Published online Feb 7, 2026. doi: 10.3748/wjg.v32.i5.115301
Revised: November 13, 2025
Accepted: December 8, 2025
Published online: February 7, 2026
Processing time: 106 Days and 8.5 Hours
Chemoresistance significantly limits the therapeutic efficacy of neoadjuvant chemotherapy (NACT) in advanced gastric cancer (AGC). There is an urgent need to identify robust biomarkers predictive of NACT response and to elucidate the molecular mechanisms that drive resistance. In this study, we systematically assess whether intercellular adhesion molecule 2 (ICAM2) predicts NACT res
To investigate the predictive significance and mechanistic role of ICAM2 in mediating 5-fluorouracil (5-FU) resistance in gastric cancer (GC).
Real-time PCR, Western blotting, enzyme-linked immunosorbent assay, and immunohistochemistry were conducted to assess alterations in ICAM2 expression between 5-FU-sensitive and -resistant GC cells as well as in AGC patient samples. Cytotoxicity assays, colony formation, flow cytometry, analyses of apoptosis-re
Low ICAM2 expression correlated significantly with poor NACT response, advanced tumor stage, worse differentiation, and reduced overall survival and disease-free survival in AGC patients. Pre-NACT serum ICAM2 demon
Our study highlights the clinical impact of ICAM2 downregulation predicting poor outcome and NACT response in AGC patients, and reveals a novel ICAM2/TGF-β/Smad/SP1/PTN signaling mediating 5-FU resistance in GC.
Core Tip: Intercellular adhesion molecule 2 (ICAM2) is both a predictive biomarker and a mechanistic mediator of chemoresistance in advanced gastric cancer. Low serum and tissue ICAM2 identify neoadjuvant nonresponders and predict shorter survival. Functionally, ICAM2 loss increases 5-fluorouracil (5-FU) resistance by impairing caspase-dependent apoptosis and remodeling an immunosuppressive, M2-biased macrophage milieu. Mechanistically, ICAM2 loss activates TGF-β/Smad signaling, which upregulates the transcription factor SP1; SP1 then directly transactivates pleiotrophin (PTN). Targeting TGF-β suppresses PTN and restores 5-FU sensitivity in preclinical models, positioning the ICAM2/TGF-β/Smad/SP1/PTN axis as a clinically actionable pathway.
- Citation: Tang XC, Chen ZJ, Chen CY, Qiu J, Huang JT, Tan RC, Li WY, Chen H, Yang ZL. ICAM2 loss drives 5-fluorouracil resistance via TGF-β/Smad/SP1/PTN-dependent apoptosis evasion and macrophage remodeling in gastric cancer. World J Gastroenterol 2026; 32(5): 115301
- URL: https://www.wjgnet.com/1007-9327/full/v32/i5/115301.htm
- DOI: https://dx.doi.org/10.3748/wjg.v32.i5.115301
Gastric cancer (GC) is the fifth most commonly diagnosed cancer and the fifth leading cause of cancer-related deaths globally[1], with approximately 71% of patients diagnosed at advanced clinical stages (cTNM II-III)[2]. The standard treatment for advanced GC (AGC) is 5-fluorouracil (5-FU)-based neoadjuvant chemotherapy (NACT) followed by radical surgery[3]. However, many AGC patients are resistant to NACT and experience disease progression. The lack of reliable biomarkers to predict NACT efficacy and the development of resistance to 5-FU are major challenges to successful treatment[4]. Therefore, identifying biomarkers for NACT response and understanding the mechanisms of 5-FU resistance are crucial for improving survival in AGC patients.
Intercellular adhesion molecule 2 (ICAM2), a type I transmembrane glycoprotein, plays a critical role in various tumor-related processes, particularly in immune surveillance and the modulation of the tumor microenvironment (TME)[5,6]. For example, overexpression of ICAM2 in pancreatic cancer cells enhances γδ T cell binding and cytotoxicity in vitro[7]. Similarly, adenoviral-mediated ICAM2 expression in tumor cells promotes natural killer cell adhesion and activation, which in turn reduces peritoneal metastasis in GC[8]. Beyond its role in immune regulation, recent studies have suggested that ICAM2 may act as a tumor suppressor, influencing key tumor characteristics such as proliferation[9], migration[10] and radioresistance[11]. These findings highlight ICAM2's dual role in both immune modulation and tumor progression. However, despite its significance in these processes, the exact biological effects and molecular mechanisms of ICAM2 in GC chemoresistance remain underexplored.
The TGF-β signaling pathway is a key mediator of chemoresistance[12]. It promotes drug resistance by regulating factors involved in metabolism[13], apoptosis[14], and the TME[15]. Targeting TGF-β signaling has shown promise in overcoming chemoresistance in various cancers, including GC[16]. Therefore, discoveries of novel molecules capable of interfering with aberrant TGF-β signaling activity may offer new therapeutic strategies to combat chemoresistance in GC.
Pleiotrophin (PTN), a secreted heparin-binding growth factor, regulates key processes such as cell proliferation, apoptosis, differentiation, and angiogenesis[17]. Recent studies have shown that PTN plays a pivotal role in the deve
In this study, we found that reduced ICAM2 expression in serum and tissue samples from AGC patients correlated with poor NACT response and worse prognosis. ROC analysis further validated serum ICAM2 levels as a reliable predictor of NACT response in AGC patients. Functional studies showed that ICAM2 knockdown enhanced 5-FU resistance by inhibiting cell apoptosis and promoting an M2 macrophage-dependent immunosuppressive microenvironment via TGF-β/Smad/SP1 signaling and PTN upregulation. These findings highlight ICAM2 as a predictive biomarker for NACT response and suggest that targeting ICAM2/TGF-β/Smad/SP1/PTN axis may improve chemotherapy efficacy and patient outcomes.
Human AGC tissue and plasma samples were collected at the Sixth Affiliated Hospital of Sun Yat-sen University. All diagnoses of GC were confirmed histopathologically. Patients underwent 4-6 cycles of 5-FU-based NACT followed by surgical resection. Treatment regimens and follow-up adhered to institutional protocols aligned with National Comprehensive Cancer Network guidelines, as previously reported[22]. Tumor regression after NACT was assessed using the Ryan tumor regression grade (TRG) system, classifying responses from grade 0 (complete response) to grade 3 (poor or no response)[22]. Patients with TRG scores of 0 or 1 were defined as NACT-sensitive, whereas those with scores of 2 or 3 were categorized as resistant[22].
For tissue-based analyses, 36 pre-NACT core needle biopsies and 93 post-NACT surgical specimens were obtained to evaluate the association between ICAM2 expression and chemoresistance in AGC. For plasma analyses, samples from 85 AGC patients before NACT and 50 patients after NACT were collected between April 2013 and December 2018 for the training cohort, aiming to assess the predictive value of circulating ICAM2 levels. An independent validation cohort included plasma samples from 95 AGC patients collected prior to NACT between January 2019 and July 2022 to confirm the utility of serum ICAM2 as a predictive biomarker for NACT response.
Human GC cell lines were sourced from the cell bank of the Chinese Academy of Sciences (Shanghai, China). Cells were maintained in RPMI-1640 medium (Gibco, United States) supplemented with 100 mL/L fetal bovine serum and 10 mL/L penicillin-streptomycin at 37 °C under a humidified atmosphere containing 50 mL/L CO2. Cell line identity was verified by short tandem repeat profiling, performed in compliance with the standards established by the cell bank. All cell cultures were routinely tested to exclude mycoplasma contamination. Galunisertib (LY2157299; 10 µM, MedChemExpress, United States) and TGF-β1 protein (10 ng/mL, MedChemExpress, United States) were introduced to the medium to inhibit or activate TGF-β signaling, respectively. To inhibit SP1 transcriptional activity, GC cells were treated with Plicamycin (MedChemExpress, United States) at concentrations of 0 nM, 50 nM, and 100 nM for 48 hours.
Paraffin-embedded GC tissues were first dewaxed in xylene and then passed through descending ethanol concentrations for rehydration. Heat-induced epitope retrieval was performed in pH 6.0 sodium citrate buffer using microwave treatment. Endogenous peroxidase activity was blocked with 30 mL/L hydrogen peroxide (H2O2), and nonspecific sites were subsequently blocked with normal goat serum (ZSGB-BIO, China). Primary antibodies detailed in Supplementary Table 1 were applied to the sections and allowed to incubate at 4 °C overnight to ensure adequate antigen-antibody binding. After washing, sections were treated with horseradish peroxidase (HRP)-conjugated secondary antibodies and visualized using 3,3′-diaminobenzidine substrate. Nuclei were counterstained with hematoxylin. Images were captured using a slide scan system (Shenzhen Shengqiang Technology Co., Ltd., China). Protein expression of ICAM2 was semi-quantitatively assessed by integrating staining intensity and the proportion of positive cells. Intensity was scored from 0 to 3 (0: Negative; 1: Weak; 2: Moderate; 3: Strong), while the percentage of positive cells was rated as 1 (1%-25%), 2 (26%-50%), 3 (51%-75%), or 4 (76%-100%). The overall immunohistochemical score was obtained by multiplying the intensity grade by the percentage of stained cells.
Total RNA was extracted from GC cell lines using TRIzol reagent (Invitrogen, United States) in accordance with the manufacturer’s protocol. RNA concentration and purity were measured by spectrophotometry (A260/A280 ratio). Subsequently, 1 µg RNA was reverse-transcribed into complementary DNA using a PrimeScript RT reagent kit (Applied Biosystems, United States). Quantitative real-time PCR (RT-PCR) was performed with SYBR Green Master Mix (Applied Biosystems, United States) on a QuantStudio system. Each reaction was run in triplicate, and primer sequences are presented in Supplementary Table 2.
Specific short hairpin RNAs (shRNAs) targeting ICAM2 and overexpression plasmids PCDH-3xFlag-ICAM2 (NM_001099789.2) were designed and synthesized by Hanyi Biotechnology (Guangzhou, China) for lentiviral vector construction. The sequences of ICAM2 shRNAs were as follows: ICAM2-sh1: 5′-AGACCTCTCTAGATAAGATTC-3′ and ICAM2-sh3: 5′-CTGACACTGCAACCCACTTTG-3′. Lentiviral transduction of GC cells was performed according to established protocols[23], and polyclonal populations stably expressing green fluorescent protein were selected for subsequent analyses.
