Published online Aug 21, 2025. doi: 10.3748/wjg.v31.i31.109948
Revised: June 28, 2025
Accepted: July 25, 2025
Published online: August 21, 2025
Processing time: 83 Days and 18.9 Hours
Gastrointestinal diseases have complex etiologies and clinical presentations. An accurate diagnosis requires physicians to integrate diverse information, including medical history, laboratory test results, and imaging findings. Existing artificial intelligence-assisted diagnostic tools are limited to single-modality information, resulting in recommendations that are often incomplete and may be associated with clinical or legal risks.
To develop and evaluate a collaborative multimodal large language model (LLM) framework for clinical decision-making in digestive diseases.
In this observational study, DeepGut, a multimodal LLM collaborative diagnostic framework, was developed to integrate four distinct large models into a four-tiered structure. The framework sequentially accomplishes multimodal infor
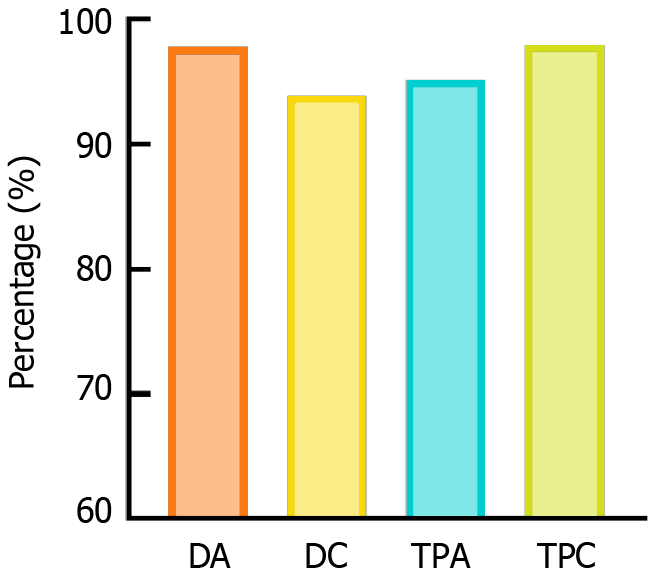
The diagnostic and treatment recommendations generated by the DeepGut framework achieved exceptional performance, with a diagnostic accuracy of 97.8%, diagnostic completeness of 93.9%, treatment plan accuracy of 95.2%, and treatment plan completeness of 98.0%, significantly surpassing the capabilities of single-modal LLM-based diagnostic tools. Experts evaluating the framework commended the completeness, relevance, and logical coherence of its outputs. However, the collaborative multimodal LLM approach resulted in increased input and output token counts, leading to higher computational costs and extended diagnostic times.
The framework achieves successful integration of multimodal diagnostic data, demonstrating enhanced performance enabled by multimodal LLM collaboration, which opens new horizons for the clinical application of artificial intelligence-assisted technology.
Core Tip: This study introduces DeepGut, a multimodal large language model (LLM) collaborative framework designed to assist in diagnostic processes by integrating multiple LLMs to extract and fuse multimodal clinical data such as medical history, laboratory tests, and imaging results. DeepGut significantly improves the diagnostic accuracy and comprehensiveness of gastrointestinal diseases compared with single-modal tools, as evidenced by expert validation. However, the framework’s higher token consumption by LLMs increases the operational costs, highlighting a key area for future optimization efforts.
- Citation: Wan XH, Liu MX, Zhang Y, Kou GJ, Xu LQ, Liu H, Yang XY, Zuo XL, Li YQ. DeepGut: A collaborative multimodal large language model framework for digestive disease assisted diagnosis and treatment. World J Gastroenterol 2025; 31(31): 109948
- URL: https://www.wjgnet.com/1007-9327/full/v31/i31/109948.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i31.109948
With rapid socioeconomic development and significant lifestyle changes, the global prevalence of digestive diseases such as inflammatory bowel disease, irritable bowel syndrome, and colorectal cancer, has shown an upward trend[1,2]. These diseases are characterized by complex etiologies and diverse clinical manifestations, requiring comprehensive assessment of multidimensional information, including past medical history, imaging studies [computed tomography (CT), magnetic resonance imaging, or ultrasound], endoscopic findings, and pathological results, to achieve an accurate diagnosis. Even in top-tier hospitals, physicians with heavy workloads may miss a key detail in the patient’s medical history or are influenced by an anchoring bias when integrating multidimensional information. Critically, China is facing a relative shortage of experienced gastroenterologists[3], resulting in limited access to care, longer waiting times, and considerable physical, psychological, and financial burdens for patients[4,5].
The rapid development of artificial intelligence (AI)-assisted diagnostic technologies has provided new solutions to these challenges[6-8]. Tong et al[9] employed convolutional neural networks and random forest models to differentiate clinically similar conditions, achieving high diagnostic accuracy. Zhou et al[10] developed the deep learning model CRCNet, trained on 460000 colonoscopy images to enable automatic optical diagnosis of colorectal cancer lesions. The model achieved an area under the precision-recall curve exceeding 0.99 on a dataset comprising 12179 patient samples.
