Published online Dec 15, 2001. doi: 10.3748/wjg.v7.i6.841
Revised: September 19, 2001
Accepted: October 20, 2001
Published online: December 15, 2001
AIM: To clone the cDNA of UGT1A9 from a Chinese human liver and establish the Chinese hamster lung (CHL) cell line expressing human UGT1A9.
METHODS: cDNA of UGT1A9 was transcripted from mRNA by reverse transcriptase-ploymerase chain reaction, and was cloned into the pGEM-T vector which was amplified in the host bacteric E. coli DH5α. The inserted fragment, verified by DNA sequencing, was subcloned into the Hind III/Not I site of a mammalian expression vector pREP9 to construct the plasmid termed pREP9-UGT1A9. CHL cells were transfected with the resultant recombinants, pREP9-UGT1A9, and selected by G418 (400 mg•L¯¹) for one month. The surviving clone (CHL-UGT1A9) was harvested as a pool and sub-cultured in medium containing G418 to obtain samples for UGT1A9 assays. The enzyme activity of CHL-UGT1A9 towards propranolol in S9 protein of the cell was determined by HPL C.
RESULTS: The sequence of the cDNA segment cloned, which was 1666 bp in length, was id entical to that released by GeneBank (GenBank accession number: AF056188) in co ding region. The recombinant constructed, pREP9-UGT1A9, contains the entire coding region, along with 18 bp of the 5’ and 55 bp of the 3’ untranslated region of the UGT1A9 cDNA, respectively. The cell lines established expressed the protein of UGT1A9, and the enzyme activity towards propranolol in S9 protein was found to be 101 ± 24 pmol•min-1•mg-1 protein (n = 3), but was not detectable in parental CHL cells.
CONCLUSION: The cDNA of UGT1A9 was successfully cloned from a Chinese human liver and transfected into CHL cells. The CHL-UGT1A9 cell lines established efficiently expressed the protein of UGT1A9 for the further enzyme study of drug glucuronidation.
- Citation: Li X, Yu YN, Zhu GJ, Qian YL. Cloning of UGT1A9 cDNA from liver tissues and its expression in CHL cells. World J Gastroenterol 2001; 7(6): 841-845
- URL: https://www.wjgnet.com/1007-9327/full/v7/i6/841.htm
- DOI: https://dx.doi.org/10.3748/wjg.v7.i6.841
Most organisms are exposed to a range of lipophilic compounds and converted them into excretable hydrophilic compounds. This metabolism of foreign compounds (xenobiotics) can be divided into two phases. For phase I metabolism, a reactive group is mostly introduced into the xenobiotic molecule. These reactions are mainly catalyzed by the cytochrome P450 monooxygenase system which consists of cytochrome P450s (CYPs) and cytochrome P450 reductase (CPR). For phase II metabolism, the reactive metabolite is conjugated to small, hydrophilic endogenous molecules such as glucuronic acid. The conjugation of this cofactor to xenobiotics is catalyzed by UDP-glucuronosyltransferases (UGTs). Since xenobiotic metabolizing enzymes have to catalyze the metabolism of structurally very diverse substrates, the various enzyme systems (e.g. CYPs and UGTs) comprise several isozymes that differ in their catalytic properties. The members of a given enzymesystem have been grouped into families and subfamilies based on sequence homologies. In UGTs, two enzyme families termed UGT1 and UGT2 have been described.
The UGT1 locus is highly conserved between species[1]. UGT1A is a subfamily of U GT1 gene complex that is located at chromosome 2q37. UGT1A subfamily is encoded by tandem individual promoters and their first exons are linked by differential splicing to four common exons. As one of the isoforms, UGT1A9, is mainly expressed in liver. UGT1A9 can be induced by polycyclic aromatic hydrocarbons (PAHs), and therefore the drug glucuronidation catalyzed by UGT1A9 will be increased in cigarette smokers who inhale PAHs[2].
Human hepatic UDP-glucuronosyltransferases (UGT) is a family of microsomal enzymes that catalyze the glucuronidation of many important drugs, xenobiotics and endogenous compounds. Attempts to characterize the microsomal enzymes by conventional purification technique are often frustrated due to its instability. UGT isoenzyme expressed by cells is a useful toolfor characterizing UGT’s function. The cDNA cloning of UGTs from various sources (rabbit, rat, monkey, human beings, etc.) and their expression in cell lines were widely used for the gene characterization and function study of UGT isoforms[3-9]. In order to study tie drug metabolisms by UGTs, the cDNA encoding UGT1A9 was cloned from human liver and expressed in Chinese hamster lung (CHL) cell line in this study. The enzyme expressed was extracted and its activity was assayed with a substrate of propranolol which is a nonselective β-adrenergic blocking agent and can be used widely clinically[10].