Additionally, small interfering RNA targeting PTN (5′-AGGCAAGAAACAGGAGAAGAT-3′) and overexpression plasmids pcDNA3.1-3xFlag-SP1 (NM_138473.3) and pcDNA3.1-3xFlag-PTN (NM_002825.7) were procured from IGEbio (Guangzhou, China). Transfection of siRNAs was conducted using LipofectaminTM RNAiMAX (Thermo Fisher Scientific, United States), while plasmid transfection employed LipofectaminTM 3000 (Thermo Fisher Scientific), following the manufacturers’ instructions.
Cell lysates were prepared using a commercial lysis buffer (Thermo Fisher Scientific, United States), and protein concentrations were quantified with a BCA assay (Beyotime, China). Equivalent amounts of total protein were separated by SDS-PAGE and transferred onto PVDF membranes (Millipore, United States). The membranes were blocked in 50 g/L non-fat milk prepared with TBST buffer for 1 hour at room temperature, followed by overnight incubation at 4 °C with primary antibodies against GAPDH (Proteintech, China), ICAM2 (Cell Signaling Technology, United States), Smad2, phospho-Smad2 (Ser467), Smad3, phospho-Smad3 (Ser423/425), phospho-Smad2 (Ser465), cleaved PARP, cleaved caspase-3 p17, Bax, Bad (all from ZENBIO, China), SP1, PTN, CD80, CD86, Arginase-1 (all from Proteintech, China), Ki67 (Servicebio, China), and CD163, CD206 (Abcam, United Kingdom). Following washes, membranes were incubated for 1 hour at room temperature with HRP-conjugated goat anti-mouse or goat anti-rabbit secondary antibodies (Proteintech, China). Signals were detected using an enhanced chemiluminescence reagent (Meilunbio, China). Detailed information on all primary antibodies is provided in Supplementary Table 1.
Serum concentrations of ICAM2 were determined by a quantitative sandwich enzyme-linked immunosorbent assay (ELISA) kit specific for human ICAM2 (FineTest, China) according to the manufacturer’s guidelines. Briefly, each serum sample was diluted 1:2 in sample diluent. Standards and diluted samples (100 μL per well) were loaded onto a 96-well microplate pre-coated with anti-ICAM2 antibody and incubated for 90 minutes at 37 °C. Wells were washed, followed by sequential incubation with biotin-conjugated anti-ICAM2 detection antibody for 60 minutes, HRP-Streptavidin for 30 minutes, and 3,3',5,5'-tetramethylbenzidine substrate for 10-20 minutes at 37 °C in the dark. After addition of the stop solution, absorbance was measured at 450 nm using a microplate reader. Standard curves were established with serially diluted recombinant ICAM2 protein (0.313-20 ng/mL), from which sample concentrations were extrapolated. Each assay was performed in triplicate to ensure reproducibility.
For determination of half-maximal inhibitory concentration (IC50), GC cells were seeded into 96-well plates at a density of 4 × 103 cells per well and allowed to adhere overnight. Cells were then exposed to escalating concentrations of 5-FU (Meilunbio, China) for 48 hours under standard culture conditions (37 °C, 50 mL/L CO2). Cell viability was determined using the CCK-8 assay (APExBIO, United States) as per the manufacturer’s protocol. Absorbance was measured at 450 nm, and IC50 values were calculated using GraphPad Prism 8 software.
GC cells (2 × 104 per well) were seeded into 6-well plates and cultured for 10 days in the presence of 5-FU at the indicated concentrations. Formed colonies were fixed with 40 g/L paraformaldehyde for 15 minutes, stained with 10 g/L crystal violet solution for 5 minutes, and washed gently with distilled water. The number of colonies was counted and analyzed for statistical significance.
Apoptotic cells were detected using the Annexin V-APC/7-AAD Apoptosis Detection Kit (MultiSciences Biotech, China) following the manufacturer’s instructions and analyzed by flow cytometry. In brief, cells were harvested, washed with cold phosphate-buffered saline (PBS), and stained with Annexin V-APC and 7-AAD in binding buffer for 5 min at room temperature in the dark. Samples were analyzed on a flow cytometer (Beckman Coulter, United States), and apoptotic cell populations were quantified using CytExpert Version 2.4 (Beckman Coulter, United States).
Human THP-1 monocytes (cell bank of the Chinese Academy of Sciences, China) were induced to differentiate into M0 macrophages by exposure to 100 ng/mL phorbol 12-myristate 13-acetate (PMA; Meilunbio, China) for 24 hours at 37 °C in RPMI-1640 medium containing 100 mL/L fetal bovine serum. Following differentiation, cells were washed twice with PBS and cultured in fresh RPMI-1640 medium without PMA for an additional 24 hours to stabilize into an M0 phenotype.
For indirect co-culture assays, transwell chambers (6-well format, 0.4 μm pore size; Corning, United States) were used. GC cells were seeded into the upper compartment, while differentiated M0 macrophages were plated in the lower chamber. After 48 hours of co-culture, tumor-associated macrophages (TAMs) previously exposed to either ICAM2-overexpressing or ICAM2-silenced GC cells were harvested for downstream analyses.
To evaluate the effects of TAMs with distinct phenotypes on GC cell behavior, these pre-conditioned macrophages were subsequently transferred to the lower chamber of new transwell systems, and wild type GC cells were seeded into the upper compartment. After an additional 48 hours of co-culture, macrophages were removed, and GC cells were collected for subsequent experimental assays.
GC cells were treated with 5-FU (2 μg/mL) for 48 hours, with three biological replicates per group. Total RNA was extracted using TRIzol reagent (Invitrogen, United States), and RNA integrity and purity were verified using a NanoDrop spectrophotometer and Agilent 2100 bioanalyzer. Sequencing libraries were generated and then processed for 150-bp paired-end sequencing using the Illumina NovaSeq 6000 platform (IGEbio, China). Bioinformatic analyses were performed using established RNA-seq pipelines (HISAT2 and DESeq2) to identify differentially expressed genes (DEGs).
Nuclear and cytoplasmic fractions were extracted using the Nuclear and Cytoplasmic Extraction Reagents Kit (Thermo Fisher Scientific, United States) in accordance with the manufacturer’s protocol. GC cells were washed with cold PBS, lysed in cytoplasmic extraction buffers, and centrifuged at 16000 × g for 5 minutes at 4 °C to collect cytoplasmic proteins. The nuclear pellets were resuspended in nuclear extraction buffer, incubated on ice with intermittent vortexing, and centrifuged at 16000 × g for 10 minutes at 4 °C. Nuclear protein concentrations were measured by BCA assay. Fraction purity was verified by Western blotting for histone H3 (nuclear marker) and GAPDH (cytoplasmic marker).
Cells were harvested and lysed on ice in Pierce IP Lysis Buffer (Thermo Fisher Scientific, United States), supplemented with protease and phosphatase inhibitor cocktails (Servicebio, China). The lysates were clarified by centrifugation at 12000 × g for 15 minutes at 4 °C, and protein concentrations were determined using the BCA Protein Assay Kit (Thermo Fisher Scientific, United States). For pre-clearing, equal amounts of total protein were incubated with Protein A/G Magnetic Beads (MedChemExpress, United States) for 4 hours at 4 °C with gentle rotation. The pre-cleared lysates were subsequently incubated overnight at 4 °C with either normal rabbit IgG (Proteintech, China) as a negative control or anti-SP1 antibody (Proteintech, China). Immune complexes were captured by adding fresh Protein A/G Magnetic Beads and incubating for an additional 4 hours at 4 °C. Beads were washed three times with IP lysis buffer to reduce nonspecific binding. Protein complexes were dissociated by boiling in 1 × SDS loading buffer (100 °C, 10 minutes) and examined by SDS-PAGE followed by immunoblotting.
The PTN promoter region (-2000 to +100 bp relative to the transcription start site) was amplified from human genomic DNA and cloned into the pGL3-basic luciferase reporter vector. Additionally, a series of truncated promoter constructs with varying upstream lengths, as well as site-directed mutant constructs targeting predicted SP1 binding sites, were generated by PCR amplification or site-directed mutagenesis. All constructs were synthesized, cloned, and sequence-verified by IGEbio (Guangzhou, China). For reporter assays, GC cells were seeded into 6-well plates at a density of 2 × 105 cells per well and transiently co-transfected with 2 µg of the PTN promoter reporter plasmid and 20 ng of pRL-TK Renilla luciferase plasmid using Lipofectamine 3000 reagent (Invitrogen, United States) following the manufacturer’s protocol. After 48 hours, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Meilunbio, China). Firefly luciferase activity was normalized to Renilla luciferase activity to control for transfection efficiency.
Chromatin immunoprecipitation (ChIP) was conducted using the Magna ChIPTM A/G kit (Merck, Germany) in accordance with the manufacturer’s instructions. Fixation was performed using 10 g/L formaldehyde for 10 minutes at room temperature, after which cross-linking was stopped with 125 mmol/L glycine and the cells were lysed. The chromatin was fragmented by sonication into 200-500 bp DNA segments, which were verified by agarose gel electrophoresis. Immunoprecipitation was carried out overnight at 4 °C using anti-SP1 antibody (Proteintech, China) or normal IgG as a negative control, followed by capture with protein A/G magnetic beads. DNA was purified and analyzed by quantitative PCR with primers specific for the PTN promoter region (PTN-site2-F: GCCTCTTGGTTCAAATCCGTTT; PTN-site2-R: AATGGAGAATGGGAGGGATGAGA). Data were normalized to input DNA and expressed as fold enrichment over IgG control.
Female BALB/c nude mice (Guangdong Yaokang Biotechnology, China) were randomly divided into two main groups (24 mice and 25 mice). One group was inoculated subcutaneously with Vector/ICAM2 MGC803 cells (1 × 106 cells in 125 µL sterile PBS per mouse), and the other group with Shcon/ICAM2-sh1 MKN45 cells (2 × 106 cells in 125 µL sterile PBS per mouse). When tumors reached approximately 100 mm3, the first group (24 mice) was further divided into four subgroups (n = 6 each) and treated with: (1) Vector + vehicle (DMSO); (2) Vector + 5-FU (20 mg/kg, i.p., every two days); (3) ICAM2-OE + vehicle; or (4) ICAM2-OE + 5-FU (20 mg/kg, i.p., every two days). The second group (25 mice) was divided into five subgroups (n = 5 each) and received: (1) Shcon + vehicle (DMSO); (2) Shcon + 5-FU (20 mg/kg, i.p., every two days); (3) ICAM2-sh1 + vehicle; (4) ICAM2-sh1 + 5-FU (20 mg/kg, i.p., every two days); or (5) ICAM2-sh1 + 5-FU (20 mg/kg, i.p., every two days) + LY2157299 (100 mg/kg, oral gavage twice daily). Tumor volumes were calculated using the formula (length × width2)/2. Mice were monitored every three days, and after euthanasia, tumors were excised, weighed, and paraffin-embedded for subsequent analyses. All animal procedures were reviewed and approved by the Laboratory Animal Ethics Committee of the Sixth Affiliated Hospital of Sun Yat-sen University.