Large language models (LLMs) have demonstrated considerable potential in the field of diagnostic support, owing to their versatility, strong text comprehension capabilities, ease of interaction[11-13], and emerging capability in image recognition[14]. Patel et al[15] demonstrated that LLMs significantly outperformed traditional natural language processing methods in extracting chief complaints and patient-reported outcomes from patients with inflammatory bowel disease. Chizhikova et al[16] utilized pretrained language models to process colorectal cancer radiology reports and automatically generated tumor, node, and metastasis staging labels to improve diagnostic efficiency and accuracy. Ferber et al[17] systematically evaluated vision-enabled generative pre-trained LLMs in cancer image processing, demonstrating their capability to classify colorectal cancer histological subtypes with a performance comparable to that of human pathologists. Zhang et al[18] evaluated the effectiveness of various commercial LLMs for patient education on inflammatory bowel disease. These findings demonstrated that the models can offer effective guidance to patients. However, existing studies have predominantly focused on single-modal data analysis (e.g., isolated imaging or pathological examination) or single-model applications. This fragmented analytical approach risks overreliance on specific test results while neglecting comprehensive clinical presentation, significantly limiting its clinical utility.
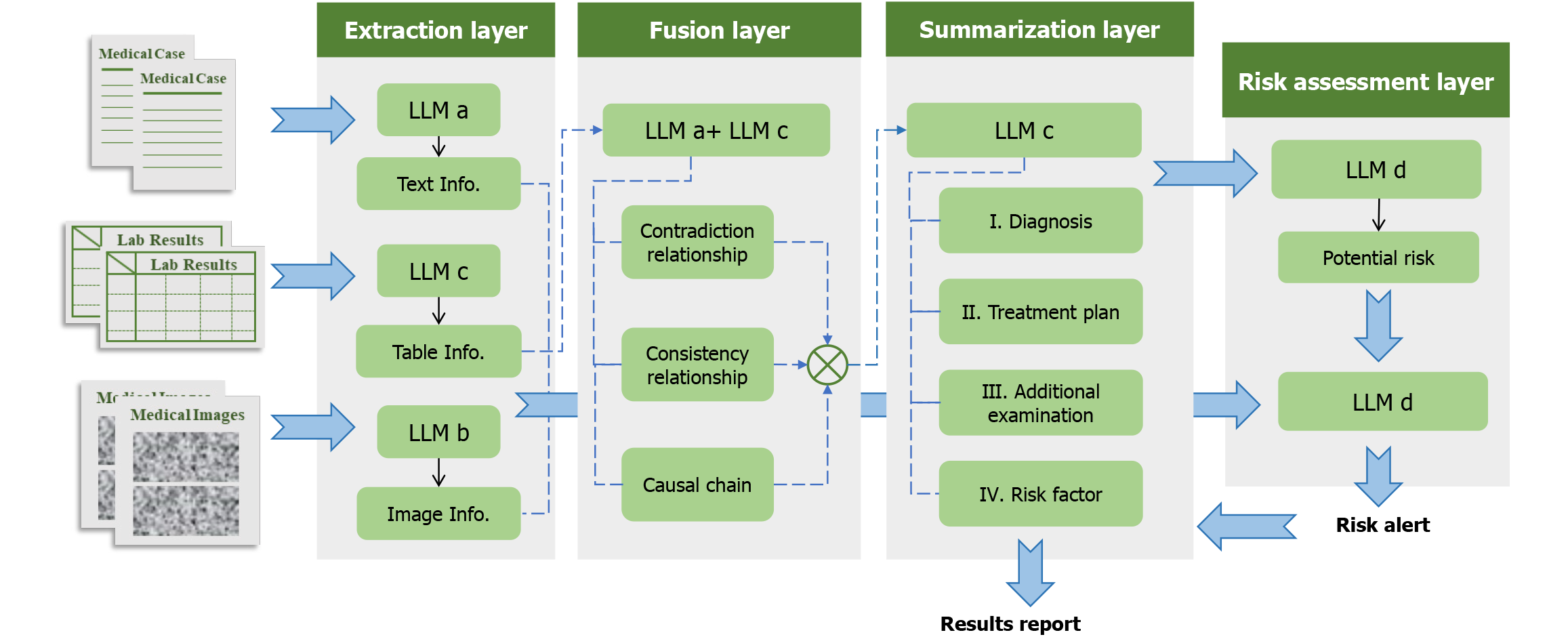
To address these limitations, we developed DeepGut, an intelligent diagnostic system for digestive diseases. By orchestrating multiple multimodal LLMs, our system extracts and synthesizes diagnostic information from medical histories, examination reports, and imaging results, leveraging the specialized capabilities of each LLM. This enables comprehensive assistance throughout the diagnostic, therapeutic, and risk management continuum. The system comprises four layers: Extraction, fusion, summarization, and risk assessment. The extraction layer identifies and retrieves key features and information from diverse data sources. The fusion layer integrates the extracted information to construct diagnostic clues and logical reasoning chains. Based on the output from the fusion layer, the summarization layer generates the final results, including diagnostic conclusions and treatment plans. Finally, the risk assessment layer detects the potential risk factors from the extracted information and evaluates the final outputs for clinical and legal risks. An evaluation of DeepGut outputs in 20 real-world clinical cases demonstrated that the system achieved more than 95% accuracy and completeness in its diagnostic and treatment recommendations. Assessments by five experienced gastroenterologists indicated that the model could effectively integrate multimodal clinical data and construct logical reasoning chains for disease diagnosis.
