Published online Dec 15, 2003. doi: 10.3748/wjg.v9.i12.2732
Revised: June 13, 2003
Accepted: July 24, 2003
Published online: December 15, 2003
AIM: Human cytochrome P-450 2E1 (CYP2E1) takes part in the biotransformation of ethanol, acetone, many small-molecule substrates and volatile anesthetics. CYP2E1 is involved in chemical activation of many carcinogens, procarcinogens, and toxicants. To assess the metabolic and toxicological characteristics of CYP2E1, we cloned CYP2E1 cDNA and established a HepG2 cell line stably expressing recombinant CYP 2E1.
METHODS: Human CYP2E1 cDNA was amplified with reverse transcription-polymerase chain reaction (RT-PCR) from total RNAs extracted from human liver and cloned into pGEM-T vector. The cDNA segment was identified by DNA sequencing and subcloned into a mammalian expression vector pREP9. A transgenic cell line was established by transfecting the recombinant plasmid of pREP9-CYP2E1 to HepG2 cells. The expression of CYP2E1 mRNA was validated by RT-PCR. The enzyme activity of CYP2E1 catalyzing oxidation of 4-nitrophenol in postmitochondrial supernate (S9) fraction of the cells was determined by spectrophotometry. The metabolic activation of HepG2-CYP2E1 cells was assayed by N-nitrosodiethylamine (NDEA) cytotoxicity and micronucleus test.
RESULTS: The cloned CYP2E1 cDNA segment was identical to that reported by Umeno et al (GenBank access No. J02843). HepG2-CYP2E1 cells expressed CYP2E1 mRNA and had 4-nitrophenol hydroxylase activity (0.162 ± 0.025 nmol·min-1·mg-1 S9 protein), which were undetectable in parent HepG2 cells. HepG2-CYP2E1 cells increased the cytotoxicity and micronucleus rate of NDEA in comparison with those of HepG2 cells.
CONCLUSION: The cDNA of human CYP2E1 can be successfully cloned, and a cell line, HepG2-CYP2E1, which can efficiently express mRNA and has CYP2E1 activity, is established. The cell line is useful for testing the cytotoxicity, mutagenicity and metabolism of xenobiotics, which may possibly be activated or metabolized by CYP2E1.
- Citation: Zhuge J, Luo Y, Yu YN. Heterologous expression of human cytochrome P450 2E1 in HepG2 cell line. World J Gastroenterol 2003; 9(12): 2732-2736
- URL: https://www.wjgnet.com/1007-9327/full/v9/i12/2732.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i12.2732
Cytochrome P450 2E1 (CYP2E1) is the only member of the CYP2E subfamily in humans. Approximately 7% of the liver CYP content consists of CYP2E1, although individual variation of the level of hepatic CYP2E1 expression can be existed by an order of magnitude. CYP2E1 is also expressed in a number of extrahepatic tissues including the lungs[1] and brain[2]. CYP2E1 takes part in the biotransformation of ethanol, acetone, and many small-molecule substrates such as halogenated hydrocarbons (1,1,1-trichlorethane, 1,2-dichloropropane, carbon tetrachloride, chloroform, ethylene dibromide, ethylene dichloride, halothane, methylchloride, methylene dichloride, vinylchloride and trichloroethylene, most of which are hepatotoxic), acetaldehyde, benzene, and styrene. It is known for its ability to metabolize volatile anesthetics such as halothane, enflurane, isoflurane, and sevoflurane, acetaminophen, phenacetin and chlorzoxazone. Another group of CYP2E1 substrates are nitrosamines. CYP2E1 is involved in chemical activation of many carcinogens, procarcinogens, and toxicants[3-5].
Genetically engineered mammalian cells expressing CYP subtypes have provided new tools for investigations of the metabolism and CYP-mediated metabolic activation of chemicals. The stable expression system of CYP in cells has made it possible to evaluate the relative risk of a chemical in toxicological testing in vitro[6,7]. Human CYP1A1[8], CYP2B6[8], CYP2A6[9], CYP3A4[10], CYP2C9[11], CYP2C18[12] and a phase II metabolism enzyme UDP-glucuronosyltransferase, UGT1A9[13] have been stably expressed in Chinese hamster lung CHL cells in our laboratory. Among the human hepatic cell lines, HepG2 derived from a human liver tumor has been characterized to retain many xenobiotic-metabolizing activities as compared to fibroblasts. Therefore, HepG2 cell is useful in prediction of the metabolism and cytotoxicity of chemicals in human liver[14]. But it does not produce significant amounts of CYP[15,16]. Yoshitomi et al[17] have established in HepG2 cells stable expression of a series of human CYP subtypes, such as CYP1A1, CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1 and CYP3A4.