Apoptotic cells were detected with a TRITC-based one-step TUNEL assay kit (KeyGen Biotech, China) following established protocols reported in earlier studies[23].
All statistical analyses were performed using GraphPad Prism version 8 (GraphPad Software, San Diego, CA, United States). Quantitative data were expressed as mean ± SD. Comparisons between two groups were conducted using unpaired Student’s t-test, while comparisons among multiple groups were analyzed by one-way ANOVA, with multiple comparisons based on Dunnett’s post hoc test. Receiver operating characteristic (ROC) curve analysis was performed to evaluate the diagnostic value of selected biomarkers. Kaplan-Meier survival analysis, assessed by the log-rank test, was used to determine the prognostic significance of biomarker expression. Univariate and multivariate analyses were further conducted to identify independent predictors of overall survival (OS) and disease-free survival (DFS). Statistical significance was defined at two-sided thresholds of P < 0.05, P < 0.01, P < 0.001, and P < 0.0001.
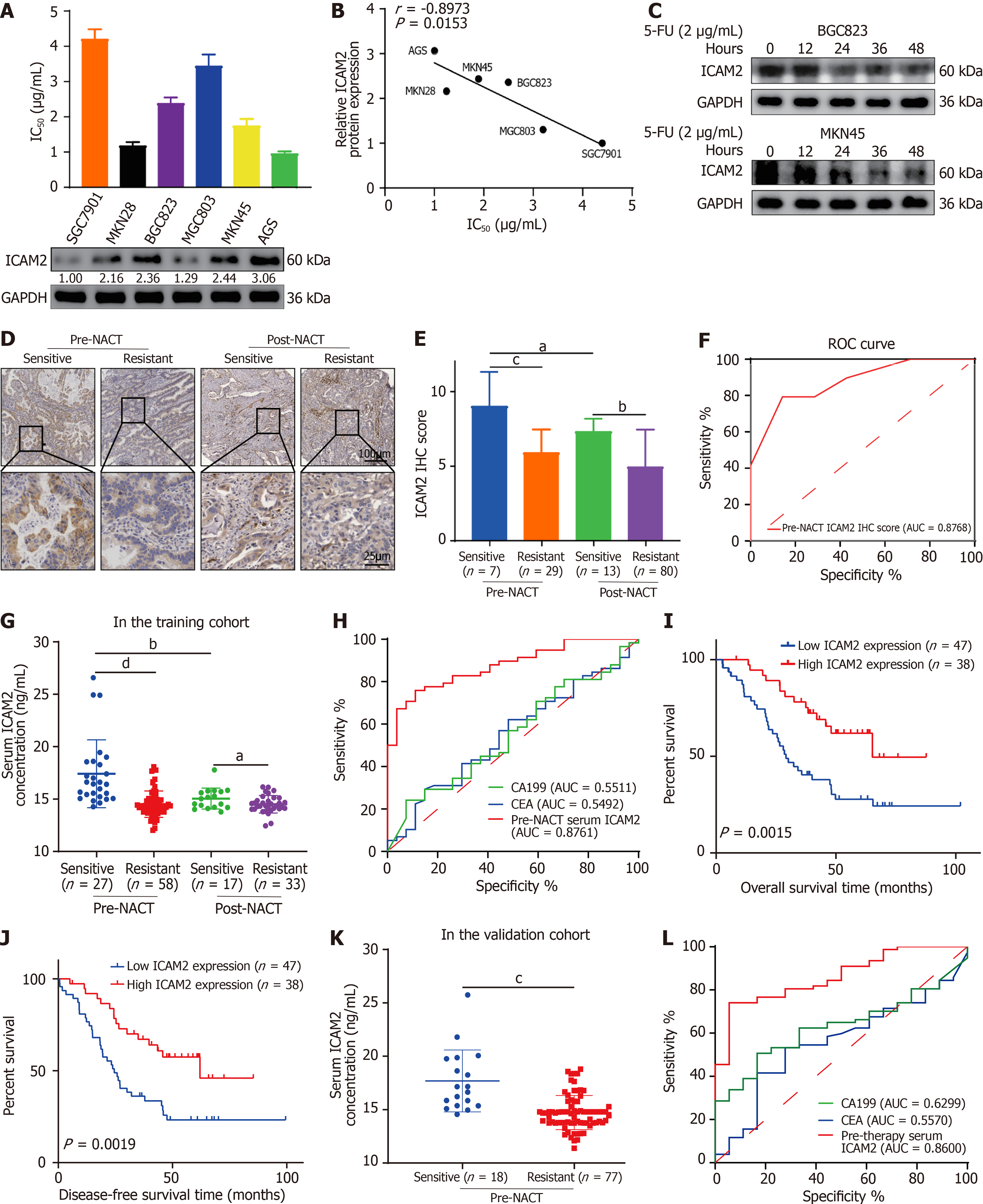
To investigate the role of ICAM2 in 5-FU resistance, we first analyzed ICAM2 expression in seven GC cell lines with varying 5-FU sensitivities. ICAM2 expression inversely correlated with the IC50 values for 5-FU (Figure 1A and B). In addition, 5-FU-resistant SGC7901 cells exhibited reduced ICAM2 levels compared to the parental line (Supplementary Figure 1A), and ICAM2 expression decreased over time in 5-FU-treated GC cells (Figure 1C), suggesting its involvement in both primary and acquired resistance.
To validate these findings in clinical samples, we performed immunohistochemistry (IHC) analysis on tumor tissues from 36 pre- and 93 post-NACT AGC patients. ICAM2 expression was significantly lower in 5-FU-resistant patients both before and after treatment (Figure 1D and E). Notably, post-NACT ICAM2 levels were further reduced compared to pre-treatment levels, suggesting a role for ICAM2 in both primary and acquired chemoresistance. ROC curve analysis demonstrated that pre-NACT ICAM2 levels could reliably distinguish between chemo-sensitive and chemo-resistant patients (Figure 1F).
In the training cohort, ELISA analysis of serum ICAM2 in AGC patients showed lower levels in 5-FU-resistant cases at both pre- and post-NACT stages, with significant reductions after treatment (Figure 1G). ROC analysis of pre-NACT ICAM2 levels demonstrated strong predictive value for NACT response (area under the curve = 0.876) with 75.86% sensitivity and 88.89% specificity (Figure 1H). Using a cut-off value of 14.96 ug/mL, patients were stratified into low and high ICAM2 groups. Lower ICAM2 levels were associated with poorer tumor differentiation, advanced TNM stage, and chemoresistance (Table 1), as well as worse OS and DFS (Figure 1I and J). Multivariate analysis confirmed ICAM2 as an independent predictor of OS and DFS (Table 2).
| Variables | Cases | Low serum ICAM2 (n = 47) | High serum ICAM2 (n = 38) | P value |
| TRG-Ryan | < 0.001 | |||
| 0 | 16 | 2 (4) | 14 (37) | |
| 1 | 11 | 1 (2) | 10 (26) | |
| 2 | 43 | 33 (70) | 10 (26) | |
| 3 | 15 | 11 (24) | 4 (11) | |
| Chemotherapy | < 0.001 | |||
| Sensitive | 27 | 3 (6) | 24 (63) | |
| Resistant | 58 | 44 (94) | 14 (37) | |
| Age (years) | 0.241 | |||
| < 60 | 50 | 25 (53) | 25 (66) | |
| ≥ 60 | 35 | 22 (47) | 13 (34) | |
| Sex | 0.483 | |||
| Male | 64 | 34 (72) | 30 (79) | |
| Female | 21 | 13 (28) | 8 (21) | |
| CEA (ng/mL) | 0.271 | |||
| ≤ 5 | 61 | 36 (77) | 25 (66) | |
| > 5 | 24 | 11 (23) | 13 (34) | |
| CA19-9 (U/mL) | 0.879 | |||
| ≤ 37 | 71 | 39 (83) | 32 (84) | |
| > 37 | 14 | 8 (17) | 6 (16) | |
| Tumor differentiation | 0.008 | |||
| High and moderate | 36 | 14 (30) | 22 (59) | |
| Poor | 47 | 32 (70) | 15 (41) | |
| Advanced TNM stage | 0.041 | |||
| II | 41 | 18 (38) | 23 (61) | |
| III | 44 | 29 (62) | 15 (39) | |
| Tumor location | 0.613 | |||
| Esophagogastric junction | 23 | 12 (26) | 11 (29) | |
| Pylorus/corpus | 29 | 18 (39) | 11 (29) | |
| Antrum | 32 | 16 (35) | 16 (42) |
| Variables | OS (univariate analysis) | OS (multivariate analysis) | DFS (univariate analysis) | DFS (multivariate analysis) | ||||||||
| HR | 95%CI | P value | HR | 95%CI | P value | HR | 95%CI | P value | HR | 95%CI | P value | |
| ICAM2 in serum (high vs low) | 0.378 | 0.202-0.707 | 0.002 | 0.441 | 0.234-0.828 | 0.011 | 0.403 | 0.222-0.730 | 0.003 | 0.446 | 0.245-0.811 | 0.008 |
| Age (years; ≥ 60 vs < 60) | 1.429 | 0.806-2.532 | 0.222 | 1.450 | 0.831-2.531 | 0.191 | ||||||
| Gender (male vs female) | 0.824 | 0.435-1.562 | 0.553 | 1.004 | 0.534-1.887 | 0.991 | ||||||
| CEA (ng/mL; > 5 vs ≤ 5) | 0.788 | 0.416-1.492 | 0.465 | 0.660 | 0.351-1.242 | 0.198 | ||||||
| CA19-9 (U/mL; > 37 vs ≤ 37) | 1.954 | 0.995-3.838 | 0.052 | 1.751 | 0.896-3.421 | 0.101 | ||||||
| Differentiation (high and moderate vs poor) | 0.588 | 0.323-1.067 | 0.081 | 0.688 | 0.388-1.221 | 0.201 | ||||||
| Advanced TNM stage (III vs II) | 3.298 | 1.763-6.170 | < 0.001 | 2.938 | 1.562-5.528 | 0.001 | 3.226 | 1.776-5.860 | < 0.001 | 2.977 | 1.644-5.462 | < 0.001 |
In the independent validation cohort, pre-NACT serum ICAM2 levels significantly differed between chemo-sensitive and chemo-resistant patients (Figure 1K), with ROC analysis confirming its ability to distinguish between the two groups (Figure 1L). These findings emphasize the potential of ICAM2 as a predictive biomarker for NACT response and prognosis in AGC patients, with lower ICAM2 levels linked to 5-FU resistance and adverse clinical outcomes.