Human liver tissue was obtained from a surgical specimen of Chinese and stored at -80 °C until use. The total RNA was isolated with TRIzol reagent (Gibco Corp, USA)
cDNA was transcr ipted from mRNA by revere transcriptase polymerase chain reaction (RT-PCR). Five μg of the total RNA and 2 μg of random primer (SANGON, Shanghai) in deionized water containing DEPC (1 g•L¯¹) were denatured at 65 °C for 15 min, then 4 μL 5 × reverse transcriptase buffer, 3 μL 10 mmol•L¯¹ dNTP, 1 μL M-MuLV reverse transcriptase (200 U) (Fermentas) and essential deionized water containing DEPC (1 g•L¯¹) were added to have the total volume of 20 μL. The reaction was performed at 25 °C for 10 min, then 42 °C for 1 h, and 70 °C for 10 min to inactivate the reverse transcriptase. The product was finally held at 4 °C. Two μL of the reactant was mixed with 2 μL of 10 mmol•L¯¹ dNTP, 30 pmol of PCR primers and 3.5 U of DNA ploymerase (Perkin-Elmer Corp). The total volume of 100 μL was reached by adding deionized water. Two 26 mer oligonucleotides as PCR primers were designed according to the DNA sequence of UGT1A9 (GenBank accession no. AF056188). The sense oligonucleo tides corresponding to base positions 1 to 26 was 5’-CTAAGCTTCAGTTCTCTGATGGCTTG-3’ with a restriction site of Hind III, and the anti-sense one, corresponding to the bases position from 1641 to 1666, was 5’-GTTGGAAATGCCTAGGGAATGGTTC-3’. The poly merase chain reaction (PCR) was performed at 94 °C 2 min, then 94 °C 15 s, 60.1 °C 30 s and 72 °C 2 min for 31 cycles, and 72 °C for 10 min. The product was finally held at 4 °C. An agrose gel electrophoresis was carried out with 10 μL of the P CR solution to check the 1666 bp DNA amplified.
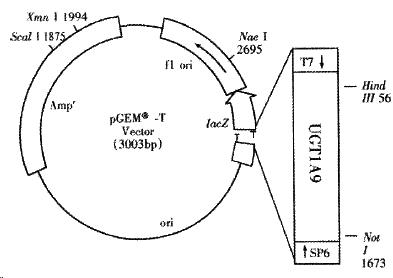
The PCR product of about 1.5 kb was isolated and ligated with pGEM-T (Promega) vector by T4 DNA ligase (Fermentas). E. coli DH5a was transformed with the resulted recombinants pGEM-UGT1A9[11] and the positive bacteria colonies were screened by ampicillin resistant and blue-white screening with X-gal and IPTG. The cDNA of UGT1A9 subcloned in pGEM-T was sequenced on both strands by dideoxy chain-termination method marked with BigDye with primers of T7 and SP6 promoters and a specific primer of 5’-CAAGTATCGTGTTGTTCGC-3’. The termination products were resolved and detected using an automated DNA sequencer (Perkin-Elmer-ABI Prism 310, Foster City, CA).
The Hind III-Not I fragment of the human UGT1A9 cDNA cleaved from the selected and amplified recombinant pGEM-UGT1A9 by Hind III and Not I digestion was purified by agarose electrophoresis and cloned directly into a unique Hind III-Not I site within the multicloning site of the mammalian expression vector pREP9 (Invitrogen, San Diego, CA) with T4 ligase.
Chinese hamster lung (CHL) cells were transfected with the resultant recombinants, pREP9-UGT1A9, using a calcium phosphate method[12]. After 24 h incubation at 37 °C, the culture was rinsed and re-fed with fresh growth medium. Seventy-two hours after transfection, the culture was split and then selected in the culture medium containing the neomycin analogue G418 (Gibco BRL, MD) (400 mg•L¯¹). The selective medium was changed every 3-4 d to remove dead cells andallow the growth of resistant colonies. After 1 mo, surviving clonies (termed C HL-UGT1A9) were harvested as a pool and propagated in medium containing G418.