In this study, human CYP2E1 cDNA was amplified by reverse transcription-polymerase chain reaction (RT-PCR), and a transgenic cell line HepG2-CYP2E1 stably expressing CYP2E1 was established to assess the metabolic and toxicological characteristics of CYP2E1.
Restriction endonucleases, Moloney murine leukemia virus (M-MuLV) reverse transcriptase were supplied by MBI Fermentas AB, Lithuania. PCR primers, DNA sequence primers, random hexamer primers, and dNTPs were synthesized or supplied by Shanghai Sangon Biotechnology Corp. Expand fidelity PCR system and NADPH were from Roche Molecular Biochemicals. DNA sequencing kit was purchased from Perkin-Elmer Co. The TRIzol reagent, G418, Dulbecco’s modified Eagle’s medium (DMEM) and newborn bovine calf sera were from Gibco. Diethyl pyrocarbonate (DEPC), MTT, and N-nitrosodiethylamine (NDEA) were purchased from Sigma Chemical Co. T4 DNA ligase and pGEM-T vector system were from Promega. 4-nitrophenol and p-nitrocatechol were from Tokyo Kasei Kogyo Co. Ltd, Japan. Other chemical reagents used were of analytical purity from the commercial sources.
Cloning of human CYP2E1 cDNA from human liver Total RNA was extracted from a surgical specimen of human liver with TRIzol reagent according to the manufacture’s instructions. RT-PCR amplifications were described before, using Expand fidelity PCR system[11]. Two specific 27 mer oligonucleotide PCR primers were designed according to the mRNA sequence of CYP2E1 reported by Song et al[18] (GenBank access No. J02625). The sense primer corresponding to base position -8 to 19 was 5’-AGGGTACCATGTCTGCCCTCGGAGTGA-3’, with a restriction site of Kpn I (underlined). The anti-sense primer, corresponding to the base position from 1507 to 1534, was 5’-ACAATTTGAAAGCTTGTTTGAAAGCGG-3’, with a restriction site of Hind III (underlined). The anticipated PCR product was 1 542 bp in length. PCR was performed at 94 °C for 2 min, then 35 cycles at 94 °C for 60 s, at 62 °C for 60 s, at 72 °C for 2 min, and extension at 72 °C for 10 min. An aliquot (10 μL) from PCR was subjected to electrophoresis in a 1% agarose gel stained with ethidium bromide.
Construction of recombinant pGEM-CYP2E1 and sequencing of CYP2E1 cDNA The PCR products were ligated with a pGEM-T vector, and transformed to E. coli DH5α. CYP2E1 cDNA cloned in pGEM-T was sequenced by Perkin-Elmer-ABI Prism 310 automated DNA sequencer with primers of T7 and SP6 promoters and two specific primers of 2E1 m1 5’-GCATCTCTTGCCTATCCTT-3’ (1042-1024), and 2E1 m2 5’-ATGGACCTACCTGGAAGGACAT-3’ (353-374).
Construction of pREP9 based expression plasmid for CYP2E1Kpn I/Hind III fragment having the total span of human CYP2E1 cDNA and correctly deduced amino acids sequence in pGEM-CYP2E1 were subcloned to a mammalian expression vector pREP9 (Invitrogen). The recombinant was transformed to E. coli Top 10, screened by ampicillin resistant, and identified by restriction mapping.
Transfection and selection[11,19] HepG2 cells grown in DMEM containing 10%(V/V) new born calf sera were transfected with the resultant recombinant plasmid, pREP9-CYP2E1, using a modified calcium phosphate method. The culture was split and then selected in the culture medium containing the neomycin analogue G418 (400 mg·L-1). A transgenic cell line named HepG2-CYP2E1 was established.
