Published online Jul 7, 2025. doi: 10.3748/wjg.v31.i25.107893
Revised: May 20, 2025
Accepted: June 9, 2025
Published online: July 7, 2025
Processing time: 95 Days and 13.9 Hours
Colorectal cancer (CRC) is a leading cause of cancer-related mortality worldwide, primarily due to tumor heterogeneity and treatment resistance. The leucine-rich repeat-containing protein 19 (LRRC19) has been linked to immune regulation and tumor suppression, yet its specific role in CRC remains poorly understood.
To investigate the tumor-suppressive role of LRRC19 in CRC, focusing on cell cycle, immune microenvironment, and chemotherapy response.
Bioinformatics analyses of Gene Expression Omnibus and The Cancer Genome Atlas databases identified differentially expressed genes in CRC. LRRC19 exp
LRRC19 expression was significantly downregulated in CRC and associated with poor prognosis. Overexpression of LRRC19 inhibited CRC cell proliferation, induced G0/G1 phase arrest, and suppressed tumor growth in vivo. Mechanistically, LRRC19 suppressed CDK6 transcription by downregulating E2F1, leading to cell cycle arrest. Additionally, LRRC19 promoted immune cell infiltration, particularly B cells and CD4+ T cells, while decreasing immunosuppressive cells. LRRC19 also sensitized CRC cells to OXA, enhancing chemotherapy efficacy.
LRRC19 suppresses CRC by targeting the CDK6/E2F1 axis, modulating the immune microenvironment, and enhancing chemotherapy sensitivity, making it a promising therapeutic target for precision medicine in CRC.
Core Tip: Colorectal cancer (CRC) remains a major global health challenge due to tumor heterogeneity and treatment resistance. Leucine-rich repeat-containing protein 19 (LRRC19) is identified as a novel tumor suppressor in CRC, inhibiting the cyclin-dependent kinase 6/E2F1 axis to induce cell cycle arrest, remodeling the immune microenvironment by enhancing immune cell infiltration, and improving chemotherapy sensitivity to oxaliplatin. These findings provide new insights into the molecular mechanisms of CRC progression and suggest LRRC19 as a promising therapeutic target for precision medicine in CRC management.
- Citation: Huang SS, Chen W, Vaishnani DK, Huang LJ, Li JZ, Huang SR, Li YZ, Xie QP. Leucine-rich repeat-containing protein 19 suppresses colorectal cancer by targeting cyclin-dependent kinase 6/E2F1 and remodeling the immune microenvironment. World J Gastroenterol 2025; 31(25): 107893
- URL: https://www.wjgnet.com/1007-9327/full/v31/i25/107893.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i25.107893
Colorectal cancer (CRC) remains a major global health challenge, ranking third in cancer incidence (9.6% of new cases) and second in mortality (9.3% of cancer deaths) worldwide, with approximately 1.93 million new diagnoses and 0.90 million fatalities reported in 2022 according to GLOBOCAN statistics[1]. In recent years, the incidence and mortality of CRC in China are also on the rise[2]. The molecular pathogenesis of CRC involves a dynamic interplay between multistep genetic alterations and tumor microenvironment (TME) reprogramming. While therapeutic advances targeting cell cycle checkpoints [e.g., cyclin-dependent kinase (CDK4/CDK6) inhibitors] and immune checkpoint blockade [anti-pro
Functional transmembrane receptors (FTRs) play pivotal roles in cellular signaling and immunometabolic reprogramming[5,6]. As specialized pattern recognition receptors (PRRs), these molecules initiate intracellular signaling cascades through pathogen-associated molecular pattern recognition, orchestrating immune responses against pathogens and tumor progression[7,8]. Leucine-rich repeat-containing 19 (LRRC19), a recently identified PRR located at chro
Our work identified LRRC19 as a context-dependent tumor suppressor in CRC, uniquely targeting the CDK6/E2F1 axis (a previously unreported mechanism) and reprogramming immune cell infiltration. This dual mechanism, com
Logged into the Gene Expression Omnibus (GEO) database and conducted a search using the keywords "colorectal cancer and normal tissue", focusing primarily on comparative studies between CRC and normal tissues, and excluding other types of CRC datasets. Ultimately, the GSE41328 and GSE23878 datasets, both containing gene expression data for CRC tissues and adjacent normal tissues, were selected[17]. Initially, rigorous quality control and preprocessing were conducted on the raw data, which included removing low-quality samples (filtering criteria: Intersample correlation coefficient < 0.8 or outlier detection), background correction, quantile normalization (using rate-monotonic analysis algorithm), and batch effect correction (with the ComBat method) to ensure data reliability. Principal component analysis (PCA) was performed separately on these two datasets. Subsequently, differential expression analysis was performed using the R package DESeq2 and Limma, with a significance threshold set at adjusted P < 0.01 (Benjamini-Hochberg correction) and a fold change (FC) threshold of |log2FC| > 1.6 to screen for significantly differentially expressed genes (DEGs). Volcano plots were drawn using ggplot2, where the X-axis represents log2FC (logarithmic FC in gene exp
The expression matrix of DEGs was further extracted, and a hierarchical clustering heatmap was generated using the pheatmap package. The data were Z-score-standardized to display the expression patterns of genes across samples. The ggVennDiagram package was used to draw a Venn diagram comparing DEGs between GSE41328 and GSE23878, identifying common and unique gene sets. The common DEGs were imported into the STRING database to construct a protein-protein interaction (PPI) network, with a confidence threshold set at ≥ 0.400[18]. Cytoscape (v3.9.1) was used for network visualization, including hiding unconnected nodes to simplify the network, applying the MCODE plugin to identify core functional modules, setting node size based on connectivity, and color mapping based on log2FC values. Based on literature screening, 19 FTR-related genes were cross-analyzed with 33 candidate genes in the PPI network, and the overlap between the two gene sets was quantified using the UpSetR package to create an interactive Venn diagram.
A systematic analysis of the regulatory network associated with the LRRC19 gene was conducted using bioinformatics methods. Firstly, potential microRNAs (miRNAs) interacting with LRRC19 were predicted using the authoritative databases TargetScan, miRDB, and miRanda. The prediction results were integrated and deduplicated to obtain a com
To further uncover the biological functions of LRRC19, co-expression genes significantly correlated (both positively and negatively) with LRRC19 expression were mined using the UALCAN database[19]. These genes were subjected to Gene Ontology (GO) functional annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. To clearly present the analysis results, a multi-layer network diagram was used for visualization: The innermost layer displayed KEGG pathway enrichment results, while the outer layers showed GO functional enrichment results, thereby revealing the biological processes and signaling pathways that LRRC19 may be involved in. All analysis results were compiled into a detailed analytical report[20,21]. Using the GSE41328 dataset, a single-gene GSEA enrichment analysis was conducted on LRRC19 to explore its potential biological functions.
This study conducted an in-depth analysis based on The Cancer Genome Atlas database, examining RNA sequencing data and clinical follow-up information of patients with CRC[22]. It explored the correlation between LRRC19 gene expression levels and disease-free interval (DFI), compared the differences in disease-specific survival (DSS) between the two groups of patients, and tested the statistical significance of LRRC19 gene expression levels in overall survival (OS). Additionally, survival analysis was performed on the progression-free interval. Moreover, a thorough analysis of LRRC19 gene expression levels in CRC samples from the TCGA database was conducted using the UALCAN database, including expression differences among patients of various age groups, sex comparisons, LRRC19 expression level contrasts in different histological subtypes (normal, adenocarcinoma, mucinous adenocarcinoma), classification comparisons based on lymph node metastasis status, LRRC19 gene expression level differences among different races, analysis of LRRC19 gene expression levels at various stages of colon adenocarcinoma (COAD), and classification comparisons based on tumor protein p53 (TP53) mutation status. Furthermore, the study investigated the differences in LRRC19 gene exp
The drug sensitivity of LRRC19 was analyzed based on the Genomics of Drug Sensitivity in Cancer database to identify candidate drugs associated with it[23]. The protein structure of LRRC19 was obtained from the Protein Data Bank database, and the molecular structures of the drugs were downloaded from the PubChem database[24,25]. Subsequently, the CB-Dock2 online molecular docking platform was used to predict the binding mode and binding energy between the drugs and LRRC19[26]. The docking results were evaluated using binding energy (kcal/mol) as the criterion, with lower binding energy indicating a more stable binding between the drug and the target protein. Using the GSE290028 dataset, which includes gene expression profiles of 13 patients with CRC insensitive to chemotherapy and 9 sensitive patients, we compared the expression differences of the LRRC19 gene between the two groups.
