Published online Jul 14, 2025. doi: 10.3748/wjg.v31.i26.109500
Revised: June 5, 2025
Accepted: June 30, 2025
Published online: July 14, 2025
Processing time: 59 Days and 5.2 Hours
Exosomal microRNAs (miRNAs) have emerged as promising biomarkers for cancer diagnosis due to their stability, tumor specificity, and accessibility. How
To investigate the diagnostic potential of serum exosomal miRNAs in metastatic pancreatic cancer.
A total of 36 patients were enrolled, comprising 8 patients in the discovery phase (4 with metastatic and 4 with non-metastatic pancreatic cancer) and 28 in the validation cohort (15 non-metastatic and 13 metastatic cases). Exosomes were isolated using the exoEasy Maxi Kit and characterized by transmission electron microscopy, nanoparticle tracking analysis, and western blotting. High-throu
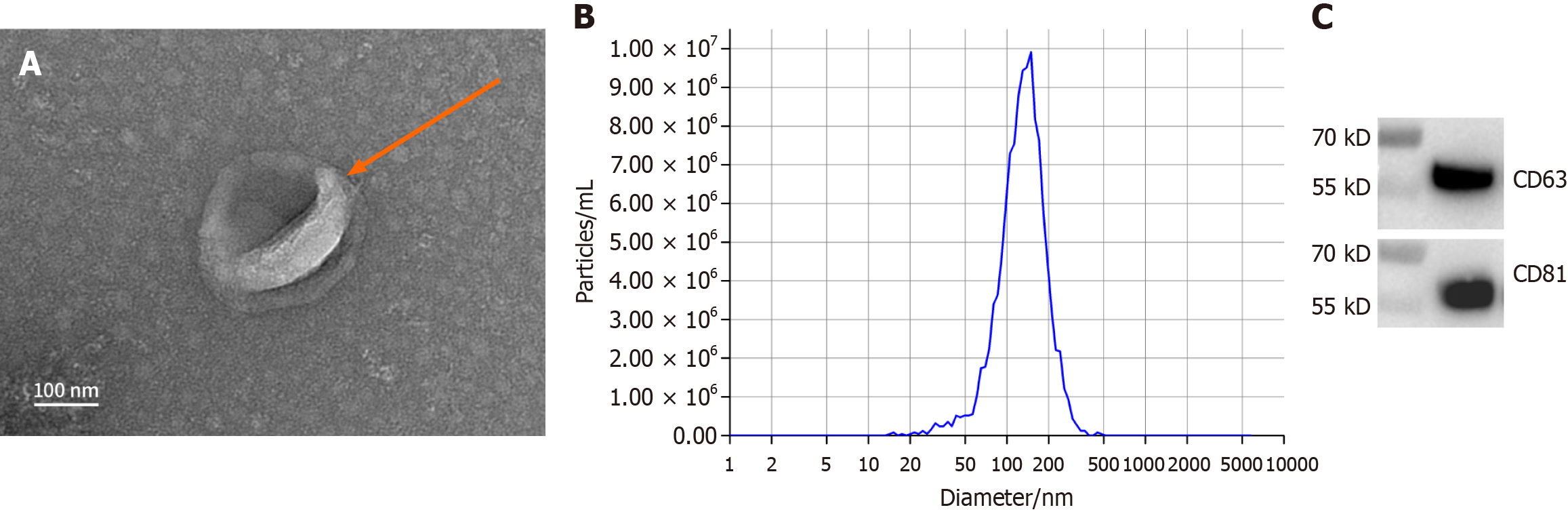
Transmission electron microscopy revealed that the isolated exosomes were predominantly round or oval with well-defined membrane boundaries. Nano
The elevated expression of serum exosomal hsa-let-7f-5p in metastatic pancreatic cancer suggests its potential as a non-invasive biomarker for distinguishing metastatic from non-metastatic disease.
Core Tip: Pancreatic cancer has a poor prognosis largely due to late-stage diagnosis and high metastatic potential. Exosomes, particularly exosome-derived microRNAs, have emerged as promising non-invasive biomarkers. This study identifies hsa-let-7f-5p as significantly upregulated in serum exosomes of metastatic pancreatic cancer patients. Validated through high-throughput sequencing and quantitative polymerase chain reaction, hsa-let-7f-5p shows potential for distinguishing metastatic from non-metastatic disease. Its target gene interactions further support its role in metastasis, offering new insights into pancreatic cancer progression and potential avenues for early detection and therapeutic intervention.
- Citation: Ren S, Song LN, Zhao R, Tian Y, Wang ZQ. Serum exosomal hsa-let-7f-5p: A potential diagnostic biomarker for metastatic pancreatic cancer detection. World J Gastroenterol 2025; 31(26): 109500
- URL: https://www.wjgnet.com/1007-9327/full/v31/i26/109500.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i26.109500
Pancreatic cancer is a highly aggressive malignancy of the digestive system and currently ranks as the third leading cause of cancer-related mortality in the United States, with its incidence and death rates continuing to rise annually[1]. It is projected that by 2030, pancreatic cancer will surpass colorectal and breast cancer to become the second leading cause of cancer-related deaths[2]. Currently, surgical resection remains the primary treatment modality; however, the overall effectiveness of available therapies, including surgery, radiotherapy, chemotherapy, and immunotherapy, remains limited. For decades, the 5-year survival rate for pancreatic cancer has remained below 5%, with a median survival time of only 6 to 10 months[3]. This poor prognosis is largely attributed to late-stage diagnosis, as most patients present with advanced disease and distant metastases at the time of detection, rendering them ineligible for curative surgical intervention. As a result, the surgical resection rate remains below 20%[4]. Given that metastasis is a critical determinant of prognosis in pancreatic cancer, elucidating the mechanisms driving metastatic progression and identifying effective therapeutic strategies are urgent priorities in the clinical management of this malignancy.