CHL-UGT1A9 cells grown in the culture medium containing G418 (400 mg•L¯¹) were rinsed with phosphate balanced solution (PBS), scraped and collected from the bottle with 11.5 g•L¯¹ KCl in aqua solution and then sonicated 3 s for 5 times with 5 s of interval break. The resulted homogenate was centrifuged at 9000 × g for 20 min and the supernatant (S9) was transferred carefully to a clean tube for assay or storage under-70 °C. The protein in S9 was determined by the same method that was used in our previous paper[13].
The UGT1A9 activities of S9 fraction were determined by the glucuronidation of propranolol. The assay was performed in a total volume of 100 μl containing final concentrations of 0.2 mmol•L¯¹ propranolol, 1 mmol·L-1 UDPGA, 1 g •L¯¹ Triton X-100, 50 μg of S9 protein in 50 mmol·L-1 Tris-HCl, 10 mmol•L¯¹ MgCl2 buffer, pH7.8 at 37 °C. The mixtures were pre-incubated and the glucuronidation was started by the addition of UDPGA and stopped after 2 h by the addition of 100 μL of methanol. The mixtures were stirred thoroughly and centrifuged a t 10000 r•min-1 for 10 min. Un-reacted propranolol in the layer of reactant was determined by HPLC and the enzyme activity was calculated according to the amount of propranolol declined after incubation.
The concentration of propranolol metabolized by S9 of CHL-UGT1A9 was assayed by the HPLC procedure[13] with modification to the mobile phase. Twenty mL of the sample was applied to a reversed phase column (Shim-pack CLC-ODS 15 cm ± 0.6 cm id, 10 μm particle size). Propranolol was monitored with a UV detector at 290 nm. The mobile phase is made up with ammonium acetate buffer (4.0 g ammonium acetate, 10 mL acetate acid and de-ionized water in 1 L)-methanol-acetonitrile (2:1:1), and to 500 mL mobile phase add 0.7 mL triethylamine as the elution modifier. The flow rate is 1.0 mL•min-1.
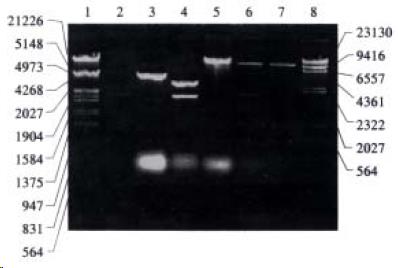
The recombinant of pGEM-UGT1A9 (Figure 1) was constructed with the human UGT1A9 inserted into the cloning site of vector pGEM-T between the promoters of T7 and SP6. Selection and identification of the recombinant was carried out by Hind III/Not I endonuclease digestion and agarose elector phoresis (Figure 2). The cloned DNA segments in selected recombinants were sequenced completely. According to the results of DNA sequencing, the cDNA in a selected recombinant was identical to the DNA sequence of UGT1A9 reported by CiottiM et al[8] (GenBank accession NO. AF056188) in the reading frame. The restriction sites of Hind III and Not I in the recombinant were used for the subcloning of insertion fragment into an expression vector.
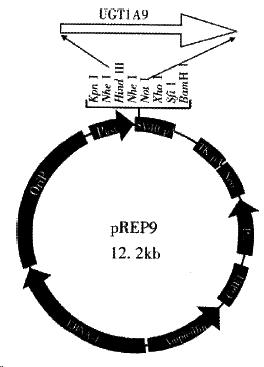
The Hind III/Not I fragment (1.5 kb) containing the complete UGT1A9 cDNA was subcloned into the Hind III/Not I site of mammalian expression vector pRE P9 (Figure 3). Selection and identification of the recombinants were carried out by Hind III/Not I endonuclease digestion and agarose electrophoresis (Figure 3). The resulting plasmid was designated as pREP9-UGT1A9 which contained the entire coding region, along with 18 bp of the 5’ and 55 bp of the 3’ untranslated region of the UGT1A9 cDNA, respectively. In addition, the neo gene of the plasmid confers the G418 resistant phenotype to CHL cells for the selection of transfected cells.