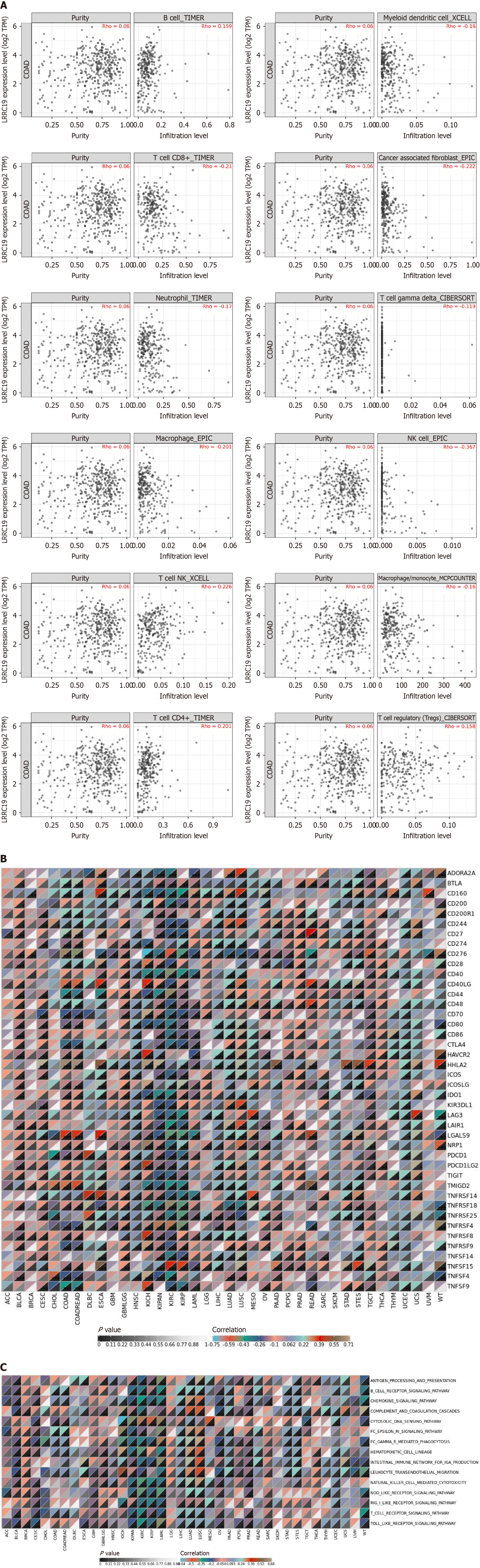
Utilizing the analytical tools provided by the Tumor Immune Estimation Resource database, we systematically investigated the correlation between the expression level of the LRRC19 gene in CRC tissues and the infiltration degree of various immune cells[27]. The specific immune cell subpopulations analyzed included B lymphocytes, cancer-associated fibroblasts (CAFs), macrophages, monocytes, myeloid dendritic cells (mDCs), neutrophils, natural killer (NK) cells, CD4+ T lymphocytes, CD8+ T lymphocytes, γδ T cells, NKT cells, and regulatory T cells (Tregs). We calculated the Spearman correlation coefficient (Rho) between LRRC19 expression level and the infiltration level of each immune cell, and assessed its statistical significance (P value). To explore the correlation of the LRRC19 gene with immune checkpoints in pan-cancer and specifically in CRC, we collected LRRC19 gene expression data and immune checkpoint-related gene expression data from multiple cancer types. Statistical methods (such as Spearman or Pearson correlation coefficients) were used to analyze the correlation between LRRC19 gene expression and immune checkpoint genes. The P value for each correlation coefficient was calculated to determine the significance level of the correlation. An analysis of the immune-inflammatory correlation of LRRC19 in pan-cancer was conducted. Based on this, we focused on the immune-inflammatory correlation of LRRC19 in CRC. By analyzing the correlation coefficients (cor) and significance levels (P) between different pathways and LRRC19 expression levels, we explored the role of LRRC19 in immune-inflammatory pathways. To conduct an in-depth analysis of the co-expression or correlation between LRRC19 and CD8 T cell ex
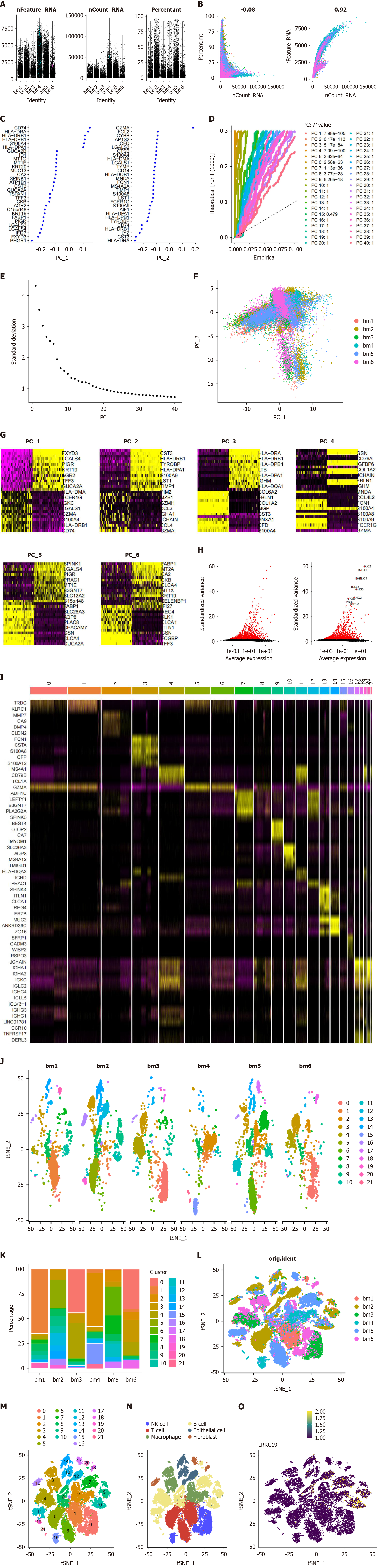
This study utilized the GSE161277 CRC single-cell dataset, randomly selecting three pairs of normal/tumor tissue samples for analysis. To mitigate potential batch effects, the proportion of mitochondrial genes in each cell was first calculated, along with the distribution of gene counts, RNA, and mitochondrial genes to assess the overall cell state and quality. Subsequently, cells were filtered based on the criteria of nFeatureRNA > 200, nCountRNA > 1000, and nCou
The LRRC19-overexpression plasmid and corresponding control vectors were purchased from Miaoling Biotechnology (Wuhan, China). The CDK6 promoter plasmid and HA-tagged vectors were constructed by Qingke Biotechnology (Beijing, China). Overexpression plasmids PCDH-CDK6 and PCDH-E2F1 were generated by our laboratory. Specific antibodies used included anti-LRRC19 (PA5-20914) from Invitrogen (Grand Island, NY, United States); antibodies against cyclin A2 (Sc-53234), cyclin B1 (Sc-245), cyclin D1 (Sc-20044), cyclin E2 (Sc-481), CDK2 (Sc-6248), CDK4 (Sc-260), CDK6 (Sc-177), E2F1 (Sc-251), SP1 (Sc-59), and ETS-1 (Sc-350) from Santa Cruz Biotechnology (Santa Cruz, CA, United States); antibodies against forkhead box P3 (FOXP3) (12829S), ELK-1 (9182S), Flag (14793S), and HA (3724S) from Cell Signaling Technology (Boston, MA, United States); and antibodies specific for glyceraldehyde-3-phosphate dehydrogenase (10494-1-AP) and α-tubulin (66031-1-Ig) from Proteintech (Chicago, IL, United States).
A total of 150 paired clinical CRC specimens and matched adjacent normal tissues were collected from the First Affiliated Hospital of Wenzhou Medical University (Zhejiang, China), with ethical approval from the Ethics Committee of Wenzhou Medical University. Comprehensive clinical information for these samples was obtained and confirmed, as detailed previously in our earlier studies. Human CRC cell lines HT29 (CBP60011; COBIOER, Nanjing, China), RKO (CBP60006; COBIOER), LoVo (CBP60032; COBIOER), SW620 (CBP60036; COBIOER), and SW480 (CBP60019; COBIOER) were cultured in RPMI-1640 medium (11875-093; Sigma, St. Louis, MO, United States). The HCT116 cell line (CBP60028; COBIOER) was maintained in McCoy’s 5A medium (PM150710; Procell Life Science & Technology Company, Wuhan, China). All cells were cultured at 37 °C in a humidified incubator containing 5% CO2.
Total RNA was extracted from cells using TRIzol reagent (15596018; Invitrogen). Complementary DNA was synthesized using a reverse transcription kit (RR037A; Takara Bio, Shiga, Japan) according to the manufacturer’s instructions. Quantitative PCR (qPCR) was conducted to determine mRNA expression levels using specific primers: LRRC19- forward: 5’-TGC TAT CAA GTG CCC AGT ATG-3’; LRRC19-reverse: 5’-TTT CTG CCT CAT GCT CTT CC-3’. CDK6-forward: 5’-GGT ACA GAG CAC CCG AAG TC-3’; CDK6-reverse: 5’-CTC CTG GGA GTC CAA TCA CG-3’. All experiments were performed according to standardized protocols. Additionally, total RNA was isolated using Qiagen miRNA extraction kits (Hilden, Germany), reverse-transcribed, and subjected to qPCR to assess the expression of miRNAs related to LRRC19 regulation.
Immunohistochemistry staining was performed to evaluate LRRC19 protein expression in 150 paired clinical CRC specimens. Tissue samples were fixed, dehydrated, embedded in paraffin, and sectioned. Sections underwent hema
Cells were seeded into 96-well plates until adherence, starved in medium containing 0.1% fetal bovine serum (FBS) for 12 hours, and subsequently cultured in medium containing 10% FBS for the indicated durations. After incubation, the medium was discarded, and an ATP-based reagent mixed with phosphate-buffered saline was added to each well. Plates were shaken gently, incubated for 10 min, and analyzed using a Centro LB 960 Luminometer (Berthold Technologies, Bad Wildbad, Germany).
For anchorage-independent growth assays, cells were mixed with an upper layer of agarose gel placed over a pre-solidified lower agarose layer in 6-well plates and cultured at 37 °C with 5% CO2. Colonies containing more than 32 cells were photographed under a microscope, counted, and analyzed.
For cell cycle analyses, cells seeded at 30%-40% confluence in 6-well plates were starved (0.1% FBS) and then cultured in complete medium (10% FBS) for 12 hours. Cells were harvested, fixed overnight in pre-chilled 70% ethanol at 4 °C, stained with propidium iodide (PI; 360 μL) and RNase A (40 μL), and incubated at room temperature for 30 minutes. Cell cycle distributions were analyzed using CytoFLEX flow cytometry (Beckman Coulter, San Diego, CA, United States).
Cells were lysed in lysis buffer, and protein concentrations in supernatants were measured using NanoDrop One (Thermo Fisher Scientific, Waltham, MA, United States). Proteins were separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred onto polyvinylidene fluoride membranes, blocked in 5% skim milk, incubated with primary and secondary antibodies, and developed using chemiluminescence reagents. Protein bands were visualized and analyzed with a phosphor imaging scanner (Typhoon FLA 7000; GE Healthcare, Chicago, IL, United States).
CRC cells (6000 cells/well) were seeded into 96-well plates, starved in medium containing 0.1% FBS for 12 hours, and subsequently treated with various concentrations of palbociclib diluted in medium containing 10% FBS for 48 hours. Cell viability was assessed using the ATP-based method as previously described.
All animal experiments were approved by the Animal Ethics Committee of Wenzhou Medical University. Twelve female BALB/c nude mice purchased from Jiangsu Jinyao Biotechnology (Nanjing, China) were housed under specific pathogen-free conditions in the Animal Facility at Wenzhou Medical University. After acclimation (approximately 1 week), mice were randomly divided into two groups and marked individually with ear tags. HCT116 (Vector) or HCT116 (LRRC19-Flag) cells were subcutaneously injected into the mice. Tumors were allowed to grow for approximately 3 weeks, after which mice were euthanized, and tumors were excised, photographed, and weighed.
Cells stably expressing the E2F1 promoter-luciferase reporter were seeded in 96-well plates (8 × 10³ cells/well). When cells reached 70%-80% confluence, cells were transfected with TK plasmids and either wild-type or mutant CDK6 promoter plasmids. After 24 hours, cells were lysed in passive lysis buffer, incubated at room temperature for 15 minutes, and luciferase activity was measured using Dual Luciferase Reporter Assay System reagents (Promega, Madison, WI, United States) on a luminometer (Wallac 1420 Victor2; PerkinElmer, Waltham, MA, United States). Promoter activity was quantified based on relative luciferase units.
Chromatin immunoprecipitation (ChIP) was performed using the SimpleChIP® Enzymatic Chromatin IP Kit (Catalog #9003; Cell Signaling Technology, Danvers, MA, United States) following the manufacturer’s protocol. Briefly, HCT116 cells (4 × 106 per sample) were crosslinked with 1% formaldehyde for 10 minutes at room temperature, followed by chromatin fragmentation to 200-500 base pairs using enzymatic digestion. IP was carried out overnight at 4 °C with 2 μg of either normal rabbit immunoglobulin G (IgG) or E2F1-specific antibody. After reversing crosslinks and purifying DNA, quantitative PCR was performed using primers targeting the CDK6 promoter region. Amplified DNA fragments were resolved via 2% agarose gel electrophoresis, stained with ethidium bromide (0.5 μg/mL), and visualized under ultraviolet transillumination.