DeepGut is a multimodal, large-model, collaborative diagnosis, and treatment assistance framework for digestive diseases. The framework incorporates four models (designated Models A, B, C, and D) specializing in text analysis, image analysis, information summarization, and logical discrimination. The model specifications, parameters, and other parameters are listed in Supplementary Table 1. Of note, these models are not fixed, and alternative models with comparable functionalities may be substituted. The DeepGut framework comprises four distinct layers: Extraction, fusion, summarization, and risk assessment. The detailed structure and workflow of this framework is shown in Figure 1. Furthermore, crafting a precise and concise prompt is essential for effectively leveraging LLMs.
The extraction layer extracted the key clinical information related to the condition from multimodal data, including medical records, test reports (tables), and various diagnostic imaging results. This process summarized and condensed multiple diagnostic materials in a unified and concise format. Model A was used for textual information processing. Because medical records typically contain lengthy texts with important information often scattered, the LLM must have sufficient context length to effectively understand all key information in the records. During this process, Model A was instructed to extract and summarize diagnosis- and treatment-related information without altering the original meaning of the text.
Model B was used to extract diagnostic results from imaging modalities such as CT and endoscopic images. As multimodal integration often weakens the comprehension ability of the model, prompts for this stage require Model B to provide relatively superficial descriptions of the imaging results, offering accurate depictions and the most direct conclusions. The tabular test results were processed using Model C. Examination reports exist in tabular form and are stored as PDFs or image files. They first undergo optical character recognition processing to convert them into a markdown format, ensuring that the model can recognize and understand the relationships between different rows and columns, as well as the information contained in parameters at different positions. Due to the highly specialized structure of diagnostic result tables, where traditional optical character recognition performs poorly, in this study, tabular information was first processed by Model B to complete the conversion to the markdown format. Because tables contain dense information with complex interrelationships, the model requires strong logical reasoning and deep-thinking capabilities. Prompts require the model to perform appropriate logical deductions based on tabular information combined with knowledge from training to accurately summarize key information and conclusions.
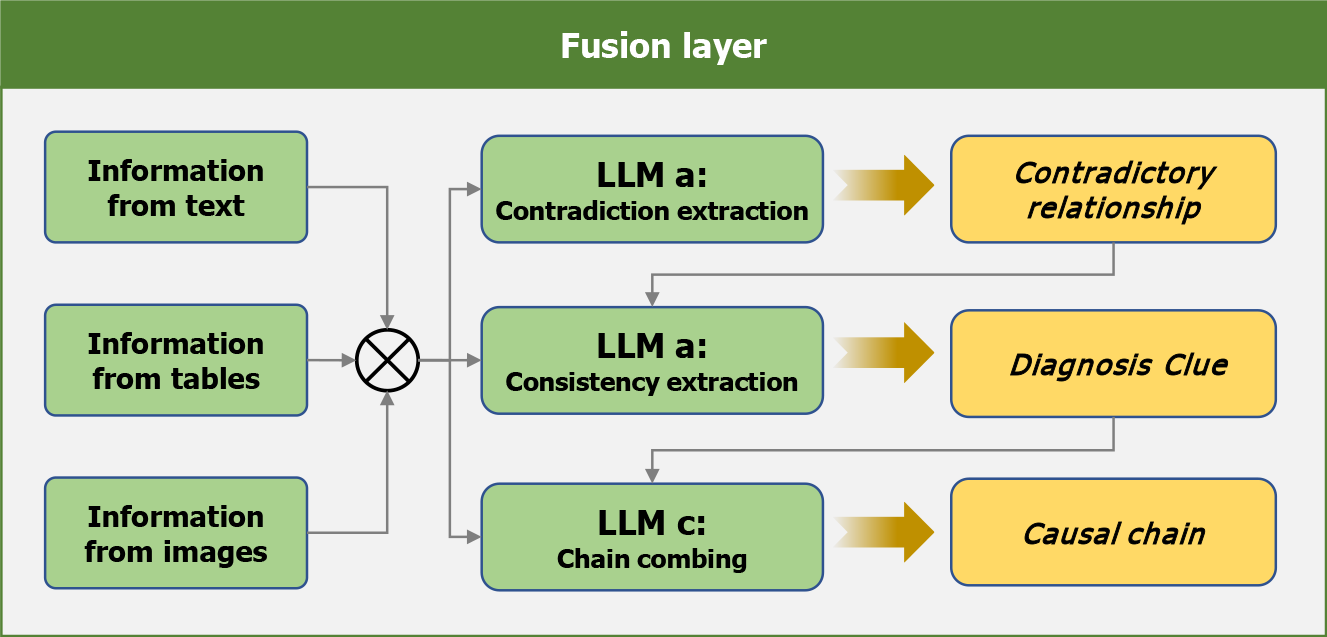
The fusion layer integrated and analyzed the information extracted from multimodal results, categorizing relationships into three types-consistent, contradictory, and sequential-while identifying potential clinical implications, as shown in Figure 2. For example, a contradiction may exist between a patient’s self-reported esophageal gastric heterotopia during admission and endoscopic findings showing a normal esophageal mucosa with a smooth surface, normal color, and no congestion, edema, ulcers, or stenosis, suggesting possible disease progression. Additionally, consistency among admission records noting recurrent melena and hematemesis, along with laboratory reports indicating reduced protein levels, may suggest active gastrointestinal bleeding.