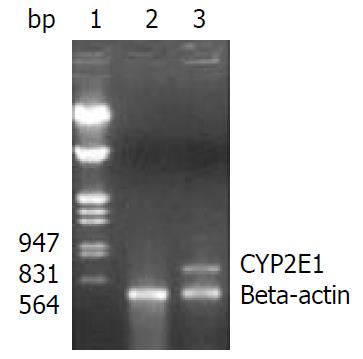
Detection of CYP2E1 mRNA expression by RT-PCR Total RNA was prepared from G-418-resistant clones by TRIzol reagent. RT-PCR was performed as described before[11], using two primers 2E1 m1 and 2E1 m2 (200 nmol·L-1), their sequence was described in above, with 20 nmol·L-1 primers of beta-actin as an internal control. The sense and anti-sense primers used for PCR amplification of beta-actin (GenBank access No. NM_001101) were 5’-TCCCTGGAGAAGAGCTACGA-3’ (776-795) and 5’-CAAGAAAGGGTGTAACGCAAC-3’ (1217-1237), respectively. PCR was performed at 94 °C for 2 min, then 35 cycles at 94 °C for 30 s, at 62 °C for 30 s, at 72 °C for 30 s, and extension at 72 °C for 7 min. The anticipated beta-actin PCR product was 462 bp in length and that of CYP2E1 was 690 bp in length. An aliquot (10 μL) from PCR was subjected to electrophoresis in a 1.5% agarose gel stained with ethidium bromide.
Preparation of postmitochondrial supernate (S9) of HepG2-CYP2E1 The procedure of preparation of S9 fraction was described before[11]. The protein in S9 was determined by the Lowry’s method, with bovine serum albumin as standard.
4-nitrophenol hydroxylase assays[20-22] CYP2E1 4-nitrophenol hydroxylase activity of S9 was determined by spectrophotometry, 0.5 mL incubations contained 0.25 mg S9 protein, 0.2 mmol·L-1 4-nitrophenol in 0.1 mmol·L-1 potassium phosphate buffer (pH 6.8), reactions were initiated with 1 mmol·L-1 NADPH and carried out in air at 37 °C for 60 min. Reactions were terminated by adding 0.25 mL of 0.6 mmol·L-1 perchloric acid and centrifuged at 10000×g for 5 min to remove protein. The supernatant was mixed with 1/10 volume of 10 mol·L-1 NaOH. The absorbance of clarified supernatants was measured at 510 nm, and the amount of product was quantitated using a standard curve generated by adding known amounts of p-nitrocatechol to incubations without NADPH.
Cytotoxicity assay[23,24] HepG2 and HepG2-CYP2E1 cells were seeded in 96 well cell culture plates at a density of 1 × 104 cells each well and incubated overnight. The medium was discarded, and a new medium containing NDEA 0, 1, 10, 100 mmol·L-1 was added to respective wells, with 6 duplications at each concentration. 72 h later, the medium was discarded and 20 μL of 50 g·L-1 MTT in PBS was added to each well. The MTT was discarded 4 h later and 100 μL dimethylsulfoxide was added. After formazan was dissolved, the absorbance was read at 570 nm with 630 nm as reference on the microtiter plate reader. Relative survival was represented as the relative toxicity to the control culture without NDEA. The significance of difference of relative survival between HepG2-CYP2E1 and HepG2 cells was analyzed by Student’s t test.
Micronucleus test[25-28] 1 × 105 HepG2 and HepG2-CYP2E1 cells were seeded in each 100 mL culture bottle and cultured overnight. The medium was discarded and 0, 5, 10, 20 mol·L-1 NDEA in 4 mL serum-free medium was applied to respective bottles and incubated for 4 h. Subsequently the cells were washed twice in PBS and incubated for 20 h with 4 mL completed medium containing 3 mg·L-1 of cytochalasin B. Then the cells were washed twice in PBS and harvested in 2 mL PBS and fixed with 0.4 mL fixed solution (methanol: acetic acid 3:1) for 30 min. The cells were centrifugated, the PBS was discarded and pelleted cells were refixed with 2 mL of fixed solution for 30 min. The cells were centrifugated, refixed and centrifugated again, and then dropped onto slides, dried in the air and stained with 10% Giemsa solution. The scoring criteria for binucleated cells and micronucleus (MN) were based on a report of HUman MicronNucleus project[27]. The frequency of MN formation was expressed as per thousand of binucleated cells with MN. The significance of difference of MN rate between HepG2-CYP2E1 and HepG2 cells was analyzed by checking the tables for determining the statistical significance of mutation frequencies[28].