Exosomes are nanoscale extracellular vesicles secreted following the fusion of multivesicular bodies with the plasma membrane and contain various bioactive molecules, including RNA, DNA fragments, and proteins[5]. Among these components, exosomal microRNAs (miRNAs), comprising both long and short chains, have garnered considerable attention. It has been reported that exosomes derived from various cell types and tissues harbor a total of 4934 distinct miRNAs[5]. These miRNAs play key roles in diverse biological processes such as stem cell differentiation, hematopoiesis, organogenesis, and particularly in tumor development and metastasis. Due to their tumor specificity, stability, and ease of preservation, exosomal miRNAs have emerged as promising biomarkers for cancer diagnosis. They hold potential for early tumor detection, treatment monitoring, and prognosis assessment[6-8]. Luo et al[9] reported that exosomes derived from peritoneal lavage fluid in gastric cancer contain abundant miRNAs associated with peritoneal metastasis. Among them, hsa-let-7g-3p showed potential as a predictive biomarker for peritoneal dissemination, chemotherapeutic response. Similarly, Masamune et al[10] demonstrated that exosomes secreted by pancreatic cancer cells can activate the phosphatidylinositol 3-kinase/protein kinase B and mitogen-activated protein kinases signaling pathways in pancreatic stellate cells, thereby enhancing the invasiveness of cancer cells and promoting tumor progression. However, the diagnostic potential of specific exosomal miRNAs such as hsa-let-7f-5p in metastatic pancreatic cancer has not been thoroughly investigated. This study aims to investigate the diagnostic potential of serum exosomal hsa-let-7f-5p in metastatic pancreatic cancer by analyzing its differential expression and validating serum exosomal miRNAs in pancreatic cancer patients with and without distant metastasis.
Instruments and equipment: Bench-top high-speed centrifuge (Eppendorf, Germany); ultra-low temperature freezer (Thermo Fisher Scientific, MA, United States); ultracentrifuge (Hitachi, Japan); transmission electron microscope (Hitachi, Japan); polymerase chain reaction machine (Bio-Rad, CA, United States); Agilent 2100 Bioanalyzer (Agilent Technologies, CA, United States). Reagents: ExoEasy Maxi Kit (QIAGEN, Germany); Agilent Small RNA Reagents Part I (Agilent Technologies, CA, United States); Agilent RNA 6000 Pico Reagents Part I (Agilent Technologies, CA, United States); XE Buffer (QIAGEN, Germany); QIAGEN RNA Lysis Solution (QIAGEN, Germany); TaqMan probes (Thermo Fisher Scientific, MA, United States).
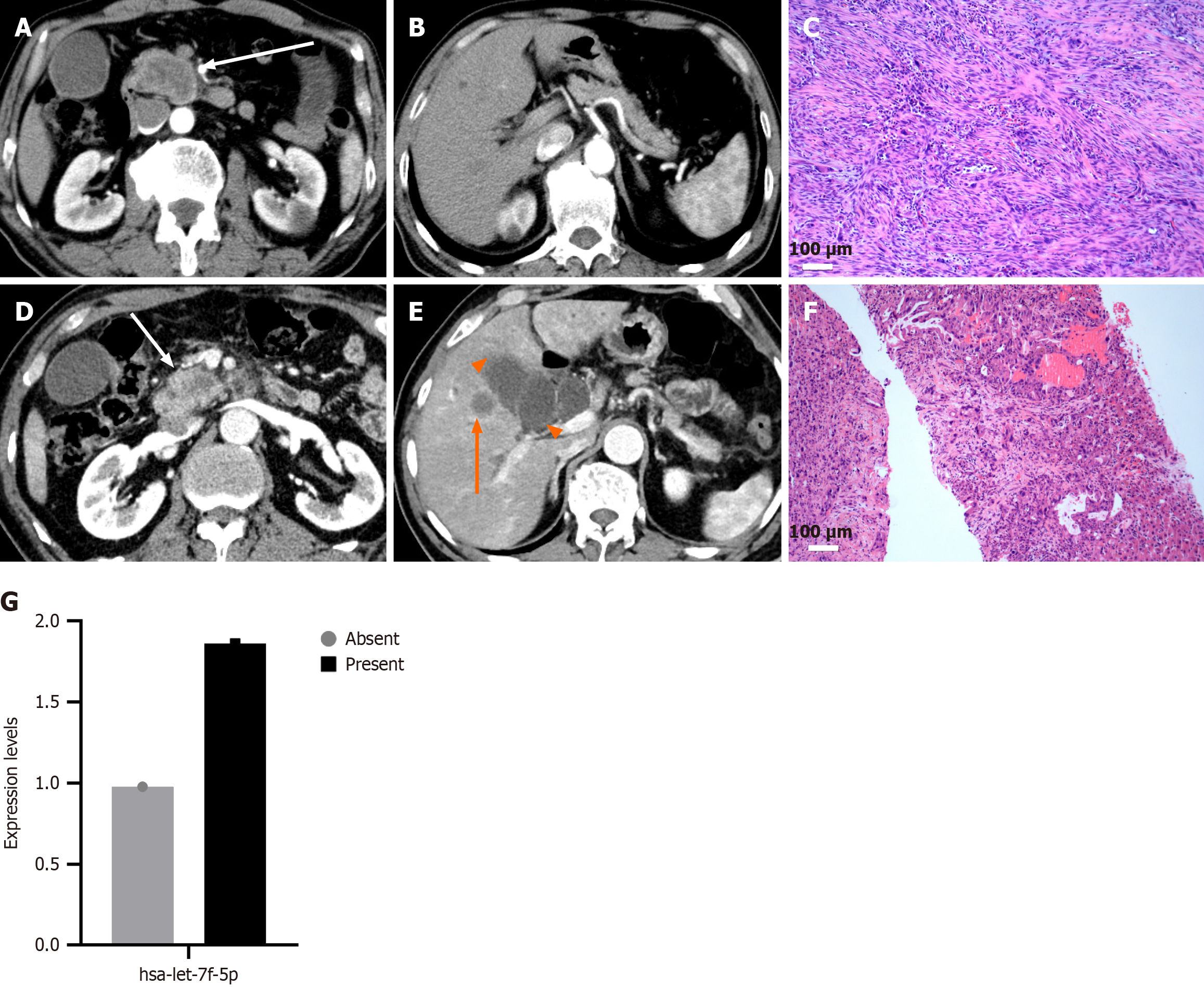
A total of 36 primary pancreatic cancer patients diagnosed by surgical pathology or biopsy between January 2019 and January 2020 at Affiliated Hospital of Nanjing University of Chinese Medicine were enrolled in this study. Among them, 19 patients had no metastasis, and 17 presented with distant metastases involving sites such as the liver, adrenal glands, mesenteric nodules, spleen, duodenum, and lungs. Prior to serum sample collection, none of the patients had undergone curative surgery, chemotherapy, or immunotherapy. All patients received abdominal computed tomography scans, including both plain and contrast-enhanced imaging, before initiating treatment.