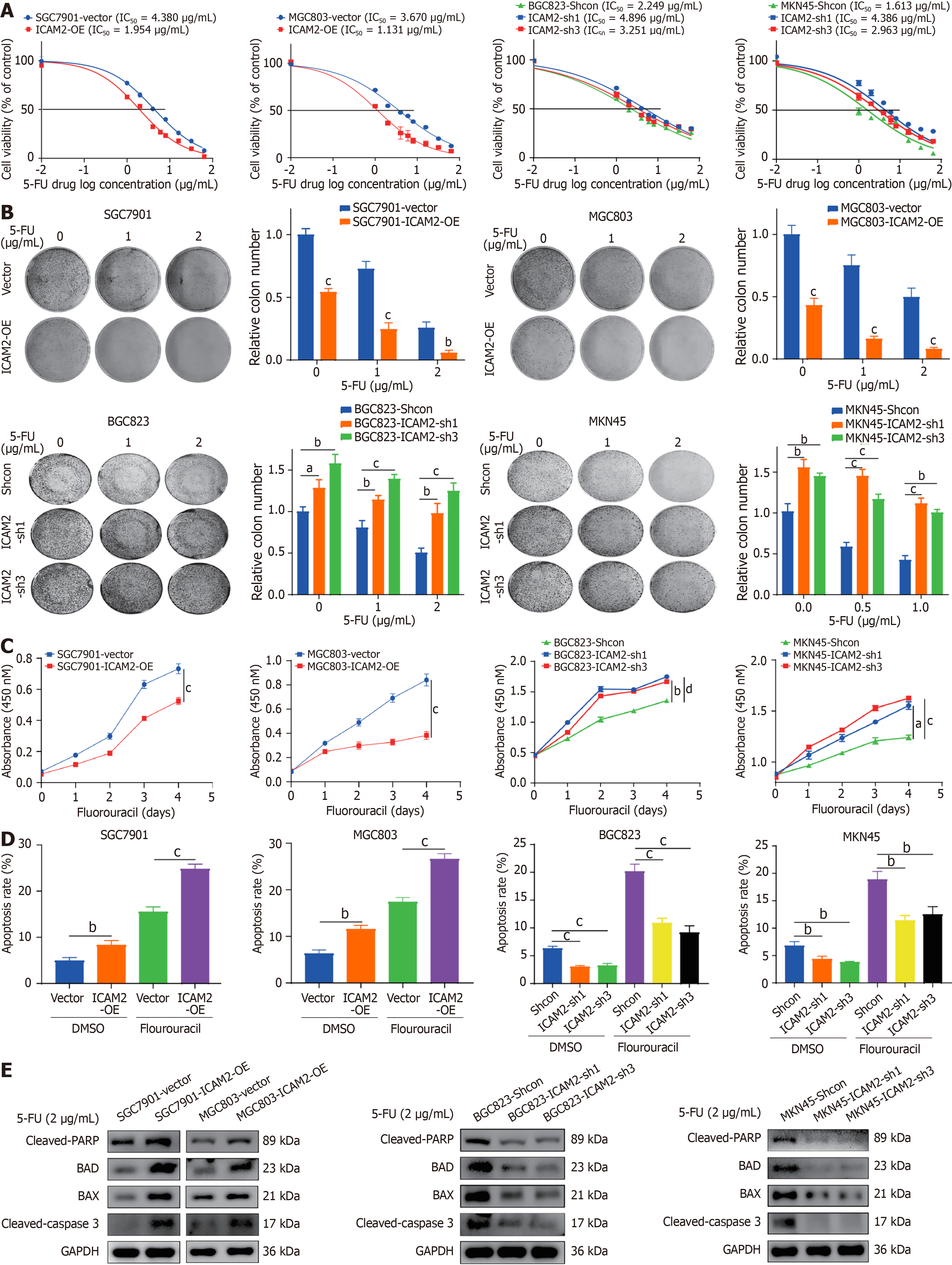
To explore ICAM2's role in GC cell chemoresistance, stable ICAM2 overexpression and knockdown GC cells were generated, with transduction efficiency confirmed by RT-PCR and western blotting (Supplementary Figure 1B). Cytotoxicity assays showed that ICAM2 overexpression significantly decreased the IC50 of 5-FU, while ICAM2 knock
Since many chemotherapy agents induce cytotoxicity through apoptosis, we assessed the relationship between ICAM2 and apoptosis. Overexpression of ICAM2 enhanced apoptosis in 5-FU-treated GC cells, while silencing ICAM2 inhibited apoptosis (Figure 2D, Supplementary Figure 1D). Analysis of apoptosis-related proteins revealed increased levels of cleaved caspase-3, cleaved PARP, Bax, and Bad following ICAM2 overexpression, while ICAM2 knockdown led to reduced levels of these pro-apoptotic markers (Figure 2E). These findings strongly support the model that ICAM2 knockdown inhibits caspase-dependent apoptosis and enhances 5-FU resistance in GC cells.
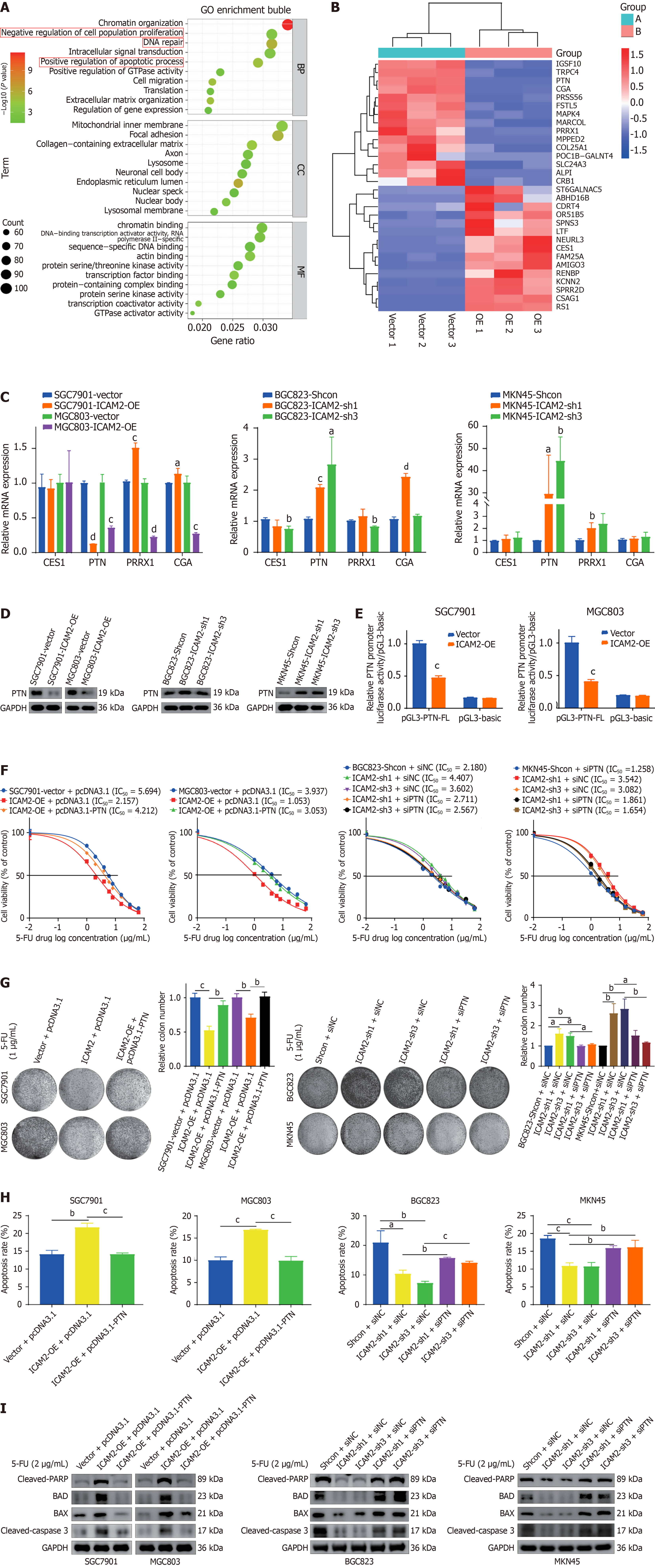
To identify the downstream effectors involved in ICAM2-mediated chemoresistance in GC, RNA sequencing was performed on 5-FU-treated GC cells overexpressing ICAM2. This analysis revealed significant changes in gene expre
To clarify PTN’s role in ICAM2-mediated chemoresistance, PTN was overexpressed in ICAM2-overexpressing GC cells and silenced in ICAM2-knockdown cells to assess its impact on resistance phenotypes (Supplementary Figure 2B). Knockdown of PTN reversed the resistance of ICAM2-silenced cells to 5-FU, as demonstrated by cytotoxicity, colony formation, flow cytometry, and apoptosis-related protein analyses. Conversely, overexpression of PTN in ICAM2-overexpressing cells increased 5-FU resistance (Figure 3F-I and Supplementary Figure 2C and D). These results highlight PTN as a key mediator of ICAM2-associated chemoresistance in GC cells.