Specific CRC cell lines, including HCT116 (Vector), HCT116 (LRRC19), HT29 (Vector), and HT29 (LRRC19), were treated with various concentrations of oxaliplatin (OXA) (O9512; Sigma-Aldrich) (0, 5, 10, 20, 40, and 80 μM) for 48 hours to assess drug sensitivity. Western blot analysis was performed to detect changes in LRRC19 expression. Morphological changes indicative of apoptosis or cytotoxicity induced by OXA treatment were observed using an inverted microscope.
All analyses were performed in R (v4.2.1). Data are presented as the mean ± SD. Differences between groups were assessed using Student’s t-test or analysis of variance, and survival outcomes were analyzed via Kaplan-Meier curves (log-rank test). Two-tailed P < 0.05 was considered statistically significant.
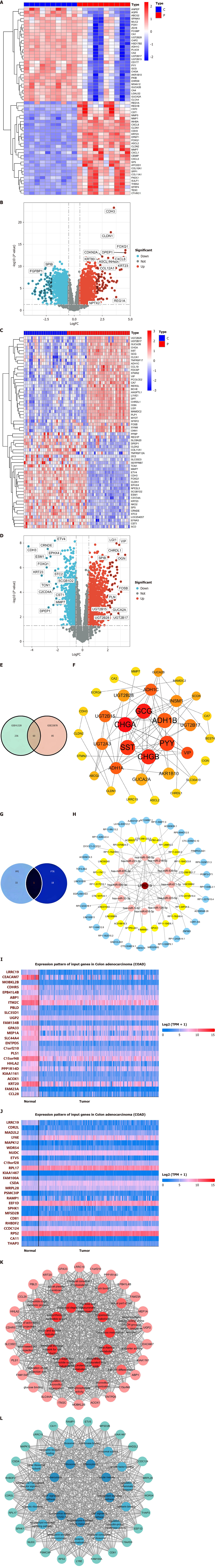
The PCA plot clearly revealed the distribution of samples from the two datasets within the principal component space, intuitively unveiling their similarities and potential batch effects (Supplementary Figure 1). Through an in-depth analysis of the CRC GSE41328 dataset, a group of genes with significant expression differences was successfully identified. The analysis results revealed a clear bidirectional regulatory pattern among these DEGs: Several genes, including P-cadherin, claudin-11, and FOXQ1, showed a significant upregulation trend, while genes such as fibroblast growth factor-binding protein 1, SPIB, and keratin 80 exhibited marked downregulation. Additionally, a large number of other genes also showed varying degrees of changes in expression levels. To systematically present these important findings, a multidimensional visualization analysis map was constructed. Figure 1A visually represents the heterogeneity of gene exp
Figure 1E reveals 60 common DEGs shared between the GSE41328 and GSE23878 datasets. Figure 1F presents a streamlined PPI network, where core genes such as chromogranin A, somatostatin, and glycogen phosphorylase B exhibit significant direct interactions with other genes, forming multiple tightly connected clusters. This suggests their potential cooperative roles in the same biological processes. Figure 1G identifies LRRC19 as the sole overlapping gene between transmembrane receptor-related genes and the PPI network, indicating its critical involvement in both biological pathways. Integrated analysis (Figure 1H) suggests that miRNAs (e.g., hsa-miR-186-5p, hsa-miR-223-3p) and lncRNAs
Figure 1I and J display genes positively and negatively correlated with LRRC19, respectively. KEGG enrichment analysis (Figure 1K) reveals that LRRC19 is closely associated with various metabolic pathways, including alpha-linolenic acid metabolism, unsaturated fatty acid biosynthesis, sugar metabolism (e.g., galactose, pyruvate, pentose, and glucuronate conversions), as well as the intestinal immune network for IgA production. GO analysis indicates that the gene is involved in regulating cellular processes related to structural components like cell protrusion size and microvillus organization, and possesses molecular functions such as guanosine diphosphate phosphatase and oxidoreductase. These findings suggest that LRRC19 plays a key role in cytoskeleton construction, metabolic regulation (particularly lipid and carbohydrate metabolism), and signal transduction, providing important clues for functional studies and the exploration of disease association mechanisms. Figure 1L presents the KEGG and GO enrichment analysis results for genes negatively correlated with LRRC19. The KEGG analysis shows that the LRRC19 gene is significantly associated with pathways such as "Ribosome", "Sphingolipid signaling pathway", and "Tuberculosis". GO analysis indicates that the gene primarily participates in the biological processes of "ribonucleoprotein complex biogenesis" and "translation", is enriched in ribosomal structures in the cellular component, and is involved in molecular functions related to "ribosome structure composition" and "growth factor binding". These discoveries suggest that LRRC19 may be involved in a variety of biological functions through the regulation of protein synthesis and metabolic processes, providing directions for further investigation into its molecular mechanisms. LRRC19 may have functioned by regulating multiple biological processes. The upregulated pathways were primarily involved in metabolic processes, including ascorbate and aldarate metabolism, other glycan degradation, pentose and glucuronate interconversions, proximal tubule bicarbonate reabsorption, and retinol metabolism. Conversely, the downregulated pathways were mainly associated with diseases and protein synthesis and degradation, such as CRC, ribosome, eukaryotic ribosome biogenesis, RNA degradation, and ubiquitin-mediated proteolysis (Supplementary Figure 2).
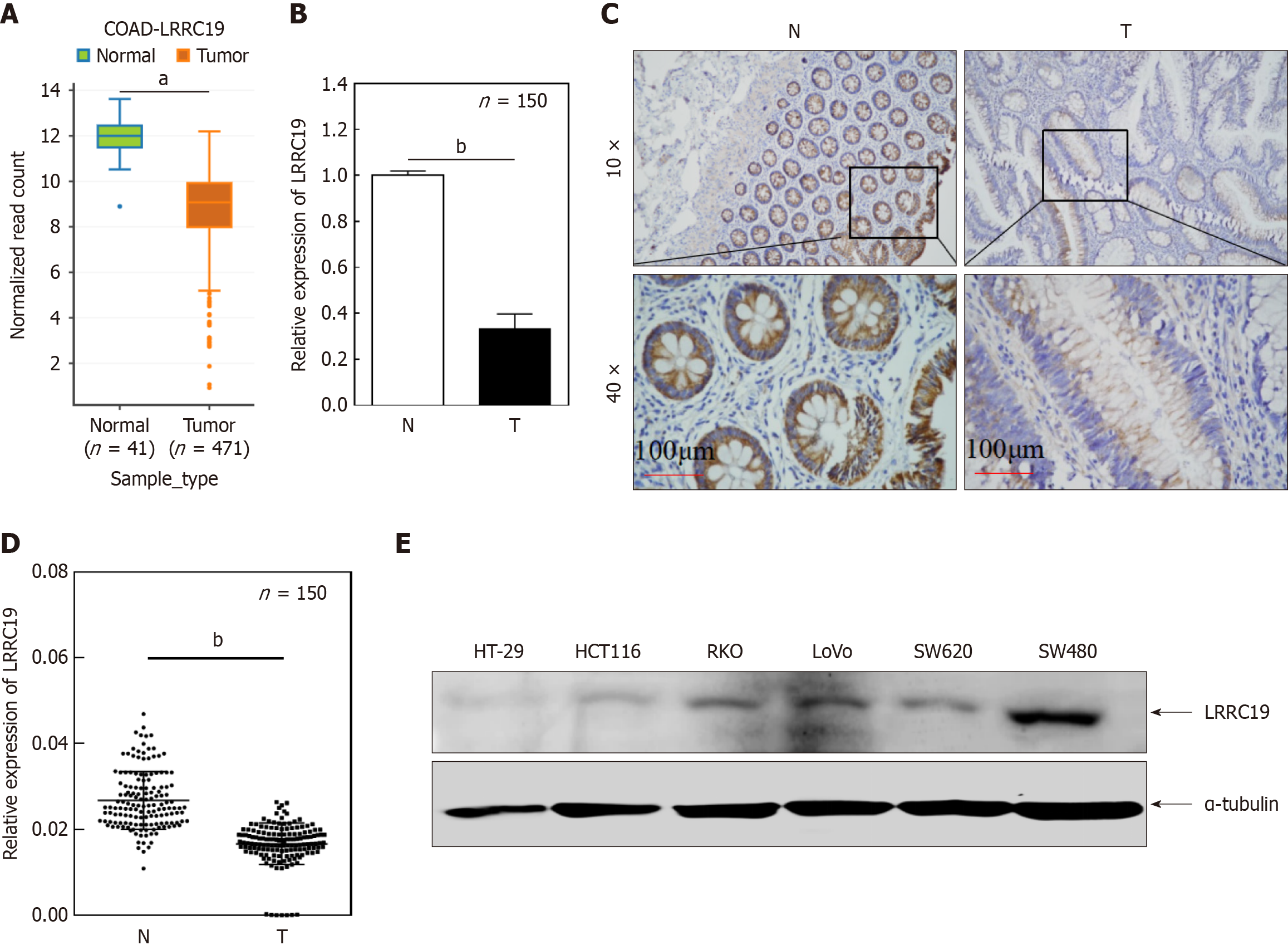
As shown in Figure 2A, the expression of LRRC19 in CRC tissues is significantly lower than in normal tissues (P < 0.01). This finding was validated in our independent clinical cohort (n = 150, P < 0.05; Figure 2B) and immunohistochemical results (P < 0.05; Figure 2C and D). Obvious brown granule deposition in the cytoplasm was observed in normal tissues, whereas the staining signal was significantly reduced in cancer tissues. Moreover, the expression of LRRC19 in metastatic CRC cell lines (HT29, HCT116, RKO, LoVo, and SW620) was significantly decreased compared to SW480 (P < 0.05; Figure 2E), with stable α-tubulin expression, suggesting that low LRRC19 expression may be associated with CRC metastasis.