Blood samples were collected from enrolled patients in the early morning following an overnight fast. Blood was drawn into red-capped coagulation tubes, labeled, and allowed to clot at 4 °C, 4000 rpm, for two cycles of 10 minutes each. Following centrifugation, 1 mL of the serum supernatant was carefully transferred into 2 mL cryovials, with each sample divided into two aliquots. These aliquots were stored in cryoboxes at -80 °C until further analysis. Patients were classified into two groups: Those with non-metastatic pancreatic cancer and those with metastatic pancreatic cancer. This study was approved by the Ethics Committee of Affiliated Hospital of Nanjing University of Chinese Medicine, and all patients provided written informed consent.
Following the manufacturer’s protocol, serum samples from 4 patients with localized pancreatic cancer and 4 patients with metastatic pancreatic cancer were used for exosome isolation using the exoEasy Maxi Kit (Catalog No. 76064, QIAGEN, Germany). The isolated exosomes were characterized by transmission electron microscopy (TEM) using the FEI Tecnai G2 Spirit BioTwin (Thermo Fisher Scientific, MA, United States), nanoparticle tracking analysis (NTA) with Zetaview (Particle Metrix, Germany), and western blotting (WB). Total RNA was subsequently extracted from the exosomes using the miRNeasy Serum/Plasma Kit (Catalog No. 217184, QIAGEN, Germany). RNA concentration and integrity were evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies, CA, United States) and the NanoDrop-2000 UV-Vis spectrophotometer (Thermo Fisher Scientific, MA, United States).
To ensure the accuracy and reproducibility of miRNA quantification by reverse transcription-quantitative polymerase chain reaction (RT-qPCR), the expression levels of target miRNAs were normalized using appropriate reference RNAs. Among the candidates, hsa-miR-486-5p and hsa-miR-423-5p exhibited stable and consistent expression across all samples and were therefore used as internal controls. The average expression of these reference miRNAs was used to calculate the relative expression levels of the target miRNAs. Differentially expressed miRNAs were identified based on a fold change (FC) ≥ 1.5 and a P value < 0.05. The potential biological functions of the selected miRNAs were predicted using miRDB, miWalk, miRTarBase, and TargetScan databases.
Fifteen patients with non-metastatic pancreatic cancer and thirteen patients with metastatic pancreatic cancer were included in the subsequent RT-qPCR validation phase. Exosomes were isolated from serum samples following the previously described protocol, and the expression levels of target miRNAs were quantified using TaqMan probe-based qRT-PCR. The qRT-PCR validation procedure included the following steps: (1) Selection of target miRNAs for validation; (2) Design of specific probes and primers; (3) Synthesis of probes and primers; (4) Preparation of the qRT-PCR reaction mixture; (5) Optimization of reaction parameters; and (6) Repetition of experiments to ensure reproducibility and to obtain reliable amplification curve data.
Differential expression of serum exosomal miRNAs between non-metastatic and metastatic pancreatic cancer patients was assessed by comparing their respective miRNA expression levels. For sequencing data, expression levels were quantified using DESeq software based on a negative binomial distribution model. Differentially expressed miRNAs were identified using the criteria of FC ≥ 1.5 and P value < 0.05. For qRT-PCR validation, statistical analyses were performed using SPSS version 24.0. Normality of the data was assessed using the Shapiro-Wilk test, followed by Levene’s test for homogeneity of variances. When data were normally distributed with equal variances, Student’s t-test was used; if variances were unequal, Welch’s t-test was applied. For non-normally distributed data, the Mann-Whitney U test was employed. A two-tailed P value < 0.05 was considered statistically significant.
A total of 36 patients with primary pancreatic cancer were included in this study, comprising 17 with metastatic disease and 19 with localized disease. Patient demographics are summarized in Table 1. The mean age was 64.32 ± 8.00 years in the metastatic group and 61.88 ± 8.92 years in the localized group, with no statistically significant difference (P = 0.394). Gender distribution was also comparable between the two groups (P = 0.345), with males accounting for 58.8% of the metastatic group and 73.7% of the localized group. No significant differences were observed in clinical symptoms, including abdominal pain, abdominal bloating or diarrhea, yellow urine or icterus, marasmus, or asymptomatic presentation (all P > 0.05). As expected, all patients in the metastatic group were classified as stage IV, while those in the localized group were classified as stages I-III, reflecting a statistically significant difference in staging between the groups (P < 0.001).
