Published online Dec 28, 2024. doi: 10.3748/wjg.v30.i48.5174
Revised: September 26, 2024
Accepted: November 8, 2024
Published online: December 28, 2024
Processing time: 161 Days and 21.8 Hours
Hepatocellular carcinoma (HCC) is a prevalent and aggressive tumor. Sorafenib is the first-line treatment for patients with advanced HCC, but resistance to sorafenib has become a significant challenge in this therapy. Cancer stem cells play a crucial role in sorafenib resistance in HCC. Our previous study revealed that the long non-coding RNA (lncRNA) KIF9-AS1 is an oncogenic gene in HCC. However, the role of KIF9-AS1 in drug resistance and cancer stemness in HCC remains unclear. Herein, we aimed to investigate the function and mechanism of the lncRNA KIF9-AS1 in cancer stemness and drug resistance in HCC.
To describe the role of the lncRNA KIF9-AS1 in cancer stemness and drug resistance in HCC and elucidate the underlying mechanism.
Tumor tissue and adjacent non-cancerous tissue samples were collected from HCC patients. Sphere formation was quantified via a tumor sphere assay. Cell viability, proliferation, and apoptosis were evaluated via Cell Counting Kit-8, flow cytometry, and colony formation assays, respectively. The interactions between the lncRNA KIF9-AS1 and its downstream targets were confirmed via RNA immunoprecipitation and coimmunoprecipitation. The tumorigenic role of KIF9-AS1 was validated in a mouse model.
Compared with that in normal controls, the expression of the lncRNA KIF9-AS1 was upregulated in HCC tissues. Knockdown of KIF9-AS1 inhibited stemness and attenuated sorafenib resistance in HCC cells. Mechanistically, N6-methyladenosine modification mediated by methyltransferase-like 3/insulin-like growth factor 2 mRNA-binding protein 1 stabilized and increased the expression of KIF9-AS1. Additionally, KIF9-AS1 increased the stability and expression of short stature homeobox 2 by promoting ubiquitin-specific peptidase 1-induced deubiquitination. Furthermore, depletion of KIF9-AS1 alleviated sorafenib resistance in a xenograft mouse model of HCC.
The N6-methyladenosine-modified lncRNA KIF9-AS1 promoted stemness and sorafenib resistance in HCC by upregulating short stature homeobox 2 expression.
Core Tip: Hepatocellular carcinoma (HCC) is a highly aggressive tumor with a poor prognosis. Our previous investigation revealed that knockdown of the long non-coding RNA KIF9-AS1 suppressed HCC progression. However, the role of KIF9-AS1 in cancer stemness and drug resistance in HCC remains unclear. This study revealed that N6-methyladenosine modification of KIF9-AS1 promotes stemness and sorafenib resistance in HCC by upregulating short stature homeobox 2 expression. This finding provides new insights into the role of KIF9-AS1 in HCC pathogenesis and highlights its potential as a biomarker and therapeutic target.
- Citation: Yu Y, Lu XH, Mu JS, Meng JY, Sun JS, Chen HX, Yan Y, Meng K. N6-methyladenosine-modified long non-coding RNA KIF9-AS1 promotes stemness and sorafenib resistance in hepatocellular carcinoma by upregulating SHOX2 expression. World J Gastroenterol 2024; 30(48): 5174-5190
- URL: https://www.wjgnet.com/1007-9327/full/v30/i48/5174.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i48.5174
Hepatocellular carcinoma (HCC), the most common type of primary liver cancer, is a highly aggressive tumor associated with a poor prognosis[1]. According to recent data, approximately 841000 cases of HCC are newly diagnosed every year globally, and the mortality rate is high[2]. Although sorafenib is the first-line treatment for advanced-stage HCC patients, resistance to sorafenib remains a major challenge in HCC therapy[3]. Cancer stem cells serve as key regulators of sorafenib resistance and tumor growth and recurrence in HCC[4]. Hence, it is crucial to explore the mechanisms of cancer cell stemness and sorafenib resistance in HCC to identify potential therapeutic targets.
Long non-coding RNAs (lncRNAs) have emerged as vital regulators of various cellular processes, and dysregulation of these lncRNAs contributes to the development of drug resistance in tumors. For example, lncRNA metastasis-associated lung adenocarcinoma transcript 1 is upregulated in HCC, where it mediates doxorubicin resistance through the microRNA-3129-5p (miR-3129-5p)/neuro-oncological ventral antigen 1 axis[5]. Moreover, Zhang et al[6] demonstrated that increased expression of the lncRNA small nucleolar RNA host gene 3 promoted invasion, epithelial-mesenchymal transition, and sorafenib resistance in HCC cells. Our previous study revealed that deletion of the lncRNA KIF9-AS1 suppressed cell proliferation and migration while promoting cell death in HCC[7]. A study by Jin et al[8] demonstrated that KIF9-AS1 promoted sorafenib resistance in renal cell carcinoma cells. However, research on the role of KIF9-AS1 in drug resistance in HCC is limited. N6-methyladenosine (m6A) modification reportedly influences RNA maturation, stability, and translational regulation[9]. A previous study revealed that methyltransferase-like 3 (METTL3)-mediated m6A methylation led to a decrease in the lncRNA maternally expressed gene 3 levels, resulting in increases in the viability, migratory ability, and invasive ability of HCC cells[10]. Furthermore, METTL3-dependent m6A methylation increased the expression of the lncRNA NIFK antisense RNA 1 (NIFK-AS1) in HCC, whereas depletion of NIFK-AS1 sensitized HCC cells to sorafenib and inhibited HCC progression[11]. Another study revealed that insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) increased the expression of the lncRNA MIR4435-2HG in HCC cells via m6A methylation, which increased the stem cell properties and proliferation of HCC cells[12]. Sequence-based RNA adenosine methylation site predictor is a useful tool that is primarily used to predict m6A modification sites on RNA sequences. Notably, the presence of m6A modification sites in KIF9-AS1 was predicted using sequence-based RNA adenosine methylation site predictor. Additionally, RM2Target predictions indicate that METTL3 and IGF2BP1 are methyltransferase recognition proteins involved in the m6A modification of KIF9-AS1. Recent evidence has shown that inhibition of METTL3-mediated m6A methylation reduces resistance to lenvatinib in HCC cells[13]. Shi et al[14] reported that increased methylation of IGF2BP1 promoted HEG1 stabilization, leading to the exacerbation of oxaliplatin resistance in HCC. These studies highlight the roles of METTL3 and IGF2BP1 as key players in drug resistance in HCC.
Several studies have focused on the downstream regulatory mechanisms of KIF9-AS1 in tumors. Numerous studies have shown that short stature homeobox 2 (SHOX2) functions as an oncogene associated with tumor progression and metastasis in bladder cancer[15], renal cancer[16] and glioma[17]. Emerging evidence shows that SHOX2 is upregulated in HCC patients and is associated with tumor recurrence[18]. Additionally, SHOX2 has been reported to increase drug resistance in lung cancer[19]. However, the functional role of SHOX2 in cancer stemness has rarely been described in previous studies. Ubiquitin-specific peptidase 1 (USP1) reportedly promotes tumorigenesis in various cancers, including breast cancer[20] and myeloma[21], by regulating protein deubiquitination. Consistently, high USP1 expression predicts poor overall survival in HCC patients[22]. Moreover, suppression of USP1 increased the sensitivity of cancer cells to platinum[23]. USP1 inhibits the K11-linked polyubiquitination of WW domain-containing transcription regulator 1, thereby increasing the stability of WW domain-containing transcription regulator 1 in HCC[24]. SHOX2 has been predicted to be a substrate for USP1 ubiquitination via UbiBrowser, and the RNA-Protein Interaction Prediction tool predicted a high score for the binding of KIF9-AS1 to USP1 (random forest classifier: 0.75; support vector machine classifier: 0.93). However, whether KIF9-AS1 regulates the expression of SHOX2 via USP1 has not been determined.
Considering all the available data, we hypothesized that METTL3 stabilizes the lncRNA KIF9-AS1 through an m6A-IGF2BP1-dependent mechanism, resulting in the upregulation of KIF9-AS1 expression. Additionally, we hypothesized that KIF9-AS1 enhances cancer stemness and sorafenib resistance in HCC by promoting USP1-mediated deubiquitination of SHOX2. These hypotheses suggest that targeting this pathway might be a promising therapeutic strategy for treating HCC.
In this study, cancer tissues and adjacent non-cancerous tissues were obtained from 20 HCC patients for analysis. The inclusion criteria were as follows: (1) A primary diagnosis of HCC; and (2) No prior chemotherapy or radiotherapy. Patients with other types of malignant tumors were excluded. The Medical Ethics Committee of the Fifth Medical Center of the Chinese People’s Liberation Army General Hospital approved this study (license KY-2024-5-75-1), and all participants provided written informed consent.
The HCC cell lines Huh-7 (American Type Culture Collection, United States) and Huh-7/R (Bio-129595, Biobw, China) were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum and 100 μg/mL penicillin-streptomycin (Thermo Fisher Scientific, United States). The cells were then cultured at 37 °C with 5% CO2 for 24 hours. In subsequent experiments, resistant cells (Huh-7/R) were generated via constant treatment with 10 μM sorafenib.