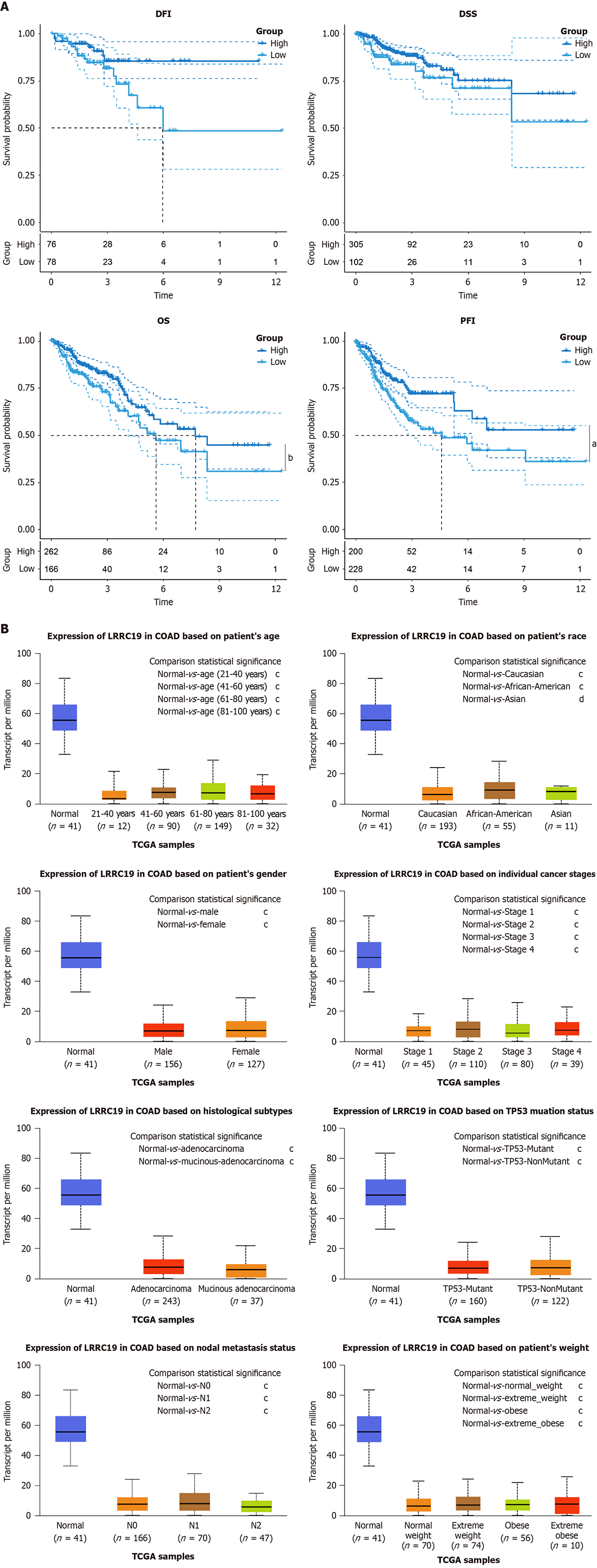
Figure 3A shows the results of survival analysis, indicating that although there were no statistically significant differences in DFI (P = 0.082) and DSS (P = 0.092) between the high LRRC19 expression group and the low expression group, the OS was significantly reduced (P < 0.05), and the probability of survival was markedly decreased (P < 0.01). Figure 3B illustrates the differential expression characteristics of LRRC19 in CRC: The LRRC19 expression level in normal tissues (n = 41) was significantly higher than that in tumor tissues across various comparisons, including sex (male n = 156: P < 1 × 10-12; female n = 127: P < 1 × 10-12), pathological type (adenocarcinoma/mucinous adenocarcinoma: P < 1 × 10-12), lymph node metastasis status (N0/N1/N2: All P < 1 × 10-12), ethnicity (Caucasian P < 1 × 10-12; African American P < 1 × 10-12; Asian P < 1 × 10-7), clinical stage (stages I-IV: All P < 1 × 10-12), TP53 mutation status (mutated group n = 160/non-mutated group n = 122: All P < 1 × 10-12), and different body mass index categories (all P < 1 × 10-12).
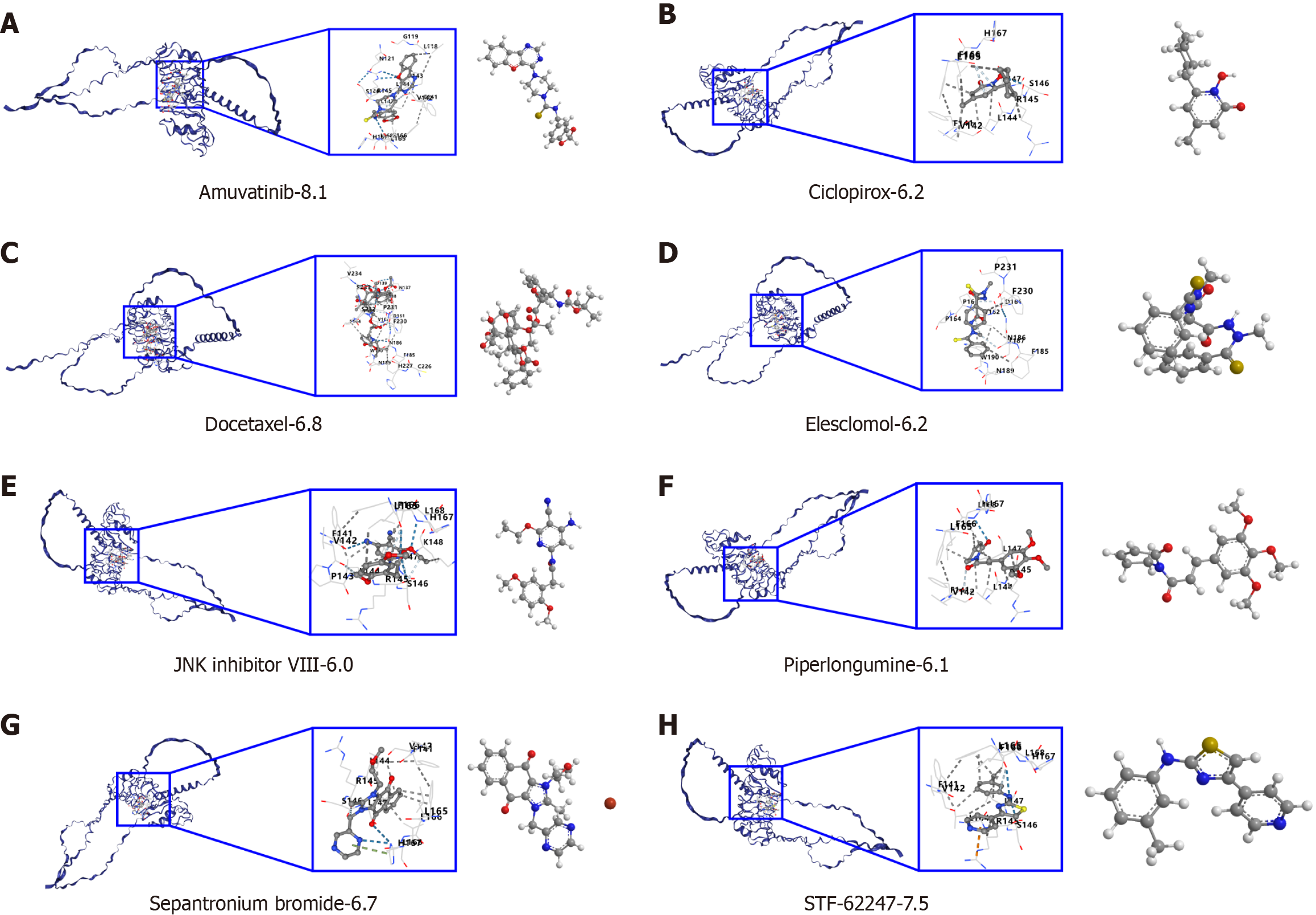
Figure 4 shows that a variety of drugs have high binding affinity to LRRC19, with amuvatinib exhibiting the lowest binding energy (-8.1 kcal/mol), suggesting it may have the strongest inhibitory effect. STF-62247 (-7.5 kcal/mol) and Docetaxel (-6.8 kcal/mol) also demonstrate good binding capabilities. The remaining drugs, such as Ciclopirox (-6.2 kcal/mol), Elesclomol (-6.2 kcal/mol), c-Jun N-terminal kinase inhibitor VIII (-6.0 kcal/mol), Piperlongumine (-6.1 kcal/mol), and Sepantronium bromide (-6.7 kcal/mol), all have binding energies below -6.0 kcal/mol, indicating they may have stable interactions with LRRC19. LRRC19 was significantly overexpressed in the chemoresistant group (P < 0.01), supporting the association between high expression of LRRC19 and chemoresistance in CRC, and suggesting its potential as a biomarker for predicting OXA response or as a target for therapeutic intervention (Supplementary Figure 3). Future studies can further validate the inhibitory effects and mechanisms of these drugs through in vitro experiments.
Figure 5A shows that the expression level of LRRC19 was significantly correlated with the degree of tumor infiltration by various immune cells in CRC. Specifically, LRRC19 expression was positively correlated with the infiltration levels of B cells (Rho = 0.159, P < 0.01), T cells/NK cells (Rho = 0.226, P < 0.001), and CD4+ T cells (Rho = 0.201, P < 0.001); whereas it showed a significant negative correlation with the infiltration levels of macrophages (Rho = -0.201, P < 0.001), mDCs (Rho = -0.16, P < 0.01), γδ T cells (Rho = -0.119, P < 0.05), neutrophils (Rho = -0.17, P < 0.01), NK cells (Rho = -0.367, P < 1 × 10-7), and CD8+ T cells (Rho = -0.21, P < 0.001). Notably, the infiltration level of Tregs showed a weak positive correlation with LRRC19 expression (Rho = 0.158, P < 0.01). These results suggest that LRRC19 may influence the tumor immune response by regulating the infiltration of specific immune cells in the TME. Figure 5B reveals that in the pan-cancer analysis, the LRRC19 gene showed significant correlation with multiple immune checkpoint genes. Particularly in CRC, the correlation between the LRRC19 gene and several immune checkpoint genes was particularly significant, such as LRRC19's significant positive correlation with CD40 LG (r = 0.338, P < 1 × 10-12), LGALS9 (r = 0.376, P < 1 × 10-12), TMIGD2 (r = 0.357, P < 1 × 10-12) genes, and significant negative correlation with CD44 (r = -0.341, P < 1 × 10-12), CTLA4 (r = -0.286, P < 1 × 10-7) genes. Figure 5C shows that in the pan-cancer analysis, LRRC19 exhibited a significant correlation with various immune-inflammatory pathways, which is particularly pronounced in CRC. Specifically, the expression level of LRRC19 was significantly positively correlated with the B cell receptor signaling pathway (cor = 0.199, P < 0.001), chemokine signaling pathway (cor = 0.107, P < 0.05), FC epsilon RI signaling pathway (cor = 0.231, P < 0.001), FC gamma R-mediated phagocytosis (cor = 0.118, P < 0.05), leukocyte transendothelial migration (cor = 0.102, P < 0.05), and T cell receptor signaling pathway (cor = 0.101, P < 0.05). These findings suggest that LRRC19 may play a significant role in the regulation of the tumor immune microenvironment. The significant positive correlation between LRRC19 and TNFRSF14, with a correlation coefficient of 0.61, indicated that LRRC19 might influence CD8 T cell exhaustion through the TNFRSF14 pathway. Additionally, LRRC19 was positively correlated with TOX, PRF1, GZMB, CTLA4, HAVCR2, LAG3, TIGIT, BTLA, ETV7, IFNG, TNF, PDCD1, HNF1A, and NR4A1, although these correlations were weaker. Conversely, LRRC19 was negatively correlated with TIGIT, HNF1A, CD274, PDCD1 LG2, and ICOSLG, suggesting an antagonistic rela