This process was completed in three rounds. First, surface-level contradictory information was summarized to assess diagnostic validity. The prompts instructed the LLM to comprehensively analyze all the results and make direct asse
The summarization layer served as the final stage of the diagnostic process. In this phase, the LLM (Model C) was required to generate conclusions based on the outputs of the fusion layer, specifically addressing four aspects: Diagnostic results, treatment plans, recommended supplementary tests, and prognostic analysis. Additionally, important supplementary information provided by physicians was incorporated as input to reinforce critical clinical values and highlight potential treatment contraindications (such as patient allergies to specific medications). Model C was used to perform the task. Because the outputs at this stage serve as crucial reference information for physicians, the prompts emphasize the need for LLM to produce outputs in formats that facilitate easy reading and comprehension by doctors.
Risk assessment was a critical step across all diagnostic stages. Following each conclusion from the extraction layer, the risk assessment Model D was tasked with identifying potential risk factors (such as patient allergies). These identified risk factors were then combined with the fusion layer outputs and input into the risk assessment model. The model evaluated both medical and legal risks to determine whether diagnostic results contained any Inappropriate treatment methods potentially leading to medical accidents or doctor-patient disputes. An effective risk assessment requires substantial medical and legal expertise. Context length is also an important consideration to enable a comprehensive analysis of all results. The outputs from the risk assessment layer are directly accessible to physicians, who use them to evaluate the reasonableness of the summarization layer’s conclusions. If physicians reject the results, the risk assessment outputs are fed back into the summarization layer as constraints, prompting the regeneration of conclusions that actively mitigate known risks. This process can be repeated iteratively until the physicians are satisfied with the final conclusions.
The evaluation dataset comprised 20 carefully selected clinical cases representing our one-year patient cohort. These de-identified case records included admission records, laboratory tests, imaging results, endoscopic images, and other relevant clinical data, with all original diagnostic information and treatment histories removed. The model outputs were evaluated through both objective metrics and expert subjective assessments. Objective evaluation comprised four key metrics: Diagnostic accuracy, diagnostic completeness, treatment plan accuracy, and treatment plan completeness. Diagnostic accuracy was calculated as the ratio of LLM-generated diagnoses matching expert conclusions to the total LLM diagnoses. Diagnostic completeness was measured as the ratio of correct LLM diagnoses to the total expert-identified diagnoses. Similarly, treatment plan accuracy was defined as the ratio of LLM-generated treatment plans matching expert recommendations to total LLM plans, whereas treatment plan completeness was defined as the ratio of correct LLM treatment plans to total expert-recommended plans. These metrics collectively evaluate both the reliability (accuracy) and comprehensiveness (completeness) of LLM outputs.
For the expert evaluation, five gastroenterology specialists independently scored the LLM outputs on five dimensions: Accuracy, completeness, relevance, logicality, and overall performance. The five participating gastroenterologists were rigorously selected based on their extensive clinical expertise, each possessing over 10 years of specialized practice and research experience in inflammatory bowel disease. All evaluations used a standardized 5-point scale (1 = poor to 5 = excellent).
Data recording and management were conducted using Microsoft Excel 2021 (Microsoft Corporation, Redmond, WA, United States). Data analysis and graphical representation were performed using GraphPad Prism (version 9.0.0; GraphPad Software, Inc., San Diego, CA, United States). For each subjective evaluation metric, central tendencies and distributions were described using statistical measures such as the median and interquartile range. Graphical presentations were used to provide a clear and comprehensive data visualization.
The objective evaluation results are presented as a histogram in Figure 3. The DeepGut framework achieved a diagnostic accuracy of 97.8%, diagnostic completeness of 93.9%, treatment plan accuracy of 95.2%, and treatment plan completeness of 98.0%. In contrast, the conventional single-modal AI tools recorded scores of 52.9%, 45.9%, 68.1%, and 60.8%, respectively. These findings demonstrate that the DeepGut framework significantly outperforms traditional single-LLM-based assistant methods. Furthermore, the evaluation metrics indicated that DeepGut adopted a more conservative approach for diagnosing complex cases, frequently recommending additional examinations rather than providing definitive conclusions. For example, in the case of patient 2, DeepGut suggested an abdominal CT enhancement scan to further assess a strip-like soft tissue density shadow near the gastroduodenal wall, despite expert gastroenterologists being able to confirm the diagnosis based on existing imaging. This conservative strategy arises from the system’s decision-making framework, in which prompt engineering explicitly restricts the model from drawing conclusions that are not supported by evidence. Although this constraint effectively reduces hallucinations and enhances output reliability, it may limit the capacity of the model to make clinically valid determinations that rely on professional judgment.
