Published online Oct 14, 2024. doi: 10.3748/wjg.v30.i38.4175
Revised: August 29, 2024
Accepted: September 12, 2024
Published online: October 14, 2024
Processing time: 117 Days and 4.4 Hours
As a research hotspot in the field of molecular biology, N6-methyladenosine (m6A) modification has made progress in the treatment of colorectal cancer (CRC), leukemia and other cancers. Numerous studies have demonstrated that the tumour microenvironment (TME) regulates the level of m6A modification in the host and activates a series of complex epigenetic signalling pathways through interactions with CRC cells, thus affecting the progression and prognosis of CRC. However, with the diversity in the composition of TME factors, this action is reci
Core Tip: This review summarizes the interactions of N6-methyladenosine (m6A) modification in colorectal cancer (CRC) with a variety of the tumour microenvironment factors such as metabolism, hypoxia, inflammation, and immunity and supports the idea that intestinal flora can influence the progression of CRC by regulating the level of m6A modification. Additionally, this review also summarizes the clinical applications of m6A modifications in CRC and suggests possible future research directions.
- Citation: Jiang TQ, Wang H, Cheng WX, Xie C. Modulation of host N6-methyladenosine modification by gut microbiota in colorectal cancer. World J Gastroenterol 2024; 30(38): 4175-4193
- URL: https://www.wjgnet.com/1007-9327/full/v30/i38/4175.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i38.4175
Colorectal cancer (CRC) is the third most common malignant tumour worldwide, and its onset is associated with factors such as lifestyle and dietary habits. It is widely distributed in regions such as Asia and Latin America[1-3]. In recent years, the incidence of CRC has been steadily increasing, with a trend towards a younger age of onset[4]. The pathological mechanisms primarily involve chromosomal instability and mutations in oncogenes, tumour