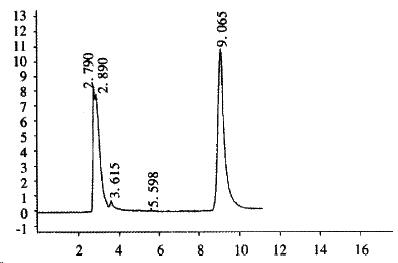
CHL cells were transfected with pREP9-UGT1A9, and selected with G418 (400 mg•L¯¹). The surviving clone was propagated and the cell line termed CHL-UGT1A9 was established. The preparation S9 was prepared from CHL-UGT1A9 cells harvested for UGT1A9 activity assay by HPLC. Figure 4 shows the typical elution of propranolol in incubation solution. The UGT enzyme activity towards propranolol in S9 protein was found to be 101 ± 24 pmol•min-1•mg-1 (n = 3), but was not detectable in parental CHL cells.
UGTs are involved in the conjugation of UDP-glucuronic acid (UDPGA) to a variety of chemicals, drugs, and endogenous compounds. The elimination of hydrophobic chemicals from cells is aided by their conversion to water-soluble glucuronides. UGTs are closely relatied to the system of cytochrome P450 monooxygenase, and involved in the transportation of carrier and the passage of drugs through cell phospholipid bilayer. In most cases, the lipophilic compounds are converted by phase I metabolism to the substrate for glucuronidation by obtaining an essential function (such as carbon, nitrogen, sulfur and oxygen), but in many cases xenobiotics and endogenous substances can also be glucuronidated by UGTs without the phase I metabolism. The xenobiotic metabolizing cytochrome P450 monooxygenase system and the UGTs reside mainly in the endoplasmic reticulum. However, CYPs and the CPR are localized on the cytosolic side of the endoplasmic reticulum, which the UGTs are localized on its luminal side[14]. UGTs are latent enzymes, needing activation (in general by detergents) to express its maximal activity.
Numerous reports revealed that glucuronidation is a major pathway involved in the metabolism of drugs, exogeno us, and numerous endogenous compounds such as bile acids and steroid hormones. E ach UGTs family or subfamily has its own substrates but the substrate spectrum are partly overlapped. UGT1 has substrates such as thyroid hormone[15], SN-38[16], bilirubin[17,18], opioids, bile acids, fatty acids, retinoids, ciprofibrate, furosemide, dilunisa, catechol estrogens, coumarins, flavonoids, anthraquinones, EM-652 (an active antiestrogen)[19] and phenolic compounds[20]. UGT2 catalyzes substrates such as estrogens, androgens, morphine, AZT, and retinoic acid, epirubicin[16,21,22,23], etc. UGT1A9 is a member of UGT1A subfamily. The endogenous substrates for UGT1A9 are estrone, 4-hydroxyestrone, ethinylestradiol, retinoic acids, etc., and exogenous substrates include propofol, propranolol, paracetamol, S-naproxen, ketoprofen, ibuprofen, entacapone, some mutagenic arylamines, etc.[2,24-26]. UGT1A9 was found to have regioselectivity on the glucuronidation of hydroxyl group of carbohydrate-containing drugs[27].
UGTs are expressed extensively in organs and tissues, and they may play a key role in the regulation of the level and action of steroid hormones in steroid target tissues. Organs that express UGTs include liver, kidney, gastrointestinal tract[28-29], olfactory [30], jejunum, ileum[31], prostate[32-33], colon[34]. UGT1A9 is mainly expressed in liver, and also expressedin steroid targets[35] and colon[34].
UGTs are inducible enzymes. In most cases this induction is due to increased transcription of the corresponding genes but sometimes it is also due to an improved stability of proteins. The pattern of enzymes affected is dependent on the inducing agent. Usually, phenobarbital induces mainly enzymes within UGT2 family, and methylcholanthrene induces enzymes belonging to the UGT1 family[28]. Other chemicals that induce UGTs include aryl hydrocarbon receptor ligands or oltipraz[36], flavonoid chrysin[37], and t-butylhydroquinone and 2,3,7,8-tetrachlorodibenzo-p-dioxin[38], etc. UGT1A9 can be induced by polycyclic aromatic hydro carbons (PAHs)[39]. On the other hand, UGTs can also be inhibited, for example by uridine diphosphate[40], and N-glycosylation is involved in the functional properties of UDP-glucuronosyltransferase enzymes[41].