GenePharma (Shanghai, China) synthesized a USP1-overexpressing vector (pcDNA3.1-USP1); small hairpin RNAs (shRNAs) targeting KIF9-AS1 (sh-KIF9-AS1), IGF2BP1 (sh-IGF2BP1), and METTL3 (sh-METTL3); and negative control (NC) constructs. In brief, these constructs or their corresponding NC constructs were transfected into cells with Lipo6000 reagent (Beyotime Biotechnology, China) for 48 hours.
To investigate the effect of IGF2BP1 knockdown on mRNA stability, Huh-7 and Huh-7/R cells were treated with
Cells (1.0 × 104) were seeded into 96-well plates and exposed to sorafenib at different concentrations (0, 3, 6, 9, 12 and 15 μM). After treatment for 48 hours, 10 μL of Cell Counting Kit-8 solution was added to each well, and the plates were then maintained at 37 °C for an additional hour. The optical density of the solutions at 450 nm was measured with a scanning microplate reader (Bio-Rad, United States), and the IC50 values of the cells were calculated.
After digestion and centrifugation, Huh-7 cells were washed twice with phosphate-buffered saline and plated into 24-well plates (500 cells/well) supplemented with antibiotics. The plates were then incubated at 37 °C and 5% CO2. After 10 days, the number of spheres was counted under a microscope.
In brief, a total of 5 × 103 Huh-7 cells and Huh-7/R cells were plated in 6-well plates and incubated for 14 days. After fixation with glutaraldehyde, the cells were stained with 1% crystal violet solution for 15 minutes. The colonies were photographed and counted using ImageJ (NIH, United States).
To assess cell apoptosis, the cells were digested with trypsin (without EDTA). The cells were rinsed with phosphate-buffered saline and resuspended in Annexin V binding buffer. The cells were then exposed to a mixture of 5 μL of Annexin V-FITC and 5 μL of propidium iodide in the dark at room temperature for 15 minutes. Finally, cell apoptosis was measured using a FSCAN flow cytometer (BD Biosciences, United States).
Polyadenylated mRNA was purified from total RNA using the GenElute™ mRNA Miniprep Kit (Sigma-Aldrich, Germany). The m6A level was assessed with the EpiQuik m6A RNA Methylation Quantification Kit (EpiQuik, United States). For this assay, 200 ng of extracted RNA was added to each well of a 96-well microplate and incubated with the prepared antibody solution. The optical density of each well at 450 nm was measured with a microplate reader (Bio-Tek, United States).
Briefly, 10 μg of mRNA was fragmented into oligonucleotides using RNA fragmentation reagents (Invitrogen) in a thermomixer. Magna ChIP Protein A/G Magnetic Beads were treated with 5 μg of m6A-specific antibody at 4 °C for 2 hours in the presence of immunoprecipitation buffer. The mixture was subsequently incubated with methylated RNA immunoprecipitation (RIP) reaction reagents at 4 °C for 1 hour. The m6A-modified RNA was then eluted and purified for further analysis via qRT-PCR.
Cells were subjected to an RIP assay using the Magna RIP Kit (Millipore, United States). Magnetic beads coated with 5 mg of normal antibodies against IGF2BP1 (ab313422, Abcam) or USP1 (LSC288484, LsBio) were incubated with the cell lysates at 4 °C overnight. Immunoglobulin G (IgG) was used as a NC. The beads containing the immunoprecipitated RNA-protein complex were treated with proteinase K to eliminate proteins. The RNA coprecipitated with the complex was detected via qRT-PCR, and the results were normalized to the input.
Cells were suspended in immunoprecipitation lysis buffer and incubated with specific antibodies against SHOX2 (#MBS6004620), ubiquitin (#MBS2118113) and USP1 (LSC288484, LsBio) at 4 °C overnight. Next, the lysate was mixed with magnetic beads conjugated with protein A/G. Finally, the antibody-protein complexes were collected for immunoblotting.
Twelve male BALB/c nude mice (6-8 weeks, 25-30 g) were purchased from SJA Laboratory Animal Company (China) and housed in a light-controlled cage at 24 °C. The mice were allocated into two groups, the experimental group and the control group. The experimental group received subcutaneous implants of Huh-7/R cells transfected with stable KIF9-AS1-knockdown shRNA (5 × 106 cells/mouse), whereas the control group was injected with normal saline. After tumor implantation, the tumor volume was measured every 7 days. Sorafenib was administered orally (p.o.) twice a week beginning on day 7 at a dose of 30 mg/kg body weight. After 4 weeks, the tumor tissues were dissected, and the weights were recorded. This study was approved by the Institutional Animal Care Committee of the Fifth Medical Center of the Chinese People’s Liberation Army General Hospital (license IACUC-2019-0025).
In brief, tissue samples were fixed in 10% formalin, embedded in paraffin and sectioned. The sections were subsequently treated with 3% H2O2 and incubated overnight at 4 °C with an anti-Ki67 antibody (ab279653; Abcam), followed by incubation with rabbit anti-mouse IgG H&L (HRP) (ab6728) at 37 °C for 30 minutes. Finally, the slides were stained with diaminobenzidine and examined with a Nikon Eclipse Ti2 confocal microscope (Nikon, Japan).
RNA extraction was performed with TRIzol reagent (Invitrogen, United States). The reverse transcription of mRNA into cDNA was conducted with the PrimeScript RT Master Mix (Takara Bio, Japan). The quantification of mRNA expression was performed on an ABI 7500 System (Applied Biosystems, United States) with TB Green® Premix Ex Taq™ II (Takara, Japan). The sequences of the PCR amplification primers used are provided in Table 1. The 2-ΔΔCt method was used to calculate the fold change in the expression of the target genes, with GAPDH serving as the internal control.
| Genes | Sequence (5’-3’) |
| KIF9-AS1 F | TCCCTGCTGAAGTGACAGTG |
| KIF9-AS1 R | ACCCCAACCCTCTCTGTTCT |
| SHOX2 F | GCCTTCATGCGAGAGGAACT |
| SHOX2 R | CGCCTGAACCTGCTGAAATG |
| PD-L1 F | ACCACCACCAATTCCAAGAG |
| PD-L1 R | GATGGCTCCCAGAATTACCA |
| GAPDH F | CCAGGTGGTCTCCTCTGA |
| GAPDH R | GCTGTAGCCAAATCGTTGT |
Protein extraction was performed, and protein expression was evaluated using a BCA Protein Assay Kit (Beyotime, China). The samples were separated by 8%-10% polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes (Bio-Rad, United States). After blocking with 5% defatted dry milk, the membranes were incubated overnight at 4 °C with primary antibodies against CD44 (ab243894, 1/1000), CD133 (ab284389, 1/1000), epithelial cell adhesion molecule (EpCAM, ab282457, 1/1000), SHOX2 (ab55740, 1 μg/mL), USP1 (ab227551, 1/500), METTL3 (ab240595, 1/2000), IGF2BP1 (ab313422, 1/1000) and GAPDH (ab9485, 1/2500, Abcam). The samples were then incubated with secondary antibodies (ab96879 or ab6940, Abcam) at 37 °C for 1 hour. The bands were visualized and analyzed with a SuperSignal West Pico chemiluminescence system (Pierce, Inc., United States) and ImageJ software (NIH).
All the data are expressed as the means ± SDs; statistical analysis and figure generation were performed with SPSS 20.0 (IBM, United States). Statistical comparisons were performed via Student’s t test. Kaplan-Meier curves were generated and log-rank tests were performed to analyze the prognostic significance of KIF9-AS1 for HCC patients. A P value less than 0.05 was considered statistically significant.
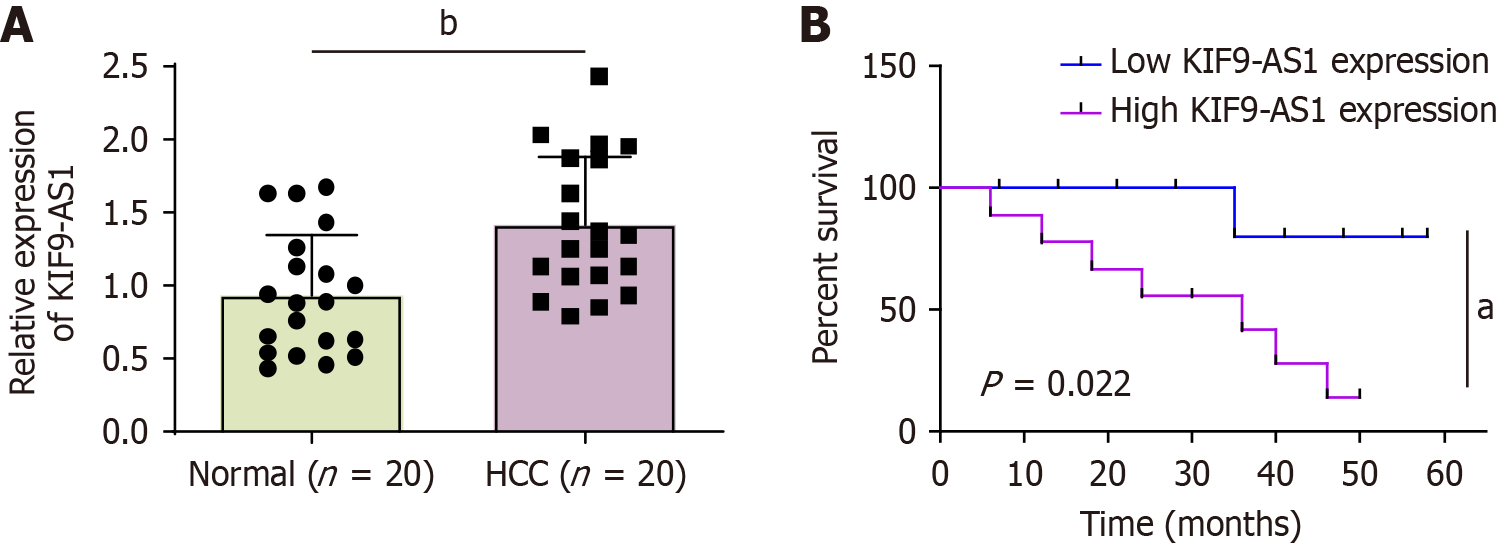
The results of this study confirmed that the expression of the lncRNA KIF9-AS1 was elevated in HCC patients (Figure 1A). Additionally, the upregulation of KIF9-AS1 demonstrated a notable correlation with poor overall survival in HCC patients (Figure 1B). The relationships between the clinical characteristics of HCC patients and KIF9-AS1 expression are shown in Table 2. Notably, high KIF9-AS1 expression in HCC patients was strongly associated with advanced tumor-node-metastasis stage and lymph node metastasis, whereas no significant relationships were found with sex, age, or tumor size. Hence, these data suggested that KIF9-AS1 promoted HCC progression.