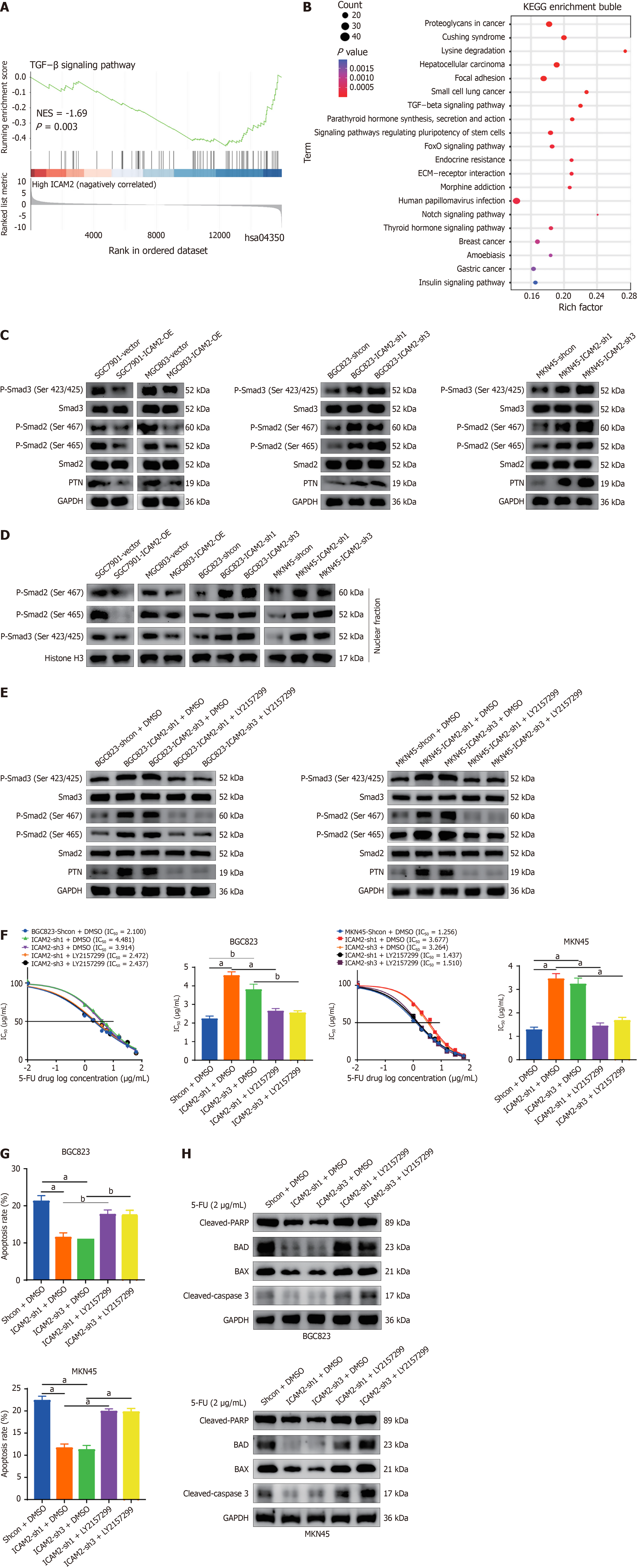
To investigate how ICAM2 knockdown induces PTN expression and confers 5-FU resistance in GC, we performed gene set enrichment analysis on RNA-seq data obtained from 5-FU-treated GC cells. The TGF-β signaling pathway emerged as one of the most significantly enriched pathways (Figure 4A). Additionally, DEGs identified after ICAM2 overexpression were highly enriched in the TGF-β pathway, as shown by KEGG analysis (Figure 4B). To investigate the involvement of TGF-β signaling in the regulation of ICAM2-mediated PTN expression, we assessed the activation of TGF-β signaling intermediaries, specifically Smad2/3. Overexpression of ICAM2 resulted in significant reductions in P-Smad2/3 and PTN protein levels, without affecting total Smad2/3 expression (Figure 4C). In contrast, ICAM2 silencing led to increased P-Smad2/3 and PTN levels. Further Western blot analysis of nuclear extracts confirmed that ICAM2 overexpression inhibited, while silencing ICAM2 enhanced, the accumulation of P-Smad2/3 in the GC cell nuclei (Figure 4D). To determine whether TGF-β signaling mediates the upregulation of PTN induced by ICAM2 loss, GC cells with ICAM2 knockdown were treated with the TGF-β receptor inhibitor galunisertib (LY2157299). Pharmacological blockade of TGF-β signaling completely abrogated the increase in PTN expression caused by ICAM2 depletion (Figure 4E), implicating TGF-β pathway activation as a critical driver of PTN upregulation following ICAM2 silencing.
Moreover, to further explore the role of TGF-β signaling in ICAM2-driven chemoresistance, LY2157299 treatment was administered to ICAM2-deficient GC cells. Inhibition of TGF-β signaling effectively restored 5-FU sensitivity, as evidenced by decreased IC50 values, increased apoptosis rates, and upregulation of pro-apoptotic proteins (Figure 4F-H and Supplementary Figure 3A). Collectively, these findings suggest that ICAM2 silencing promotes chemoresistance in GC by activating TGF-β signaling and inducing PTN expression.
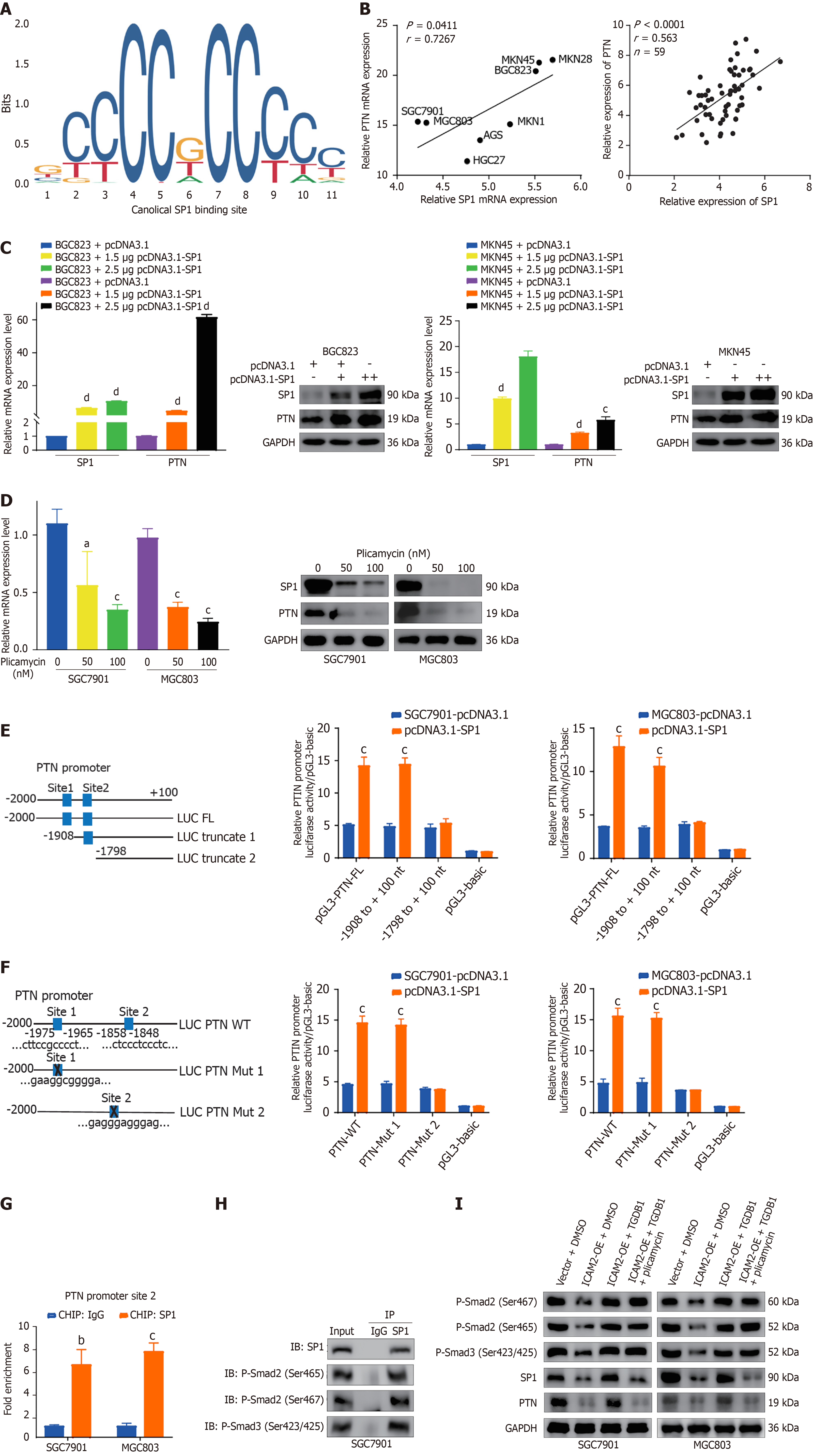
To investigate the transcription factors responsible for PTN upregulation in GC cells, we analyzed data from the TRRUST and JASPAR databases, which predicted SP1 as a potential regulator of PTN. Two SP1-binding sites (sites 1 and 2) within the PTN promoter were identified, both showing predicted scores greater than 10 (Figure 5A and Supplementary Table
To determine whether SP1 directly binds to the PTN promoter, we constructed full-length and truncated PTN promoter plasmids (Figure 5E). In luciferase reporter assays, SP1 overexpression enhanced the activity of the full-length PTN promoter (pGL3-PTN-FL) and the truncated version (pGL3-PTN-truncated 1), but not the other truncated version (pGL3-PTN-truncated 2; Figure 5E). Mutation of binding site 1 did not abolish the ability of SP1 overexpression to enhance PTN promoter luciferase activity, whereas mutation of binding site 2 completely abrogated this effect, indicating that site 2 is essential for SP1-driven transcriptional activation of PTN (Figure 5F). ChIP assays confirmed SP1 binding to site 2 of the PTN promoter (Figure 5G), located between nucleotides -1858 and -1848, thereby establishing SP1 as a direct regulator of PTN expression.