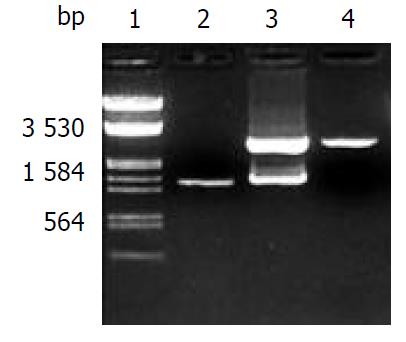
The recombinant of pGEM-CYP2E1 was constructed with the human CYP2E1 cDNA inserted into the cloning site of pGEM-T vector. Selection and identification of the recombinant were carried out by Kpn I/Hind III endonuclease digestion and agarose gel electrophoresis (Figure 1). The cloned cDNA segment was sequenced completely. In comparison with the cDNA sequence reported by Song et al (GenBank access No. J02625), there was only one base difference in cloned cDNA, 105 T→C, while the encoding amino acid was 35G. But there was no sequence difference as compared with that reported by Umeno et al[29] (GenBank access No. J02843).
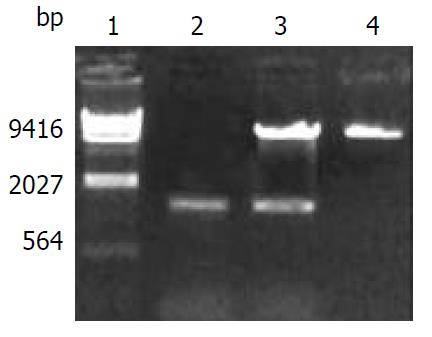
The Kpn I/Hind III fragment (1.54 kb) containing complete CYP2E1 cDNA was subcloned into the Kpn I/Hind III site of mammalian expression vector pREP9. Selection and identification of the recombinant were carried out by Kpn I/ Hind III endonuclease digestion and agarose gel electrophoresis (Figure 2). The resulting plasmid was designated as pREP9-CYP2E1 and contained the entire coding region, along with 8 bp of the 5’and 52 bp of the 3’ untranslation region of CYP2E1 cDNA, respectively.
HepG2 cells were transfected with pREP9-CYP2E1, and selected with G418. The surviving clones were propagated and a cell line termed HepG2-CYP2E1 was established. The expression of CYP2E1 mRNA could be detected in HepG2-CYP2E1 cells but not in HepG2 cells by RT-PCR (Figure 3). The 4-nitrophenol hydroxylase activity in S9 of HepG2-CYP2E1 cells was found 0.162 ± 0.025 nmol·min-1·mg-1 S9 protein (n = 3), but not detectable in parent HepG2 cells.
