Published online Apr 15, 2000. doi: 10.3748/wjg.v6.i2.220
Revised: December 6, 1999
Accepted: December 24, 1999
Published online: April 15, 2000
AIM: To clone core gene cDNA of Chinese hepatitis C virus (HCV) into eukaryotic expression vector cosmid pTM3 and to express HCV core antigen in HepG2 cells.
METHODS: Core gene cDNA of HCV was introduced into eukaryotice xpression vector cosmid pTM3. Using vaccinia virus/bacterio phage T7 hybrid expression system, HepG2 cells were transfected with the recombinant plasmid pTM3-Q534 by lipofectin.
RESULTS: From the transfected bacteria Top10F′, 2 pTM3-Q534 clones containing the recombinant plasmid were identified from randomly selected 10 ampicillin-resistant colonies. By reverse transcription PCR and indirect immunofluorescence technique, HCV RNA and core protein was identified in HepG2 cells transfected with the recombinant plasmid.
CONCLUSION: The construction of a recombinant plasmid and the expression of core gene cDNA of HCV in HepG2 was successful.
- Citation: Jiang RL, Lu QS, Luo KX. Cloning and expression of core gene cDNA of Chinese hepatitis C virus in cosmid pTM3. World J Gastroenterol 2000; 6(2): 220-222
- URL: https://www.wjgnet.com/1007-9327/full/v6/i2/220.htm
- DOI: https://dx.doi.org/10.3748/wjg.v6.i2.220
Although the genome of several strains of hepatitis C virus (HCV) have been cloned and sequenced, the HCV particles have not been observed so far. Currently, because of very low infection efficiency, one of the major impediments to the structural analysis of HCV genome and genetic analysis of viral replication is the lack of a reliable cell culture system permissive for HCV replication. In this study, using recombinant DNA technique, we constructed a recombinant plasmid by subcloning core gene cDNA of Chinese HCV isolated into a eukaryotic expression vector pTM3 and expressed HCV core antigen in transfected HepG2 cells by using vaccinia virus/bacteriophage T7 hybrid expression system.
Details of the plasmid construction are as follows. First, plasmid pQ534[1] containing core gene cDNA of Chinese HCV (provided by Prof. QI Zhong-Tian, Second Military Medican University) was digested partially with Pst-I and Sma I, cosmid vector pTM3 (provided by St. Mary′s Hospital, London University, England) was digested thoroughly with Pst I and Sma I, and the target gene and large fragment vector was purified by agarose gel electrophoresis. The 559 base pairs (bp) Pst I and Sma I target gene fragment was ligated with a 7418 bp Pst I and Sma I linearized plasmid vector of pTM3 by bacteriophage T4 DNA ligase overnight at 37 °C, and stored at -20 °C. The ligation product was routinely transformed into bacteria Top10F′, and was incubated in an LB plate containing ampicillin overnight at 37 °C. Ten bacterial colonies were individually transferred into 2 mL of LB medium containing ampicillin in a loosely capped 15 mL tube, and the culture was incubated overnight at 37 °C with vigorous shaking. To confirm that the culture did contain the correct plasmid, we prepared a small amount of plasmid DNA and analyzed it by digestion with restriction enzymes. Then, we propagated the transformed positive bacterial colony, prepared and purified a large amount of plasmid DNA, and stored it at -20 °C for transfections.
After the inoculum was removed, the cells were transfected with 1 mg of pTM3-Q534 plus lipofection (GIBCO-BRL) 1 mg for 4 h at 37 °C. For the control experiment, the infected cells were transfected with pTM3 instead of pTM3-Q534 for 4 h. The cells were then incubated at 37 °C for 4 h in DMEM containing 2% fetal bovine serum.
Briefly, HepG2 cells were seeded into 6-well plate and were approximately 80% confluent 24 h later. The cells were infected with the recombinant vaccinia virus vvT-7-3 (provided by St. Mary′s Hospital, London University, England) at the multiplicity of infection of 8 plaque-forming units (PFU)/cell for 1 h at 37 °C in Dulbecco′s Modified Eagle Medium (DMEM) containing 2% fetal bovine serum.
Production of core region HCV-RNA in the transfected HepG2 cells was examined by RT-PCR using primers located at the core region of the HCV genome. (First PCR : sense 5′-CCCAAACCTCAAAGAAA-3′, antisense 5′-AGCGGTATGTACCCCATG-3′; second PRC: sense 5′-CAGATCGTTGGTGGAGTT-3′, antisense 5′-GCAGCCCTCATTGCCAT-3′). RT-PCR was performed using a standard procedure described previously. In all experiments, RNA extracted from a liver specimen known to contain HCV was included as positive control and cloned HBV-DNA transfected HepG2 cells (HepG2 2.2.15 cell line) as negative control.
After infection/transfection, the medium was removed and cells were washed once with PBS and were fixed with methanol/acetone (50∶50) for 20 min at -20 °C. The methanol/acetone was removed and the cells were rinsed with PBS. For immunofluorescent assays, the fixed cells were incubated for 1 h at 37 °C with a 1 ∶50 dilution of the clinical HCV positive human serum in PBS for 5 min per wash. FITC-conjugated secondary antibody IgG (goat anti-human) were added at 1 ∶5 dilution in PBS and incubated for 1 h at 37 °C. The cells were washed for 4-5 times as above. The stained cells were observed with a Meridian-ACAS 470.
In order to satisfy the directional cloning and the correct reading frame, plasmid pQ534 was double digested with Sma I and Pst I. Since there are 2 Sma I recognition sites in the core gene cDNA of HCV, partial digestion has to be carried out, so at first, pQ534 was digested with the restriction enzyme Pst I for 1 h at 37 °C, and then digested with the second enzyme Sma I for 10 min at 37 °C. The 559 bp segment containing core gene cDNA of HCV was purified by agarose gel electrophoresis (Figure 1).