| Variables | Patients with metastatic pancreatic cancer | Patients with localized pancreatic cancer | P value |
| Age, years | 64.32 ± 8.00 | 61.88 ± 8.92 | 0.3941 |
| Gender | 0.3452 | ||
| Male | 10 (58.8) | 14 (73.7) | |
| Female | 7 (41.2) | 5 (26.3) | |
| Clinical symptoms | |||
| Abdominal pain | 9 (52.9) | 12 (63.2) | 0.5352 |
| Abdominal bloating or diarrhea | 3 (17.6) | 4 (21.1) | 0.5662 |
| Yellow urine or icterus | 6 (35.3) | 7 (41.2) | 0.7242 |
| Marasmus | 2 (11.8) | 2 (10.5) | 0.6552 |
| Asymptomatic | 6 (35.3) | 9 (47.4) | 0.4632 |
| Staging | < 0.0012 | ||
| I | 0 | 3 | |
| II | 0 | 8 | |
| III | 0 | 8 | |
| IV | 17 | 0 |
Exosome morphology was examined by TEM using the FEI Tecnai G2 Spirit BioTwin (Thermo Fisher Scientific, MA, United States), which revealed predominantly round or oval vesicles with varying sizes and well-defined membrane boundaries (Figure 1A). NTA demonstrated a primary peak particle size of approximately 138 nm, comprising about 99.2% of the total vesicle population, consistent with the typical size range of exosomes (Figure 1B). WB confirmed the presence of exosome-specific marker proteins CD63 and CD81 (Figure 1C), further validating successful exosome isolation.
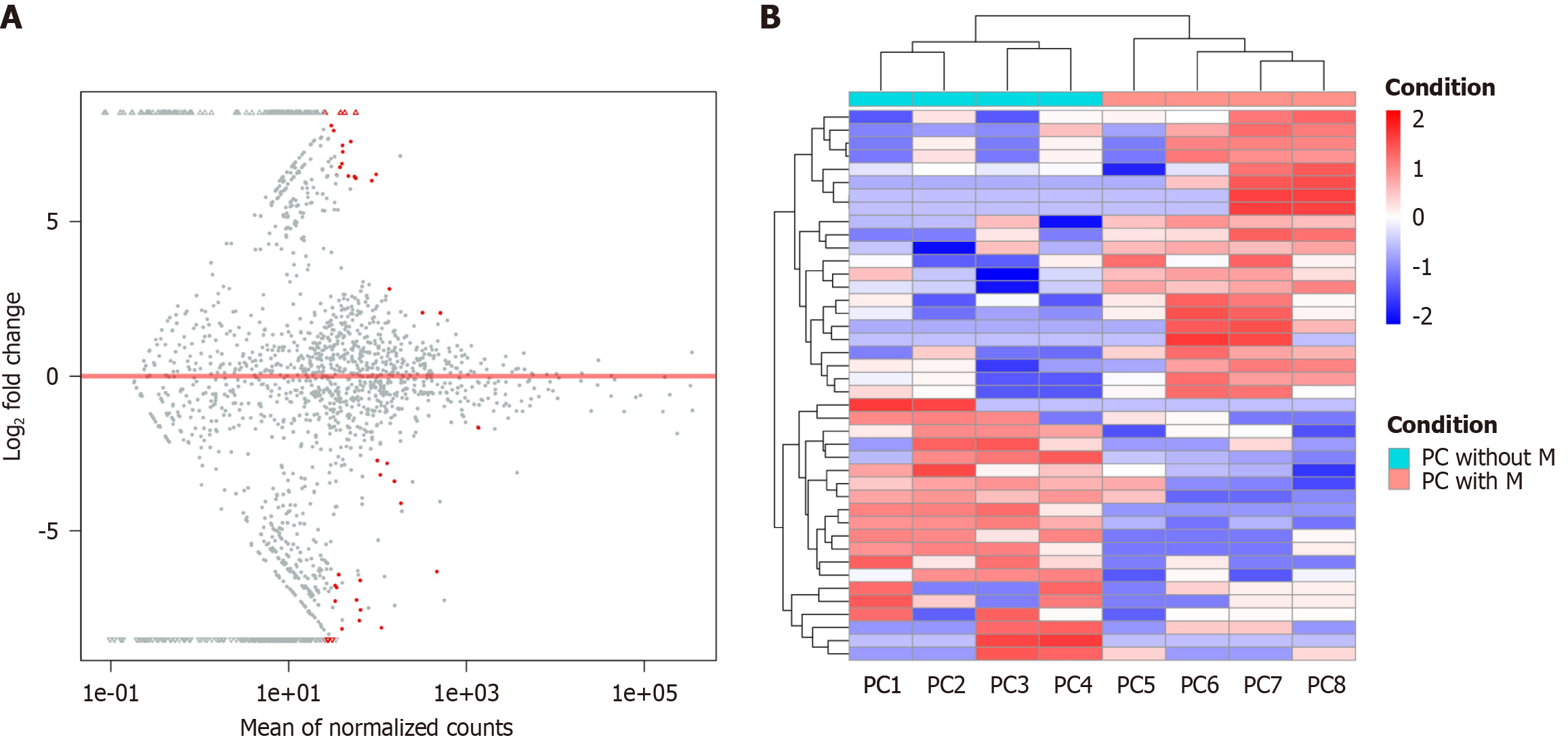
High-throughput sequencing was employed to analyze the miRNA expression profiles of serum-derived exosomes from pancreatic cancer patients with and without distant metastasis. A total of 2483 exosomal miRNAs were identified, including 845 previously reported miRNAs and 1638 newly predicted small RNAs not yet cataloged in miRBase. MiRNA expression levels were calculated using the DESeq package in R, which applies a negative binomial distribution model. Differential expression analysis was performed based on a FC threshold of ≥ 1.5 and a significance level of P < 0.05, resulting in the identification of 42 differentially expressed miRNAs between the metastatic and non-metastatic groups (Supplementary Table 1). The top eight named (previously reported) differentially expressed exosomal miRNAs are presented in Table 2. A volcano plot and hierarchical clustering heatmap comparing miRNA expression profiles between the two groups are shown in Figure 2.