Figure 6A shows that normal and tumor data partially overlap in the "nFeature_RNA" dimension, but the median was higher in the tumor group, suggesting more significant mitochondrial characteristics in their cells; the "nCount_RNA" also showed an overall higher trend in the tumor group, which may reflect an increase in metabolic activity and proliferation rate, while "percent.mt" indicated that both groups were mainly concentrated in the lower percentage range. Figure 6B indicated a weak negative correlation (r = -0.08) between mitochondrial gene proportion (percent_mt) and total mRNA molecule count (nCount_RNA), whereas nFeature_RNA and nCount_RNA showed a strong positive correlation
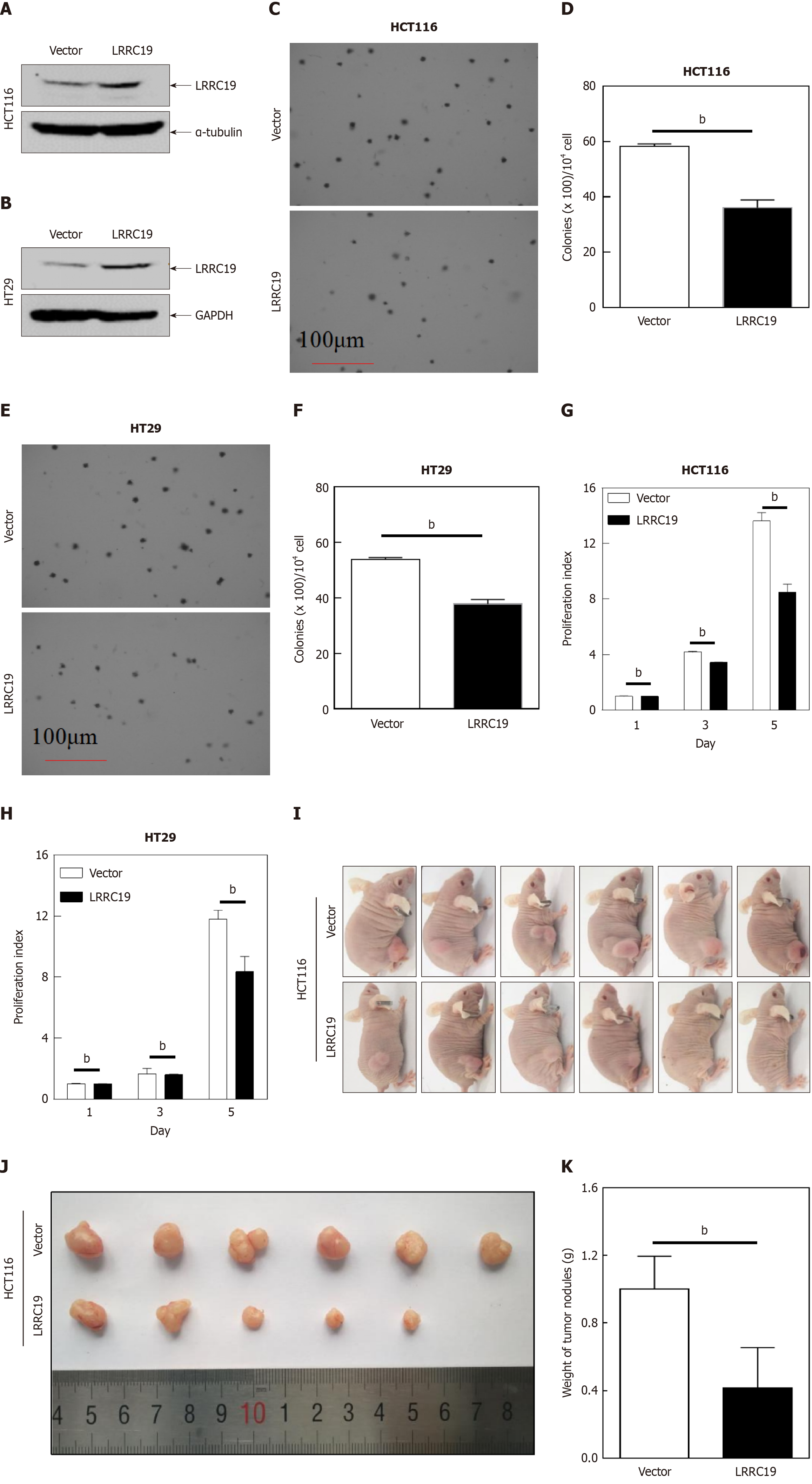
Figures 7A and B reveal that a marked enhancement of protein bands was indeed observed in the cell groups with stable overexpression of LRRC19 compared to the untreated control group or the cells transfected with the empty vector. This indicates that the target gene had been effectively expressed. Consequently, stable overexpression models of LRRC19 in two different CRC cell lines were successfully established, laying the foundation for further exploration of its role in tumor development and progression. The results showed that, compared to the vector control group, the number of colonies formed on soft agar by LRRC19-transfected HCT116 cells (Figure 7C and D) and HT29 cells (Figure 7E and F) was significantly reduced (P < 0.05), suggesting that LRRC19 effectively inhibits the non-adherent growth of these two cell types. The monolayer proliferation assay demonstrated that LRRC19 overexpression significantly suppressed cell growth, with the most pronounced effect observed on day 5. In HT-29 and HCT116 cells, the optical density values were significantly lower than the control group (P < 0.05; Figure 7G and H). The results indicate that, compared to the control group, the HCT116 cells overexpressing LRRC19 formed tumors with significantly reduced volume and weight (P < 0.05; Figure 7I-K), demonstrating that LRRC19 has a significant tumor-suppressive effect. Overall, these findings align with previous reports describing the antiproliferative role of LRRC19 in gastrointestinal malignancies, highlighting its tumor-suppressive functions mediated through intrinsic cellular mechanisms and TME regulation.
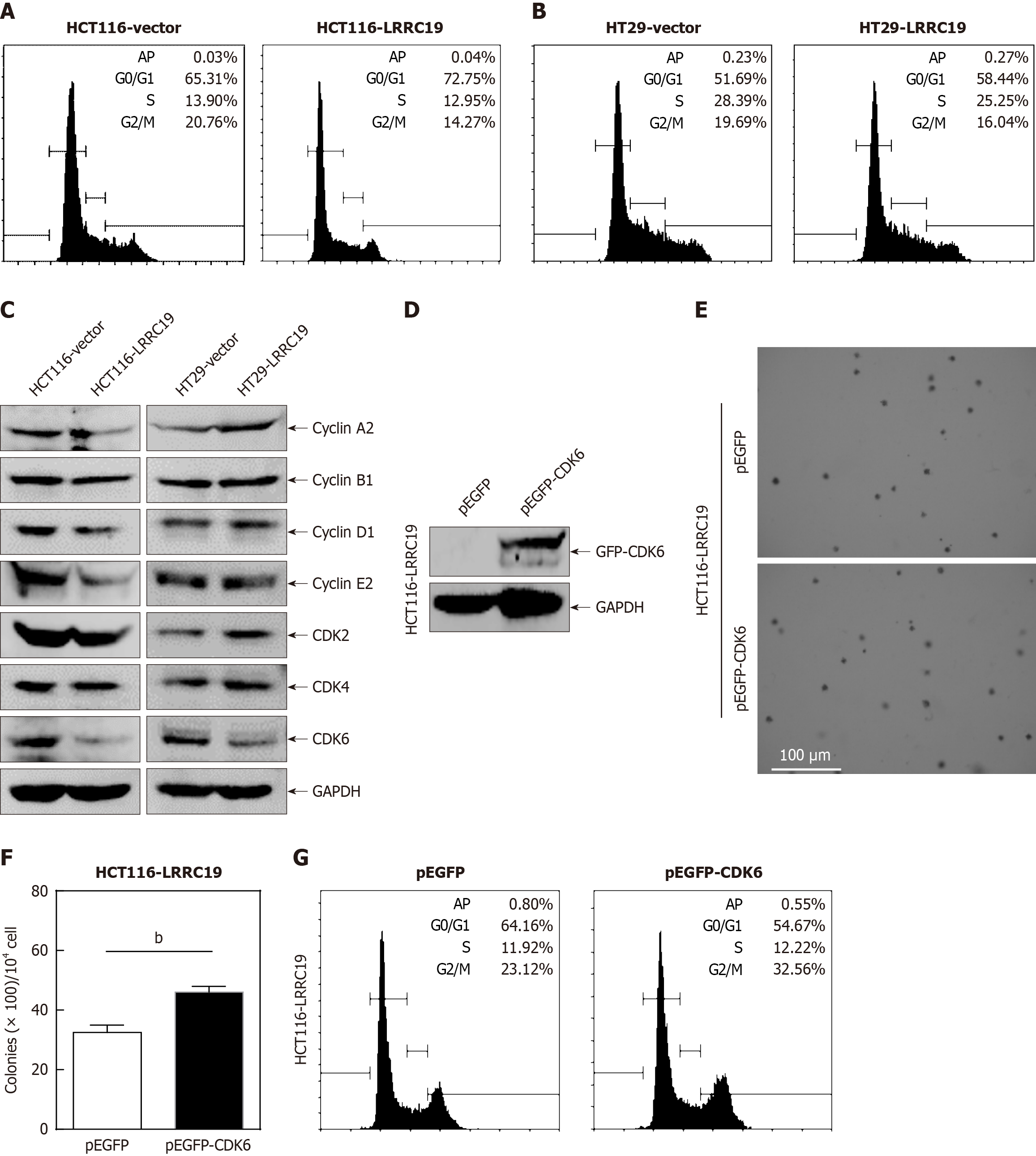
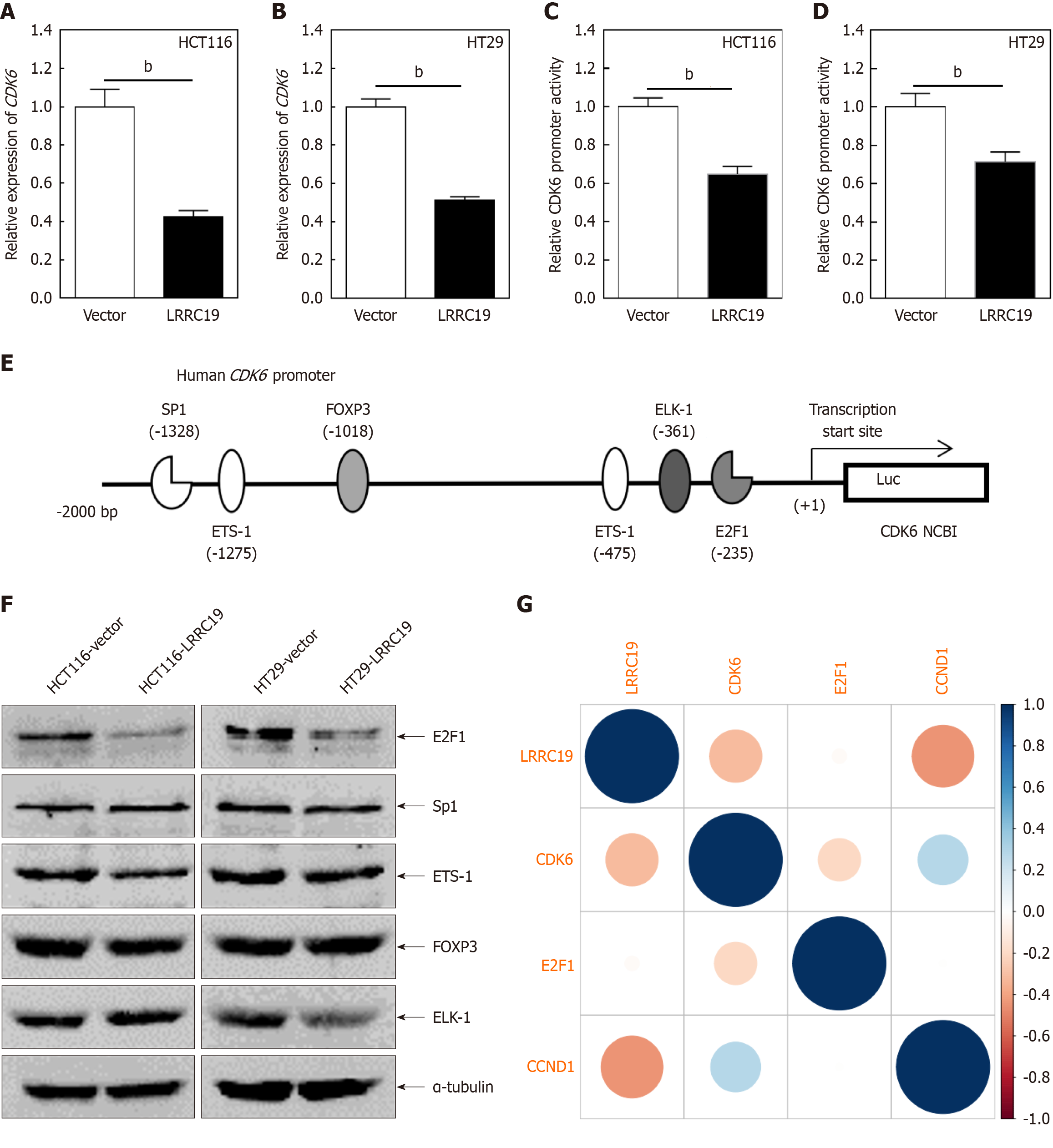
Figure 8A and B illustrates that, compared to the control, the proportion of LRRC19-overexpressing HCT116 cells in the G0/G1 phase increased from 65.31% to 72.75%, while HT29 cells increased from 51.69% to 58.44%, indicating a significant induction of cell cycle arrest in the G0/G1 phase by LRRC19 overexpression. Figure 8C demonstrates that under specific conditions, the protein level of CDK6 was significantly reduced (P < 0.05), while the expression levels of CDK2, CDK4, cyclin A2, cyclin B1, cyclin D1, and cyclin E2 remained stable. To investigate the functional involvement of CDK6 in the LRRC19-mediated suppression of CRC proliferation, we performed targeted transfection of CDK6 expression plasmids and matched control vectors into CRC cells with stable LRRC19 overexpression (Figure 8D). Figure 8E and F show that, compared to the control group overexpressing LRRC19 alone, cells simultaneously overexpressing CDK6 exhibited a significant increase in colony formation number on soft agar (P < 0.05), suggesting that CDK6 can partially restore the anchorage-independent growth capacity suppressed by LRRC19 overexpression. Additionally, flow cytometry data revealed that ectopic expression of CDK6 also promoted the release of cells from the G0/G1 phase into the S phase, thereby accelerating the progression of the entire cell cycle (P < 0.05; Figure 8G). These findings suggest that CDK6 may play a crucial role in regulating the proliferation of tumor cells.