In treatment planning, DeepGut achieved particularly high-performance metrics. By leveraging the LLM’s extensive medical knowledge base, the system consistently provided accurate and comprehensive treatment recommendations, including excellent performance in post-treatment follow-up reminders. Furthermore, as most missed diagnoses were secondary conditions that did not require immediate treatment, they had minimal impact on the overall quality of the treatment plans. The results clearly demonstrate that the medical specialization level of the LLMs significantly affects the performance of DeepGut. It should be noted that all LLMs used in this study were general-purpose models. In theory, employing LLMs that are specifically fine-tuned for medical tasks can yield better performance.
Importantly, the design of the DeepGut framework effectively accommodates the use of nonspecialized models. By incorporating contradiction analysis, diagnostic clue identification, and reasoning chain construction, the system systematically decomposes the diagnostic and treatment tasks. This structured approach enables LLMs to interpret tasks better and apply relevant knowledge, thereby achieving superior performance metrics that are challenging for general-purpose LLMs without this framework.
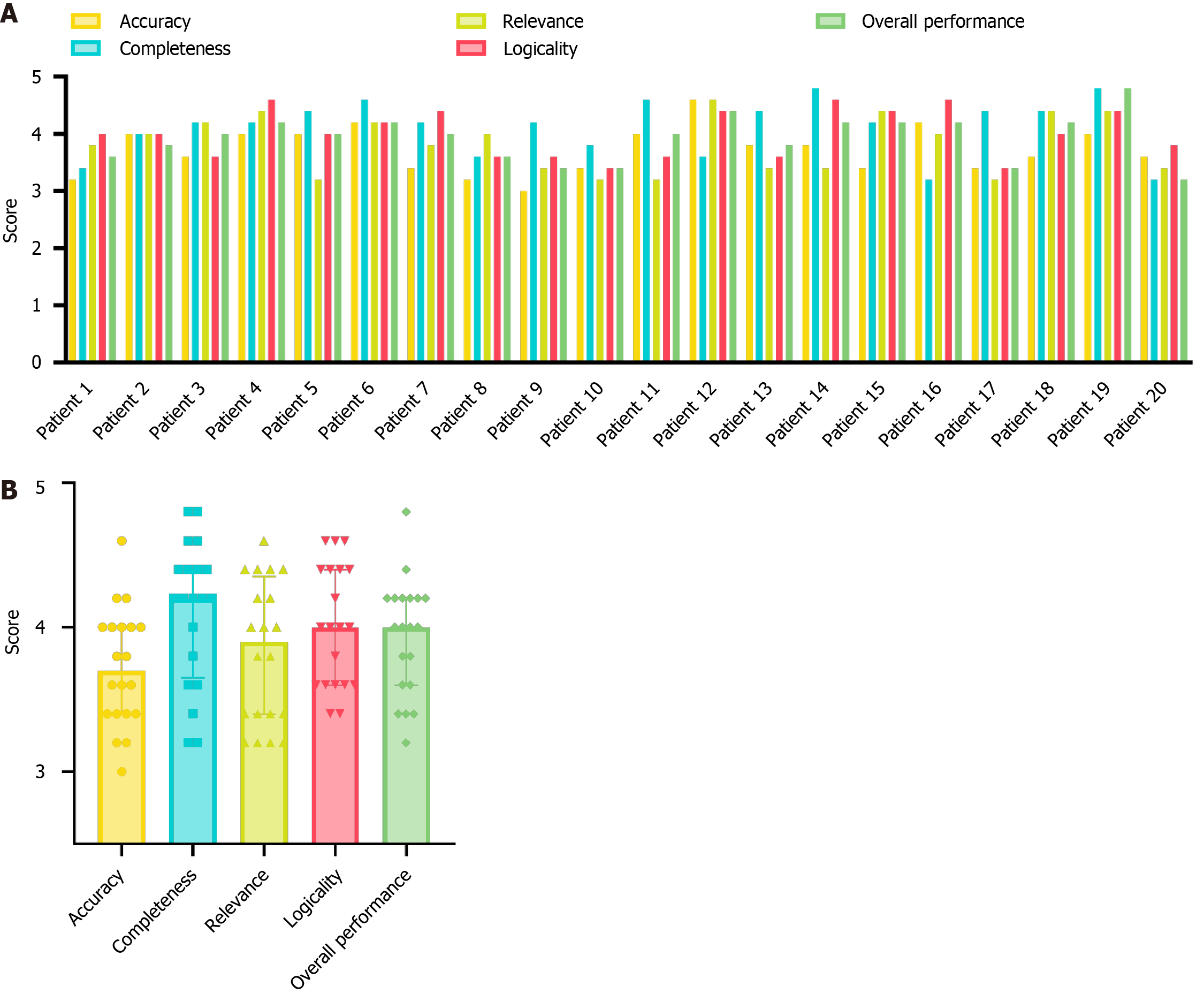
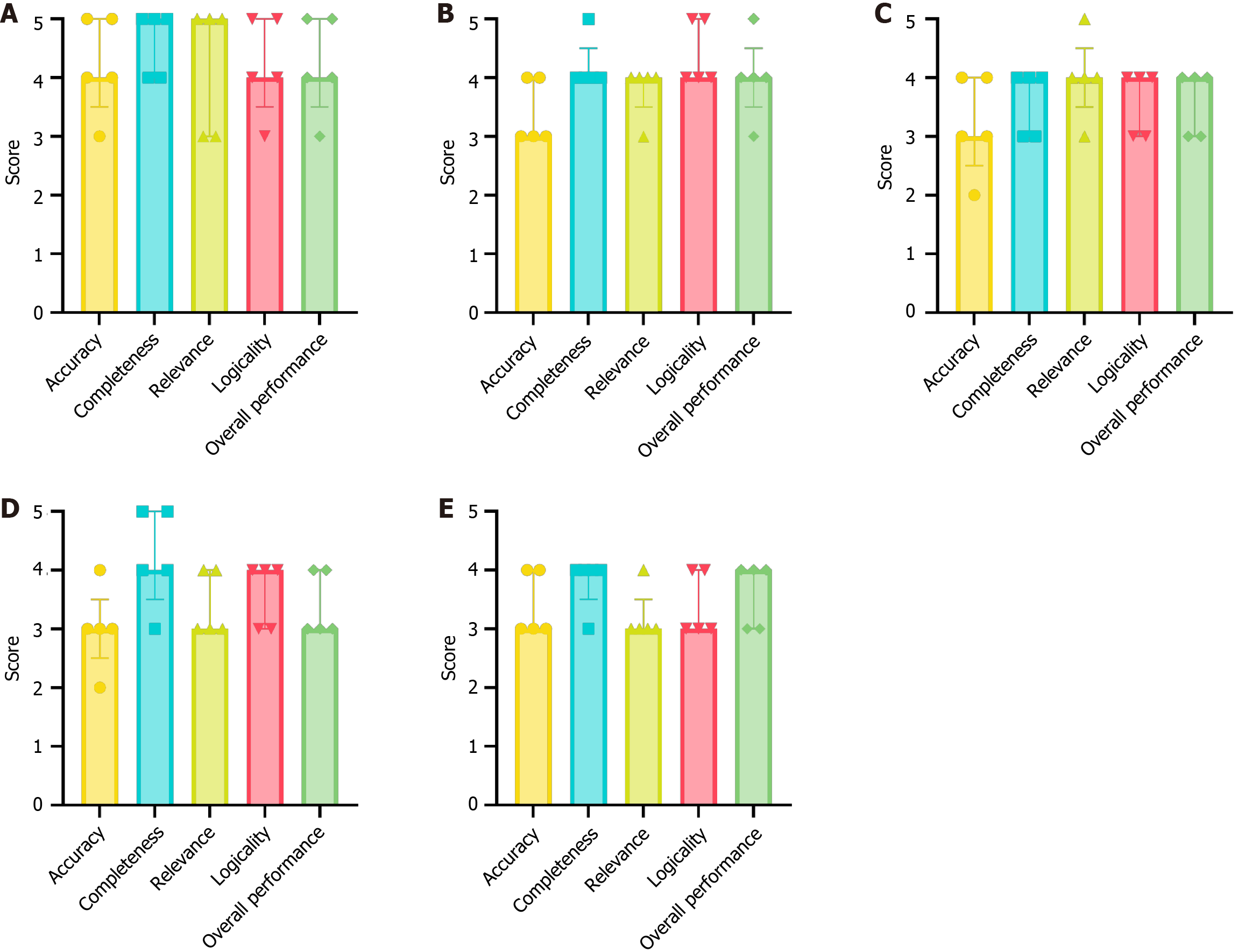
The results of the subjective evaluations are presented in Figure 4. Figure 4A illustrates the average scores across the five evaluation indicators for the 20 patients as assessed by the five expert raters. Figure 4B summarizes the statistical outcomes. Notably, nearly half of the patients (patients 2, 3, 4, 6, 12, 15, 18, and 19) received scores exceeding 3.5 for all evaluation indicators, reflecting strong expert endorsement of the medical assistant tool’s usability. In particular, the completeness, logicality, and overall performance of the case evaluations were highly rated, with 13 or more outputs achieving an average score above 4. These findings demonstrate that DeepGut effectively analyzed multimodal patient data and presented results in a clear and organized manner, indicating a high level of acceptance among expert phy