| MicroRNA | log2 (FC) | P value | microRNA_seq | Regulation |
| hsa-let-7f-5p | 7.735016707 | 0.000929468 | TGAGGTAGTAGATTGTATAGTT | Up |
| hsa-miR-150-5p | 7.14531257 | 0.007156657 | AACCCGTAGATCCGAACTTGTG | Up |
| hsa-miR-15b-5p | 1.118971956 | 0.026993311 | TCGAGGAGCTCACAGTCT | Up |
| hsa-miR-151a-3p | 1.0528602 | 0.015206751 | CAACGGAATCCCAAAAGCAGCTG | Up |
| hsa-miR-200b-3p | -0.675276934 | 0.038884084 | TAATACTGCCTGGTAATGATGA | Down |
| hsa-miR-200b-5p | -1.12850605 | 0.000277183 | CATCTTACTGGGCAGCATTGGA | Down |
| hsa-miR-203b-5p | -7.79645688 | 0.013200973 | TAGTGGTCCTAAACATTTCACA | Down |
| hsa-miR-204-3p | -8.83325841 | 0.000617642 | GCTGGGAAGGCAAAGGGACGT | Down |
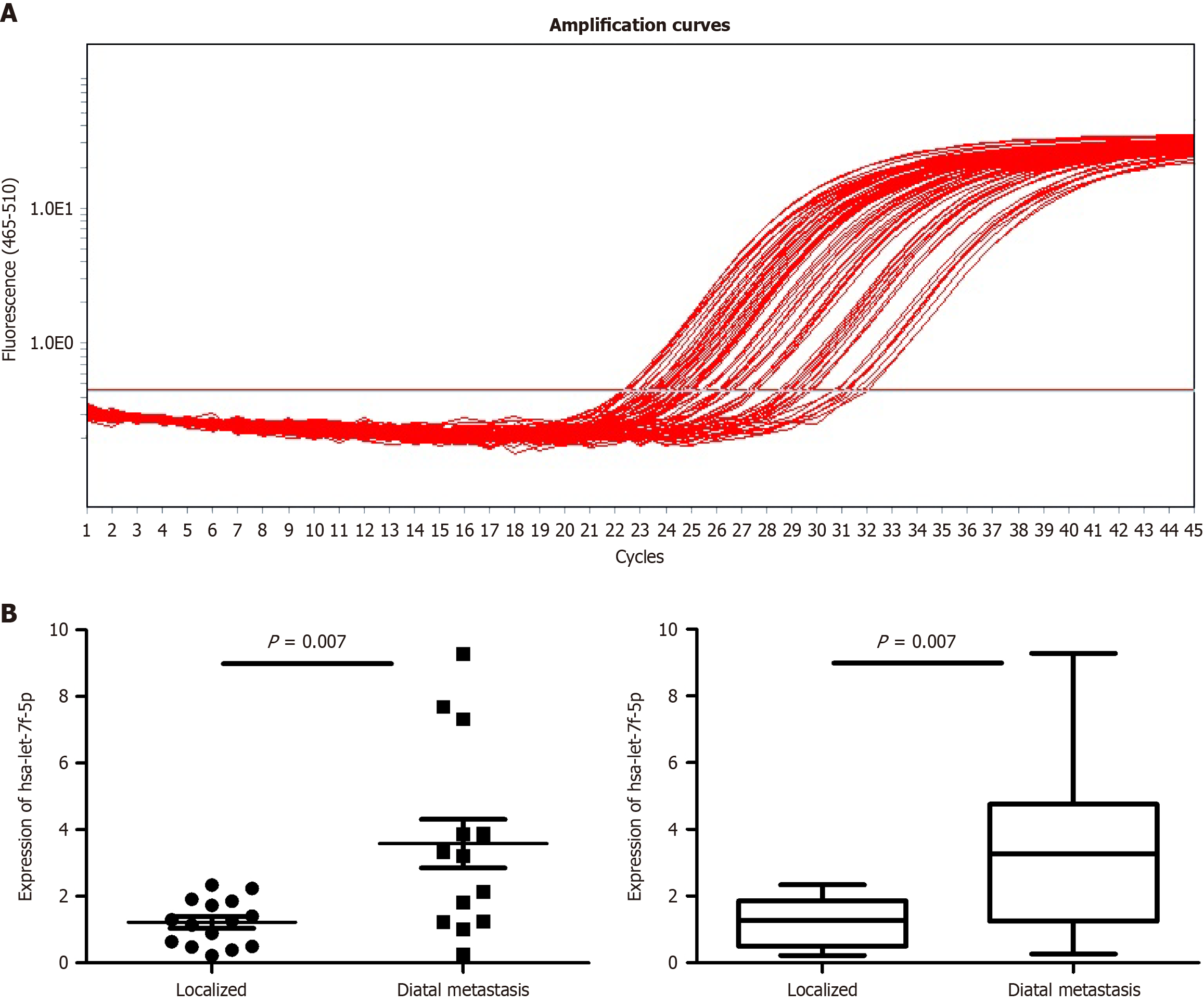
To validate the differential expression of serum exosomal miRNAs identified by high-throughput sequencing, qRT-PCR was performed on serum samples from 15 patients with localized pancreatic cancer and 13 patients with metastatic pancreatic cancer. Four candidate miRNAs (hsa-let-7f-5p, hsa-miR-150-5p, hsa-miR-15b-5p, and hsa-miR-151a-3p) were selected for validation based on their differential expression in the sequencing analysis. Among these, hsa-let-7f-5p showed significant upregulation in metastatic pancreatic cancer patients compared to the localized group (P < 0.05), consistent with the sequencing data. The relative expression levels of the validated hsa-let-7f-5p are summarized in Figure 3, which displays the qRT-PCR amplification curves (Figure 3A) and validation results (Figure 3B). These findings confirm the reliability of the sequencing results and suggest that hsa-let-7f-5p may serve as a promising biomarker for metastatic pancreatic cancer.