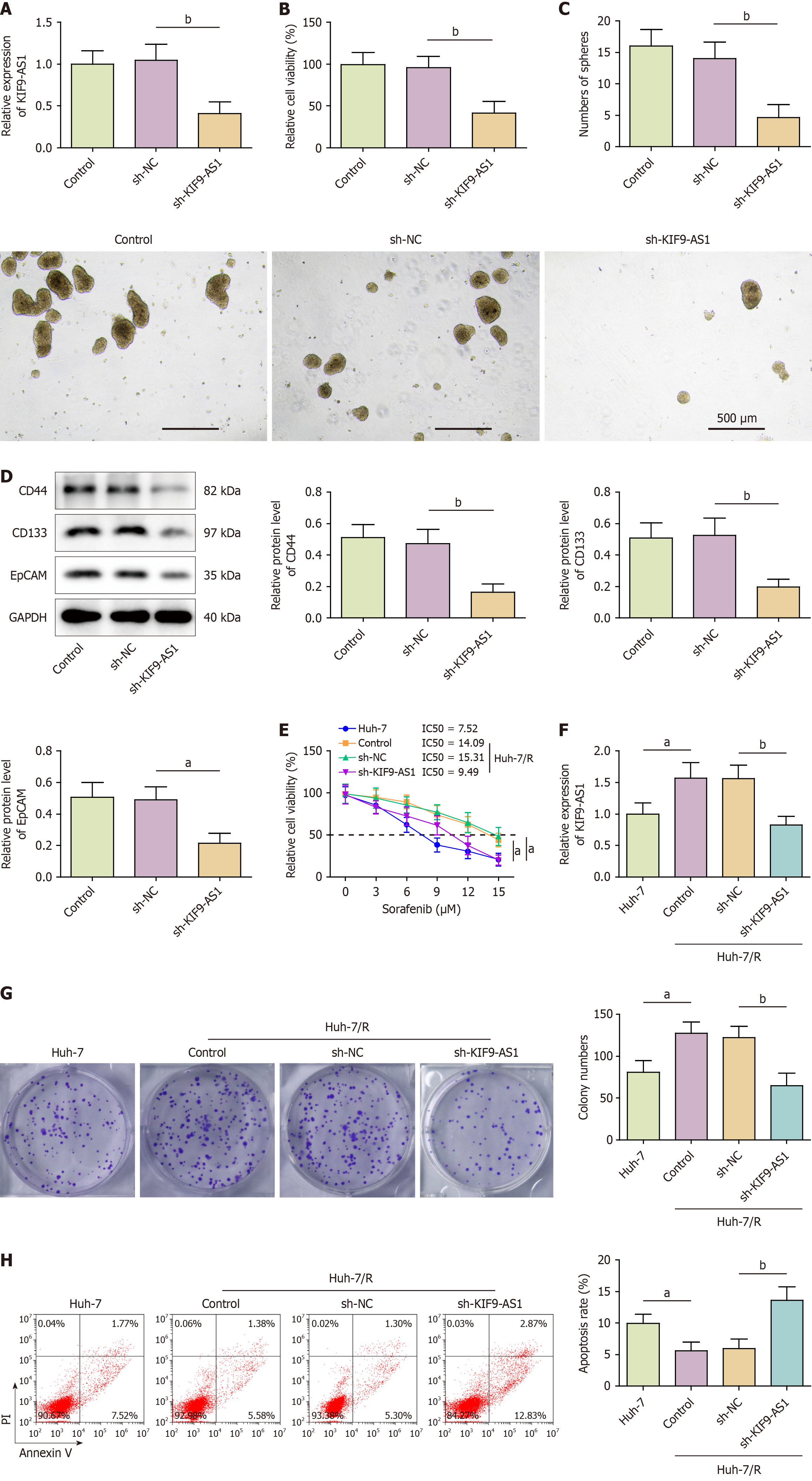
Depletion of KIF9-AS1 suppressed stemness characteristics and reduced sorafenib resistance in HCC cells. To evaluate the impact of KIF9-AS1 on HCC cell stemness, we silenced KIF9-AS1 in Huh-7 cells through transfection with sh-KIF9-AS1 (Figure 2A). Suppression of KIF9-AS1 significantly reduced Huh-7 cell viability (Figure 2B). Furthermore, suppression of KIF9-AS1 reduced the number of tumor spheres formed and the expression of stem cell markers, including CD44, CD133, and EpCAM, in Huh-7 cells (Figure 2C and D). Next, we assessed the impact of KIF9-AS1 on sorafenib resistance in HCC cells. Compared with Huh-7 cells, Huh-7/R cells had a greater IC50 value and greater expression of KIF9-AS1, whereas silencing KIF9-AS1 resulted in a reduced IC50 value (Figure 2E and F). Additionally, colony formation was accelerated and cell apoptosis was suppressed in Huh-7/R cells; these effects were reversed by downregulation of KIF9-AS1 (Figure 2G and H). Together, these findings suggested that inhibition of KIF9-AS1 suppressed stemness characteristics and attenuated sorafenib resistance in HCC cells.
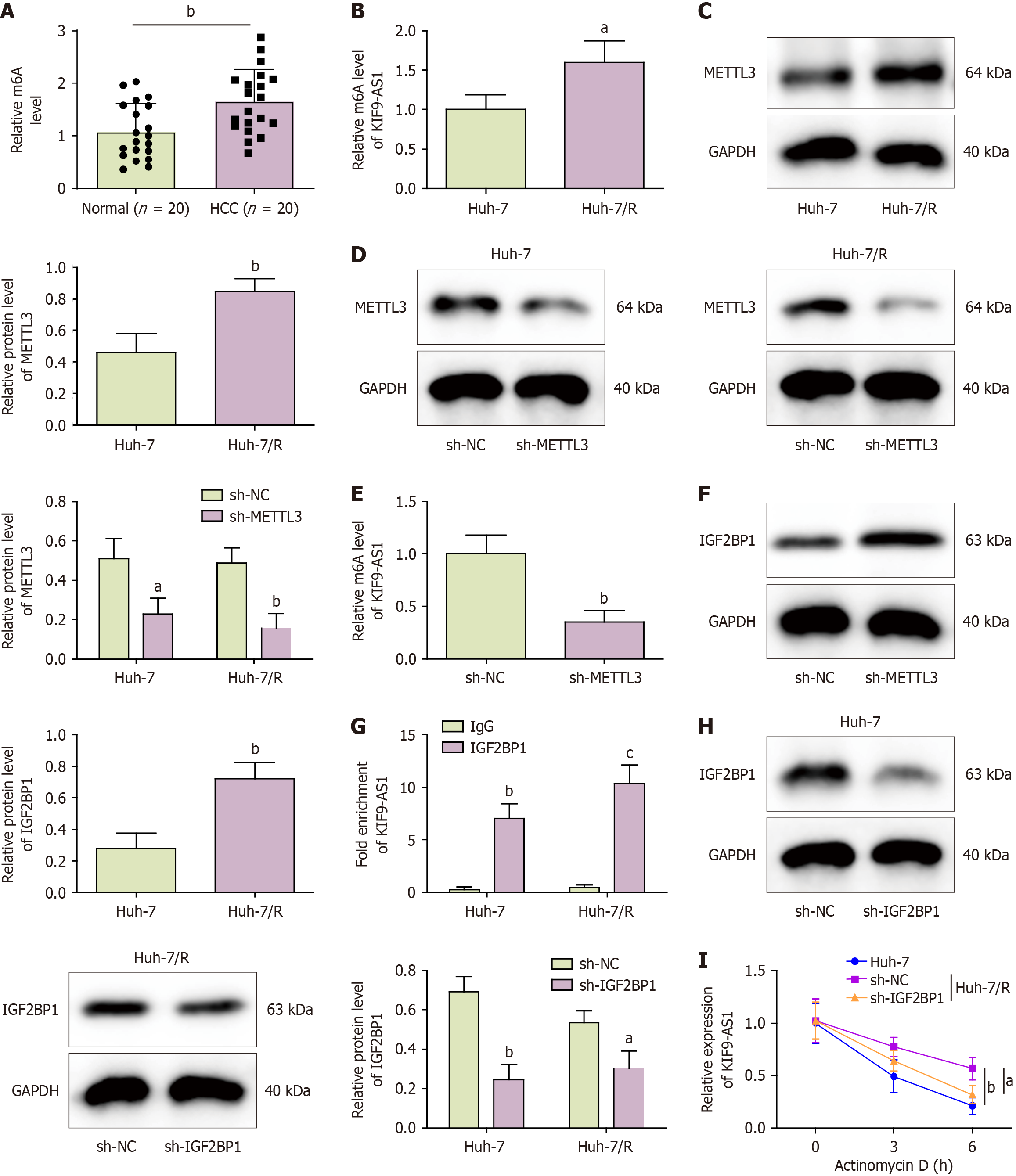
The importance of m6A modification in tumor stemness and drug resistance has gained widespread recognition. For example, YTHDF1 regulates the m6A modification of NOTCH1 mRNA to increase its stability and upregulate its expression, which facilitates stemness and drug resistance in HCC cells[25]. As depicted in Figure 3A, we observed an obvious increase in the total m6A content in tissue samples from HCC patients. Notably, the m6A level of KIF9-AS1 and the protein level of METTL3 were elevated in Huh-7/R cells compared with Huh-7 cells (Figure 3B and C). Silencing METTL3 decreased the m6A levels of KIF9-AS1 (Figure 3D and E). The data presented in Figure 3F revealed a significant increase in IGF2BP1 expression in Huh-7/R cells compared with that in Huh-7 cells. Moreover, the anti-IGF2BP1 antibody bound significantly more KIF9-AS1 than the IgG controls did in both Huh-7 cells and Huh-7/R cells (Figure 3G). Finally, IGF2BP1 was knocked down in both Huh-7 and Huh-7/R cells (Figure 3H). When both cell types were treated with actinomycin D (5 μg/mL), the stability of KIF9-AS1 in Huh-7/R cells was greater than that in Huh-7 cells; however, this stability was decreased in Huh-7/R cells upon IGF2BP1 silencing (Figure 3I). These results highlighted that METTL3 stabilized and increased KIF9-AS1 expression through an m6A-IGF2BP1-dependent mechanism.