suppressor genes, and mismatch repair-related genes caused by CpG methylation, thereby facilitating the occurrence and progression of cancer[3,5]. Due to the continuous increase in the global incidence and mortality rates of CRC in recent decades, the prevention and treatment of this disease have become increasingly crucial for human health[6,7].
RNA modifications are widely found in various types of RNA in organisms and constitute a class of epigenetic modifications at the RNA level. These modifications include N6-methyladenosine (m6A), N1-methyladenosine (m1A), N7-methylguanosine, 5-methylcytidine, 2-o-methylation, pseudouridine, and inosine[8,9]. Currently, there are many studies on m6A modification, which refers to adenylate methylation at the 6th N position in an RNA molecule[10]. This modification is enriched mainly near the termination codon, in long internal exons, in the 5' and 3' untranslated regions, and in the shared sequence RRACH (R = G/A and H = A/C/U). Currently, m6A is the most prevalent and abundant internal chemical and epigenetic modification known in eukaryotic RNA molecules[11] and is important in the regulation of RNA splicing[12], translation[13], stability[14], and DNA damage repair[15], which in turn affects cellular differentiation[16], embryonic development[17], sex determination[18], cancer occurrence[19,20] and other processes.
The CRC tumour microenvironment (TME) is composed mainly of tumour cells, blood vessels, lymphocytes, fibro
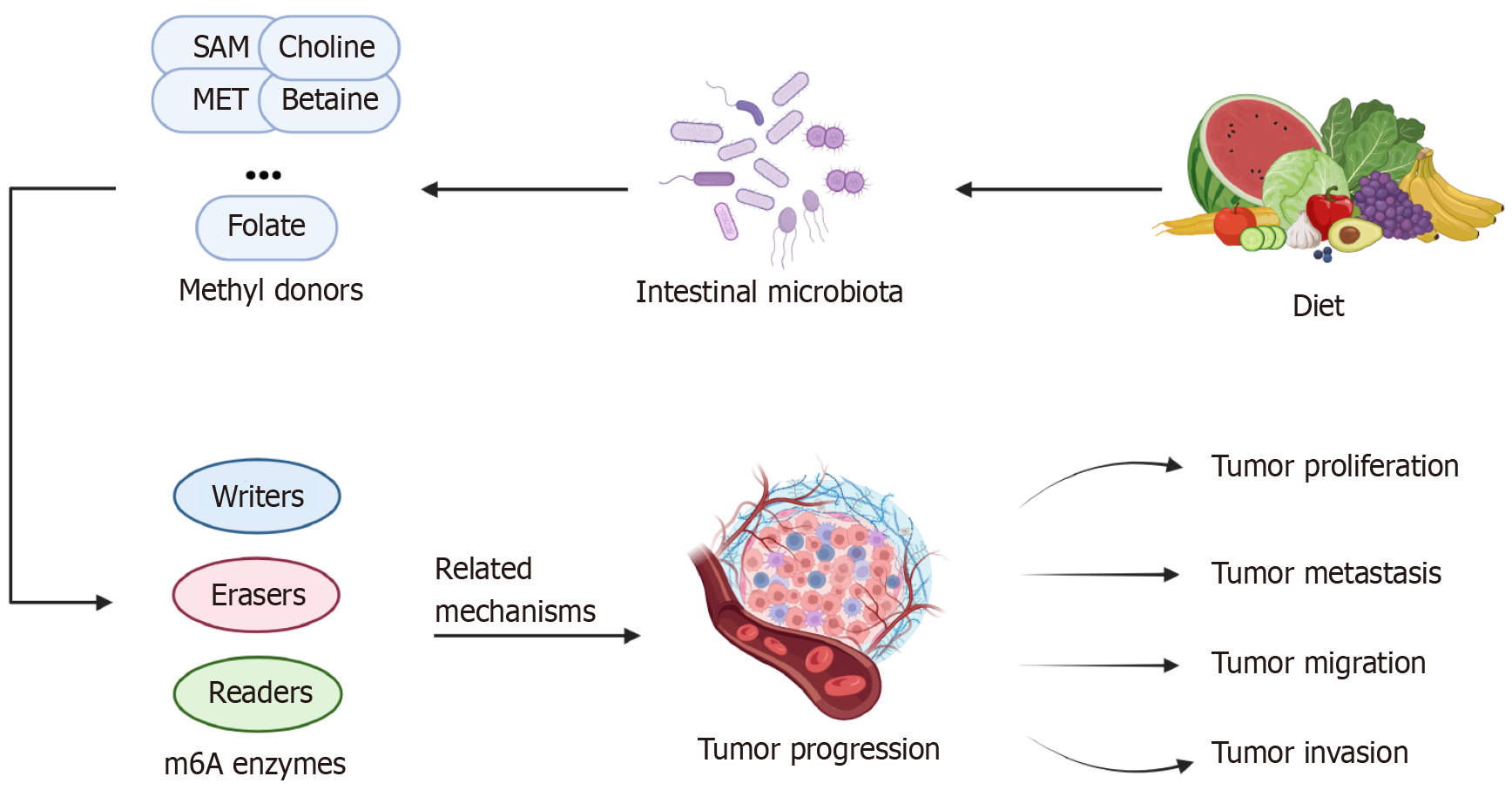
There is a strong correlation between the gut microbiota and the production and utilization of methyl donors by the body. Methyl groups are produced mainly by the single-carbon metabolic pathway and are involved in m6A RNA methylation through the folate cycle and methionine cycle pathways, and the substances that provide methyl groups are known as methyl donors. Among them, S-adenosylmethionine (SAM) is the most prominent methyl donor, and other common methyl donors