Figure 9A and B reveal that in HCT116 cells, the relative expression of CDK6 mRNA in the LRRCl9 group was sig
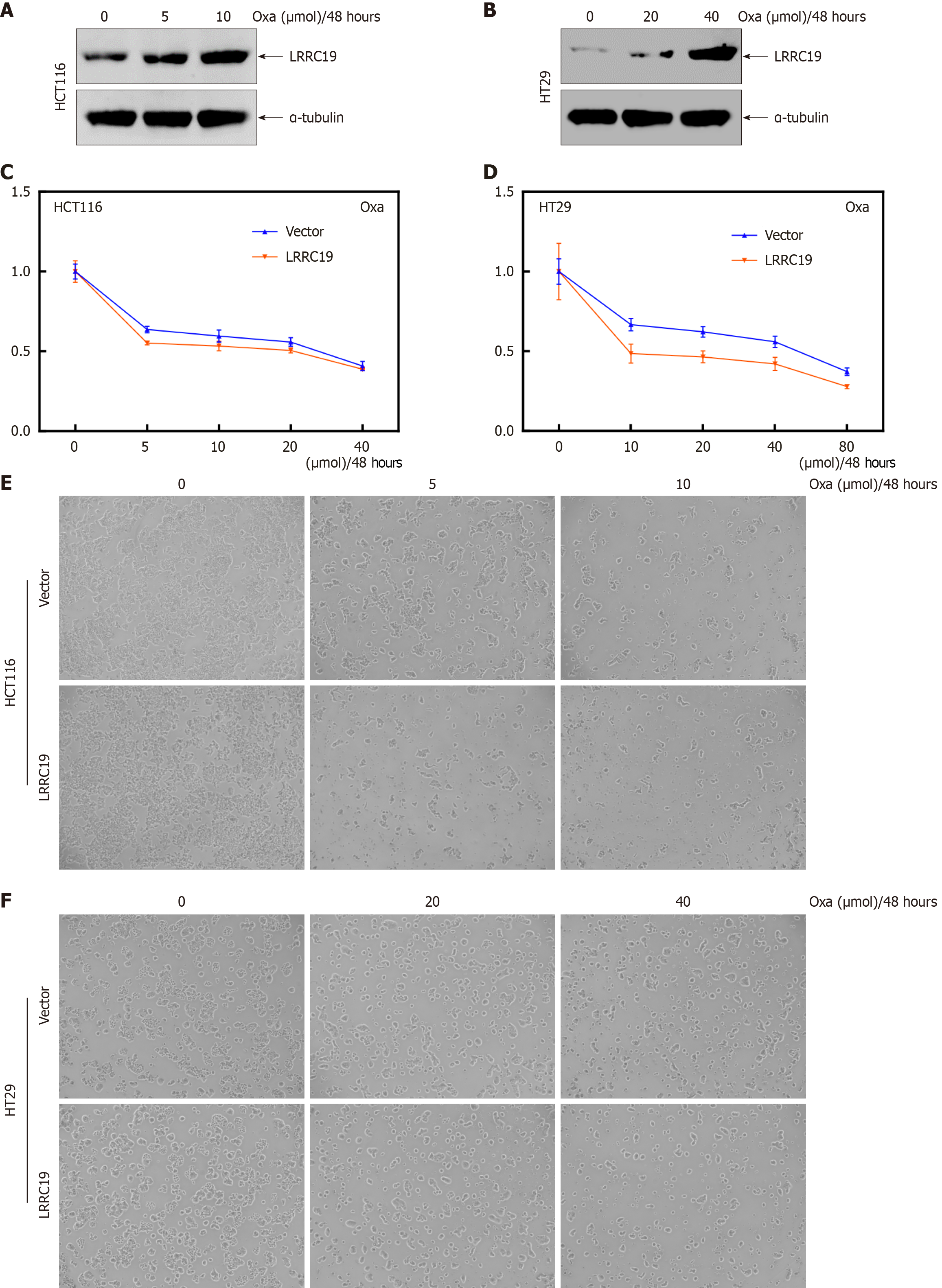
OXA is the most commonly used chemotherapeutic agent in the clinical treatment of CRC[23], and when CRC cells increase their proliferation and metastasis, they become resistant to OXA chemotherapy. Experimental investigations demonstrated that OXA treatment induced an upregulation of LRRC19 protein expression in HCT116 and HT29 cells (P < 0.05; Figure 10A and B). Functional validation revealed that LRRC19 overexpression significantly sensitized CRC cells to OXA, with IC50 values decreasing in HCT116 (P < 0.05) and HT29 (P < 0.05) after 48-hour treatment (Figure 10C-F). These findings collectively identify LRRC19 as both a predictive biomarker for OXA response and a therapeutic target to overcome chemoresistance in CRC.
Despite advancements in therapeutic strategies, CRC remains challenging to treat, particularly in metastatic disease, where the 5-year survival rate is below 15%. Identifying novel molecular targets is thus crucial for improving CRC prognosis and therapeutic efficacy. In this study, we established that LRRC19 is recurrently downregulated in CRC (Figure 2A and E). Next, we determined whether its loss correlates with aggressive clinical features. Survival analysis revealed that patients with LRRC19-low disease exhibited significantly reduced OS (P < 0.05) but comparable DFS (P = 0.092; Figure 3A). This disparity suggests that LRRC19 depletion may not drive early recurrence but exacerbates late-stage mortality through mechanisms such as immune evasion or chemoresistance (explored in Figures 4-10). LRRC19 loss promotes CRC progression via two interconnected axes: (1) Cell cycle dysregulation-LRRC19 deficiency activates CDK6/E2F1 signaling (Figures 7-9), accelerating G1/S transition and uncontrolled proliferation; and (2) Immune suppression-LRRC19 depletion recruits immunosuppressive Tregs while depleting cytotoxic CD8+ T cells (Figure 5A), fostering an immune-evasive niche. These dual pathways collectively drive poor prognosis in patients with LRRC19-low disease (Figure 3B).
While LRRC19 low expression was associated with significantly worse OS (P < 0.05), no statistical differences were observed in DFS or DFI (P > 0.05). This apparent paradox may arise from several factors. First, OS reflects cumulative mortality from both cancer progression and non-cancer causes (e.g., chemotherapy toxicity, comorbidities), whereas DFS/DFI specifically captures tumor recurrence. The lack of DFS/DFI association suggests that LRRC19 may not directly regulate early recurrence but instead influences long-term survival through mechanisms such as therapy resistance or immune exhaustion (e.g., reduced CD8+ T cell infiltration; Figure 5A). Second, our cohort’s median follow-up time (15-30 months) may be insufficient to detect DFS/DFI differences, as CRC recurrence often occurs beyond 5 years. Third, clinical heterogeneity (e.g., adjuvant therapy regimens, microsatellite instability status) could dilute DFS/DFI signals, whereas OS aggregates broader risk factors. Future studies with extended follow-up and stratified analyses (e.g., by treatment type) are needed to resolve this discrepancy.
The CDK family plays a pivotal role in cell cycle regulation, with CDK6 being a key member. CDKs regulate cyclins, thereby influencing various cell cycle processes. Abnormal activation of cell cycle-related proteins can lead to excessive cell proliferation and tumorigenesis[28]. Consequently, CDKs have become prominent targets for inhibitors in cancer therapy[29,30]. CDK4 and CDK6, in conjunction with D-type cyclins, drive the transition from the G1 to S phase through RB phosphorylation and E2F transcription factor release. Dysregulation of the CDK4/6 axis, including gene amplification, mutations, and upstream signaling pathway activation, is observed in various cancers. Unlike broad-spectrum CDK4/6 inhibitors (e.g., palbociclib, ribociclib, abemaciclib)[31,32] employed in breast cancers, LRRC19 specifically targeted CDK6 without affecting other cyclins or kinases, indicating superior target specificity. In this study, CDK6 was the only kinase downregulated in two CRC cell lines overexpressing LRRC19, while CDK4 and other cell cycle-related proteins remained unchanged. This suggests that LRRC19 specifically suppresses CDK6 expression, inhibiting the G0/G1 to S phase transition in CRC cells. This regulatory relationship positions LRRC19 as a potential novel therapeutic target for CRC.