Previous studies have demonstrated that SP1 functions as a downstream effector of TGF-β signaling, forming complexes with P-Smad2/3 to co-regulate transcriptional programs[24-26]. Given that TGF-β1 is a potent activator of this pathway and widely used to mimic TGF-β signaling activation
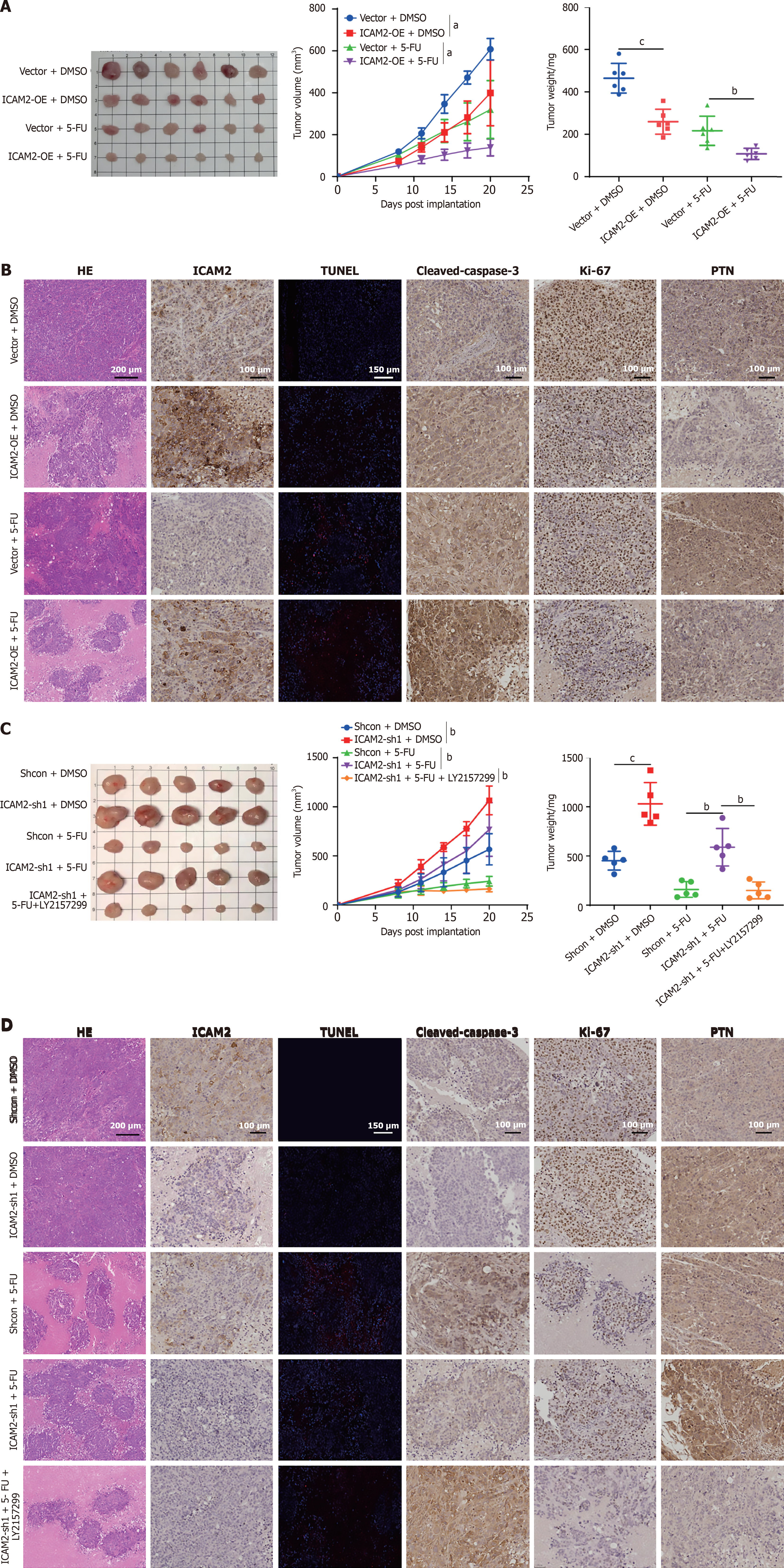
To investigate the in vivo role of ICAM2 in chemoresistance, a subcutaneous xenograft model of GC cells was established in nude mice. When tumors reached approximately 100 mm3, the mice were randomized into four groups (vector + DMSO, ICAM2-OE + DMSO, vector + 5-FU, ICAM2-OE + 5-FU), and treated with intraperitoneal injections of 5-FU or DMSO. Tumor growth was monitored every three days. The combination treatment (ICAM2-OE + 5-FU) showed significantly lower tumor growth rates and smaller tumor sizes compared to the other groups, with tumors in this group also weighing less (Figure 6A). Apoptosis rates were highest in the combination group, which also exhibited the lowest Ki-67 expression and the highest levels of cleaved caspase-3 (Figure 6B). These results confirmed that ICAM2 overexpression enhances GC tumor sensitivity to 5-FU in vivo.
Given that excessive TGF-β signaling activity in GC cells with low ICAM2 expression is critical for chemoresistance, we hypothesized that targeting the TGF-β pathway could improve chemotherapeutic efficacy in vivo. To test this, we evaluated the impact of 5-FU treatment alone or combined with the TGF-β inhibitor LY2157299 on GC xenografts (ShNC + DMSO, ICAM2-sh1 + DMSO, ShNC + 5-FU, ICAM2-sh1 + 5-FU, and ICAM2-sh1 + 5-FU + LY2157299). ICAM2 silencing increased 5-FU resistance, whereas combination treatment with 5-FU and LY2157299 significantly impaired tumor growth (Figure 6C). Both TUNEL staining and IHC analyses showed that LY2157299 enhanced tumor cell apoptosis and reduced Ki-67 staining compared to 5-FU treatment alone in xenografts with low ICAM2 levels (Figure 6D). These findings highlight the potential of targeting TGF-β signaling to overcome 5-FU resistance in GC tumors with low ICAM2 expression.
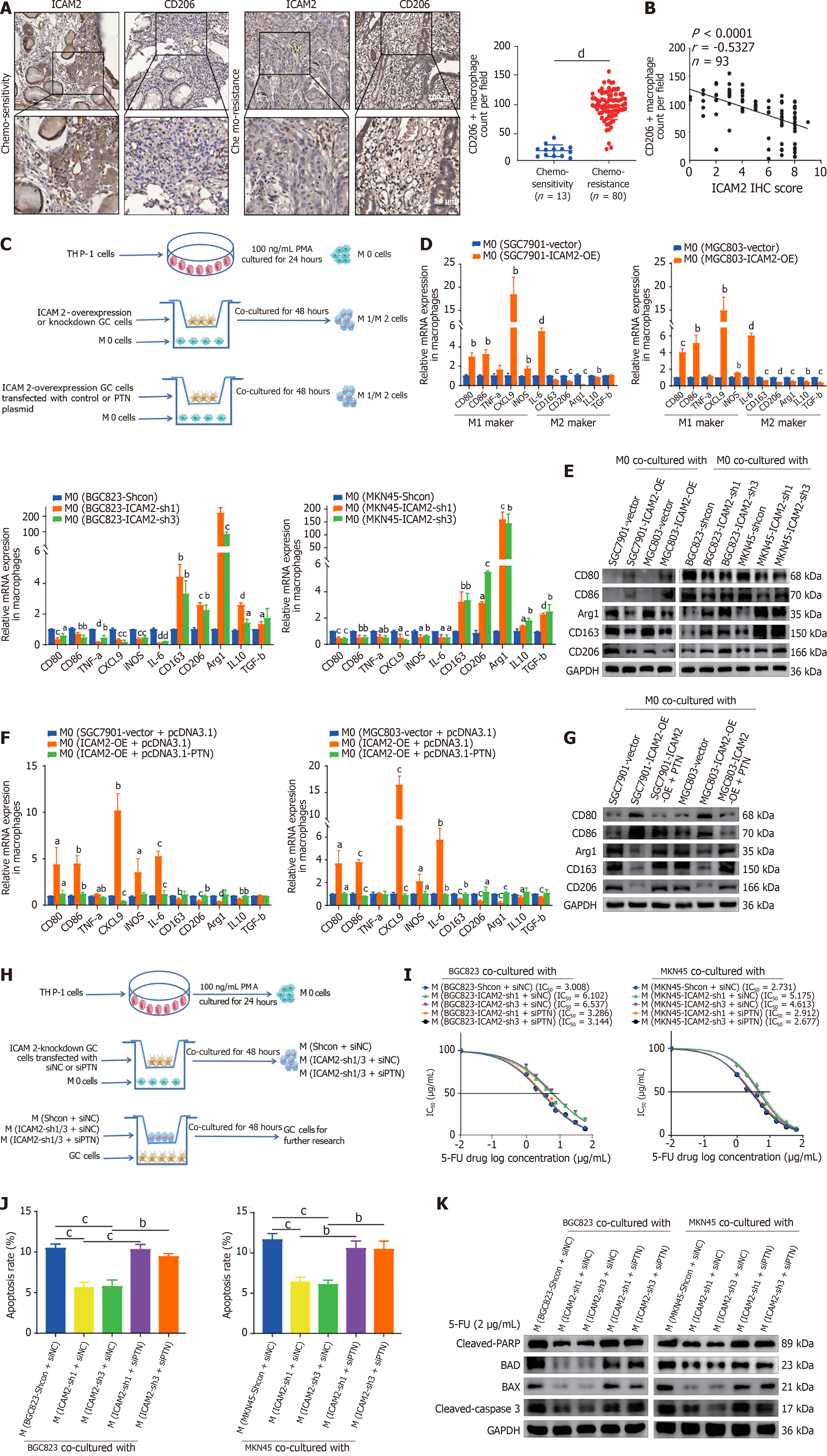
Chemoresistance is strongly influenced by interactions between tumor cells and the TME, with M2 macrophages playing a pivotal role in this process[27,28]. ICAM2 has been linked to immune surveillance and immune response modulation, but its role in regulating chemoresistance through M2 macrophage polarization in GC is unclear. Therefore, we analyzed ICAM2 expression and M2 macrophage infiltration in tumor tissues from 93 post-NACT AGC patients using IHC. M2 macrophages (CD206+ macrophages) were significantly more abundant in chemo-resistant tissues compared to chemo-sensitive ones (Figure 7A). A negative correlation between ICAM2 expression and M2 macrophage infiltration was also observed (Figure 7B), suggesting that ICAM2 downregulation may contribute to 5-FU resistance in AGC by promoting M2 macrophage polarization.
Next, we explored the effect of ICAM2 on macrophage activation through co-culture of PMA-stimulated THP-1 cells with GC cells (Figure 7C). Co-culture of M0 macrophages with ICAM2-overexpressing GC cells resulted in upregulation of M1 markers (CD80, CD86, CXCL9, iNOS, IL6) and downregulation of M2 markers (CD163, CD206, Arg1, IL10, TGF-β) at the mRNA level (Figure 7D). Conversely, co-culture with ICAM2-knockdown GC cells exhibited the opposite effect (Figure 7D). Western blotting of macrophages further demonstrated that co-culture with ICAM2-overexpressing GC cells elevated M1 markers (CD80 and CD86) while reducing M2 markers (CD163, CD206, and Arg1), whereas ICAM2 knockdown led to the opposite effect (Figure 7E). These findings suggest that ICAM2 downregulation promotes M2 macrophage polarization.