are methionine, betaine, choline, folate, cobalamin, and pyridoxine[26]. All of these methyl donors can be ingested through the diet or produced from the gut microbiota, and their metabolism is also influenced by the gut microbiota. Their metabolites are involved in the synthesis of nucleotides, proteins, and lipids in the body through epigenetic mechanisms[27]. In addition, several other B vitamins are important for these pathways, and their production and metabolism are also influenced by the gut microbiota[28]. The balance of the distribution state of the gut microbiota is important for maintaining the normal functioning of human metabolism[29].
In this review, we focus on the role of the intestinal flora, which is an important factor in TME and an important upstream factor related to m6A during CRC occurrence and development, and leads to alterations in host m6A me
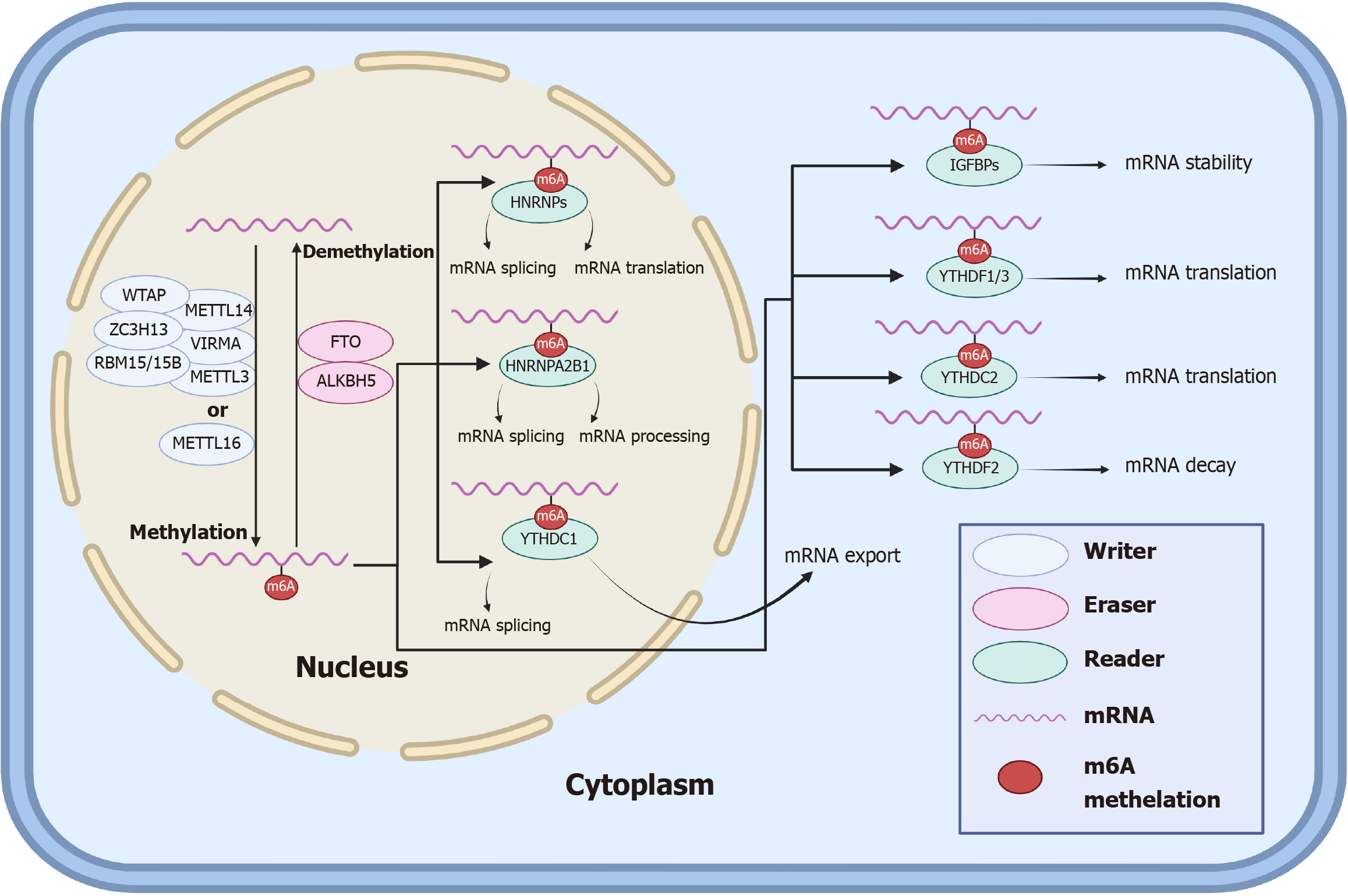
m6A, the most common RNA modification in organisms, mediates gene expression and is involved in many life processes. m6A modification is achieved through regulatory proteins such as writers, erasers, and readers (Table 1)[30-45], which can play roles in the methylation, demethylation, and recognition, respectively, of m6A on bound RNAs[46]. The specific processes by which m6A methylation regulates RNA modification in cells can be summarized as follows: In the nucleus, m6A methylation is accomplished by a complex consisting of METTL3, METTL14, WTAP, RBM15/15B, VIRMA, ZC3H13, or METTL16, and removal of the m6A modification is performed by the demethylation of erasers, including FTO and ALKBH5. After RNA methylation, the readers recognize m6A and perform posttranscriptional regulation, in which HNRNPs in the nucleus regulate mRNA splicing and translation, HNRNPA2B1 regulates mRNA splicing and processing, YTHDC1 regulates mRNA splicing, IGFBPs in the cytoplasm regulate mRNA stability, YTHDF1/3 and YTHDC2 regulate mRNA translation, and YTHDF2 regulates mRNA decay.