Notably, most experts considered the completeness of the diagnostic recommendations provided by DeepGut to be greater than its accuracy. This is because the experts generally approved of DeepGut’s tendency to “advise patients to undergo further examinations to confirm or exclude specific diseases”, considering this approach to be “more consistent with medical standards and a responsible practice towards patients”. In addition, the experts highly valued DeepGut’s detailed risk assessment features, which they regarded as exemplifying its exceptional completeness. These features support physicians in standardizing the treatment processes and mitigating potential medical disputes with patients. However, regarding diagnostic accuracy, experts observed that DeepGut’s conclusions for some conditions appeared overly definitive. They suggested that as a diagnostic aid, DeepGut should maintain a higher degree of skepticism to allow physicians to exercise their professional judgment and minimize the risk of misdiagnosis. Figure 5 presents detailed box plots of the scores provided by the experts for the five selected cases (cases 6, 7, 8, 9, and 10). Overall, the experts’ scores were consistent, with only a few exceptions. In these cases, scores for specific indicators exhibited greater variability, likely due to the subjective nature of the assessments. Notably, the overall impression scores for the five cases were high and consistent. However, the DeepGut framework involves multiple sequential model generation processes, which significantly increases the analysis time, resulting in a processing time that is approximately 4-12 times longer than that of traditional single-modality LLMs. While the experts acknowledged that this waiting time was still much shorter than the typical duration of an outpatient visit, they emphasized the need to optimize the runtime to allow physicians to receive prompts more quickly.
