubMed] |
| 4. | Bhat NK, Thompson CB, Lindsten T, June CH, Fujiwara S, Koizumi S, Fisher RJ, Papas TS. Reciprocal expression of human ETS1 and ETS2 genes during T-cell activation: regulatory role for the protooncogene ETS1. Proc Natl Acad Sci USA. 1990;87:3723-3727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 117] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 5. | Ma X, Neurath M, Gri G, Trinchieri G. Identification and characterization of a novel Ets-2-related nuclear complex implicated in the activation of the human interleukin-12 p40 gene promoter. J Biol Chem. 1997;272:10389-10395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 123] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 6. | Blumenthal SG, Aichele G, Wirth T, Czernilofsky AP, Nordheim A, Dittmer J. Regulation of the human interleukin-5 promoter by Ets transcription factors. Ets1 and Ets2, but not Elf-1, cooperate with GATA3 and HTLV-I Tax1. J Biol Chem. 1999;274:12910-12916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 82] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 7. | Sacchi N, Cheng SV, Tanzi RE, Gusella JF, Drabkin HA, Patterson D, Haines JH, Papas TS. The ETS genes on chromosome 21 are distal to the breakpoint of the acute myelogenous leukemia translocation (8; 21). Genomics. 1988;3:110-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 8. | Santoro A, Maggio A, Carbone P, Mirto S, Caronia F, Acuto S. Amplification of ETS2 oncogene in acute nonlymphoblastic leukemia with t (6; 21; 18). Cancer Genet Cytogenet. 1992;58:71-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 9. | Sapi E, Flick MB, Rodov S, Kacinski BM. Ets-2 transdominant mutant abolishes anchorage-independent growth and macrophage colony-stimulating factor-stimulated invasion by BT20 breast carcinoma cells. Cancer Res. 1998;58:1027-1033. [PubMed] |
| 10. | Sementchenko VI, Schweinfest CW, Papas TS, Watson DK. ETS2 function is required to maintain the transformed state of human prostate cancer cells. Oncogene. 1998;17:2883-2888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 57] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 11. | Zhang W, Bailey-Wilson JE, Li W, Wang X, Zhang C, Mao X, Liu Z, Zhou C, Wu M. Segregation analysis of esophageal cancer in a moderately high-incidence area of northern China. Am J Hum Genet. 2000;67:110-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 56] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 12. | Lu J, Liu Z, Xiong M, Wang Q, Wang X, Yang G, Zhao L, Qiu Z, Zhou C, Wu M. Gene expression profile changes in initiation and progression of squamous cell carcinoma of esophagus. Int J Cancer. 2001;91:288-294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (1)] |
| 13. | Sumarsono SH, Wilson TJ, Tymms MJ, Venter DJ, Corrick CM, Kola R, Lahoud MH, Papas TS, Seth A, Kola I. Down's syndrome-like skeletal abnormalities in Ets2 transgenic mice. Nature. 1996;379:534-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 134] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 14. | Neznanov N, Man AK, Yamamoto H, Hauser CA, Cardiff RD, Oshima RG. A single targeted Ets2 allele restricts development of mammary tumors in transgenic mice. Cancer Res. 1999;59:4242-4246. [PubMed] |
| 15. | Foos G, Hauser CA. Altered Ets transcription factor activity in prostate tumor cells inhibits anchorage-independent growth, survival, and invasiveness. Oncogene. 2000;19:5507-5516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 16. | Pack SD, Karkera JD, Zhuang Z, Pak ED, Balan KV, Hwu P, Park WS, Pham T, Ault DO, Glaser M. Molecular cytogenetic fingerprinting of esophageal squamous cell carcinoma by comparative genomic hybridization reveals a consistent pattern of chromosomal alterations. Genes Chromosomes Cancer. 1999;25:160-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 17. | Yen CC, Chen YJ, Chen JT, Hsia JY, Chen PM, Liu JH, Fan FS, Chiou TJ, Wang WS, Lin CH. Comparative genomic hybridization of esophageal squamous cell carcinoma: correlations between chromosomal aberrations and disease progression/prognosis. Cancer. 2001;92:2769-2777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 18. | Roth MJ, Hu N, Emmert-Buck MR, Wang QH, Dawsey SM, Li G, Guo WJ, Zhang YZ, Taylor PR. Genetic progression and heterogeneity associated with the development of esophageal squamous cell carcinoma. Cancer Res. 2001;61:4098-4104. [PubMed] |
| 19. | Mayama T, Nishihira T, Satomi S, Horii A. [Frequent loss of heterozygosity on the long arm of chromosome 21 in human esophageal, squamous cell carcinoma]. Gan To Kagaku Ryoho. 1998;25 Suppl 3:459-463. [PubMed] |
| 20. | Mayama T, Fukushige S, Shineha R, Nishihira T, Satomi S, Horii A. Frequent loss of copy number on the long arm of chromosome 21 in human esophageal squamous cell carcinoma. Int J Oncol. 2000;17:245-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
Edited By Wu XN