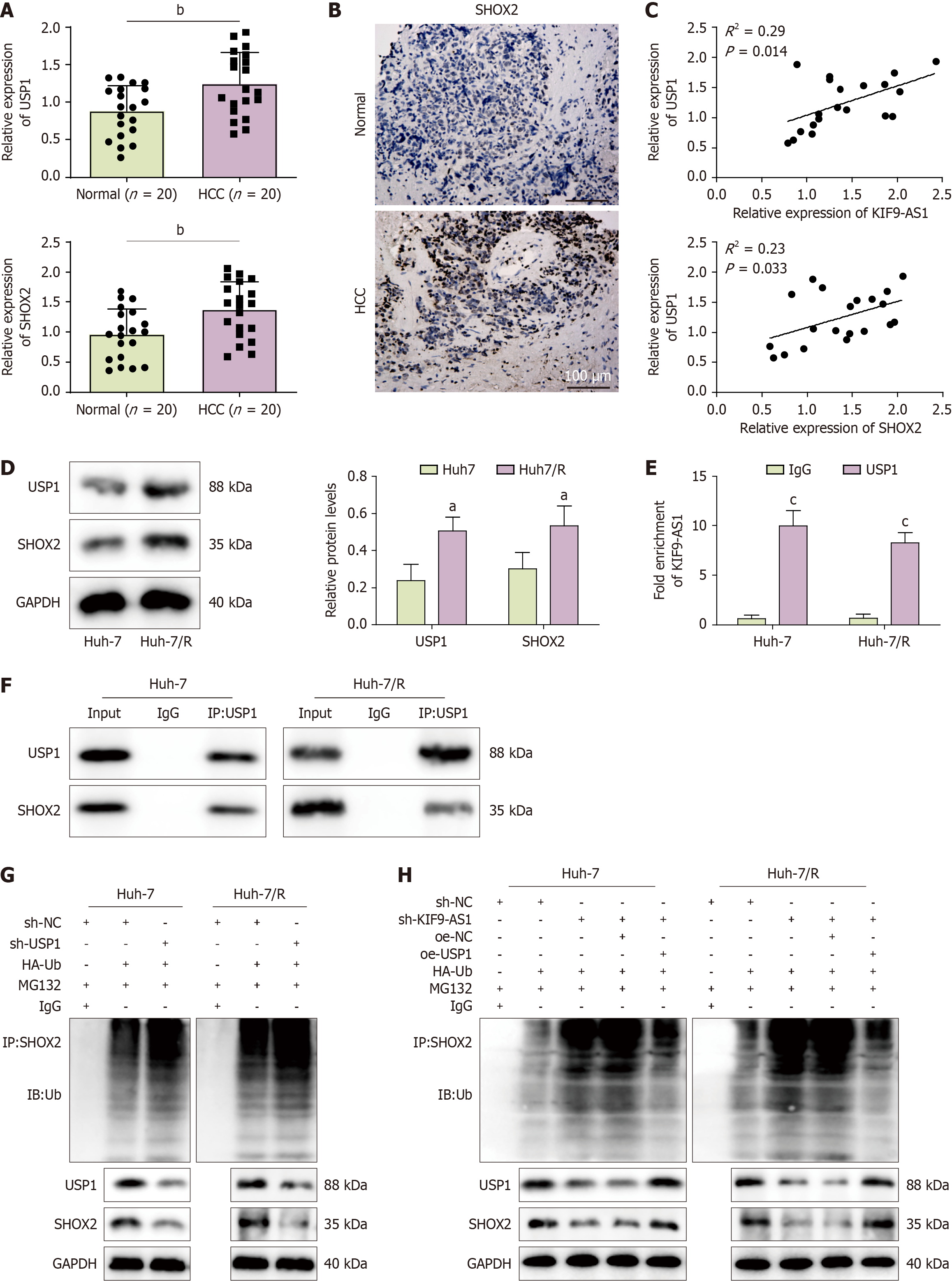
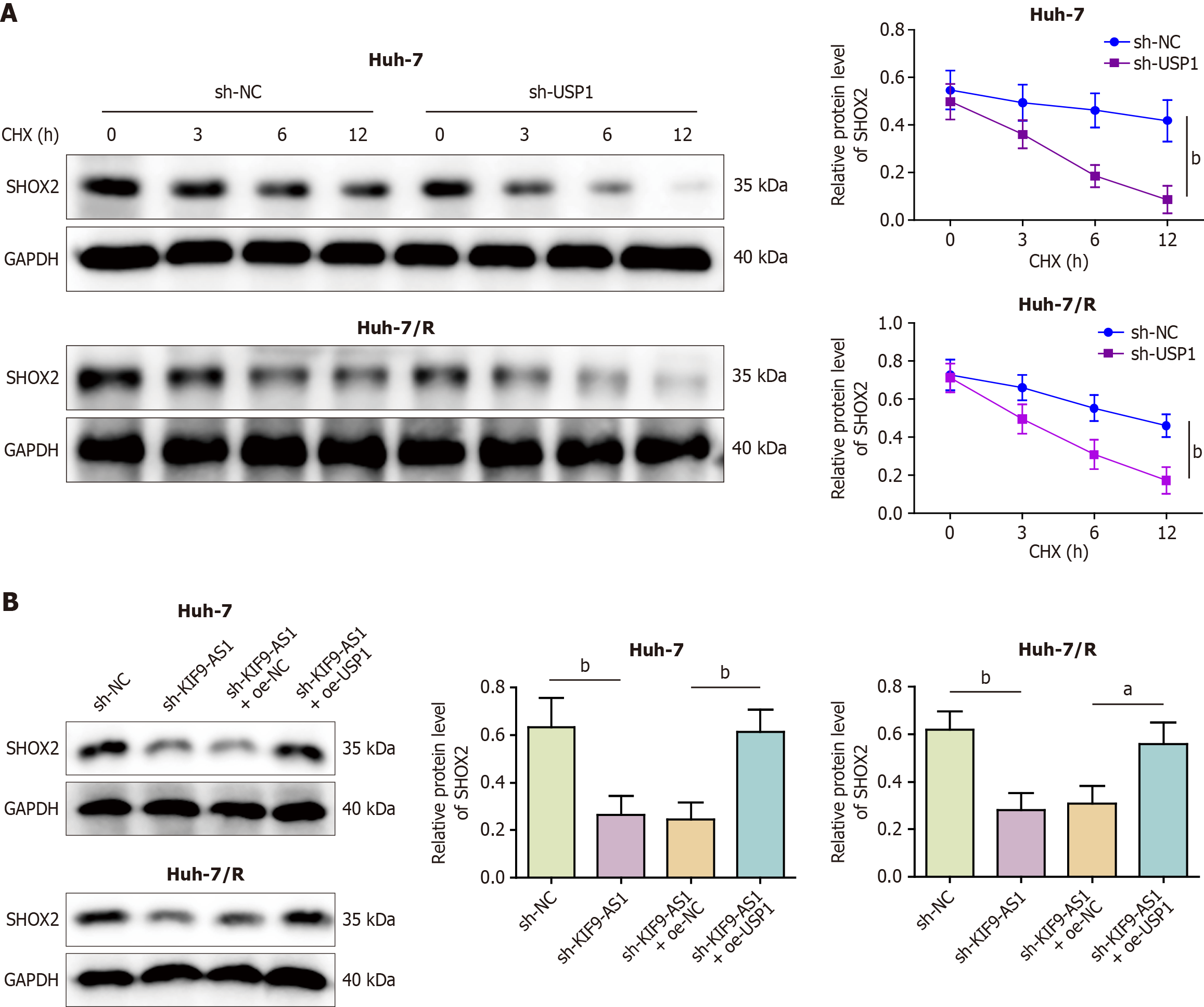
We further explored the downstream mechanism of KIF9-AS1 in HCC. As shown in Figure 4A-C, the levels of USP1 and SHOX2 were increased in HCC tissues, and positive correlations were observed between the levels of KIF9-AS1 and USP1 and between the levels of USP1 and SHOX2. Compared with those in Huh-7 cells, the expression levels of USP1 and SHOX2 were upregulated in Huh-7/R cells (Figure 4D). UbiBrowser predicted SHOX2 as a substrate for USP1 ubiquitination, and RNA-Protein Interaction Prediction predicted a high score for the binding of KIF9-AS1 to USP1 (random forest classifier: 0.75; support vector machine classifier: 0.93). Consistent with this prediction, the RIP and coimmunoprecipitation results revealed that KIF9-AS1 was enriched by the anti-USP1 antibody and that USP1 efficiently coimmunoprecipitated with SHOX2 in both cell lines (Figure 4E and F). Next, we knocked down USP1 and observed an increase in the level of SHOX2 ubiquitination, leading to reduced SHOX2 protein expression (Figure 4G). Moreover, downregulation of KIF9-AS1 led to increased SHOX2 ubiquitination and decreased SHOX2 protein expression, whereas simultaneous knockdown of KIF9-AS1 and overexpression of USP1 repressed SHOX2 ubiquitination and increased SHOX2 protein expression (Figure 4H). USP1 knockdown promoted degradation of the SHOX2 protein after cycloheximide treatment (Figure 5A). Overall, the results revealed that KIF9-AS1 knockdown inhibited the protein expression of SHOX2, whereas the overexpression of USP1 increased SHOX2 protein expression, which reversed the effect of silencing KIF9-AS1 (Figure 5B). In conclusion, KIF9-AS1 increased the stability and expression of SHOX2 by promoting USP1-mediated deubiquitination.