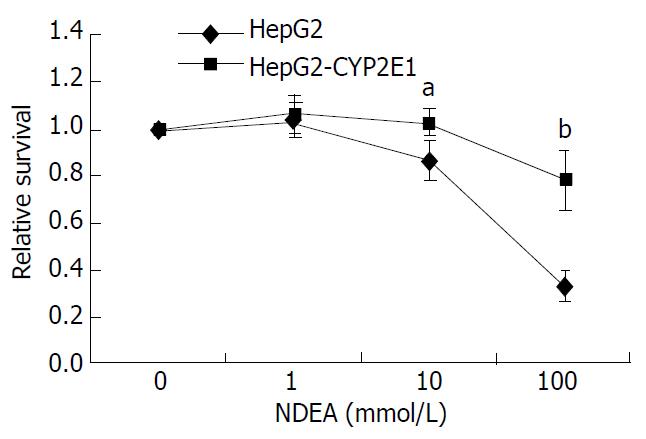
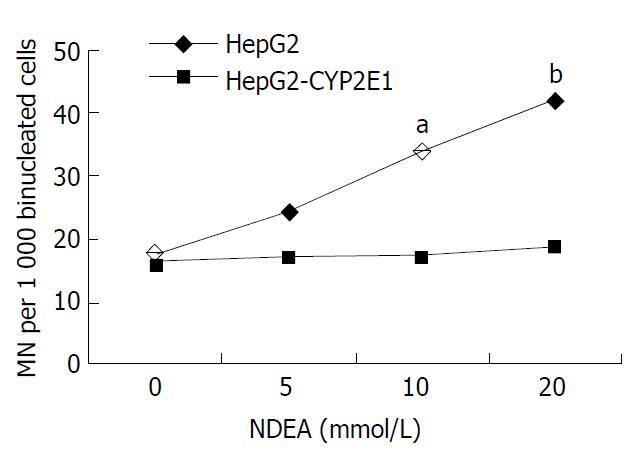
Cells were exposed to various concentrations of NDEA. The relative survival rate of HepG2-CYP2E1 cells was lower than that of HepG2 cells in 10 and 100 mmol·L-1 NDEA (P < 0.05 and P < 0.01, respectively), as shown in Figure 4. The MN rate of HepG2-CYP2E1 was higher than that of HepG2 cells in 10 and 20 mmol·L-1 NDEA (P < 0.05 and P < 0.01 respectively) as shown in Figure 5.
Human CYP2E1 gene is located on the chromosome 10q24.3-qter. Up to date, seven CYP2E1 alleles have been identified (see: CYP2E1 alleles nomenclature at: http://www.imm.ki.se/CYPalleles/cyp2e1.htm). Only 3 alleles have nucleotide substitute, resulting in amino acid change. CYP2E1*1 is the wild type of human CYP2E1. CYP2E1*2 has a 1 168 G→A point mutation in exon 2 causing an R76H amino acid substitution, and CYP2E1*3 has a 10 059 G→A base substitution in exon 8 yielding a V389I amino acid exchange. The corresponding CYP2E1 cDNAs were expressed in COS-1 cells by Hu et al[30]. The cellular levels of CYP2E1 mRNA, protein, and the rate of chlorzoxazone hydroxylation were monitored. CYP2E1*3 cDNA variant was indistinguishable from the wild type cDNA on all variables investigated, whereas CYP2E1*2 cDNA, although yielding similar amounts of mRNA, only caused 37% of the protein expression and 36% of the catalytic activity compared with the wild type cDNA. Complete screening by single-stranded conformation polymorphism of the three populations revealed that these variant alleles were rare. Human CYP2E1 gene was functionally well conserved compared with other CYP enzymes active in drug metabolism, which suggested an important endogenous function in humans[30]. CYP2E1*4 has a 4 804 G→A point mutation in exon 4, resulting in V179I amino acid exchange. No significant difference in kinetic constants for chlorzoxazone hydroxylation between mutant and wild type was observed by expression of the wild type and mutated full length cDNAs in lymphoblastoid cells[31]. Our laboratory has once cloned a CYP2E1 cDNA (GenBank access No. AF182276), which has two point mutations, i.e. 105 T→C, no amino change 35G, and 704G→T, and can result in V235A amino acid exchange. This cDNA was expressed in Chinese hamster lung CHL cells. We could not detect the N-nitrosodimethylamine demethylase activity in transgenic cells (data not shown). According to the homology modelling of human CYP2E1 based on the CYP2C5 crystal structure, the substrate recognition site (SRS) 1 was located at codon 100-118, SRS 2 at 200-211, SRS3 at 236-241, SRS4 at 291-305, SRS5 at 361-370, SRS6 at 470-480. The point mutations of CYP2E1*2, *3, *4 were not located on the SRS. The V235A amino acid exchange in our formerly cloned CYP2E1 was just at the front of SRS3. This might influence SRS3 and reduce the enzyme activity. Fortunately, this time we cloned a wild type CYP2E1.
It has been found that polymorphism of CYP2E1 gene is significant for inter-individual differences in toxicity of its substrates[33], and has some effect on the development of gastric cancer[34] and colorectal cancer[35].