| Types | Regulators | Functions | Ref. |
| METTL3 | Catalyses m6A modification | [30,31] | |
| METTL14 | Provides structural support and recognizes target RNAs | [30,31] | |
| WTAP | Contributes to the orientation of MTC | [32,33] | |
| VIRMA/KIAA1429 | Recruits m6A complexes to specialized RNA sites | [32,33] | |
| m6A writers | ZC3H13 | Bridges WTAP to Nito | [32,33] |
| RBM15 | Binds to the m6A complex and recruits to specialized RNA sites | [32,33] | |
| CBLL1/HAKAI | Contributes to the stabilization of MTC | [32,34] | |
| METTL16 | Catalyses m6A modification | [35] | |
| FTO | Affects RNA splicing stabilization and deletes m6A modifications | [36] | |
| m6A erasers | ALKBH5 | Regulates RNA export and splicing, and deletes m6A modifications | [36] |
| IGF2BPs | Enhances mRNA stability and translation | [37] | |
| YTHDC1 | Mediates RNA splicing and export | [38] | |
| YTHDC2 | Enhances target RNA translation and reduces RNA abundance | [39] | |
| YTHDF1 | Enhances mRNA translation | [40] | |
| YTHDF2 | Promotes mRNA degradation | [40] | |
| m6A readers | YTHDF3 | Synergizes with YTHDF1 and YTHDF2 to enhance translation and degradation | [40] |
| ELF3 | Enhances mRNA translation | [41] | |
| FMR1 | Promotes mRNA degradation | [42] | |
| HNRNPs | Mediates mRNA splicing and translation | [43] | |
| HNRNPA2B1 | Mediates mRNA splicing and miRNA processing | [44] | |
| ELAL1/HuR | Improve translation efficiency and stability of mRNA | [45] |
m6A RNA modification plays both a promoting and inhibitory role in CRC tumour differentiation, angiogenesis, metastasis, and immunity[47]. In addition, m6A modification, as an epigenetic regulator, can be used to detect disease progression in a variety of solid tumours, including CRC, gastric cancer, breast cancer, liver cancer, and other diseases[48]. Due to its biological function and molecular mechanism, m6A RNA is a promising target site for CRC therapy and is valuable for further research[32].
The specific process and role of m6A in the normal human body are shown in the following schematic diagram (Figure 1).
Numerous studies have confirmed that aberrant m6A modification of cancer-related genes is closely associated with cancer progression[49]. CRC, which is closely related to the gastrointestinal tract, is the third most common cancer in the world and the second leading cause of cancer-related death[50,51]. The focus of this review is on the associations of gut flora-mediated m6A changes in CRC patients with cancer development and progression. Hypoxia, immunity, and inflammation are also discussed as components of the TME.
The onset of CRC is characterised by an invasive cancer process that originates from epithelial cells and is intricately linked to the replacement of normal tissues within the intestinal wall by cancer cells[52]. The main aetiologies of CRC include benign adenomatous malignancy and inflammatory bowel disease (IBD). This section focuses on these two aetiologies to discuss their relationship with m6A modification.
Adenoma-related CRC: The developmental trajectory of adenoma-related CRC can be succinctly described as normal mucosa–adenoma–CRC[53]. Throughout this process, numerous RNA methylation modifications, including m6A, m1A, and N2-methylguanosine, actively participate in modulating the antitumour immune function of the patient. Their regulatory role involves either fostering or inhibiting the expression of corresponding genes, thereby influencing the occurrence of CRC[54].
To date, researchers have conducted targeted experimental studies on the expression levels and potential roles of certain m6A regulators in this sequence of changes. Pan et al[55], after contrasting the differential expression levels of METTL3 in normal, adenoma, and CRC tumour tissues, reported significant increases in m6A and METTL3 Levels in adenoma and CRC tissues compared with normal tissues. Moreover, they reported that the m6A content was greater in adenomas than in CRC tissues. This observation suggests that METTL3 may exhibit increased expression during the adenoma-CRC process, thereby promoting the onset of CRC. By comparing the differences in ALKBH5 Levels between tumour tissues and normal intestinal mucosal tissues in the colon adenocarcinoma (COAD) patient database, Yang et al[56]. reported decreased expression of ALKBH5 in tumour tissues. Building upon this finding, researchers established an in vivo CRC metastasis model in nude mice, revealing that mice in the ALKBH5 overexpression group presented fewer lung metastatic nodules. These findings indicate that ALKBH5 potentially exerts an inhibitory effect on tumour invasion and metastasis[56]. In a study on colon cancer, researchers identified a potential correlation between low ALKBH5 expression and high YTHDF1 expression. This correlation may contribute to the transformation of cold tumours into hot tumours during the adenomatous lesion-COAD process, thereby promoting the occurrence of CRC. These effects are mediated through the modulation of the patient's immune environment, affecting the expression levels of immune cells such as CD4+ T cells, CD8+ T cells, NK cells, dendritic cells, and macrophages[57].