To clone and express UGTs in cells can help screen substrates that an isoenzyme is responsible. The production of a UGT enzyme protein using transgenic cell lines is a practical manner to study its function[42-43]. We report here the cloning of UGT1A9 cDNA and establishment of a CHL cell line expressing UGT1A9 from a Chinese human liver. The full-length cDNA, UGT1A9, that encodes for a human UDP-glucuronosyltransf erase protein, was isolated from a Chinese human liver total RNA. To achieve high expression levels of UGT1A9, the UGT1A9 cDNA was cloned into the eukaryotic expression vector pREP9, which we had previously used in this laboratory for the express ion of human CYP450 1A1, 2B6, 3A4, etc. in CHL cells[44-45]. The salient feature of this vector has an EBV origin of replication and nuclear antigen (EBNA-1) to allow high-copy episomal replication in mammal cell lines. The Rous sarcoma virus long terminal repeat (RSV LTR) early promoter controls the expression of the U GT1A9 cDNA. As noted under “Results", the isolated clone contains a 1592-nucleotide open reading frame flanked by 18 and 55 base pairs of 5’ and 3’ noncoding sequences, respectively. The DNA sequence in the reading code frame is identical to that reported (GenBank accession no. AF056188). The expression of a protein that catalyzed the glucuronidation of propranolol was proven in the Chinese hamster lung cells transfected with the recombinant plasmid pREP9-UGT1A9.
Conjugation with glucuronic acid is an important biotransformation pathway for a large number of clinically used drugs. In human intestinal, UGTs play an important role in the detoxification of xenobiotics compounds and, in some cases, may limit the bioavailability of therapeutic agents[20]. The deficient of a UGTs isoenzyme, m ay cause disease and clinical incident[46-47], the typical example was serious adverse events associated with chloramphenicol toxicity in neonates. Human UGTs are regulated in cases of healthy condition and exposure of harmful environmental carcinogens[48-50]. Moreover, UGT was identified as an antigenic target in a subgroup of liver- kidney microsomal auto-antibodies[51]. Hence, it is very necessary to undertake the study of functions and characteristics of UGTs. Over the last decade, some research papers were published about the usage of cloned and expressed human UGTs for the assessment of human drug conjugations and identification potential drug interactions[6-8]. However, the information gap still exists regarding the enzymatic aspects of UGTs to drugs elimination and its potential impact on therapy. More researches on the drug metabolism by UGTs are necessary for effective translation of scientific information into clinically applicable knowledge. As has been shown with the CYPs, coupling of basic and clinical science is needed to continually improve our understanding of the UGTs. Many factors are known to influence the activities of UGTs involved in drug metabolism, hence plasma clearances of glucuronidated drugs. Such factors include age (especially neonatal period), cigarette smoking, diet, certain disease states, drug therapy, ethnicity, genetics and hormonal effects. Knowledge of the profile, substrate specificities and regulation of human UGTs remains limited and consequently it is still generally not possible to predict the effects of specific environmental and genetic factors on the metabolism and pharmacokinetics of individual glucuronidated drugs. Future investigations must define the substrate specificities of the various UGTs and investigate mechanisms by which the separate isozymes are regulated. Only then will it become possible to rationalize (and predict) the alterations in pharmacokinetics and response to glucuronidated drugs in specific patient groups.