Since PTN has been implicated in tumor progression and is known to influence the TME[29], we hypothesized that it might also contribute to macrophage polarization in the context of ICAM2-mediated M2 polarization in GC. Although we did not directly measure PTN levels in cell culture supernatants following ICAM2 modulation, we performed a rescue experiment by overexpressing PTN in ICAM2-overexpressing GC cells prior to macrophage co-culture (Figure 7C). PTN overexpression partially reversed the effects of ICAM2 overexpression on macrophage polarization, increasing the expression of M2 markers (CD163, CD206, and Arg1) and reducing M1 markers (CD80 and CD86) at both mRNA and protein levels (Figure 7F and G). These results indicate that PTN may contribute to macrophage polarization in this context, although further studies are needed to confirm whether PTN acts as a direct mediator of ICAM2-regulated macrophage phenotype changes.
To further explore the impact of ICAM2-induced macrophage polarization on GC cell chemoresistance, M0 macrophages were first polarized by co-culture with ICAM2-knockdown or ICAM2-overexpressing GC cells. These polarized macrophages were then harvested and co-cultured with wild-type GC cells, whose chemoresistance was subsequently evaluated (Figure 7H). Cytotoxicity assays revealed that ICAM2 knockdown-induced macrophage polarization enhanced 5-FU resistance in GC cells, an effect that was reduced when PTN was silenced in GC cells (Figure 7I, Supplementary Figure 3D). Similarly, apoptotic induction was diminished in GC cells co-cultured with ICAM2 knockdown-induced macrophages compared to controls, with PTN silencing reversing this effect (Figure 7J, Supplementary Figure 3E). Western blotting confirmed that ICAM2 knockdown-induced macrophages led to increased anti-apoptotic proteins and reduced pro-apoptotic proteins in GC cells, with PTN knockdown reversing these changes (Figure 7K). Collectively, these data suggest that ICAM2 downregulation contributes to 5-FU resistance in GC, potentially through PTN-associated modulation of macrophage polarization, though further investigation is warranted to delineate the exact mechanisms involved.
GC remains a significant global health challenge, characterized by high morbidity and mortality, with the majority of cases diagnosed at advanced clinical stages (cTNM II-III)[30]. NACT, predominantly 5-FU-based regimens, serves as a cornerstone for the management of AGC, aiming to improve surgical resectability and patient survival[31]. However, intrinsic or acquired chemoresistance considerably limits therapeutic efficacy, leading to poor outcomes in a substantial proportion of patients[32]. Thus, there is an urgent clinical need to identify reliable biomarkers predictive of chemotherapeutic response and to uncover the intricate molecular pathways driving chemoresistance in AGC. In this study, we established ICAM2 as a robust and clinically relevant biomarker predictive of the response to NACT in patients with AGC. Reduced ICAM2 expression in both serum and tumor tissues correlated strongly with diminished responsiveness to chemotherapy and poorer overall prognosis. The high diagnostic performance of ICAM2, validated via ROC curve analysis, highlights its potential utility as a predictive biomarker with significant clinical implications. These findings align with current clinical demands for precise biomarkers that can stratify patient treatment and facilitate tailored therapeutic interventions.
Chemoresistance in GC involves complex interactions between intrinsic tumor cell mechanisms and extrinsic modulation by the TME[33]. A critical hallmark of chemo-resistant cancer cells is their ability to evade apoptosis induced by chemotherapeutic agents[34]. Our results convincingly demonstrate that ICAM2 downregulation significantly inhibits caspase-dependent apoptosis following exposure to 5-FU, marked by decreased cleavage of caspase-3 and PARP, and reduced expression of the pro-apoptotic proteins Bax and Bad. These findings confirm apoptosis resistance as a fundamental mechanism by which reduced ICAM2 expression promotes intrinsic tumor cell chemoresistance.
Beyond intrinsic mechanisms, a novel and compelling aspect of our study is the identification of ICAM2 as a key regulator of immune cell function within the GC microenvironment, specifically through modulation of macrophage polarization. We observed a pronounced shift towards the immunosuppressive M2 macrophage phenotype following ICAM2 downregulation in GC cells. M2 macrophages are well-characterized as key players in creating an immunosuppressive milieu that fosters tumor cell survival and chemotherapeutic resistance by secreting pro-survival cytokines and growth factors[35,36]. Consistent with these reports, our co-culture experiments demonstrated that M2 macrophages induced by ICAM2 deficiency significantly promote GC cell resistance to 5-FU, thereby providing direct experimental evidence linking ICAM2 loss to immune-mediated chemoresistance.
Mechanistically, our data causally support a critical ICAM2-regulated signaling pathway that connects tumor-intrinsic and immune-mediated chemoresistance. Our data indicate that decreased ICAM2 expression activates the TGF-β/Smad/SP1 signaling cascade, resulting in increased production of PTN, a growth factor previously implicated in promoting tumor cell survival and immune evasion[37,38]. Elevated PTN expression, driven by this axis, was found to simultaneously reinforce apoptotic resistance in tumor cells and promote M2 macrophage polarization, thus exacerbating chemoresistance through dual, complementary mechanisms. In addition, we cannot exclude the possibility that ICAM2 loss also influences the TME. Specifically, reduced ICAM2 expression may promote M2 macrophage polarization and increase their secretion of TGF-β, thereby generating a feed-forward loop that amplifies TGF-β/Smad signaling and indirectly contributes to 5-FU resistance. Crucially, our preclinical studies demonstrated that targeted inhibition of the TGF-β pathway using LY2157299, a selective TGF-β receptor I inhibitor, robustly reversed ICAM2-mediated chemoresistance both in vitro and in vivo. These findings support emerging therapeutic strategies combining TGF-β pathway inhibitors with standard chemotherapy to effectively overcome chemoresistance in AGC patients exhibiting low ICAM2 expression. Of note, other ICAM family members such as ICAM1 have also been implicated in chemotherapy resistance, particularly to cisplatin and docetaxel, through context-dependent mechanisms involving cytokine signaling and STAT3 activation[39,40]. However, our data indicate that the ICAM2/TGF-β/Smad/SP1/PTN axis specifically mediates 5-FU resistance in GC, suggesting a distinct, ICAM2-linked regulatory pathway.
Despite the strengths of our study, several limitations should be acknowledged. Although our data suggest that ICAM2 downregulation activates the TGF-β/Smad/SP1 pathway and elevates PTN expression, we have not yet defined the precise molecular link between ICAM2 and TGF-β signaling. Moreover, while our findings demonstrate that PTN ex
In conclusion, this study identifies ICAM2 as a novel and clinically significant biomarker predictive of NACT response and patient prognosis in AGC, while unveiling its pivotal role in governing chemoresistance through apoptosis evasion and immunosuppressive reprogramming of the TME mediated by M2 macrophage polarization. Mechanistically, we delineate an ICAM2/TGF-β/Smad/SP1/PTN signaling axis that orchestrates these processes, underscoring its potential as a therapeutic target. Notably, pharmacological inhibition of this pathway restores chemosensitivity, establishing ICAM2 as both a predictive indicator and a viable therapeutic node for precision treatment in GC.
We gratefully acknowledge the support provided by the National Key Clinical Discipline.
| 1. | Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5690] [Cited by in RCA: 12562] [Article Influence: 6281.0] [Reference Citation Analysis (6)] |
| 2. | Gastric Cancer Association; China Anti-Cancer Association. [Chinese expert consensus on perioperative treatment of locally advanced gastric cancer (2021 version)]. Zhonghua Wei Chang Wai Ke Za Zhi. 2021;24:741-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 3. | Wang FH, Zhang XT, Tang L, Wu Q, Cai MY, Li YF, Qu XJ, Qiu H, Zhang YJ, Ying JE, Zhang J, Sun LY, Lin RB, Wang C, Liu H, Qiu MZ, Guan WL, Rao SX, Ji JF, Xin Y, Sheng WQ, Xu HM, Zhou ZW, Zhou AP, Jin J, Yuan XL, Bi F, Liu TS, Liang H, Zhang YQ, Li GX, Liang J, Liu BR, Shen L, Li J, Xu RH. The Chinese Society of Clinical Oncology (CSCO): Clinical guidelines for the diagnosis and treatment of gastric cancer, 2023. Cancer Commun (Lond). 2024;44:127-172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 212] [Article Influence: 106.0] [Reference Citation Analysis (0)] |
| 4. | Guo T, Tang XH, Gao XY, Zhou Y, Jin B, Deng ZQ, Hu Y, Xing XF, Li ZY, Ji JF. A liquid biopsy signature of circulating exosome-derived mRNAs, miRNAs and lncRNAs predict therapeutic efficacy to neoadjuvant chemotherapy in patients with advanced gastric cancer. Mol Cancer. 2022;21:216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 52] [Reference Citation Analysis (0)] |