This study aimed to develop and evaluate a multimodal LLM collaborative diagnostic framework designed to overcome the limitations of traditional single-modality approaches and enhance clinical decision-making for physicians. The superiority of this framework is evident in its ability to integrate diverse data sources, leverage the complementary strengths of multiple LLMs, and incorporate robust evaluations and regeneration mechanisms. The framework excelled in integrating multimodal information, addressing a key limitation of traditional AI-assisted medical methods, which often rely on a single modality and fail to comprehensively incorporate all diagnostic data, potentially leading to misdiagnoses or missed diagnoses[19,20]. In contrast, the proposed framework employed a text summarization model (Model A), image recognition model (Model B), and tabular recognition model (Model C) within the extraction layer to systematically extract and unify information from diverse sources, including textual case descriptions, physician-provided supplementary information, tabular diagnostic indicators, and imaging results.
The formulation of medical conclusions is a complex process involving a series of subtasks including induction, analysis, reasoning, and evaluation[21]. A three-round information fusion process in the fusion layer of DeepGut resolves contradictions, identifies consistencies, and establishes logical connections among diagnostic data. This approach not only filters out erroneous information, but also identifies diagnostic clues and constructs reasoning chains, providing a comprehensive and accurate foundation for subsequent diagnostic conclusions. This robust integration process was the primary contributor to the high accuracy and completeness of the framework.
In addition, the framework maintained high compatibility, theoretically allowing any LLM with relevant medical knowledge to be integrated and function under the coordination of the DeepGut framework. However, this places stringent demands on the prompt design for LLMs, requiring the use of restrictive wording to ensure they adhere to an objective and cautious diagnostic approach, preventing some general LLMs from generating “hallucinations” on complex medical issues. However, this limits the full potential of LLMs, as they cannot provide direct and definitive answers to all questions. Fine-tuning large models on medical datasets could theoretically alleviate this issue; however, more significant improvements will depend on advancements in LLM technology. The introduction of high-performance models is expected to considerably enhance the performance and reliability of DeepGut.
Subjective expert evaluation indicated a high level of acceptance among physicians of DeepGut as a diagnostic aid. One expert noted, “it comprehensively considers various test results from multiple patients and provides thorough theoretical support and reasoning for each conclusion, making its output trustworthy and helpful in verifying its own diagnostic judgments”. This suggests that the physicians’ acceptance of AI-assisted tools largely depends on how well they fit the medical process. Compared to general LLMs, which often fail to provide the desired responses, developing AI-assisted tools that are practical, accurate, easily understood, and can genuinely assist physicians in their medical practice is a valuable and promising research direction. However, this multimodal collaborative approach has several limitations. The repeated input of information into the models inevitably increases the number of input and output tokens, thereby increasing operational costs. Moreover, the models in different layers cannot operate in parallel; the sequential generation time of the models across layers increases, prolonging the overall diagnostic time.
The DeepGut framework represents a sophisticated approach for enhancing the diagnosis and treatment of gas
This study constitutes an exploratory effort to advance diagnostic performance through multi-model collaboration, demonstrating that the strategic integration of LLMs can significantly enhance their efficacy. By leveraging the diverse modalities and complementary strengths of LLMs, DeepGut fosters synergistic effects, thereby enriching the clinical applications of these models. However, the collaborative approach resulted in increased token counts, leading to longer generation times and increased computational costs. Future research should focus on refining the input methods to ensure that critical information is concisely and comprehensively delivered to each LLM layer, thereby reducing token waste and improving efficiency.