| 1. | Li Q, Lamb G, Tukey RH. Characterization of the UDP-glucuronosyltransferase 1A locus in lagomorphs: evidence for duplication of the UGT1A6 gene. Mol Pharmacol. 2000;58:89-97. [PubMed] |
| 2. | Bock KW, Gschaidmeier H, Heel H, Lehmköster T, Münzel PA, Bock-Hennig BS. Functions and transcriptional regulation of PAH-inducible human UDP-glucuronosyltransferases. Drug Metab Rev. 1999;31:411-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 38] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 3. | Barbier O, Bélanger A, Hum DW. Cloning and characterization of a simian UDP-glucuronosyltransferase enzyme UGT2B20, a novel C19 steroid-conjugating protein. Biochem J. 1999;337:567-574. [PubMed] |
| 4. | Jedlitschky G, Cassidy AJ, Sales M, Pratt N, Burchell B. Cloning and characterization of a novel human olfactory UDP-glucuronosyltransferase. Biochem J. 1999;340:837-843. [PubMed] |
| 5. | Cheng Z, Radominska-Pandya A, Tephly TR. Cloning and expression of human UDP-glucuronosyltransferase (UGT) 1A8. Arch Biochem Biophys. 1998;356:301-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 87] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 6. | Ethell BT, Beaumont K, Rance DJ, Burchell B. Use of cloned and expressed human UDP-glucuronosyltransferases for the assessment of human drug conjugation and identification of potential drug interactions. Drug Metab Dispos. 2001;29:48-53. [PubMed] |
| 7. | Lautala P, Ethell BT, Taskinen J, Burchell B. The specificity of glucuronidation of entacapone and tolcapone by recombinant human UDP-glucuronosyltransferases. Drug Metab Dispos. 2000;28:1385-1389. [PubMed] |
| 8. | Ciotti M, Lakshmi VM, Basu N, Davis BB, Owens IS, Zenser TV. Glucuronidation of benzidine and its metabolites by cDNA-expressed human UDP-glucuronosyltransferases and pH stability of glucuronides. Carcinogenesis. 1999;20:1963-1969. [PubMed] |
| 9. | Ren Q, Murphy SE, Zheng Z, Lazarus P. O-Glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) by human UDP-glucuronosyltransferases 2B7 and 1A9. Drug Metab Dispos. 2000;28:1352-1360. [PubMed] |
| 10. | Li XS, Shen DM, Zou JZ, Liu CA, Zhang L. Low dose propranolol in combination with ligustrazine for prevention of recurrent esophageal varices bleeding: a randomly controlled experimental and clinical study. Shijie Huaren Xiaohua Zazhi. 2000;8:135-138. |
| 11. | Jin DY, Li MF. Molecular Cloning, A Laboratory Manual. 2nd ed. Translating from: Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, A L aboratory Manual. 2nd ed. New York: Cold Spring Harbor Laboratory Press. Beijing: Science Press 1999; 34-74. |
| 12. | Jin DY, Li MF. Molecular Cloning, A Laboratory Manual. 2nd ed. Translating from: Sambrook J, Fritsch EF, Maniatis T. Molecul ar Clo ning, A Laboratory Manual. 2nd ed. New York: Cold Spring Harbor Laboratory Press. Beijing: Science Press 1999; 792-793. |
| 13. | Li X, Zeng S. Stereoselective propranolol metabolism in two drug induced rat hepatic microsomes. World J Gastroenterol. 2000;6:74-78. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 9] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 14. | McLaughlin L, Burchell B, Pritchard M, Wolf CR, Friedberg T. Treatment of mammalian cells with the endoplasmic reticulum-proliferator compactin strongly induces recombinant and endogenous xenobiotic metabolizing enzymes and 3-hydroxy-3-methylglutaryl-CoA reductase in vitro. J Cell Sci. 1999;112:515-523. [PubMed] |
| 15. | Findlay KA, Kaptein E, Visser TJ, Burchell B. Characterization of the uridine diphosphate-glucuronosyltransferase-catalyzing thyroid hormone glucuronidation in man. J Clin Endocrinol Metab. 2000;85:2879-2883. [PubMed] |
| 16. | Innocenti F, Iyer L, Ramírez J, Green MD, Ratain MJ. Epirubicin glucuronidation is catalyzed by human UDP-glucuronosyltransferase 2B7. Drug Metab Dispos. 2001;29:686-692. [PubMed] |
| 17. | Bernard P, Goudonnet H, Artur Y, Desvergne B, Wahli W. Activation of the mouse TATA-less and human TATA-containing UDP-glucuronosyltransferase 1A1 promoters by hepatocyte nuclear factor 1. Mol Pharmacol. 1999;56:526-536. [PubMed] |