E2F1, a critical transcription factor, plays a central role in cell cycle regulation, DNA damage repair, and apoptosis[33,34]. By interacting with cell cycle regulators like Rb protein, E2F1 modulates the expression of multiple genes, driving cell proliferation[33-37]. Aberrant activation of E2F1 often leads to excessive cell proliferation and tumor progression[38,39]. Involvement of E2F1 in nuclear transcription factor Y subunit beta-E2F1-checkpoint kinase 1 signaling has been reported to contribute to OXA resistance in CRC[40]. Many drugs exert anti-CRC effects by affecting E2F1 expression. For example, downregulation of E2F1 by HGF and downregulation of E2F1 by inhibition of HSP90 in CRC cells can enhance the efficacy of 5-FU[41]. Nano-liposome-encapsulated small interfering RNA and others can downregulate E2F1, inhibit CRC cell growth, or enhance chemotherapy effect[42]. Our study showed that overexpression of LRRC19 inhibited CDK6 transcription by downregulating E2F1, although the exact binding site between LRRC19 and E2F1 remains unknown. Our study confirmed that LRRC19 regulates two genes with important biological functions. Clinicians may consider deve
By integrating predictive results from TargetScan, miRDB, and miRanda databases, we identified several miRNAs (e.g., hsa-miR-186-5p, hsa-miR-223-3p) potentially targeting LRRC19. miRNAs are known to regulate gene expression post-transcriptionally by binding to the 3' untranslated region (3'-UTR) of target mRNAs, thereby suppressing their stability or translation efficiency[43]. We hypothesize that these miRNAs may interact with the 3’-UTR of LRRC19 to modulate its expression levels. Notably, miR-186-5p[44] and miR-223-3p[45] have been implicated in various cancers, suggesting their potential involvement in CRC pathogenesis through LRRC19 regulation. Using the spongeScan tool, we further analyzed lncRNAs that could serve as competing endogenous RNAs for these miRNAs. Several lncRNAs, including AC005614.3, DYX1C1-CCPG1, and LA16C60D12.2, were predicted to form intricate regulatory networks with the identified miRNAs. LncRNAs typically function as "molecular sponges" to sequester miRNAs, thereby alleviating their inhibitory effects on target mRNAs[46].
The significant negative correlations between LRRC19 and immune checkpoint molecules CTLA4 and CD274/PD-L1 suggest that elevated LRRC19 expression may counteract T cell dysfunction by downregulating these inhibitory signals, thereby enhancing anti-tumor immunity[47,48]. This observation aligns with LRRC19-associated enrichment of plasma cells and activated dendritic cells in the TME. Plasma cells mediate humoral immunity through antibody secretion, while activated dendritic cells serve as central orchestrators of antigen presentation, potentially synergizing to amplify adaptive immune responses. The elevated TIDE scores (indicative of immune escape potential) in patients with LRRC19-low disease further corroborate its role as an immunostimulatory molecule[49,50]. Notably, LRRC19 expression negatively correlated with immunosuppressive cell populations, including M0/M1/M2 macrophages and Tregs, implying its capacity to restrict the recruitment or polarization of these cells, thereby enhancing TME immunogenicity[51]. Intri
Chemoresistance remains a major obstacle in cancer therapy[54]. While our computational models identified LRRC19-binding drugs with potential therapeutic relevance (Figure 3), experimental validation (e.g., surface plasmon resonance, dose-response assays) is required to confirm binding kinetics and functional efficacy. Future studies will address this limitation. OXA is a cornerstone therapy in CRC, widely used in metastatic first-line. Our study found that LRRC19 overexpression significantly increased the sensitivity of CRC cells to OXA. This finding underscores LRRC19’s critical role in overcoming tumor immune evasion. Mechanistically, chemotherapy-induced upregulation of LRRC19 may reverse EMT or inhibit pro-survival signaling pathways, thereby overcoming drug resistance. These results provide a theoretical basis for optimizing chemotherapy regimens based on LRRC19 expression. These concentrations align with established in vitro models of CRC chemoresistance. Clinically, OXA is typically delivered via intravenous infusion at 85 mg/m² every 2 weeks (equivalent to approximately 1.5-3 μM plasma concentration). While our in vitro doses are higher than clinical plasma levels, this accounts for the shorter exposure time (48 hours vs sustained circulation in vivo) and compensates for reduced drug penetration in monolayer cultures. Future studies will validate these findings in patient-derived organoids and murine xenograft models with clinically relevant dosing schedules.
Although this study elucidates the multifaceted functions of LRRC19, its specific ligands and downstream signaling pathways require further investigation. For instance, does LRRC19 directly regulate anti-tumor immune responses by recruiting immune cells such as T cells? Additionally, how does the lncRNA/miRNA regulatory network influence LRRC19 expression? Future research should integrate single-cell sequencing and organoid models to explore LRRC19’s dynamic role in the TME and develop small-molecule agonists or gene therapy strategies targeting LRRC19.
This work establishes LRRC19 as a pleiotropic tumor suppressor in CRC, bridging cell cycle control, immune microenvironment remodeling, and chemotherapeutic response. Its dual mechanisms and stromal specificity position LRRC19 as both a predictive biomarker and a novel therapeutic target for precision oncology approaches. Clinical translation should prioritize combinatorial regimens integrating LRRC19 modulation with existing immunotherapies or chemotherapy protocols.
| 1. | Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5690] [Cited by in RCA: 12644] [Article Influence: 6322.0] [Reference Citation Analysis (6)] |
| 2. | Li N, Lu B, Luo C, Cai J, Lu M, Zhang Y, Chen H, Dai M. Incidence, mortality, survival, risk factor and screening of colorectal cancer: A comparison among China, Europe, and northern America. Cancer Lett. 2021;522:255-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 287] [Article Influence: 57.4] [Reference Citation Analysis (0)] |
| 3. | Waldner MJ, Neurath MF. TGFβ and the Tumor Microenvironment in Colorectal Cancer. Cells. 2023;12:1139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 29] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 4. | Wang F, Ruan DY, Xu RH. Challenges and opportunities in oncology drug development and clinical research in China. Cell. 2024;187:1578-1583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 5. | Carroll SL, Pasare C, Barton GM. Control of adaptive immunity by pattern recognition receptors. Immunity. 2024;57:632-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 72] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 6. | Kumar V, Stewart Iv JH. Pattern-Recognition Receptors and Immunometabolic Reprogramming: What We Know and What to Explore. J Innate Immun. 2024;16:295-323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 7. | Man SM, Jenkins BJ. Context-dependent functions of pattern recognition receptors in cancer. Nat Rev Cancer. 2022;22:397-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 104] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 8. | Li D, Wu M. Pattern recognition receptors in health and diseases. Signal Transduct Target Ther. 2021;6:291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 414] [Cited by in RCA: 1146] [Article Influence: 229.2] [Reference Citation Analysis (0)] |
| 9. | Ng A, Xavier RJ. Leucine-rich repeat (LRR) proteins: integrators of pattern recognition and signaling in immunity. Autophagy. 2011;7:1082-1084. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 159] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 10. | Sun J, Wang Z, Wang X. Suppression of LRRC19 promotes cutaneous wound healing in pressure ulcers in mice. Organogenesis. 2018;14:13-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 11. | Wang YJ, Liu M, Jiang HY, Yu YW. Downregulation of LRRC19 Is Associated with Poor Prognosis in Colorectal Cancer. J Oncol. 2022;2022:5848823. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 12. | Dong Y, Meng F, Wang J, Wei J, Zhang K, Qin S, Li M, Wang F, Wang B, Liu T, Zhong W, Cao H. Desulfovibrio vulgaris flagellin exacerbates colorectal cancer through activating LRRC19/TRAF6/TAK1 pathway. Gut Microbes. 2025;17:2446376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 13. | Rajeshkumar NV, Tan AC, De Oliveira E, Womack C, Wombwell H, Morgan S, Warren MV, Walker J, Green TP, Jimeno A, Messersmith WA, Hidalgo M. Antitumor effects and biomarkers of activity of AZD0530, a Src inhibitor, in pancreatic cancer. Clin Cancer Res. 2009;15:4138-4146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 70] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 14. | Xin S, Su J, Li R, Cao Q, Wang H, Wei Z, Wang C, Zhang C. Identification of a risk model for prognostic and therapeutic prediction in renal cell carcinoma based on infiltrating M0 cells. Sci Rep. 2024;14:13390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 15. | Xie R, Gu Y, Li M, Li L, Yang Y, Sun Y, Zhou B, Liu T, Wang S, Liu W, Yang R, Su X, Zhong W, Wang B, Cao H. Desulfovibrio vulgaris interacts with novel gut epithelial immune receptor LRRC19 and exacerbates colitis. Microbiome. 2024;12:4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 39] [Reference Citation Analysis (0)] |
| 16. | Huang G, Zheng Y, Zhang N, Huang G, Zhang W, Li Q, Ren X. Desulfovibrio vulgaris caused gut inflammation and aggravated DSS-induced colitis in C57BL/6 mice model. Gut Pathog. 2024;16:39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 28] [Reference Citation Analysis (0)] |
| 17. | Xiao H, Wang K, Li D, Wang K, Yu M. Evaluation of FGFR1 as a diagnostic biomarker for ovarian cancer using TCGA and GEO datasets. PeerJ. 2021;9:e10817. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 18. | Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, Gable AL, Fang T, Doncheva NT, Pyysalo S, Bork P, Jensen LJ, von Mering C. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638-D646. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1815] [Cited by in RCA: 5147] [Article Influence: 1715.7] [Reference Citation Analysis (0)] |
| 19. | Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne U, Creighton CJ, Varambally S. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18-27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 1641] [Article Influence: 410.3] [Reference Citation Analysis (0)] |