| 5. | Staunton DE, Dustin ML, Springer TA. Functional cloning of ICAM-2, a cell adhesion ligand for LFA-1 homologous to ICAM-1. Nature. 1989;339:61-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 576] [Cited by in RCA: 571] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 6. | Perez OD, Kinoshita S, Hitoshi Y, Payan DG, Kitamura T, Nolan GP, Lorens JB. Activation of the PKB/AKT pathway by ICAM-2. Immunity. 2002;16:51-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 102] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 7. | Liu Z, Guo B, Lopez RD. Expression of intercellular adhesion molecule (ICAM)-1 or ICAM-2 is critical in determining sensitivity of pancreatic cancer cells to cytolysis by human gammadelta-T cells: implications in the design of gammadelta-T-cell-based immunotherapies for pancreatic cancer. J Gastroenterol Hepatol. 2009;24:900-911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 8. | Tanaka H, Yashiro M, Sunami T, Sakate Y, Kosaka K, Hirakawa K. ICAM-2 gene therapy for peritoneal dissemination of scirrhous gastric carcinoma. Clin Cancer Res. 2004;10:4885-4892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 27] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 9. | Tang X, Huang J, Jiang Y, Qiu J, Li T, Li W, Chen Z, Huang Z, Yu X, Yang T, Ji X, Tan R, Lv L, Yang Z, Chen H. Intercellular adhesion molecule 2 as a novel prospective tumor suppressor induced by ERG promotes ubiquitination-mediated radixin degradation to inhibit gastric cancer tumorigenicity and metastasis. J Transl Med. 2023;21:670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 10. | Yoon KJ, Phelps DA, Bush RA, Remack JS, Billups CA, Khoury JD. ICAM-2 expression mediates a membrane-actin link, confers a nonmetastatic phenotype and reflects favorable tumor stage or histology in neuroblastoma. PLoS One. 2008;3:e3629. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 11. | Ishigami T, Uzawa K, Fushimi K, Saito K, Kato Y, Nakashima D, Higo M, Kouzu Y, Bukawa H, Kawata T, Ito H, Tanzawa H. Inhibition of ICAM2 induces radiosensitization in oral squamous cell carcinoma cells. Br J Cancer. 2008;98:1357-1365. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 12. | Shin DH, Choi M, Han C, Kim SS. Targeting TGF-β-Smad2/3-JNK1-mediated SIRT1 activity overcomes the chemoresistance of KRAS mutation lung cancer. Exp Mol Med. 2025;57:2022-2039. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 13. | Xu Y, Li Y, Chen X, Xiang F, Deng Y, Li Z, Wei D. TGF-β protects osteosarcoma cells from chemotherapeutic cytotoxicity in a SDH/HIF1α dependent manner. BMC Cancer. 2021;21:1200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 14. | Carrillo-Gálvez AB, Quintero JE, Rodríguez R, Menéndez ST, Victoria González M, Blanco-Lorenzo V, Allonca E, de Araújo Farias V, González-Correa JE, Erill-Sagalés N, Martínez-Zubiaurre I, Hellevik T, Sánchez-Hernández S, Muñoz P, Zurita F, Martín F, Rodríguez-Manzaneque JC, Anderson P. GARP promotes the proliferation and therapeutic resistance of bone sarcoma cancer cells through the activation of TGF-β. Cell Death Dis. 2020;11:985. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 15. | Chen TW, Hung WZ, Chiang SF, Chen WT, Ke TW, Liang JA, Huang CY, Yang PC, Huang KC, Chao KSC. Dual inhibition of TGFβ signaling and CSF1/CSF1R reprograms tumor-infiltrating macrophages and improves response to chemotherapy via suppressing PD-L1. Cancer Lett. 2022;543:215795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 16. | He W, Liang B, Wang C, Li S, Zhao Y, Huang Q, Liu Z, Yao Z, Wu Q, Liao W, Zhang S, Liu Y, Xiang Y, Liu J, Shi M. MSC-regulated lncRNA MACC1-AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene. 2019;38:4637-4654. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 246] [Article Influence: 35.1] [Reference Citation Analysis (0)] |
| 17. | Wang X. Pleiotrophin: Activity and mechanism. Adv Clin Chem. 2020;98:51-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 18. | Hu J, Hu Y, Zhang X, Zhang J, Zhou Y, Wang X, Wu W, Chen J, Han Y. Tenacissoside G reverses paclitaxel resistance by inhibiting Src/PTN/P-gp signaling axis activation in ovarian cancer cells. J Nat Med. 2025;79:621-638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 19. | Huang P, Ouyang DJ, Chang S, Li MY, Li L, Li QY, Zeng R, Zou QY, Su J, Zhao P, Pei L, Yi WJ. Chemotherapy-driven increases in the CDKN1A/PTN/PTPRZ1 axis promote chemoresistance by activating the NF-κB pathway in breast cancer cells. Cell Commun Signal. 2018;16:92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 20. | Sun X, Tian C, Zhang H, Han K, Zhou M, Gan Z, Zhu H, Min D. Long noncoding RNA OIP5-AS1 mediates resistance to doxorubicin by regulating miR-137-3p/PTN axis in osteosarcoma. Biomed Pharmacother. 2020;128:110201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 21. | Wu D, Liu L, Yan X, Wang C, Wang Y, Han K, Lin S, Gan Z, Min D. Pleiotrophin promotes chemoresistance to doxorubicin in osteosarcoma by upregulating P-glycoprotein. Oncotarget. 2017;8:63857-63870. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 22. | Ajani JA, D'Amico TA, Bentrem DJ, Chao J, Cooke D, Corvera C, Das P, Enzinger PC, Enzler T, Fanta P, Farjah F, Gerdes H, Gibson MK, Hochwald S, Hofstetter WL, Ilson DH, Keswani RN, Kim S, Kleinberg LR, Klempner SJ, Lacy J, Ly QP, Matkowskyj KA, McNamara M, Mulcahy MF, Outlaw D, Park H, Perry KA, Pimiento J, Poultsides GA, Reznik S, Roses RE, Strong VE, Su S, Wang HL, Wiesner G, Willett CG, Yakoub D, Yoon H, McMillian N, Pluchino LA. Gastric Cancer, Version 2.2022, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2022;20:167-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 1163] [Article Influence: 290.8] [Reference Citation Analysis (0)] |
| 23. | Zhou J, Li T, Chen H, Jiang Y, Zhao Y, Huang J, Chen Z, Tang X, Huang Z, Yang Z. ADAMTS10 inhibits aggressiveness via JAK/STAT/c-MYC pathway and reprograms macrophage to create an anti-malignant microenvironment in gastric cancer. Gastric Cancer. 2022;25:1002-1016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 27] [Reference Citation Analysis (0)] |
| 24. | Zhu HF, Liu YP, Liu DL, Ma YD, Hu ZY, Wang XY, Gu CS, Zhong Y, Long T, Kan HP, Li ZG. Role of TGFβ3-Smads-Sp1 axis in DcR3-mediated immune escape of hepatocellular carcinoma. Oncogenesis. 2019;8:43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 25. | Jungert K, Buck A, Buchholz M, Wagner M, Adler G, Gress TM, Ellenrieder V. Smad-Sp1 complexes mediate TGFbeta-induced early transcription of oncogenic Smad7 in pancreatic cancer cells. Carcinogenesis. 2006;27:2392-2401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 48] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 26. | Jungert K, Buck A, von Wichert G, Adler G, König A, Buchholz M, Gress TM, Ellenrieder V. Sp1 is required for transforming growth factor-beta-induced mesenchymal transition and migration in pancreatic cancer cells. Cancer Res. 2007;67:1563-1570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 91] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 27. | Chen C, Jin L, Wan H, Liu H, Zhang S, Shen G, Gong J, Zhu Y. TIPE3 promotes drug resistance in colorectal cancer by enhancing autophagy via the USP19/Beclin1 pathway. Cell Death Discov. 2025;11:202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 28. | Li H, Luo F, Jiang X, Zhang W, Xiang T, Pan Q, Cai L, Zhao J, Weng D, Li Y, Dai Y, Sun F, Yang C, Huang Y, Yang J, Tang Y, Han Y, He M, Zhang Y, Song L, Xia JC. CircITGB6 promotes ovarian cancer cisplatin resistance by resetting tumor-associated macrophage polarization toward the M2 phenotype. J Immunother Cancer. 2022;10:e004029. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 121] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 29. | Shen D, Podolnikova NP, Yakubenko VP, Ardell CL, Balabiyev A, Ugarova TP, Wang X. Pleiotrophin, a multifunctional cytokine and growth factor, induces leukocyte responses through the integrin Mac-1. J Biol Chem. 2017;292:18848-18861. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 30. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 68525] [Article Influence: 13705.0] [Reference Citation Analysis (201)] |
| 31. | Wagner AD, Syn NL, Moehler M, Grothe W, Yong WP, Tai BC, Ho J, Unverzagt S. Chemotherapy for advanced gastric cancer. Cochrane Database Syst Rev. 2017;8:CD004064. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 444] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 32. | Alsina M, Arrazubi V, Diez M, Tabernero J. Current developments in gastric cancer: from molecular profiling to treatment strategy. Nat Rev Gastroenterol Hepatol. 2023;20:155-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 227] [Article Influence: 75.7] [Reference Citation Analysis (1)] |
| 33. | Wang Y, Zhang J, Shi H, Wang M, Yu D, Fu M, Qian Y, Zhang X, Ji R, Wang S, Gu J, Zhang X. M2 Tumor-Associated Macrophages-Derived Exosomal MALAT1 Promotes Glycolysis and Gastric Cancer Progression. Adv Sci (Weinh). 2024;11:e2309298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 48] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 34. | Tang X, Liang Y, Sun G, He Q, Hou Z, Jiang X, Gao P, Qu H. Upregulation of CRABP2 by TET1-mediated DNA hydroxymethylation attenuates mitochondrial apoptosis and promotes oxaliplatin resistance in gastric cancer. Cell Death Dis. 2022;13:848. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 32] [Reference Citation Analysis (0)] |
| 35. | Umeda H, Shigeyasu K, Takahashi T, Moriwake K, Kondo Y, Yoshida K, Takeda S, Yano S, Matsumi Y, Kishimoto H, Fuji T, Yasui K, Yamamoto H, Takagi K, Kayano M, Michiue H, Nakamura K, Mori Y, Teraishi F, Tazawa H, Umeda Y, Kagawa S, Goel A, Fujiwara T. ADAR1-high tumor-associated macrophages induce drug resistance and are therapeutic targets in colorectal cancer. Mol Cancer. 2025;24:116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 36. | Yang T, Deng Z, Xu L, Li X, Yang T, Qian Y, Lu Y, Tian L, Yao W, Wang J. Macrophages-aPKC(ɩ)-CCL5 Feedback Loop Modulates the Progression and Chemoresistance in Cholangiocarcinoma. J Exp Clin Cancer Res. 2022;41:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 42] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 37. | Ganguly D, Schmidt MO, Coleman M, Ngo TC, Sorrelle N, Dominguez ATA, Murimwa GZ, Toombs JE, Lewis C, Fang YV, Valdes-Mora F, Gallego-Ortega D, Wellstein A, Brekken RA. Pleiotrophin drives a prometastatic immune niche in breast cancer. J Exp Med. 2023;220:e20220610. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 38. | Zhang M, Zhou K, Wang Z, Liu T, Stevens LE, Lynce F, Chen WY, Peng S, Xie Y, Zhai D, Chen Q, Shi Y, Shi H, Yuan Z, Li X, Xu J, Cai Z, Guo J, Shao N, Lin Y. A Subpopulation of Luminal Progenitors Secretes Pleiotrophin to Promote Angiogenesis and Metastasis in Inflammatory Breast Cancer. Cancer Res. 2024;84:1781-1798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 39. | Hsieh CY, Lin CC, Huang YW, Chen JH, Tsou YA, Chang LC, Fan CC, Lin CY, Chang WC. Macrophage secretory IL-1β promotes docetaxel resistance in head and neck squamous carcinoma via SOD2/CAT-ICAM1 signaling. JCI Insight. 2022;7:e157285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 35] [Reference Citation Analysis (0)] |
| 40. | Tinajero-Rodríguez JM, Ramírez-Vidal L, Becerril-Rico J, Alvarado-Ortiz E, Romero-Rodríguez DP, López-Casillas F, Hernández-Sotelo D, Fernández-Ramírez F, Contreras-Paredes A, Ortiz-Sánchez E. ICAM1 (CD54) Contributes to the Metastatic Capacity of Gastric Cancer Stem Cells. Int J Mol Sci. 2024;25:8865. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/