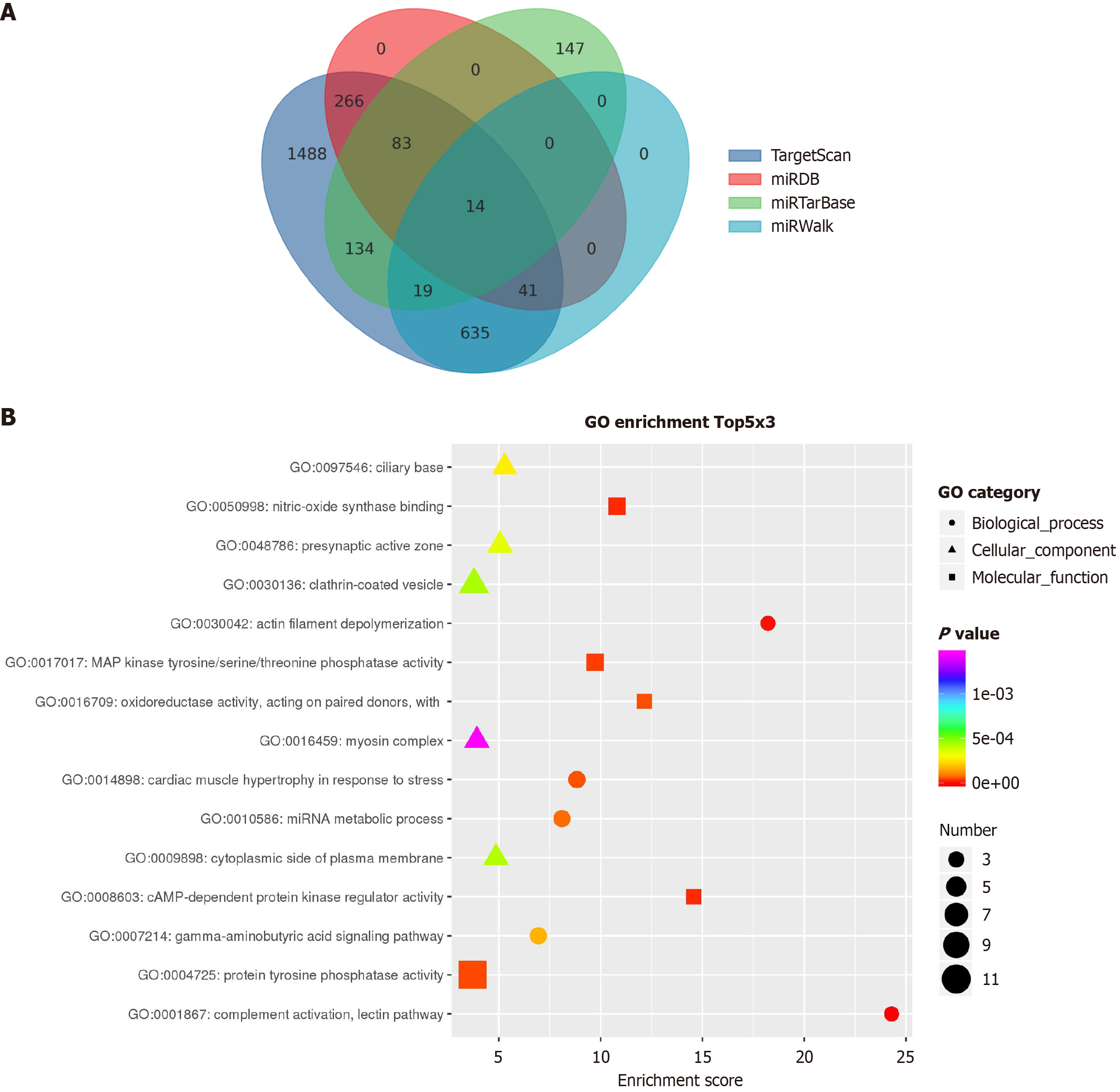
To explore the underlying molecular mechanisms, target genes of hsa-let-7f-5p were predicted using multiple databases, including miRDB, miWalk, miRTarBase, and TargetScan. After integrating the results, a total of 14 target genes were identified, including several genes associated with pancreatic cancer metastasis. Notably, several of these genes are implicated in pancreatic cancer metastasis, including nerve growth factor (NGF), cyclin-dependent kinase inhibitor 1A
Pancreatic cancer is an aggressive, highly malignant neoplasm characterized by rapid progression and early metastatic propensity. Due to this aggressive biology, dissemination to distant organs frequently occurs at initial stages, thereby complicating both diagnosis and clinical management. This propensity for early metastasis is one of the key factors contributing to its poor prognosis and high mortality rate. The most common metastatic pattern in pancreatic cancer is local invasion; however, both lymphatic and hematogenous dissemination also occur. Pancreatic cancer frequently metastasizes to regional organs, with the liver representing the most frequent metastatic site. Other common sites include the stomach, adrenal glands, kidneys, gallbladder, and duodenum[11,12]. Lymphatic metastasis commonly disseminates to distant sites including the pelvis, lungs, or brain, whereas hematogenous metastasis frequently involves osseous tissue. Consequently, identifying serum exosomal miRNAs directly correlated with distant metastatic status in pancreatic cancer could significantly enhance clinical detection of occult or micrometastatic lesions at early stages. This would facilitate timely therapeutic intervention, enable optimized treatment planning, and ultimately improve patient prognosis and quality of life.
In this study, high-throughput sequencing of serum exosomes from metastatic and non-metastatic pancreatic cancer patients identified differentially expressed miRNAs. TEM revealed exosomes with spherical or elliptical morphology, heterogeneous sizes, and defined membrane boundaries. NTA demonstrated a predominant particle size peak at approximately 138 nm (representing approximately 99.2% of samples), consistent with typical exosomal dimensions. WB confirmed expression of exosome-specific markers CD63 and CD8. Sequencing analysis showed elevated expression levels of hsa-let-7f-5p, hsa-miR-150-5p, hsa-miR-15b-5p, and hsa-miR-151a-3p in metastatic vs non-metastatic groups. Subsequent TaqMan probe-based RT-qPCR confirmed significant upregulation of hsa-let-7f-5p in metastatic patients. These findings suggest serum exosomal hsa-let-7f-5p may serve as a novel non-invasive biomarker for detecting metastatic pancreatic cancer.