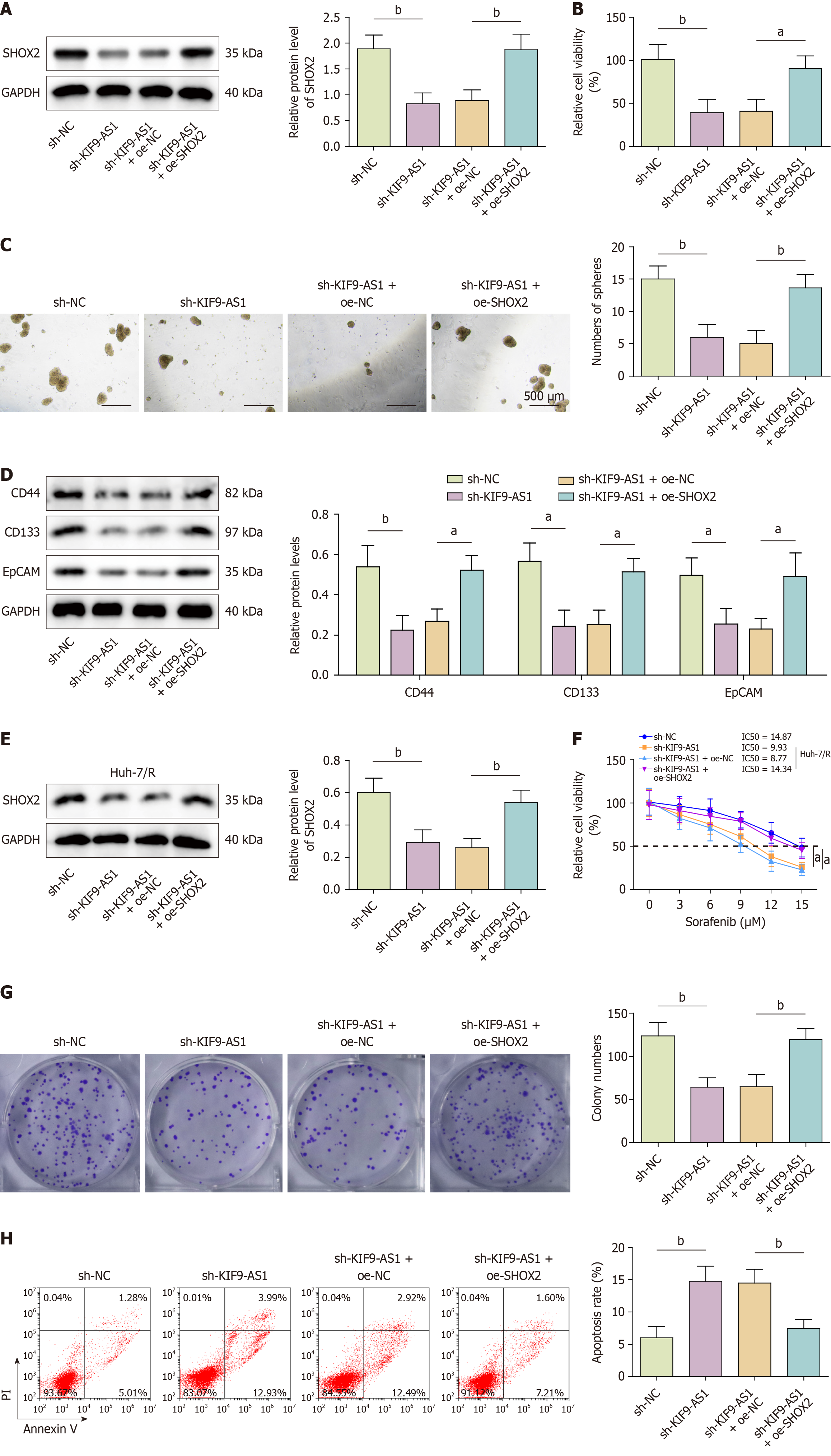
Further investigations were conducted to explore the function of the KIF9-AS1/SHOX2 axis in mediating stemness and sorafenib resistance in HCC cells. Silencing of KIF9-AS1 decreased SHOX2 protein levels and cell viability, which were reversed by SHOX2 overexpression (Figure 6A and B). Co-silencing of KIF9-AS1 and SHOX2 overexpression resulted in increased formation of tumor spheres and expression of stemness markers (CD44, CD133, EpCAM) (Figure 6C and D). These results suggested that increased levels of SHOX2 mitigated the suppressive effects of sh-KIF9-AS1 on tumor sphere formation and stemness marker expression in Huh-7 cells. Similar trends were observed in Huh-7/R cells. Knockdown of KIF9-AS1 significantly reduced SHOX2 expression, whereas co-transfection with OE-SHOX2 restored its expression (Figure 6E). Furthermore, SHOX2 overexpression increased the IC50 value and the number of colonies formed and inhibited cell apoptosis, effectively reversing the impacts of KIF9-AS1 knockdown in Huh-7/R cells (Figure 6F-H). Thus, KIF9-AS1 promoted stemness and sorafenib resistance in HCC cells through the regulation of SHOX2.