The expression of CYP2E1 mRNA in HepG2 cells was validated by RT-PCR. The commonly used CYP2E1 probe substrates were chlorzoxazone[36,37] and 4-nitrophenol[38]. In this research, we used 4-nitrophenol 2-hydroxylase activity to evaluate the expression of CYP2E1, and the 4-nitrophenol hydroxylase activity of HepG2-CYP2E1 cells was found to be 0.162 ± 0.025 nmol·min-1·mg-1 S9 protein, a little lower than those of HepG2-CYP2E1 E43 and E47 cells (0.19 and 0.34 nmol·min-1·mg-1 of microsome, respectively)[39], much lower than that of human liver (1.91 ± 0.28 nmol·min-1·mg-1of microsome)[40].
The most frequently used genotoxicity test in mammals is the micronucleus test, which provides a simple and rapid indirect measure of induced structural and numerical chromosome aberrations and is a scientific and regulatory assay accepted by supranational authorities such as the Organization for Economic Cooperation and Development (OECD), International Conference on Harmonization (ICH) and European Union (EU). NDEA could induce early experimental hepatocellular carcinomas[41] and esophageal neoplasms[42]. The metabolic activation of NDEA was mediated mainly by CYP2A6 and CYP2E1[43]. This study has shown that NDEA can decrease the relative survival rate of HepG2-CYP2E1 cells and increase the MN rate in binucleated cells as compared with HepG2 cells.
cDNA of human CYP2E1 was successfully cloned and a cell line, HepG2-CYP2E1, efficiently expressing mRNA and having the CYP2E1 enzymatic activities, was established. The cell line is useful for testing the cytotoxicity, mutagenicity and metabolism of xenobiotics and drugs, which may possibly be activated by CYP2E1.
| 1. | Hukkanen J, Pelkonen O, Hakkola J, Raunio H. Expression and regulation of xenobiotic-metabolizing cytochrome P450 (CYP) enzymes in human lung. Crit Rev Toxicol. 2002;32:391-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 235] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 2. | Upadhya SC, Tirumalai PS, Boyd MR, Mori T, Ravindranath V. Cytochrome P4502E (CYP2E) in brain: constitutive expression, induction by ethanol and localization by fluorescence in situ hybridization. Arch Biochem Biophys. 2000;373:23-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 95] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 3. | Lieber CS. Cytochrome P-4502E1: its physiological and pathological role. Physiol Rev. 1997;77:517-544. [PubMed] |
| 4. | Tanaka E, Terada M, Misawa S. Cytochrome P450 2E1: its clinical and toxicological role. J Clin Pharm Ther. 2000;25:165-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 145] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 5. | Anzenbacher P, Anzenbacherová E. Cytochromes P450 and metabolism of xenobiotics. Cell Mol Life Sci. 2001;58:737-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 686] [Cited by in RCA: 648] [Article Influence: 25.9] [Reference Citation Analysis (1)] |
| 6. | Sawada M, Kamataki T. Genetically engineered cells stably expressing cytochrome P450 and their application to mutagen assays. Mutat Res. 1998;411:19-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 7. | Crespi CL, Miller VP. The use of heterologously expressed drug metabolizing enzymes--state of the art and prospects for the future. Pharmacol Ther. 1999;84:121-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 107] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 8. | Wu J, Dong H, Cai Z, Yu Y. Stable expression of human cytochrome CYP2B6 and CYP1A1 in Chinese hamster CHL cells: their use in micronucleus assays. Chin Med Sci J. 1997;12:148-155. [PubMed] |
| 9. | Yan LQ, Yu YN, Zhuge J, Xie HY. Cloning of human cytochrome P450 2A6 cDNA and its expression in mammalian cells. Zhongguo Yaolixue Yu Dulixue Zazhi. 2000;14:31-35. |
| 10. | Chen Q, Wu J, Yu Y. [Establishment of transgenic cell line CHL-3A4 and its metabolic activation]. Zhonghua Yufang Yixue Zazhi. 1998;32:281-284. [PubMed] |
| 11. | Zhu GJ, Yu YN, Li X, Qian YL. Cloning of cytochrome P-450 2C9 cDNA from human liver and its expression in CHL cells. World J Gastroenterol. 2002;8:318-322. [PubMed] |
| 12. | Zhu-Ge J, Yu YN, Qian YL, Li X. Establishment of a transgenic cell line stably expressing human cytochrome P450 2C18 and identification of a CYP2C18 clone with exon 5 missing. World J Gastroenterol. 2002;8:888-892. [PubMed] |