| 1. | Cheng ZY, Gao Y, Mao F, Lin H, Jiang YY, Xu TL, Sun C, Xin L, Li ZS, Wan R, Zhou MG, Wang LW; China Gastrointestinal Health Expert Group (CGHEG). Construction and results of a comprehensive index for gastrointestinal health monitoring in China: a nationwide study. Lancet Reg Health West Pac. 2023;38:100810. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 2. | Chen Y, Chen T, Fang JY. Burden of gastrointestinal cancers in China from 1990 to 2019 and projection through 2029. Cancer Lett. 2023;560:216127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 39] [Reference Citation Analysis (0)] |
| 3. | Xiao Y, Zhang Z, Xu CM, Yu JY, Chen TT, Jia SW, Du N, Zhu SY, Wang JH. Evolution of physician resources in China (2003-2021): quantity, quality, structure, and geographic distribution. Hum Resour Health. 2025;23:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (1)] |
| 4. | Yu Q, Zhu C, Feng S, Xu L, Hu S, Chen H, Chen H, Yao S, Wang X, Chen Y. Economic Burden and Health Care Access for Patients With Inflammatory Bowel Diseases in China: Web-Based Survey Study. J Med Internet Res. 2021;23:e20629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 5. | Gong S, Zhang Y, Wang Y, Yang X, Cheng B, Song Z, Liu X. Study on the burden of digestive diseases among Chinese residents in the 21st century. Front Public Health. 2023;11:1314122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 6. | Busnatu Ș, Niculescu AG, Bolocan A, Petrescu GED, Păduraru DN, Năstasă I, Lupușoru M, Geantă M, Andronic O, Grumezescu AM, Martins H. Clinical Applications of Artificial Intelligence-An Updated Overview. J Clin Med. 2022;11:2265. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 75] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 7. | Wu SY, Wang JJ, Guo QQ, Lan H, Zhang JJ, Wang L, Janne E, Luo X, Wang Q, Song Y, Mathew JL, Xun YQ, Yang N, Lee MS, Chen YL. Application of artificial intelligence in clinical diagnosis and treatment: an overview of systematic reviews. Intell Med. 2022;2:88-96. [DOI] [Full Text] |
| 8. | Elemento O, Leslie C, Lundin J, Tourassi G. Artificial intelligence in cancer research, diagnosis and therapy. Nat Rev Cancer. 2021;21:747-752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 143] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 9. | Tong Y, Lu K, Yang Y, Li J, Lin Y, Wu D, Yang A, Li Y, Yu S, Qian J. Can natural language processing help differentiate inflammatory intestinal diseases in China? Models applying random forest and convolutional neural network approaches. BMC Med Inform Decis Mak. 2020;20:248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 10. | Zhou D, Tian F, Tian X, Sun L, Huang X, Zhao F, Zhou N, Chen Z, Zhang Q, Yang M, Yang Y, Guo X, Li Z, Liu J, Wang J, Wang J, Wang B, Zhang G, Sun B, Zhang W, Kong D, Chen K, Li X. Diagnostic evaluation of a deep learning model for optical diagnosis of colorectal cancer. Nat Commun. 2020;11:2961. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 11. | Kim JK, Chua M, Rickard M, Lorenzo A. ChatGPT and large language model (LLM) chatbots: The current state of acceptability and a proposal for guidelines on utilization in academic medicine. J Pediatr Urol. 2023;19:598-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 71] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 12. | Laupichler MC, Rother JF, Grunwald Kadow IC, Ahmadi S, Raupach T. Large Language Models in Medical Education: Comparing ChatGPT- to Human-Generated Exam Questions. Acad Med. 2024;99:508-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 50] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 13. | Thirunavukarasu AJ, Ting DSJ, Elangovan K, Gutierrez L, Tan TF, Ting DSW. Large language models in medicine. Nat Med. 2023;29:1930-1940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 1290] [Article Influence: 430.0] [Reference Citation Analysis (0)] |
| 14. | Tian D, Jiang S, Zhang L, Lu X, Xu Y. The role of large language models in medical image processing: a narrative review. Quant Imaging Med Surg. 2024;14:1108-1121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 40] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 15. | Patel PV, Davis C, Ralbovsky A, Tinoco D, Williams CYK, Slatter S, Naderalvojoud B, Rosen MJ, Hernandez-Boussard T, Rudrapatna V. Large Language Models Outperform Traditional Natural Language Processing Methods in Extracting Patient-Reported Outcomes in Inflammatory Bowel Disease. Gastro Hep Adv. 2025;4:100563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 16. | Chizhikova M, López-Úbeda P, Martín-Noguerol T, Díaz-Galiano MC, Ureña-López LA, Luna A, Martín-Valdivia MT. Automatic TNM staging of colorectal cancer radiology reports using pre-trained language models. Comput Methods Programs Biomed. 2025;259:108515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 17. | Ferber D, Wölflein G, Wiest IC, Ligero M, Sainath S, Ghaffari Laleh N, El Nahhas OSM, Müller-Franzes G, Jäger D, Truhn D, Kather JN. In-context learning enables multimodal large language models to classify cancer pathology images. Nat Commun. 2024;15:10104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 43] [Reference Citation Analysis (0)] |
| 18. | Zhang Y, Wan XH, Kong QZ, Liu H, Liu J, Guo J, Yang XY, Zuo XL, Li YQ. Evaluating large language models as patient education tools for inflammatory bowel disease: A comparative study. World J Gastroenterol. 2025;31:102090. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (8)] |
| 19. | Zhou L, Zhang Z, Chen YC, Zhao ZY, Yin XD, Jiang HB. A Deep Learning-Based Radiomics Model for Differentiating Benign and Malignant Renal Tumors. Transl Oncol. 2019;12:292-300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 92] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 20. | Suganyadevi S, Seethalakshmi V, Balasamy K. A review on deep learning in medical image analysis. Int J Multimed Inf Retr. 2022;11:19-38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 138] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 21. | Corazza GR, Lenti MV, Howdle PD. Diagnostic reasoning in internal medicine: a practical reappraisal. Intern Emerg Med. 2021;16:273-279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/