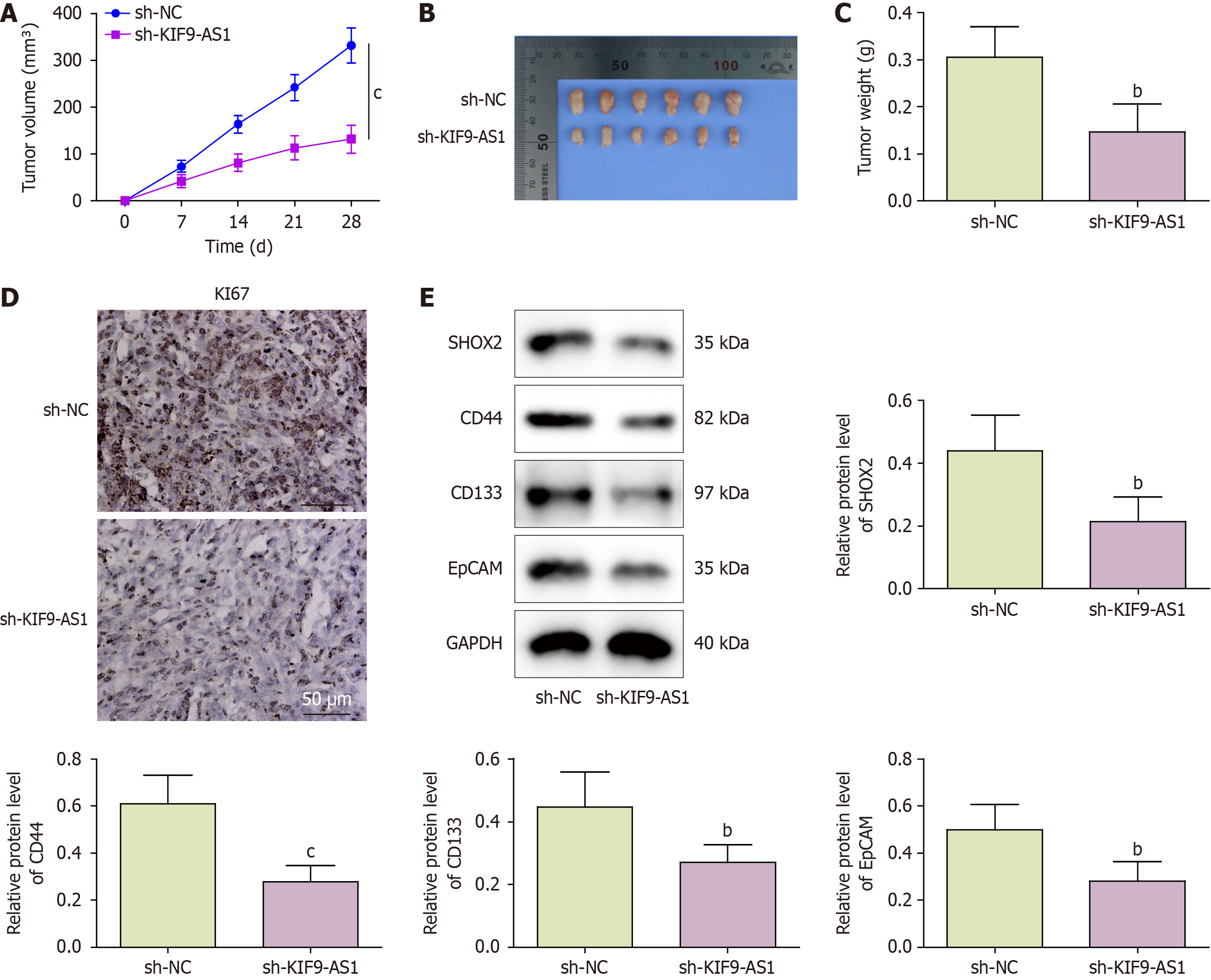
We validated the role of KIF9-AS1 in regulating the stemness and sorafenib resistance of HCC cells in an HCC xenograft mouse model. The mice were subcutaneously implanted with Huh-7/R cells transfected with sh-KIF9-AS1 and orally administered sorafenib. sh-KIF9-AS1 treatment resulted in significant reductions in both tumor volume and weight (Figure 7A-C). In addition, depletion of KIF9-AS1 downregulated the expression of Ki67, SHOX2, and stemness markers (CD44, CD133, and EpCAM) in HCC model mice (Figure 7D and E). These findings suggested that the suppression of KIF9-AS1 alleviated sorafenib resistance in HCC xenograft model mice.
Studies have shown that tumor stem cells play crucial roles in drug resistance, cell renewal and proliferation, differentiation ability, and sphere formation ability[26,27]. Despite the widespread use of sorafenib as a first-line treatment for HCC, the emergence of sorafenib resistance remains a significant challenge in HCC treatment[28]. This study demonstrated that the m6A-modified lncRNA KIF9-AS1 promoted stemness and sorafenib resistance in HCC by upregulating SHOX2 expression.
Differentially expressed lncRNAs in HCC are associated with prognosis and sensitivity to therapy[29]. For example, the lncRNA AC007639.1 increased drug resistance in HCC cells and accelerated tumor growth in HCC mouse models[30]. Additionally, increased levels of the lncRNA nuclear paraspeckle assembly transcript 1 are correlated with poor prognosis in HCC patients, and a molecular mechanism study revealed that nuclear paraspeckle assembly transcript 1 decreases sorafenib resistance in HCC by targeting the miR-149-5p/AKT serine/threonine kinase 1 axis[31]. Our previous study revealed that KIF9-AS1 is upregulated in HCC tumor tissues and that deletion of this lncRNA suppressed cell proliferation and migration but accelerated cell death in HCC cells[7]. In the present study, we confirmed that KIF9-AS1 expression was increased in HCC patient tissues and that high expression of KIF9-AS1 predicted poor clinical outcomes in patients. These results suggested the oncogenic potential of KIF9-AS1 in the development of HCC. To date, the regulatory role of KIF9-AS1 in drug resistance in tumors has rarely been discussed. Our previous study revealed that the upregulation of KIF9-AS1 accelerated cell proliferation, suppressed cell apoptosis, and enhanced sorafenib resistance in renal cell carcinoma cells[8]. Consistent with these findings, the present study revealed that knocking down KIF9-AS1 alleviated resistance to sorafenib, suppressed cell proliferation and promoted cell death in HCC cells. As previously reported, lncRNAs can be targeted to combat chemoresistance mediated by cancer stem cells, suggesting that stemness and therapeutic resistance are linked by lncRNAs[32]. However, the functional role of KIF9-AS1 in stemness in HCC has not been thoroughly elucidated. Notably, our study demonstrated for the first time that KIF9-AS1 promoted the expression of stemness marker genes in sorafenib-resistant HCC cells, which is consistent with previous studies. For example, the lncRNA DPPA2 upstream binding RNA increased cancer stemness marker expression and amplified chemoresistance in HCC through the E2F transcription factor 1/cellular inhibitor of PP2A axis[33]. These results suggest the potential of KIF9-AS1 as a promising therapeutic target for HCC in the future.
Recent evidence indicates that m6A modification can mediate RNA metabolism and gene expression, influencing the malignant phenotypes and drug resistance of HCC[34]. A study by Wu et al[10] demonstrated that m6A modification increased lncRNA NIFK-AS1 expression in HCC, thus promoting resistance to sorafenib and tumor development. Another study revealed that METTL14 modulates m6A modification, stabilization and upregulation of the lncRNA MIR155 host gene, thereby facilitating immune escape in HCC cells[35]. Bioinformatics analyses have predicted m6A modification sites in KIF9-AS1. However, the role of m6A-modified KIF9-AS1 in HCC has not been clarified. Our findings provide evidence that m6A modification of KIF9-AS1 is significantly enhanced in sorafenib-resistant HCC cells. METTL3 and IGF2BP1 are considered regulators of m6A methylation; the RM2Target database also predicts that these proteins regulate the m6A modification of KIF9-AS1. However, regulation of the METTL3/IGF2BP1 axis by m6A-modified KIF9-AS1 has not been reported in HCC. A previous study revealed that METTL3 mediated m6A modification and upregulation of the expression of the lncRNA LINC00958, leading to lipogenesis in HCC and tumor progression[36]. Liu et al[37] reported that METTL3, an m6A writer, facilitates m6A modification and the upregulation of the expression of the lncRNA glucosylceramidase beta pseudogene 1, increasing the migratory and invasive capabilities of HCC cells. Our findings revealed that METTL3 expression was elevated in HCC cells and that silencing METTL3 suppressed m6A modification of KIF9-AS1. Cai et al[38] reported that RNA binding motif protein 15 promoted the proliferation and invasion of HCC cells through m6A methylation of YES1 via an IGF2BP1-dependent mechanism. This study also revealed that silencing IGF2BP1 decreased the stability of KIF9-AS1 in HCC cells. Our study confirmed that METTL3 stabilized and promoted KIF9-AS1 expression in HCC cells in an m6A-IGF2BP1-dependent manner. These results indicate that the METTL3/IGF2BP1 axis regulates m6A modification and KIF9-AS1 expression in HCC.
Finally, we explored the downstream mechanism of KIF9-AS1 in HCC. As demonstrated in a study by Yang et al[18], the upregulation of SHOX2 was positively correlated with tumor recurrence and advanced tumor-node-metastasis stage, while silencing SHOX2 obviously suppressed the proliferative and invasive abilities of HCC cells. Consistently, our findings revealed that SHOX2 overexpression enhanced the stemness and sorafenib resistance of HCC cells. According to the bioinformatics analysis, USP1 is predicted to interact with SHOX2 and KIF9-AS1. The ubiquitination function of USP1 plays an essential regulatory role in various types of cancers. Reportedly, an increase in USP1 can lead to the deubiquitination of tumor-related proteins, thereby increasing protein stability and function in tumor cells[39]. Another study demonstrated that USP1 suppression decreased tumor stemness and sensitized HCC cells to doxycycline by increasing the ubiquitylation of proliferating cell nuclear antigen[40]. Accordingly, we observed that USP1 suppression induced an increase in SHOX2 ubiquitination, leading to the downregulation of SHOX2 expression in HCC cells. Moreover, we found that KIF9-AS1 increased SHOX2 stability and expression by promoting deubiquitination via USP1. The functional mechanism of SHOX2 has been implicated in different cancers. For example, SHOX2 facilitates the oncogenic behavior of prostate cancer cells by blocking the Hippo-Yes-associated protein pathway through the activation of nephronophthisis type 4 transcription[41]. Teng et al[42] reported that SHOX2 accelerated breast cancer metastasis by activating WASF3 transcription in cooperation with signal transducer and activator of transcription 3 (STAT3)[42]. Notably, Yes-associated protein expression was found to be associated with vascular invasion, stemness and epithelial-mesenchymal transition in HCC cells[43]. Furthermore, glycochenodeoxycholic acid-induced activation of the STAT3 signaling pathway enhances chemoresistance and stemness in HCC[44]. Another study revealed that inhibition of the NANOG/STAT3 pathway increased sensitivity to sorafenib treatment in HCC[45]. Therefore, we speculate that SHOX2 might exert its functional effects on stemness and sorafenib resistance in HCC by mediating downstream pathways or proteins.