In recent years, miRNA research has garnered significant attention due to their roles in tumor cell proliferation, invasion, and metastasis[13]. The Let-7 family, in particular, has been associated with the initiation and progression of various malignancies, with studies reporting its involvement in regulating cancer cell proliferation, invasion, metastasis, and apoptosis[14,15]. Di Fazio et al[16] investigated hsa-let-7f-5p expression in typical and atypical pulmonary carcinoids, demonstrating overexpression in most lesions. They further identified differential downregulation of its downstream target gene HMGA2, aiding discrimination between carcinoid subtypes. Valera et al[17] reported significantly higher hsa-let-7f-5p expression in older vs younger prostate cancer patients (P = 0.02), whereas no significant difference was observed between cancer and adjacent normal tissues in younger patients (P > 0.05). In contrast, our study revealed significantly elevated hsa-let-7f-5p expression in metastatic vs non-metastatic pancreatic cancer patients.
Through bioinformatic analyses using miRDB, miWalk, miRTarBase, and TargetScan databases, we identified 14 potential downstream target genes of hsa-let-7f-5p, including NGF, CDKN1A, HMGA2, IGF2BP1, and IGF2BP3, all of which have been associated with pancreatic cancer metastasis. NGF is secreted by pancreatic cancer cells under conditions of metabolic stress, facilitating serine secretion by neurons and thereby supporting the metabolic demands of cancer cells[18,19]. Jiang et al[20] investigated the role of NGF and its receptor, tropomyosin-related kinase A (TrkA), demonstrating that NGF/TrkA is overexpressed and promotes the invasion and proliferation of pancreatic cancer cells. Furthermore, epithelial-mesenchymal transition-related genes were modulated following small interfering RNA-mediated TrkA knockdown. Additionally, previous studies have implicated HMGA2, IGF2BP1, and IGF2BP3 in the invasiveness and metastasis of pancreatic cancer[21]. Insulin-like growth factor I (IGF-I) signaling induces the epithelial-mesenchymal transition program and contributes to metastasis and drug resistance across various tumor subtypes. Preclinical studies have shown that targeting the IGF-I receptor produces promising antitumor effects[22]. Taken together, these findings support our hypothesis that hsa-let-7f-5p may promote pancreatic cancer cell invasiveness by regulating the expression of key genes such as HMGA2, IGF2BP1, and IGF2BP3.
In this study, we utilized high-throughput sequencing technology to identify differentially expressed miRNAs in serum exosomes of pancreatic cancer patients with metastasis. Subsequent validation via TaqMan probe-based RT-qPCR confirmed hsa-let-7f-5p as a potential biomarker, demonstrating significantly higher expression in the serum exosomes of metastatic pancreatic cancer patients compared to non-metastatic patients. Furthermore, through bioinformatic prediction of hsa-let-7f-5p downstream target genes, we identified several genes associated with pancreatic cancer metastasis, including NGF, CDKN1A, HMGA2, IGF2BP1, and IGF2BP3.
However, several limitations of this study should be acknowledged. First, a total of 42 differentially expressed miRNAs were identified, among which we selected the top four named (previously reported) upregulated miRNAs for validation. The remaining four named downregulated miRNAs and 34 novel miRNAs were not validated due to resource and time constraints. Preliminary screening and functional characterization of these miRNAs were not performed in this study. Future research is warranted to validate and explore the biological roles of these novel and downregulated miRNAs in pancreatic cancer. Second, the sample size was relatively small, and validation in larger, multi-center cohorts is necessary to confirm the robustness of hsa-let-7f-5p expression across different pancreatic cancer subgroups. Third, given the known differences in miRNA expression between serum exosomes and tumor tissues, further studies are needed to investigate the expression levels of hsa-let-7f-5p in tissue-derived exosomes from patients with metastatic pancreatic cancer. Fourth, due to the lack of available clinical follow-up data, we were unable to analyze the correlation between hsa-let-7f-5p expression and patient prognosis or treatment response. Future studies incorporating well-annotated clinical data will be essential to determine the prognostic and predictive value of this biomarker. Finally, in vitro and in vivo experiments are required to further elucidate the molecular mechanisms by which hsa-let-7f-5p may contribute to pancreatic cancer metastasis, ultimately supporting its development as a novel biomarker for the diagnosis and management of metastatic pancreatic cancer.
In conclusion, our findings indicate that serum exosomal hsa-let-7f-5p is significantly elevated in patients with metastatic pancreatic cancer, suggesting its potential as a non-invasive biomarker for the detection of metastatic disease. This elevation may reflect its biological involvement in the metastatic process. Given the stability and accessibility of exosomal miRNAs in circulation, hsa-let-7f-5p could be leveraged not only for early diagnosis but also for dynamic monitoring of disease progression and treatment response. In future clinical applications, serum exosomal hsa-let-7f-5p may be integrated into routine follow-up protocols or ambulatory monitoring systems, potentially in combination with imaging modalities, to improve the early detection and risk stratification of pancreatic cancer metastasis. This approach could provide a less invasive and more accessible alternative to traditional tissue biopsy, ultimately aiding personalized disease management.