While some researchers have conducted experiments on certain m6A regulators, the existing reports remain quite limited. Nevertheless, data analysis allows researchers to discern differences in the expression levels of common m6A regulators between tumour and nontumor tissues in patients with COAD. Ji et al[58] team performed an analysis on data from The Cancer Genome Atlas (TCGA) database and revealed that regulators such as METTL3, WTAP, METTL16, VIRMA, RBM15, YTHDC1, YTHDF1/2, IGF2BP1/2/3, and FTO were expressed at relatively high levels in COAD tumour tissues, whereas ALKBH5 was expressed at relatively low levels. However, there was no significant difference in the expression of METTL14 and YTHDC2/3. Kuai et al[59] also utilized the TCGA database and reported significant differential expression of regulatory genes associated with YTHDF1, IGF2BP1/3, and EIFB3 in both adenomas and CRC tissues compared with normal tissues (P < 0.0001). Interestingly, the mRNA expression levels governed by these four related genes were found to be significantly upregulated only in CRC, with no apparent difference in adenomas. In a comprehensive analysis of data from the TCGA, Gene Expression Atlas, and Human Protein Atlas databases, Liu et al[60] reported that, in contrast with normal tissues, CRC tumour tissues presented lower expression levels of METTL14. Notably, WTAP, METTL16, HNPNPC, and YTHDC1 are highly expressed in COAD but not in rectal adenocarcinoma[60]. In summary, the majority of m6A regulators play crucial roles in the normal mucosa-adenoma-CRC process.
In summary, during the process of normal mucosa evolving into adenoma and further progressing to CRC, the majority of associated m6A regulatory factors exhibit expression patterns in adenoma and CRC tissues that differ from those in normal tissues. Despite initial advancements in understanding the role of m6A modification in the pathological development of CRC, further targeted experiments are necessary to confirm the specific roles played by these m6A regulatory factors in this intricate process.
IBD-related CRC: IBD, which includes mainly ulcerative colitis (UC) and Crohn's disease (CD), is another major cause of CRC, and it also plays an important role in the development of CRC and influences the level of m6A modification in the host[19]. m6A methylation is closely associated with the development of IBD and the transition from IBD to CRC through the regulation of RNA metabolism and the immune cells in intestinal mucosal immunity[19]. Some researchers have analysed the interaction network between m6A genes and IBD risk genes and reported that there is a significant interaction between them, which also reaffirms that m6A methylation plays a broad regulatory role in the occurrence and development of IBD from the perspective of data analysis[26].
To date, some m6A regulatory factors have been observed in experimental studies to have clear disease-promoting or -inhibiting effects on the pathogenesis of IBD. Fang et al[61] reported that METTL3 is highly expressed in the nuclei of intestinal epithelial cells from IBD patients, which is consistent with the oncogenic effect of METTL3 during the progression of CRC. Lu et al[62] utilized colitis experiments in a mouse model and reported that the absence of METTL14 in T cells led to spontaneous colitis, resulting in features such as increased inflammatory cell infiltration and increased Th1 and Th17 cytokine levels in mice, which is consistent with the oncogenic role of METTL14 during CRC progression. Similarly, Zhang et al[63] verified that the absence of METTL14 Led to severe colitis and suggested that m6A modification could be a potential therapeutic target for IBD. Motawi et al[64] compared different m6A regulators among UC patients, CD patients and healthy volunteers and reported that METTL3 was more frequently expressed in CD patients and had good diagnostic accuracy, whereas the expression of WTAP demonstrated good discrimination between UC patients and CD patients.
Some researchers have also analysed data to compare the differences in m6A modification levels between IBD patients and the healthy controls. For example, Chen et al[65] reported that the expression of IGF2BP1 and IGF2BP2 was lower in CD tissues than in normal tissues and that the expression of IGF2BP2 was similarly lower in UC tissues than in normal tissues.
Taken together, the results of the above experimental studies and data analyses suggest that the expression levels of multiple m6A regulators in the organisms of IBD patients differ significantly from those of healthy controls and that this difference in turn correlates with the process of the transition from IBD to CRC. In addition, dysregulation of the intestinal flora is considered one of the main reasons for the disruption of the immune response in the intestines of IBD patients and is worth exploring more deeply as a link in the pathogenesis of IBD-CRC[66].
As a pivotal factor in the metabolic microenvironment, the intestinal flora plays a crucial role in the onset and progression of CRC; not only does the intestinal flora modulate CRC through bacterial surface receptors and the secretion of metabolites, but more significantly, it also engages in reciprocal interactions with m6A modifications through mechanisms such as gene expression programs[67]. This section primarily elaborates on the process by which the intestinal flora influences the progression of CRC by impacting m6A modification levels.
Currently, many studies have unequivocally demonstrated the intimate relationships between several intestinal flora, such as Fusobacterium nucleatum (F. nucleatum), enterotoxigenic Bacteroides fragilis (B. fragilis), and Escherichia coli, and the onset and progression of CRC. Additionally, other intestinal bacteria, including Enterococcus faecalis and Salmonella enterica, participate in mechanisms that promote CRC cell proliferation. Beneficial bacteria, such as Lactobacillus acidophilus, next-generation probiotics, such as Akkermansia muciniphila, and other protective bacteria, exert inhibitory effects against CRC. Several typical gut bacteria that have been described to promote or inhibit CRC development and progression are listed in Table 2[68-80].