This study still has limitations. First, we plan to collect more clinical samples to analyze the use of KIF9-AS1 as a diagnostic biomarker for HCC in future work. Second, this study revealed that m6A modification led to the differential expression of KIF9-AS1 in HCC; however, other epigenetic mechanisms, such as transcription factor expression and histone acetylation, may also be involved. Third, our findings revealed that KIF9-AS1 regulated SHOX2 expression through USP1-mediated ubiquitination. Further investigations are needed to clarify whether KIF9-AS1 influences SHOX2 expression through other factors or epigenetic modifications. Overall, our study also indicates the potential clinical application of KIF9-AS1. For example, this study might provide a theoretical foundation for the use of KIF9-AS1 as a diagnostic marker for HCC. Furthermore, KIF9-AS1 might be delivered to target sites through various delivery systems for HCC treatment.
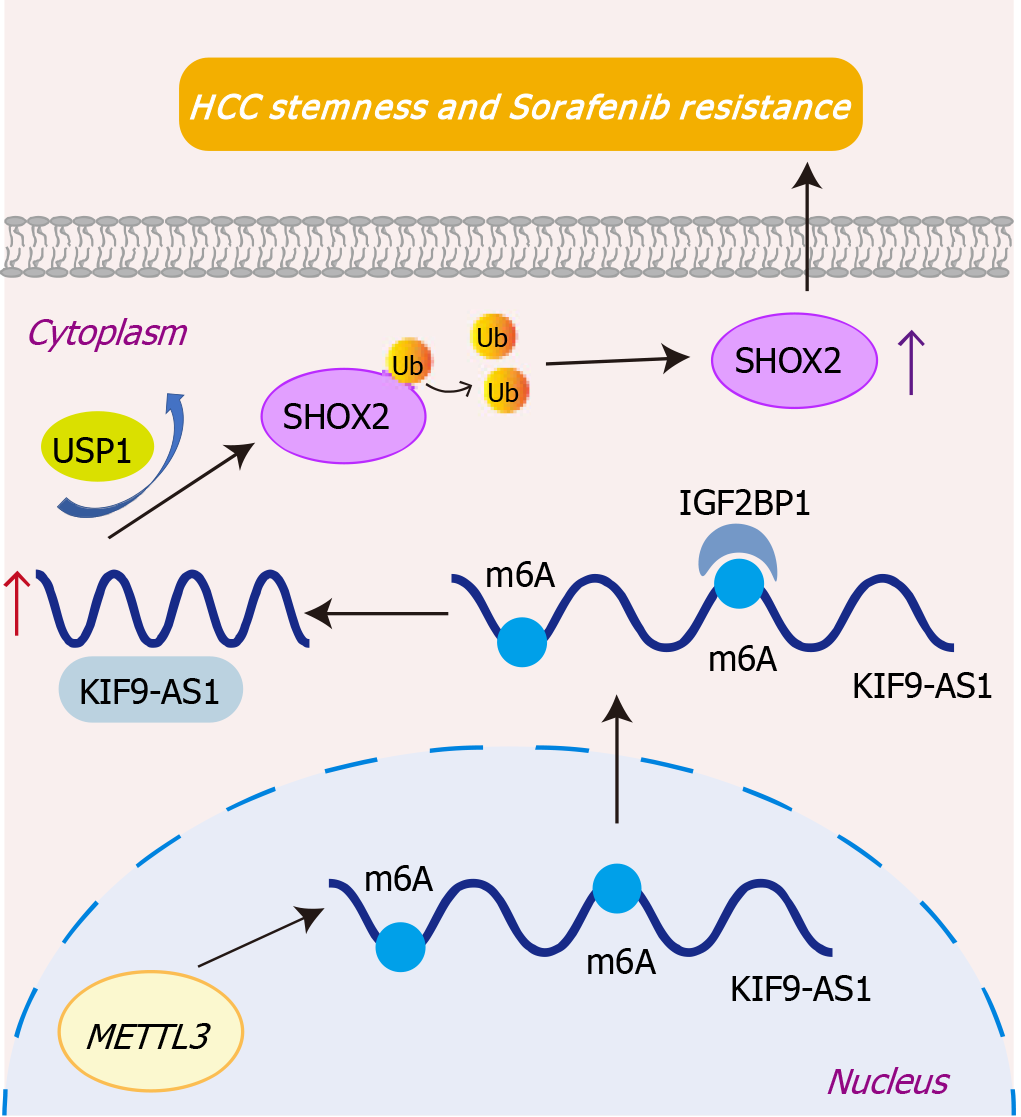
In conclusion, the present study demonstrated that the m6A-modified lncRNA KIF9-AS1 promoted stemness and sorafenib resistance in HCC through USP1-mediated deubiquitination of SHOX2 (Figure 8). Our work might offer a promising new clinical target for treating HCC.
We thank Xiao-Mei Zhang for her technical assistance.
| 1. | Chidambaranathan-Reghupaty S, Fisher PB, Sarkar D. Hepatocellular carcinoma (HCC): Epidemiology, etiology and molecular classification. Adv Cancer Res. 2021;149:1-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 330] [Cited by in RCA: 639] [Article Influence: 106.5] [Reference Citation Analysis (0)] |
| 2. | Brown ZJ, Tsilimigras DI, Ruff SM, Mohseni A, Kamel IR, Cloyd JM, Pawlik TM. Management of Hepatocellular Carcinoma: A Review. JAMA Surg. 2023;158:410-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 494] [Reference Citation Analysis (1)] |
| 3. | Huang A, Yang XR, Chung WY, Dennison AR, Zhou J. Targeted therapy for hepatocellular carcinoma. Signal Transduct Target Ther. 2020;5:146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 473] [Cited by in RCA: 549] [Article Influence: 91.5] [Reference Citation Analysis (0)] |
| 4. | Tsui YM, Chan LK, Ng IO. Cancer stemness in hepatocellular carcinoma: mechanisms and translational potential. Br J Cancer. 2020;122:1428-1440. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 93] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 5. | Cao Y, Zhang F, Wang H, Bi C, Cui J, Liu F, Pan H. LncRNA MALAT1 mediates doxorubicin resistance of hepatocellular carcinoma by regulating miR-3129-5p/Nova1 axis. Mol Cell Biochem. 2021;476:279-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 6. | Zhang PF, Wang F, Wu J, Wu Y, Huang W, Liu D, Huang XY, Zhang XM, Ke AW. LncRNA SNHG3 induces EMT and sorafenib resistance by modulating the miR-128/CD151 pathway in hepatocellular carcinoma. J Cell Physiol. 2019;234:2788-2794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 148] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 7. | Yu Y, Lu X, Yan Y, Wang Y, Meng J, Tian S, Mu J. The lncRNA KIF9-AS1 Accelerates Hepatocellular Carcinoma Growth by Recruiting DNMT1 to Promote RAI2 DNA Methylation. J Oncol. 2022;2022:3888798. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 8. | Jin Y, Huang R, Xia Y, Huang C, Qiu F, Pu J, He X, Zhao X. Long Noncoding RNA KIF9-AS1 Regulates Transforming Growth Factor-β and Autophagy Signaling to Enhance Renal Cell Carcinoma Chemoresistance via microRNA-497-5p. DNA Cell Biol. 2020;39:1096-1103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 9. | Zhu ZM, Huo FC, Zhang J, Shan HJ, Pei DS. Crosstalk between m6A modification and alternative splicing during cancer progression. Clin Transl Med. 2023;13:e1460. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 64] [Reference Citation Analysis (0)] |
| 10. | Wu J, Pang R, Li M, Chen B, Huang J, Zhu Y. m6A-Induced LncRNA MEG3 Suppresses the Proliferation, Migration and Invasion of Hepatocellular Carcinoma Cell Through miR-544b/BTG2 Signaling. Onco Targets Ther. 2021;14:3745-3755. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 11. | Chen YT, Xiang D, Zhao XY, Chu XY. Upregulation of lncRNA NIFK-AS1 in hepatocellular carcinoma by m(6)A methylation promotes disease progression and sorafenib resistance. Hum Cell. 2021;34:1800-1811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 12. | Zhu Y, Xiao B, Liu M, Chen M, Xia N, Guo H, Huang J, Liu Z, Wang F. N6-methyladenosine-modified oncofetal lncRNA MIR4435-2HG contributed to stemness features of hepatocellular carcinoma cells by regulating rRNA 2'-O methylation. Cell Mol Biol Lett. 2023;28:89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 25] [Reference Citation Analysis (0)] |
| 13. | Wang L, Yang Q, Zhou Q, Fang F, Lei K, Liu Z, Zheng G, Zhu L, Huo J, Li X, Peng S, Kuang M, Lin S, Huang M, Xu L. METTL3-m(6)A-EGFR-axis drives lenvatinib resistance in hepatocellular carcinoma. Cancer Lett. 2023;559:216122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 87] [Reference Citation Analysis (0)] |
| 14. | Shi Y, Niu Y, Yuan Y, Li K, Zhong C, Qiu Z, Li K, Lin Z, Yang Z, Zuo D, Qiu J, He W, Wang C, Liao Y, Wang G, Yuan Y, Li B. PRMT3-mediated arginine methylation of IGF2BP1 promotes oxaliplatin resistance in liver cancer. Nat Commun. 2023;14:1932. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 65] [Reference Citation Analysis (0)] |
| 15. | Zhi X, Zhou J, Tian H, Zhou R, Huang Z, Liu C. [SHOX2 promotes migration, invasion and stemness of bladder cancer cells in vitro]. Nan Fang Yi Ke Da Xue Xue Bao. 2021;41:995-1001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 16. | Zhou J, Li P, Feng J, Wu Q, You S. MiR-24-1-5p Hinders Malignant Phenotypes of Clear Cell Renal Cell Carcinoma by Targeting SHOX2. Biochem Genet. 2023;61:2004-2019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 17. | Wu X, Chen H, You C, Peng Z. A potential immunotherapeutic and prognostic biomarker for multiple tumors including glioma: SHOX2. Hereditas. 2023;160:21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 18. | Yang T, Zhang H, Cai SY, Shen YN, Yuan SX, Yang GS, Wu MC, Lu JH, Shen F. Elevated SHOX2 expression is associated with tumor recurrence of hepatocellular carcinoma. Ann Surg Oncol. 2013;20 Suppl 3:S644-S649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 19. | Li N, Zeng Y, Huang J. Signaling pathways and clinical application of RASSF1A and SHOX2 in lung cancer. J Cancer Res Clin Oncol. 2020;146:1379-1393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 20. | Niu Z, Li X, Feng S, Huang Q, Zhuang T, Yan C, Qian H, Ding Y, Zhu J, Xu W. The deubiquitinating enzyme USP1 modulates ERα and modulates breast cancer progression. J Cancer. 2020;11:6992-7000. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 21. | Das DS, Das A, Ray A, Song Y, Samur MK, Munshi NC, Chauhan D, Anderson KC. Blockade of Deubiquitylating Enzyme USP1 Inhibits DNA Repair and Triggers Apoptosis in Multiple Myeloma Cells. Clin Cancer Res. 2017;23:4280-4289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 80] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 22. | Zhao Y, Xue C, Xie Z, Ouyang X, Li L. Comprehensive analysis of ubiquitin-specific protease 1 reveals its importance in hepatocellular carcinoma. Cell Prolif. 2020;53:e12908. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 23. | Sonego M, Pellarin I, Costa A, Vinciguerra GLR, Coan M, Kraut A, D'Andrea S, Dall'Acqua A, Castillo-Tong DC, Califano D, Losito S, Spizzo R, Couté Y, Vecchione A, Belletti B, Schiappacassi M, Baldassarre G. USP1 links platinum resistance to cancer cell dissemination by regulating Snail stability. Sci Adv. 2019;5:eaav3235. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 103] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 24. | Liu D, Li Q, Zang Y, Li X, Li Z, Zhang P, Feng C, Yang P, Cui J, Sun Y, Wei T, Su P, Zhao X, Yang H, Ding Y. USP1 modulates hepatocellular carcinoma progression via the Hippo/TAZ axis. Cell Death Dis. 2023;14:264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 29] [Reference Citation Analysis (0)] |