| 18. | Kren BT, Parashar B, Bandyopadhyay P, Chowdhury NR, Chowdhury JR, Steer CJ. Correction of the UDP-glucuronosyltransferase gene defect in the gunn rat model of crigler-najjar syndrome type I with a chimeric oligonucleotide. Proc Natl Acad Sci USA. 1999;96:10349-10354. [PubMed] |
| 19. | Barbier O, Albert C, Martineau I, Vallée M, High K, Labrie F, Hum DW, Labrie C, Bélanger A. Glucuronidation of the nonsteroidal antiestrogen EM-652 (SCH 57068), by human and monkey steroid conjugating UDP-glucuronosyltransferase enzymes. Mol Pharmacol. 2001;59:636-645. [PubMed] |
| 20. | Cheng Z, Radominska-Pandya A, Tephly TR. Studies on the substrate specificity of human intestinal UDP- lucuronosyltransferases 1A8 and 1A10. Drug Metab Dispos. 1999;27:1165-1170. [PubMed] |
| 21. | Carrier JS, Turgeon D, Journault K, Hum DW, Bélanger A. Isolation and characterization of the human UGT2B7 gene. Biochem Biophys Res Commun. 2000;272:616-621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 22. | Barbier O, Bélanger A, Hum DW. Cloning and characterization of a simian UDP-glucuronosyltransferase enzyme UGT2B20, a novel C19 steroid-conjugating protein. Biochem J. 1999;337:567-574. [PubMed] |
| 23. | Coffman BL, King CD, Rios GR, Tephly TR. The glucuronidation of opioids, other xenobiotics, and androgens by human UGT2B7Y (268) and UGT2B7H (268). Drug Metab Dispos. 1998;26:73-77. [PubMed] |
| 24. | Radominska-Pandya A, Czernik PJ, Little JM, Battaglia E, Mackenzie PI. Structural and functional studies of UDP-glucuronosyltransferases. Drug Metab Rev. 1999;31:817-899. [PubMed] |
| 25. | Lautala P, Ethell BT, Taskinen J, Burchell B. The specificity of glucuronidation of entacapone and tolcapone by recombinant human UDP-glucuronosyltransferases. Drug Metab Dispos. 2000;28:1385-1389. [PubMed] |
| 26. | Yueh MF, Nguyen N, Famourzadeh M, Strassburg CP, Oda Y, Guengerich FP, Tukey RH. The contribution of UDP-glucuronosyltransferase 1A9 on CYP1A2-mediated genotoxicity by aromatic and heterocyclic amines. Carcinogenesis. 2001;22:943-950. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 44] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 27. | Pless D, Gouze JN, Senay C, Herber R, Leroy P, Barberousse V, Fournel-Gigleux S, Magdalou J. Characterization of the UDP-glucuronosyltransferases involved in the glucuronidation of an antithrombotic thioxyloside in rat and humans. Drug Metab Dispos. 1999;27:588-595. [PubMed] |
| 28. | Grams B, Harms A, Braun S, Strassburg CP, Manns MP, Obermayer-Straub P. Distribution and inducibility by 3-methylcholanthrene of family 1 UDP-glucuronosyltransferases in the rat gastrointestinal tract. Arch Biochem Biophys. 2000;377:255-265. [PubMed] |
| 29. | Kobayashi T, Tatano A, Yokota H, Onaga T, Watanabe T, Yuasa A. Small intestinal UDP-glucuronosyltransferase sheUGT1A07: partial purification and cDNA cloning from sheep small intestine. Arch Biochem Biophys. 1999;364:143-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 30. | Jedlitschky G, Cassidy AJ, Sales M, Pratt N, Burchell B. Cloning and characterization of a novel human olfactory UDP-glucuronosyltransferase. Biochem J. 1999;340:837-843. [PubMed] |
| 31. | Dong M, Owens IS, Sheen YY. Cloning and expression of human liver UDP-glucuronosyltransferase cDNA, UDPGTh2. Arch Pharm Res. 1997;20:459-464. [PubMed] |
| 32. | Bélanger G, Barbier O, Hum DW, Bélanger A. Molecular cloning, expression and characterization of a monkey steroid UDP-glucuronosyltransferase, UGT2B19, that conjugates testosterone. Eur J Biochem. 1999;260:701-708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 33. | Barbier O, Lapointe H, El Alfy M, Hum DW, Bélanger A. Cellular localization of uridine diphosphoglucuronosyltransferase 2B enzymes in the human prostate by in situ hybridization and immunohistochemistry. J Clin Endocrinol Metab. 2000;85:4819-4826. [PubMed] |