| 13. | Li X, Yu YN, Zhu GJ, Qian YL. Cloning of UGT1A9 cDNA from liver tissues and its expression in CHL cells. World J Gastroenterol. 2001;7:841-845. [PubMed] |
| 14. | Rueff J, Chiapella C, Chipman JK, Darroudi F, Silva ID, Duverger-van Bogaert M, Fonti E, Glatt HR, Isern P, Laires A. Development and validation of alternative metabolic systems for mutagenicity testing in short-term assays. Mutat Res. 1996;353:151-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 46] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 15. | Rodríguez-Antona C, Donato MT, Boobis A, Edwards RJ, Watts PS, Castell JV, Gómez-Lechón MJ. Cytochrome P450 expression in human hepatocytes and hepatoma cell lines: molecular mechanisms that determine lower expression in cultured cells. Xenobiotica. 2002;32:505-520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 298] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 16. | Jover R, Bort R, Gómez-Lechón MJ, Castell JV. Cytochrome P450 regulation by hepatocyte nuclear factor 4 in human hepatocytes: a study using adenovirus-mediated antisense targeting. Hepatology. 2001;33:668-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 167] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 17. | Yoshitomi S, Ikemoto K, Takahashi J, Miki H, Namba M, Asahi S. Establishment of the transformants expressing human cytochrome P450 subtypes in HepG2, and their applications on drug metabolism and toxicology. Toxicol In Vitro. 2001;15:245-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 85] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 18. | Song BJ, Gelboin HV, Park SS, Yang CS, Gonzalez FJ. Complementary DNA and protein sequences of ethanol-inducible rat and human cytochrome P-450s. Transcriptional and post-transcriptional regulation of the rat enzyme. J Biol Chem. 1986;261:16689-16697. [PubMed] |
| 19. | Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, A Labo-ratory Manual. 2nd ed. New York: Cold Spring Harbor Laboratory Press. 1989;6.28-6.29. |
| 20. | Lin HL, Roberts ES, Hollenberg PF. Heterologous expression of rat P450 2E1 in a mammalian cell line: in situ metabolism and cytotoxicity of N-nitrosodimethylamine. Carcinogenesis. 1998;19:321-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 0.7] [Reference Citation Analysis (1)] |
| 21. | Koop DR. Hydroxylation of p-nitrophenol by rabbit ethanol-inducible cytochrome P-450 isozyme 3a. Mol Pharmacol. 1986;29:399-404. [PubMed] |
| 22. | Tassaneeyakul W, Veronese ME, Birkett DJ, Gonzalez FJ, Miners JO. Validation of 4-nitrophenol as an in vitro substrate probe for human liver CYP2E1 using cDNA expression and microsomal kinetic techniques. Biochem Pharmacol. 1993;46:1975-1981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 87] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 23. | Zhuge J, Wang LR, Bi AH, Liu GH. Investigation of the role of Interleukin-2 and soluble Interleukin-2 receptor in the pathogen-esis of asthma. Mianyixue Zazhi. 1994;10:242-244. |
| 24. | Moskatelo D, Benjak A, Laketa V, Polanc S, Kosmrlj J, Osmak M. Cytotoxic effects of diazenes on tumor cells in vitro. Chemotherapy. 2002;48:36-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 25. | Garriott ML, Phelps JB, Hoffman WP. A protocol for the in vitro micronucleus test. I. Contributions to the development of a protocol suitable for regulatory submissions from an examination of 16 chemicals with different mechanisms of action and different levels of activity. Mutat Res. 2002;517:123-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 47] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 26. | Erexson GL, Periago MV, Spicer CS. Differential sensitivity of Chinese hamster V79 and Chinese hamster ovary (CHO) cells in the in vitro micronucleus screening assay. Mutat Res. 2001;495:75-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 27. | Fenech M, Chang WP, Kirsch-Volders M, Holland N, Bonassi S, Zeiger E. HUMN project: detailed description of the scoring criteria for the cytokinesis-block micronucleus assay using isolated human lymphocyte cultures. Mutat Res. 2003;534:65-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 951] [Cited by in RCA: 962] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 28. | Kastenbaum MA, Bowman KO. Tables for determining the statistical significance of mutation frequencies. Mutat Res. 1970;9:527-549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 779] [Cited by in RCA: 716] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 29. | Umeno M, McBride OW, Yang CS, Gelboin HV, Gonzalez FJ. Human ethanol-inducible P450IIE1: complete gene sequence, promoter characterization, chromosome mapping, and cDNA-directed expression. Biochemistry. 