From the recombinant plasmid transformed bacteria Top10F′, 2 pTM3-Q534 clones were identified from 10 randomly selected ampicillin-resistant colonies. Plasmid DNA obtained by alkaline lysis was cleaved with restriction enzyme EcoR I/Pst I, a 534 bp insertion segment was observed in 2 of the 10 colonies (Figure 2, lane 3, 4). The plasmid DNA of positive clones was digested with EcoR I, and actually only a 7970 bp band was found after electrophoresis (Figure 2, lane 5). Because an EcoR I site also existed in the core gene cDNA of HCV (9 base pairs near the 5′ terminus of core gene cDNA), if the target gene was reverse-inserted, the length between the site and the Eco-R I site of the polyclonal site of plasmid pTM3 was 550 bp, so a 550 bp Eco-R I Eco-R I fragment (7970 bp) after electrophoresis. It was confirmed that construction of core gene cDNA of HCV in cosmid vector pTM3 was successful.
Positive-strand HCV RNA was detected in HepG2 cells over a 6-week period after transfection with pTM3-Q534. Negative-strand HCV RNA was detected over a 4-week period after transfection with pTM3-Q534. However, there was no HCV-RNA specific strand found in the control cells. It was demonstrated that there was HCV replication in the HepG2 cells transfected with pTM3-Q534.
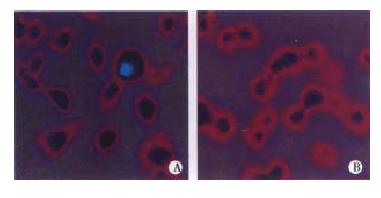
In the experimental group, some of the HepG2 cells transfected with pTM3-Q534 were positive for HCV core protein. The other cells were negative. The negative control HepG2 cells transfected with pTM3 were all negative for HCV core protein (Figure 3), and the same results were obtained in the repeated experiments of transfection and indirect immunofluorescence.
More and more studies about the expression of HCV gene in mammalian cells have been reported in the world[2,4,6,8,9], but no reports from China. In this study, we successfully cloned core gene cDNA of HCV into eukaryotic expression vector pTM3. To achieve transcription with high efficiency and sufficient translation and expression of the HCV genome, we used a vaccinia virus/bacteriophage T7 hybrid expression system[3,5,7]. In this system, T7 RNA polymerase was expressed in the cells infected with a recombinant vaccinia virus vvT-7.3, and the transfected target genes controlled under the T7 promotor sequence were transcribed with high efficiency. Thus, the core protein of HCV can be efficiently produced in the cells. Lipofectin reagent interacts spontaneously with DNA to form a lipid-DNA complex. The fusion of the complex with tissue culture cells could result in the efficient uptake and expression of the DNA. Compared to transfection methods employing calcium phosphate, a protocol using lipofectin reagent has been shown to be 50-100 fold more efficient. Cloning of core gene cDNA of HCV into eukaryotic expression vector pTM3 and expression of HCV core antigen in HepG2 cells confirmed that the recombinant plasmid pTM3-Q534 which contains core gene cDNA of HCV possesses the property of propagation in HepG2 cells. As all the results were reproducible, a solid foundation was laid for the further investigation in the expression of HCV genes in mammalian cell lines and the detection of the function of the various regions in HCV genome.
| 1. | Qi ZT, Pan W, Du P. Cloning and sequencing of core gene cDNA of Chinese hepatitis C virus. Dier Junyi Daxue Xuebao. 1992;13:301-306. |
| 2. | Selby MJ, Choo QL, Berger K, Kuo G, Glazer E, Eckart M, Lee C, Chien D, Kuo C, Houghton M. Expression, identification and subcellular localization of the proteins encoded by the hepatitis C viral genome. J Gen Virol. 1993;74:1103-1113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 163] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 3. | Mizuno M, Yamada G, Tanaka T, Shimotohno K, Takatani M, Tsuji T. Virion-like structures in HeLa G cells transfected with the full-length sequence of the hepatitis C virus genome. Gastroenterology. 1995;109:1933-1940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 4. | Yoo BJ, Selby MJ, Choe J, Suh BS, Choi SH, Joh JS, Nuovo GJ, Lee HS, Houghton M, Han JH. Transfection of a differentiated human hepatoma cell line (Huh7) with in vitro-transcribed hepatitis C virus (HCV) RNA and establishment of a long-term culture persistently infected with HCV. J Virol. 1995;69:32-38. [PubMed] |
| 5. | Fuerst TR, Niles EG, Studier FW, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122-8126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1645] [Cited by in RCA: 1788] [Article Influence: 44.7] [Reference Citation Analysis (0)] |
| 6. | Hiramatsu N, Dash S, Gerber MA. HCV cDNA transfection to HepG2 cells. J Viral Hepat. 1997;4 Suppl 1:61-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 15] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 7. | Yanagi M, Purcell RH, Emerson SU, Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci USA. 1997;94:8738-8743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 433] [Cited by in RCA: 423] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 8. | Pasquinelli C, Shoenberger JM, Chung J, Chang KM, Guidotti LG, Selby M, Berger K, Lesniewski R, Houghton M, Chisari FV. Hepatitis C virus core and E2 protein expression in transgenic mice. Hepatology. 1997;25:719-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 98] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 9. | Dash S, Halim AB, Tsuji H, Hiramatsu N, Gerber MA. Transfection of HepG2 cells with infectious hepatitis C virus genome. Am J Pathol. 1997;151:363-373. [PubMed] |
Edited by Ma JY
Project supported by the National Natural Science Foundation of China, No. 39500129