| 34. | Strassburg CP, Manns MP, Tukey RH. Expression of the UDP-glucuronosyltransferase 1A locus in human colon. Identification and characterization of the novel extrahepatic UGT1A8. J Biol Chem. 1998;273:8719-8726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 211] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 35. | Albert C, Vallée M, Beaudry G, Bélanger A, Hum DW. The monkey and human uridine diphosphate-glucuronosyltransferase UGT1A9, expressed in steroid target tissues, are estrogen-conjugating enzymes. Endocrinology. 1999;140:3292-3302. [PubMed] |
| 36. | Metz RP, Ritter JK. Transcriptional activation of the UDP-glucuronosyltransferase 1A7 gene in rat liver by aryl hydrocarbon receptor ligands and oltipraz. J Biol Chem. 1998;273:5607-5614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 37. | Walle T, Otake Y, Galijatovic A, Ritter JK, Walle UK. Induction of UDP-glucuronosyltransferase UGT1A1 by the flavonoid chrysin in the human hepatoma cell line hep G2. Drug Metab Dispos. 2000;28:1077-1082. [PubMed] |
| 38. | Münzel PA, Schmohl S, Heel H, Kälberer K, Bock-Hennig BS, Bock KW. Induction of human UDP glucuronosyltransferases (UGT1A6, UGT1A9, and UGT2B7) by t-butylhydroquinone and 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin in Caco-2 cells. Drug Metab Dispos. 1999;27:569-573. [PubMed] |
| 39. | Bock KW, Gschaidmeier H, Heel H, Lehmköster T, Münzel PA, Raschko F, Bock-Hennig B. AH receptor-controlled transcriptional regulation and function of rat and human UDP-glucuronosyltransferase isoforms. Adv Enzyme Regul. 1998;38:207-222. [PubMed] |
| 40. | Yokota H, Ando F, Iwano H, Yuasa A. Inhibitory effects of uridine diphosphate on UDP-glucuronosyltransferase. Life Sci. 1998;63:1693-1699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 41. | Barbier O, Girard C, Breton R, Bélanger A, Hum DW. N-glycosylation and residue 96 are involved in the functional properties of UDP-glucuronosyltransferase enzymes. Biochemistry. 2000;39:11540-11552. [PubMed] |
| 42. | Iwano H, Yotsumoto N, Yokota H, Yuasa A. cDNA cloning and expression of a bovine phenol UDP-glucuronosyltransferase, BovUGT1A6. Life Sci. 2001;68:2131-2139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 43. | Forsman T, Lautala P, Lundström K, Monastyrskaia K, Ouzzine M, Burchell B, Taskinen J, Ulmanen I. Production of human UDP-glucuronosyltransferases 1A6 and 1A9 using the Semliki Forest virus expression system. Life Sci. 2000;67:2473-2484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 44. | Chen Q, Wu J, Yu Y. [Establishment of transgenic cell line CHL-3A4 and its metabolic activation]. Zhonghua Yu Fang Yi Xue Za Zhi. 1998;32:281-284. [PubMed] |
| 45. | Zhuge J, Qian YL, Xie HY, Yu YN. Cloning and iden tification of human cytochrome P4501A2 cDNA. Chin J of Pharmacol Toxicol. 2000;14:315-317. |
| 46. | Court MH, Greenblatt DJ. Molecular genetic basis for deficient acetaminophen glucuronidation by cats: UGT1A6 is a pseudogene, and evidence for reduced diversity of expressed hepatic UGT1A isoforms. Pharmacogenetics. 2000;10:355-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 122] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 47. | de Wildt SN, Kearns GL, Leeder JS, van den Anker JN. Glucuronidation in humans. Pharmacogenetic and developmental aspects. Clin Pharmacokinet. 1999;36:439-452. [PubMed] |
| 48. | Choo EF, Angus PW, Morgan DJ. Effect of cirrhosis on sulphation by the isolated perfused rat liver. J Hepatol. 1999;30:498-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 49. | Strassburg CP, Strassburg A, Nguyen N, Li Q, Manns MP, Tukey RH. Regulation and function of family 1 and family 2 UDP-glucuronosyltransferase genes (UGT1A, UGT2B) in human oesophagus. Biochem J. 1999;338:489-498. [PubMed] |
| 50. | Ren Q, Murphy SE, Zheng Z, Lazarus P. O-Glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) by human UDP-glucuronosyltransferases 2B7 and 1A9. Drug Metab Dispos. 2000;28:1352-1360. [PubMed] |
| 51. | Bachrich T, Thalhammer T, Jäger W, Haslmayer P, Alihodzic B, Bakos S, Hitchman E, Senderowicz AM, Penner E. Characterization of autoantibodies against uridine-diphosphate glucuronosyltransferase in patients with inflammatory liver diseases. Hepatology. 2001;33:1053-1059. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
Edited by Ma JY