We thank all authors for their continuous and excellent support with patient data collection, imaging analysis, statistical analysis, and valuable suggestions for the article.
| 1. | Siegel RL, Kratzer TB, Giaquinto AN, Sung H, Jemal A. Cancer statistics, 2025. CA Cancer J Clin. 2025;75:10-45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 1448] [Article Influence: 1448.0] [Reference Citation Analysis (3)] |
| 2. | Xia C, Dong X, Li H, Cao M, Sun D, He S, Yang F, Yan X, Zhang S, Li N, Chen W. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J (Engl). 2022;135:584-590. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1790] [Cited by in RCA: 2490] [Article Influence: 622.5] [Reference Citation Analysis (2)] |
| 3. | Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5690] [Cited by in RCA: 12367] [Article Influence: 6183.5] [Reference Citation Analysis (6)] |
| 4. | Torphy RJ, Fujiwara Y, Schulick RD. Pancreatic cancer treatment: better, but a long way to go. Surg Today. 2020;50:1117-1125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 145] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 5. | Li J, Wang A, Guo H, Zheng W, Chen R, Miao C, Zheng D, Peng J, Wang J, Chen Z. Exosomes: innovative biomarkers leading the charge in non-invasive cancer diagnostics. Theranostics. 2025;15:5277-5311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 6. | Jain G, Das P, Ranjan P, Neha, Valderrama F, Cieza-Borrella C. Urinary extracellular vesicles miRNA-A new era of prostate cancer biomarkers. Front Genet. 2023;14:1065757. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 7. | Sha M, Kunduzi B, Froghi S, Quaglia A, Davidson B, Fusai GK. Role of circulating exosomal biomarkers and their diagnostic accuracy in pancreatic cancer. JGH Open. 2023;7:30-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 8. | Nicoletti A, Negri M, Paratore M, Vitale F, Ainora ME, Nista EC, Gasbarrini A, Zocco MA, Zileri Dal Verme L. Diagnostic and Prognostic Role of Extracellular Vesicles in Pancreatic Cancer: Current Evidence and Future Perspectives. Int J Mol Sci. 2023;24:885. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 9. | Luo J, Jiang L, He C, Shi M, Yang ZY, Shi M, Lu S, Li C, Zhang J, Yan M, Zhu ZG, Yan C. Exosomal hsa-let-7g-3p and hsa-miR-10395-3p derived from peritoneal lavage predict peritoneal metastasis and the efficacy of neoadjuvant intraperitoneal and systemic chemotherapy in patients with gastric cancer. Gastric Cancer. 2023;26:364-378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 10. | Masamune A, Yoshida N, Hamada S, Takikawa T, Nabeshima T, Shimosegawa T. Exosomes derived from pancreatic cancer cells induce activation and profibrogenic activities in pancreatic stellate cells. Biochem Biophys Res Commun. 2018;495:71-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 79] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 11. | Cao BY, Tong F, Zhang LT, Kang YX, Wu CC, Wang QQ, Yang W, Wang J. Risk factors, prognostic predictors, and nomograms for pancreatic cancer patients with initially diagnosed synchronous liver metastasis. World J Gastrointest Oncol. 2023;15:128-142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 3] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 12. | Ren S, Qian LC, Cao YY, Daniels MJ, Song LN, Tian Y, Wang ZQ. Computed tomography-based radiomics diagnostic approach for differential diagnosis between early- and late-stage pancreatic ductal adenocarcinoma. World J Gastrointest Oncol. 2024;16:1256-1267. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 11] [Cited by in RCA: 48] [Article Influence: 24.0] [Reference Citation Analysis (2)] |
| 13. | Hasani F, Masrour M, Khamaki S, Jazi K, Hosseini S, Heidarpour H, Namazee M. Diagnostic and Prognostic Accuracy of MiRNAs in Pancreatic Cancer: A Systematic Review and Meta-Analysis. J Cell Mol Med. 2025;29:e70337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 14. | Han K, Liu H, Cui J, Liu Y, Pan P. Recent strategies for electrochemical sensing detection of miRNAs in lung cancer. Anal Biochem. 2023;661:114986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 15. | Niculae AM, Dobre M, Herlea V, Manuc TE, Trandafir B, Milanesi E, Hinescu ME. Let-7 microRNAs Are Possibly Associated with Perineural Invasion in Colorectal Cancer by Targeting IGF Axis. Life (Basel). 2022;12:1638. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 16. | Di Fazio P, Maass M, Roth S, Meyer C, Grups J, Rexin P, Bartsch DK, Kirschbaum A. Expression of hsa-let-7b-5p, hsa-let-7f-5p, and hsa-miR-222-3p and their putative targets HMGA2 and CDKN1B in typical and atypical carcinoid tumors of the lung. Tumour Biol. 2017;39:1010428317728417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 17. | Valera VA, Parra-Medina R, Walter BA, Pinto P, Merino MJ. microRNA Expression Profiling in Young Prostate Cancer Patients. J Cancer. 2020;11:4106-4114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 18. | Zhang W, He R, Yang W, Zhang Y, Yuan Q, Wang J, Liu Y, Chen S, Zhang S, Zhang W, Zhu Z, Zhang J, Wang Z, Li J. Autophagic Schwann cells promote perineural invasion mediated by the NGF/ATG7 paracrine pathway in pancreatic cancer. J Exp Clin Cancer Res. 2022;41:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 62] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 19. | Wang K, Ni B, Xie Y, Li Z, Yuan L, Meng C, Zhao T, Gao S, Huang C, Wang H, Ma Y, Zhou T, Feng Y, Chang A, Yang C, Yu J, Yu W, Zang F, Zhang Y, Ji RR, Wang X, Hao J. Nociceptor neurons promote PDAC progression and cancer pain by interaction with cancer-associated fibroblasts and suppression of natural killer cells. Cell Res. 2025;35:362-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 29] [Reference Citation Analysis (0)] |
| 20. | Jiang J, Bai J, Qin T, Wang Z, Han L. NGF from pancreatic stellate cells induces pancreatic cancer proliferation and invasion by PI3K/AKT/GSK signal pathway. J Cell Mol Med. 2020;24:5901-5910. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 21. | Kugel S, Sebastián C, Fitamant J, Ross KN, Saha SK, Jain E, Gladden A, Arora KS, Kato Y, Rivera MN, Ramaswamy S, Sadreyev RI, Goren A, Deshpande V, Bardeesy N, Mostoslavsky R. SIRT6 Suppresses Pancreatic Cancer through Control of Lin28b. Cell. 2016;165:1401-1415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 223] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 22. | Li H, Batth IS, Qu X, Xu L, Song N, Wang R, Liu Y. IGF-IR signaling in epithelial to mesenchymal transition and targeting IGF-IR therapy: overview and new insights. Mol Cancer. 2017;16:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 91] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/