1988;27:9006-9013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 133] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 30. | Hu Y, Oscarson M, Johansson I, Yue QY, Dahl ML, Tabone M, Arincò S, Albano E, Ingelman-Sundberg M. Genetic polymorphism of human CYP2E1: characterization of two variant alleles. Mol Pharmacol. 1997;51:370-376. [PubMed] |
| 31. | Fairbrother KS, Grove J, de Waziers I, Steimel DT, Day CP, Crespi CL, Daly AK. Detection and characterization of novel polymorphisms in the CYP2E1 gene. Pharmacogenetics. 1998;8:543-552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 70] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 32. | Lewis DF, Lake BG, Bird MG, Loizou GD, Dickins M, Goldfarb PS. Homology modelling of human CYP2E1 based on the CYP2C5 crystal structure: investigation of enzyme-substrate and enzyme-inhibitor interactions. Toxicol In Vitro. 2003;17:93-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 33. | Bolt HM, Roos PH, Thier R. The cytochrome P-450 isoenzyme CYP2E1 in the biological processing of industrial chemicals: consequences for occupational and environmental medicine. Int Arch Occup Environ Health. 2003;76:174-185. [PubMed] |
| 34. | Cai L, Yu SZ, Zhan ZF. Cytochrome P450 2E1 genetic polymorphism and gastric cancer in Changle, Fujian Province. World J Gastroenterol. 2001;7:792-795. [PubMed] |
| 35. | Le Marchand L, Donlon T, Seifried A, Wilkens LR. Red meat intake, CYP2E1 genetic polymorphisms, and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1019-1024. [PubMed] |
| 36. | Lucas D, Ferrara R, Gonzalez E, Bodenez P, Albores A, Manno M, Berthou F. Chlorzoxazone, a selective probe for phenotyping CYP2E1 in humans. Pharmacogenetics. 1999;9:377-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 71] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 37. | Yuan R, Madani S, Wei XX, Reynolds K, Huang SM. Evaluation of cytochrome P450 probe substrates commonly used by the pharmaceutical industry to study in vitro drug interactions. Drug Metab Dispos. 2002;30:1311-1319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 254] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 38. | Spatzenegger M, Liu H, Wang Q, Debarber A, Koop DR, Halpert JR. Analysis of differential substrate selectivities of CYP2B6 and CYP2E1 by site-directed mutagenesis and molecular modeling. J Pharmacol Exp Ther. 2003;304:477-487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 62] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 39. | Chen Q, Cederbaum AI. Cytotoxicity and apoptosis produced by cytochrome P450 2E1 in Hep G2 cells. Mol Pharmacol. 1998;53:638-648. [PubMed] |
| 40. | Adas F, Berthou F, Salaün JP, Dréano Y, Amet Y. Interspecies variations in fatty acid hydroxylations involving cytochromes P450 2E1 and 4A. Toxicol Lett. 1999;110:43-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 26] [Article Influence: 1.0] [Reference Citation Analysis (1)] |
| 41. | Wang Z, Ruan YB, Guan Y, Liu SH. Expression of IGF-II in early experimental hepatocellular carcinomas and its significance in early diagnosis. World J Gastroenterol. 2003;9:267-270. [PubMed] |
| 42. | Waddell WJ. Threshold for carcinogenicity of N-nitrosodiethylamine for esophageal tumors in rats. Food Chem Toxicol. 2003;41:739-741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 43. | Fujita K, Kamataki T. Predicting the mutagenicity of tobacco-related N-nitrosamines in humans using 11 strains of Salmonella typhimurium YG7108, each coexpressing a form of human cytochrome P450 along with NADPH-cytochrome P450 reductase. Environ Mol Mutagen. 2001;38:339-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 72] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
Edited by Zhang JZ and Wang XL