| Gut microbiota | Roles | Functions | Mechanism | Ref. |
| Coriobacteriaceae | Promote | Tumorigenesis, progression↑ | CPT1A-ERK axis | [68] |
| Clostridioides difficile | Promote | Tumorigenesis↑ | Secrete toxin TcdB | [69] |
| F. nucleatum | Promote | Tumorigenesis, metastasis↑ | modulate TME | [70,71] |
| ETBF | Promote | Progression↑ | Suppress the immune responses | [72] |
| Escherichia coli | Promote | Proliferation, progression↑ | Exert toxic effects on DNA | [73] |
| Enterococcus faecalis | Promote | Tumorigenesis, progression↑ | Secrete metabolite biliverdin | [74] |
| Salmonella enterica | Promote | Tumorigenesis, proliferation↑ | Express secretory protein AvrA | [75] |
| Streptococcus bovis/gallolyticus | Promote | Tumorigenesis, progression↑ | Promote inflammatory response | [76] |
| Blautia producta | Suppress | Tumorigenesis, progression↓ | Facilitate the immune surveillance | [77] |
| Lactobacillus acidophilus, Lactobacillus rhamnosus, and Lactobacillus casei | Suppress | Migration, invasion↓ | Reduce abnormal crypt foci | [78] |
| Roseburia intestinalis | Suppress | Tumorigenesis, proliferation↓ | Produce butyrate and induce CD8+ T cells | [79] |
| Akkermansia muciniphila | Suppress | Proliferation↓ | Modulate CD8+ T cells | [80] |
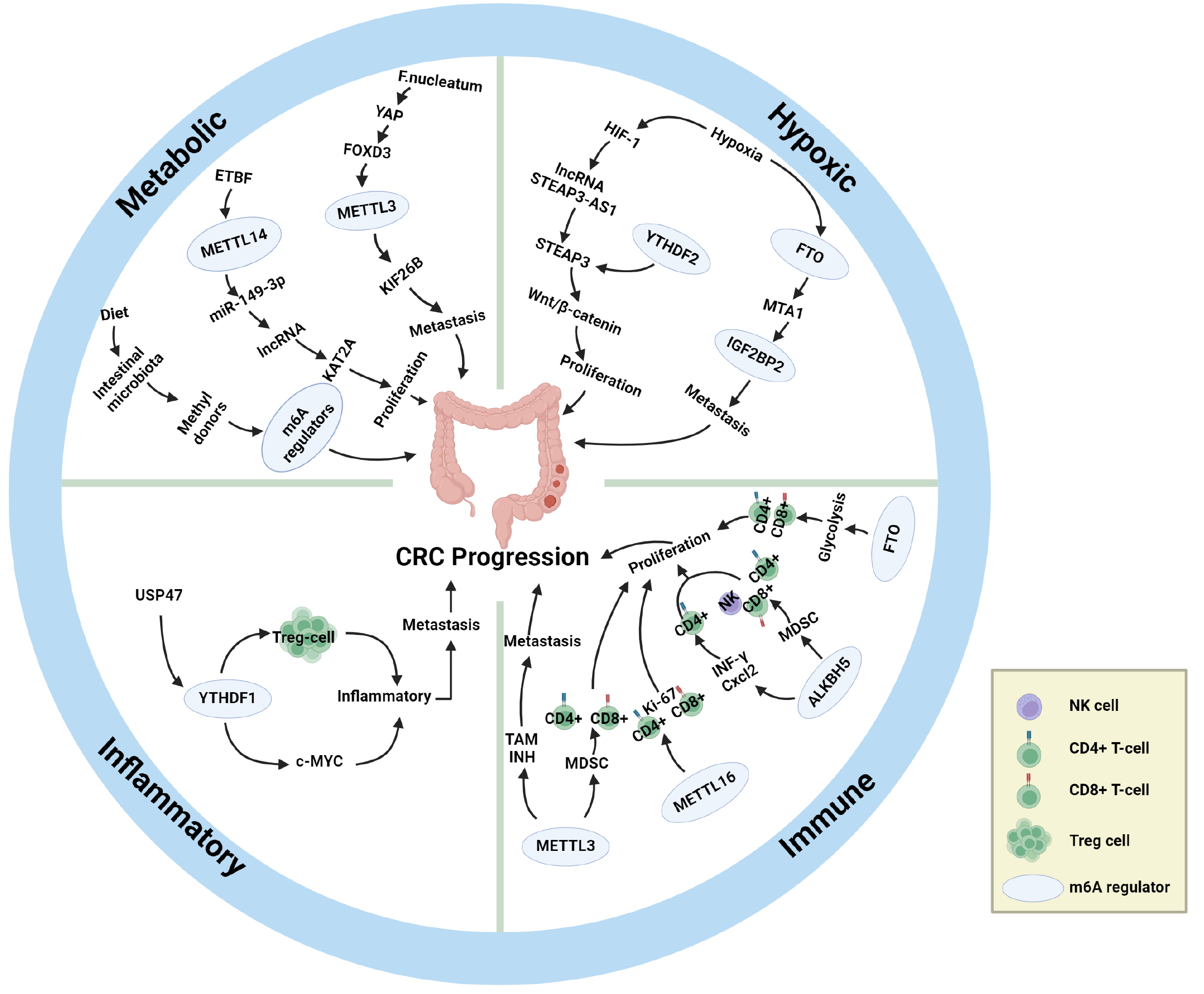
In the process of exerting the aforementioned physiological effects, the most crucial mechanism by which the intestinal flora influences the m6A modification level is as an upstream regulator of m6A modification. Through various pathways, the intestinal flora modulates the m6A modification level, thereby either promoting or inhibiting CRC. One of the more commonly observed regulatory pathways is the methyl donor pathway. Under normal circumstances, the gut microbiota participates in the generation and metabolism of numerous methyl donors, including SAM, MET, betaine, choline, and folate. Disruption of the gut microbiota due to factors such as dietary changes alters the supply of methyl donors. SAM, the most crucial methyl donor, interacts with the writers METTL3 and METTL16, and changes in SAM levels directly impact their activity. Although METT14 Lacks methyltransferase activity, it is indirectly influenced by gut microbiota regulation, as it typically forms an MTC with METTL3 to exert methylation effects. Research has confirmed that the suppression of METTL3 in CRC cells results in the inhibition of CRC. Mechanistically, METTL3 inhibits antitumour immunity and promotes CRC progression through the m6A-BHLHE41-CXCL1/CXCR2 axis[81]. One study demonstrated that METTL3 also increases the levels of m6A-modified HK2 and SLC2A1 via IGF2BP2/3, thereby promoting the progression of glycolysis and tumorigenesis in CRC[82]. There are limited findings regarding METTL16, and the current research suggests that METTL16 is highly expressed in CRC patients and that mechanistically, METTL16 can increase SOGA1 Levels through binding to IGF2BP1, with a consequent upregulation of PDK4, which promotes glycolytic me