| 20. | Wu B, Xi S. Bioinformatics analysis of differentially expressed genes and pathways in the development of cervical cancer. BMC Cancer. 2021;21:733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 21. | Gene Ontology Consortium; Aleksander SA, Balhoff J, Carbon S, Cherry JM, Drabkin HJ, Ebert D, Feuermann M, Gaudet P, Harris NL, Hill DP, Lee R, Mi H, Moxon S, Mungall CJ, Muruganugan A, Mushayahama T, Sternberg PW, Thomas PD, Van Auken K, Ramsey J, Siegele DA, Chisholm RL, Fey P, Aspromonte MC, Nugnes MV, Quaglia F, Tosatto S, Giglio M, Nadendla S, Antonazzo G, Attrill H, Dos Santos G, Marygold S, Strelets V, Tabone CJ, Thurmond J, Zhou P, Ahmed SH, Asanitthong P, Luna Buitrago D, Erdol MN, Gage MC, Ali Kadhum M, Li KYC, Long M, Michalak A, Pesala A, Pritazahra A, Saverimuttu SCC, Su R, Thurlow KE, Lovering RC, Logie C, Oliferenko S, Blake J, Christie K, Corbani L, Dolan ME, Drabkin HJ, Hill DP, Ni L, Sitnikov D, Smith C, Cuzick A, Seager J, Cooper L, Elser J, Jaiswal P, Gupta P, Jaiswal P, Naithani S, Lera-Ramirez M, Rutherford K, Wood V, De Pons JL, Dwinell MR, Hayman GT, Kaldunski ML, Kwitek AE, Laulederkind SJF, Tutaj MA, Vedi M, Wang SJ, D'Eustachio P, Aimo L, Axelsen K, Bridge A, Hyka-Nouspikel N, Morgat A, Aleksander SA, Cherry JM, Engel SR, Karra K, Miyasato SR, Nash RS, Skrzypek MS, Weng S, Wong ED, Bakker E, Berardini TZ, Reiser L, Auchincloss A, Axelsen K, Argoud-Puy G, Blatter MC, Boutet E, Breuza L, Bridge A, Casals-Casas C, Coudert E, Estreicher A, Livia Famiglietti M, Feuermann M, Gos A, Gruaz-Gumowski N, Hulo C, Hyka-Nouspikel N, Jungo F, Le Mercier P, Lieberherr D, Masson P, Morgat A, Pedruzzi I, Pourcel L, Poux S, Rivoire C, Sundaram S, Bateman A, Bowler-Barnett E, Bye-A-Jee H, Denny P, Ignatchenko A, Ishtiaq R, Lock A, Lussi Y, Magrane M, Martin MJ, Orchard S, Raposo P, Speretta E, Tyagi N, Warner K, Zaru R, Diehl AD, Lee R, Chan J, Diamantakis S, Raciti D, Zarowiecki M, Fisher M, James-Zorn C, Ponferrada V, Zorn A, Ramachandran S, Ruzicka L, Westerfield M. The Gene Ontology knowledgebase in 2023. Genetics. 2023;224:iyad031. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 998] [Cited by in RCA: 1515] [Article Influence: 505.0] [Reference Citation Analysis (0)] |
| 22. | Jamieson A, McAlpine JN. Molecular Profiling of Endometrial Cancer From TCGA to Clinical Practice. J Natl Compr Canc Netw. 2023;21:210-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 62] [Reference Citation Analysis (0)] |
| 23. | Kusch N, Schuppert A. Two-step multi-omics modelling of drug sensitivity in cancer cell lines to identify driving mechanisms. PLoS One. 2020;15:e0238961. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 24. | Karuppasamy MP, Venkateswaran S, Subbiah P. PDB-2-PBv3.0: An updated protein block database. J Bioinform Comput Biol. 2020;18:2050009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 25. | Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, Zaslavsky L, Zhang J, Bolton EE. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021;49:D1388-D1395. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2091] [Cited by in RCA: 2365] [Article Influence: 473.0] [Reference Citation Analysis (0)] |
| 26. | Liu Y, Yang X, Gan J, Chen S, Xiao ZX, Cao Y. CB-Dock2: improved protein-ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res. 2022;50:W159-W164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 939] [Article Influence: 234.8] [Reference Citation Analysis (0)] |
| 27. | Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509-W514. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2451] [Cited by in RCA: 3751] [Article Influence: 625.2] [Reference Citation Analysis (0)] |
| 28. | Nebenfuehr S, Kollmann K, Sexl V. The role of CDK6 in cancer. Int J Cancer. 2020;147:2988-2995. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 140] [Article Influence: 23.3] [Reference Citation Analysis (8)] |
| 29. | Bury M, Le Calvé B, Ferbeyre G, Blank V, Lessard F. New Insights into CDK Regulators: Novel Opportunities for Cancer Therapy. Trends Cell Biol. 2021;31:331-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 96] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 30. | Wu X, Yang X, Xiong Y, Li R, Ito T, Ahmed TA, Karoulia Z, Adamopoulos C, Wang H, Wang L, Xie L, Liu J, Ueberheide B, Aaronson SA, Chen X, Buchanan SG, Sellers WR, Jin J, Poulikakos PI. Distinct CDK6 complexes determine tumor cell response to CDK4/6 inhibitors and degraders. Nat Cancer. 2021;2:429-443. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 31. | Goel S, Bergholz JS, Zhao JJ. Targeting CDK4 and CDK6 in cancer. Nat Rev Cancer. 2022;22:356-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 347] [Article Influence: 86.8] [Reference Citation Analysis (0)] |
| 32. | Fassl A, Geng Y, Sicinski P. CDK4 and CDK6 kinases: From basic science to cancer therapy. Science. 2022;375:eabc1495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 328] [Article Influence: 82.0] [Reference Citation Analysis (4)] |
| 33. | Li J, Bi W, Lu F, Pan B, Xiong M, Nasifu L, Nie Z, He B. Prognostic role of E2F1 gene expression in human cancer: a meta-analysis. BMC Cancer. 2023;23:509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 34. | Fouad S, Hauton D, D'Angiolella V. E2F1: Cause and Consequence of DNA Replication Stress. Front Mol Biosci. 2020;7:599332. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 35. | Mandigo AC, Yuan W, Xu K, Gallagher P, Pang A, Guan YF, Shafi AA, Thangavel C, Sheehan B, Bogdan D, Paschalis A, McCann JJ, Laufer TS, Gordon N, Vasilevskaya IA, Dylgjeri E, Chand SN, Schiewer MJ, Domingo-Domenech J, Den RB, Holst J, McCue PA, de Bono JS, McNair C, Knudsen KE. RB/E2F1 as a Master Regulator of Cancer Cell Metabolism in Advanced Disease. Cancer Discov. 2021;11:2334-2353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 36. | Yu L, Qiu W, Gao Y, Sun M, Chen L, Cui Z, Zhu D, Guo P, Tang H, Luo H. JNK1 activated pRb/E2F1 and inhibited p53/p21 signaling pathway is involved in hydroquinone-induced pathway malignant transformation of TK6 cells by accelerating the cell cycle progression. Environ Toxicol. 2023;38:2344-2351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 37. | Barczak W, Carr SM, Liu G, Munro S, Nicastri A, Lee LN, Hutchings C, Ternette N, Klenerman P, Kanapin A, Samsonova A, La Thangue NB. Long non-coding RNA-derived peptides are immunogenic and drive a potent anti-tumour response. Nat Commun. 2023;14:1078. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 85] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 38. | Zheng X, Huang M, Xing L, Yang R, Wang X, Jiang R, Zhang L, Chen J. The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Mol Cancer. 2020;19:73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 321] [Article Influence: 53.5] [Reference Citation Analysis (0)] |
| 39. | Jing Z, Liu Q, He X, Jia Z, Xu Z, Yang B, Liu P. NCAPD3 enhances Warburg effect through c-myc and E2F1 and promotes the occurrence and progression of colorectal cancer. J Exp Clin Cancer Res. 2022;41:198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 109] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 40. | Gao H, Zhou F, Li R, Yuan J, Ye L. E2F1 inhibits cellular senescence and promotes oxaliplatin resistance in colorectal cancer. Ann Transl Med. 2023;11:185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 41. | Shah ZA, Nouroz F, Ejaz S, Tayyeb A. An Insight into the Role of E2F1 in Breast Cancer Progression, Drug Resistance, and Metastasis. Curr Mol Med. 2023;23:365-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 42. | Jesenko T, Kranjc Brezar S, Pisljar Z, Bozic T, Markelc B, Cazzato M, Grassi G, Cemazar M. Effective targeting of E2F1 transcription factor via siRNA gene electrotransfer in HT-29 colorectal carcinoma xenografts. Bioelectrochemistry. 2025;165:108994. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 43. | Ali Syeda Z, Langden SSS, Munkhzul C, Lee M, Song SJ. Regulatory Mechanism of MicroRNA Expression in Cancer. Int J Mol Sci. 2020;21:1723. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 502] [Cited by in RCA: 725] [Article Influence: 120.8] [Reference Citation Analysis (0)] |
| 44. | Hsu XR, Wu JE, Wu YY, Hsiao SY, Liang JL, Wu YJ, Tung CH, Huang MF, Lin MS, Yang PC, Chen YL, Hong TM. Exosomal long noncoding RNA MLETA1 promotes tumor progression and metastasis by regulating the miR-186-5p/EGFR and miR-497-5p/IGF1R axes in non-small cell lung cancer. J Exp Clin Cancer Res. 2023;42:283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 46] [Reference Citation Analysis (0)] |
| 45. | Bao H, Peng Z, Cheng X, Jian C, Li X, Shi Y, Zhu W, Hu Y, Jiang M, Song J, Fang F, Chen J, Shu X. GABA induced by sleep deprivation promotes the proliferation and migration of colon tumors through miR-223-3p endogenous pathway and exosome pathway. J Exp Clin Cancer Res. 2023;42:344. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 42] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 46. | Fontemaggi G, Turco C, Esposito G, Di Agostino S. New Molecular Mechanisms and Clinical Impact of circRNAs in Human Cancer. Cancers (Basel). 2021;13:3154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 47. | Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16:275-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1659] [Cited by in RCA: 2177] [Article Influence: 217.7] [Reference Citation Analysis (0)] |
| 48. | Zhang Y, Zheng J. Functions of Immune Checkpoint Molecules Beyond Immune Evasion. Adv Exp Med Biol. 2020;1248:201-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 177] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 49. | Oliveira G, Wu CJ. Dynamics and specificities of T cells in cancer immunotherapy. Nat Rev Cancer. 2023;23:295-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 357] [Article Influence: 119.0] [Reference Citation Analysis (0)] |
| 50. | Bruni D, Angell HK, Galon J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat Rev Cancer. 2020;20:662-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 529] [Cited by in RCA: 1157] [Article Influence: 192.8] [Reference Citation Analysis (0)] |
| 51. | Zhang Y, Tang M, Guo Q, Xu H, Yang Z, Li D. The value of erlotinib related target molecules in kidney renal cell carcinoma via bioinformatics analysis. Gene. 2022;816:146173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 52. | Zheng L, Qin S, Si W, Wang A, Xing B, Gao R, Ren X, Wang L, Wu X, Zhang J, Wu N, Zhang N, Zheng H, Ouyang H, Chen K, Bu Z, Hu X, Ji J, Zhang Z. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science. 2021;374:abe6474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 929] [Article Influence: 185.8] [Reference Citation Analysis (0)] |
| 53. | Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, Fearon D, Greten FR, Hingorani SR, Hunter T, Hynes RO, Jain RK, Janowitz T, Jorgensen C, Kimmelman AC, Kolonin MG, Maki RG, Powers RS, Puré E, Ramirez DC, Scherz-Shouval R, Sherman MH, Stewart S, Tlsty TD, Tuveson DA, Watt FM, Weaver V, Weeraratna AT, Werb Z. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer. 2020;20:174-186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1940] [Cited by in RCA: 2744] [Article Influence: 457.3] [Reference Citation Analysis (0)] |
| 54. | Boumahdi S, de Sauvage FJ. The great escape: tumour cell plasticity in resistance to targeted therapy. Nat Rev Drug Discov. 2020;19:39-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 528] [Article Influence: 88.0] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/