| 25. | Zhang X, Su T, Wu Y, Cai Y, Wang L, Liang C, Zhou L, Wang S, Li XX, Peng S, Kuang M, Yu J, Xu L. N6-Methyladenosine Reader YTHDF1 Promotes Stemness and Therapeutic Resistance in Hepatocellular Carcinoma by Enhancing NOTCH1 Expression. Cancer Res. 2024;84:827-840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 58] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 26. | Li Y, Wang Z, Ajani JA, Song S. Drug resistance and Cancer stem cells. Cell Commun Signal. 2021;19:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 238] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 27. | Huang T, Song X, Xu D, Tiek D, Goenka A, Wu B, Sastry N, Hu B, Cheng SY. Stem cell programs in cancer initiation, progression, and therapy resistance. Theranostics. 2020;10:8721-8743. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 351] [Article Influence: 58.5] [Reference Citation Analysis (0)] |
| 28. | Li Z, Gao J, Zheng S, Wang Y, Xiang X, Cheng Q, Zhu J. Therapeutic Efficacy of Sorafenib in Patients with Hepatocellular Carcinoma Recurrence After Liver Transplantation: A Systematic Review and Meta-Analysis. Turk J Gastroenterol. 2021;32:30-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 29. | Zhang G, Sun J, Zhang X. A novel Cuproptosis-related LncRNA signature to predict prognosis in hepatocellular carcinoma. Sci Rep. 2022;12:11325. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 138] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 30. | Bai Y, Ding M, Lu D, Li Y, Yao S, Wang L, Li H, Cui G, Li X, Sun X, Yang Y. Long Noncoding RNA AC007639.1 Promotes the Pathogenesis and Progression of Hepatocellular Carcinoma Through Inhibiting Apoptosis and Stimulating Chemotherapeutic Resistance. Front Oncol. 2021;11:715541. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 31. | Niu Y, Tang G, Wu X, Wu C. LncRNA NEAT1 modulates sorafenib resistance in hepatocellular carcinoma through regulating the miR-149-5p/AKT1 axis. Saudi J Gastroenterol. 2020;26:194-203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 32. | Yue J, Wu Y, Qiu L, Zhao R, Jiang M, Zhang H. LncRNAs link cancer stemness to therapy resistance. Am J Cancer Res. 2021;11:1051-1068. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 33. | Liu S, Bu X, Kan A, Luo L, Xu Y, Chen H, Lin X, Lai Z, Wen D, Huang L, Shi M. SP1-induced lncRNA DUBR promotes stemness and oxaliplatin resistance of hepatocellular carcinoma via E2F1-CIP2A feedback. Cancer Lett. 2022;528:16-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 51] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 34. | Qiu Z, Yuan X, Wang X, Liu S. Crosstalk between m6A modification and non-coding RNAs in HCC. Cell Signal. 2024;117:111076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 35. | Peng L, Pan B, Zhang X, Wang Z, Qiu J, Wang X, Tang N. Lipopolysaccharide facilitates immune escape of hepatocellular carcinoma cells via m6A modification of lncRNA MIR155HG to upregulate PD-L1 expression. Cell Biol Toxicol. 2022;38:1159-1173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 80] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 36. | Zuo X, Chen Z, Gao W, Zhang Y, Wang J, Wang J, Cao M, Cai J, Wu J, Wang X. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol. 2020;13:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 337] [Article Influence: 56.2] [Reference Citation Analysis (0)] |
| 37. | Liu R, Yin G, Tuo H, Guo Y, Zhu Y, Zhang L, Yang W, Liu Q, Wang Y. METTL3-induced lncRNA GBAP1 promotes hepatocellular carcinoma progression by activating BMP/SMAD pathway. Biol Direct. 2023;18:53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 38. | Cai X, Chen Y, Man D, Yang B, Feng X, Zhang D, Chen J, Wu J. RBM15 promotes hepatocellular carcinoma progression by regulating N6-methyladenosine modification of YES1 mRNA in an IGF2BP1-dependent manner. Cell Death Discov. 2021;7:315. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 63] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 39. | García-Santisteban I, Peters GJ, Giovannetti E, Rodríguez JA. USP1 deubiquitinase: cellular functions, regulatory mechanisms and emerging potential as target in cancer therapy. Mol Cancer. 2013;12:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 142] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 40. | Lu Z, Zhang Z, Yang M, Xiao M. Ubiquitin-specific protease 1 inhibition sensitizes hepatocellular carcinoma cells to doxorubicin by ubiquitinated proliferating cell nuclear antigen-mediated attenuation of stemness. Anticancer Drugs. 2022;33:622-631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 41. | Yang W, Chen H, Ma L, Dong J, Wei M, Xue X, Li Y, Jin Z, Xu W, Ji Z. SHOX2 promotes prostate cancer proliferation and metastasis through disruption of the Hippo-YAP pathway. iScience. 2023;26:107617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 42. | Teng Y, Loveless R, Benson EM, Sun L, Shull AY, Shay C. SHOX2 cooperates with STAT3 to promote breast cancer metastasis through the transcriptional activation of WASF3. J Exp Clin Cancer Res. 2021;40:274. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 43. | Park H, Lee Y, Lee K, Lee H, Yoo JE, Ahn S, Park YN, Kim H. The Clinicopathological Significance of YAP/TAZ Expression in Hepatocellular Carcinoma with Relation to Hypoxia and Stemness. Pathol Oncol Res. 2021;27:604600. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 44. | Shi C, Yang J, Hu L, Liao B, Qiao L, Shen W, Xie F, Zhu G. Glycochenodeoxycholic acid induces stemness and chemoresistance via the STAT3 signaling pathway in hepatocellular carcinoma cells. Aging (Albany NY). 2020;12:15546-15555. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 45. | Qiu D, Wang T, Xiong Y, Li K, Qiu X, Feng Y, Lian Q, Qin Y, Liu K, Zhang Q, Jia C. TFCP2L1 drives stemness and enhances their resistance to Sorafenib treatment by modulating the NANOG/STAT3 pathway in hepatocellular carcinoma. Oncogenesis. 2024;13:33. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/