In addition to the methyl donor pathway, some experiments have revealed the ability of the gut flora to alter CRC cell proliferation by acting directly on m6A and thereby altering CRC cell proliferation. For example, METTL3 promotes CRC progression by increasing the m6A level of CCNE1, whereas butyrate, a metabolite of the gut microbiota, reduces the expression of METTL3 and thus inhibits CRC[25]. However, the role of METTL3 in CRC remains controversial. Another study demonstrated that F. nucleatum downregulates METTL3 levels via the YAP/FOXD3 axis and that downregulated METTL3 promotes CRC metastasis by increasing KIF26B expression through a reduction in m6A levels[24]. However, a recent study demonstrated that although F. nucleatum is also enriched in CRC patients, it increases the level of METTL3 and exerts its oncogenic effects through the c-Myc pathway[85]. These reports demonstrate that the mechanism by which F. nucleatum regulates METTL3 and the corresponding m6A levels is contradictory and remains to be clarified by further studies. Mouse experiments have shown that m6A levels of METTL16 and its target mRNA Mat2a are downregulated under gut microbiota-deficient conditions[86], whereas METTL16 overexpression promotes the proliferation of CRC cells[87]. In addition, ETBF can directly promote CRC cell proliferation through METTL14-mediated m6A downregulation of miR-149-3p[88].
In summary, the level of METTL3 and the corresponding m6A content in CRC cells are still controversial, and the prevailing view is that the level of METTL3 and the corresponding m6A content are increased, but a few other studies have suggested that the level of METTL3 and the corresponding m6A content are decreased. This discrepancy may not account for bias in individual experimental cases, such as differences in experimental methodology or limitations in the selection of assay targets. More experiments of greater consistency may be necessary to support a clear conclusion and allow for further exploration of the possibility of additional regulatory mechanisms. Furthermore, available studies have confirmed that METTL16 Levels and corresponding m6A levels are elevated in CRC patients. However, METTL14 Levels and corresponding m6A levels were significantly decreased. Studies related to the regulation of m6A by the gut microbiota in the progression of CRC are beginning to progress, but more experimental data are still needed to support these findings.
Direct associations between the gut microbiota and erasers have rarely been reported, though it has been reported that FTO-deficient mice have increased levels of Helicobacter, Lactobacillus, and Porphyromonas in the intestinal tract[89]. Other upstream mechanisms have also been relatively infrequently studied, and the effects of ALKBH5 and FTO on CRC and the corresponding m6A levels are still under debate. On the one hand, some studies have indicated that high ALKBH5 expression is closely associated with poor prognosis in CRC patients and that ALKBH5 promotes CRC by binding to the downstream target AXIN2 to reduce m6A levels and subsequently suppressing the immune system through the AXIN2/Wnt/DKK1 axis[90]. On the other hand, it has also been observed that downregulation of ALKBH5 predicts a poor prognosis for CRC patients, and mechanistically ALKBH5 inhibits CRC by decreasing PHF20 m6A modification[91]. Studies on FTO have reported the two distinct findings of both promoting and inhibiting CRC progression[92-94]. After comparing the research methods and targets of the related papers, we hypothesise that ALKBH5 and FTO do not act directly on CRC tissues or cells but instead act indirectly on CRC tissues or cells by altering the m6A levels of various downstream targets, which may explain the conflicting results. In addition, studies have reported that the combination of ALKBH5 and FTO inhibits glycolysis and CRC cell proliferation and that a decrease in ALKBH5/FTO levels increases METTL3 levels while decreasing METTL14, which leads to accelerated CRC progression[95].
The mechanism by which the gut microbiota directly regulates readers has also not been reported; however, readers such as IGF2BP2 influence CRC progression via other upstream mechanisms. circEZH2/miR-133b first upregulates the level of IGF2BP2, which enhances the stability of CREB1 mRNA via IGF2BP2-mediated m6A modification, thereby promoting CRC progression[96]. YTHDF1 can inhibit antitumour immunity through the m6A-p65-CXCL1/CXCR2 axis, thereby promoting CRC progression[97]. KRT17 first degrades YTHDF2, and then YTHDF2 enhances T-cell infiltration by downregulating the level of m6A modification of CXCL10, preventing immune escape, and ultimately inhibiting CRC[98]. The lncRNA H19 binds to HNRNPA2B1 and promotes CRC metastasis by promoting epithelial-to-mesenchymal transition through the m6A/Raf-1 mRNA/Raf-ERK axis[99]. Research on the associations of the remaining readers with CRC is ongoing.
In conclusion, the gut microbiota, as an important upstream mechanism regulating m6A modification, can alter the expression levels of m6A-related enzymes and thus promote or inhibit the progression of CRC in various ways. We summarize existing upstream mechanisms regulating m6A modification levels in CRC patients and their roles in CRC progression in Table 3[100-111] (which mainly include the gut flora), and the basic process by which the intestinal flora affects m6A modification in CRC patients is shown in Figure 2 and Table 3.
| m6A regulator | Upstream mechanism | Roles | Functions | Mechanism | Ref. |
| METTL3 | Gut microbiota | Oncogene | Glycolysis, chemo-resistance↑ | METTL3↑/LDHA↑ | [100] |
| METTL3 | Gut microbiota | Oncogene | Proliferation, migration, invasion↑ | METTL3↑/m6A↑/YTHDF2↑/CRB3↓/Hippo↓ | [55] |
| METTL3 | Gut microbiota | Oncogene | Glycolysis, progression↑ | METTL3↑/m6A/GLUT1↑/mTORC1 ↑ | [101] |
| METTL3 | Gut microbiota | Oncogene | Invasion, migration↑ | METTL3↑/m6A/circ1662↑/YAP1↑/SMAD3↓ | [102] |
| METTL3 | F. nucleatum | Suppressor | Metastasis↑ | METTL3↓/m6A↓/YTHDF2/KIF26B↑ | [24] |
| METTL16 | Gut microbiota | Oncogene | Glycolysis, progression↑ | METTL14↑/IGF2BP1/SOGA1↑/PDK4↑ | [83] |
| METTL14 | ETBF | Suppressor | Proliferation↑ | METTL14↓/m6A/miR-149-3p↓/PHF5A/KAT2A | [88] |
| METTL14 | Gut microbiota | Suppressor | Proliferation, metastasis↑ | METTL14↓/m6A↓/YTHDF2↓/lncRNA XIST↑ | [103] |
| METTL14 | Gut microbiota | Suppressor | Migration, invasion, metastasis↑ | METTL14↓/m6A↓/YTHDF2/SOX4↑ | [104] |
| ALKBH5 | H3K27 | Oncogene | Glycolysis, progression↓ | ALKBH5↓/JMJD8↓/PKM2↓ | [105] |
| ALKBH5 | Oncogene | Proliferation, migration, invasion↑ | ALKBH5↑/YTHDF2/RAB5A↑ | [106] | |
| ALKBH5 | Suppressor | Radiosensitivity↑ | ALKBH5↑/YTHDF2/circAFF2↑/Cullin-NEDD8↓ | [107] | |
| ALKBH5 | Suppressor | Proliferation, migration, invasion↓ | ALKBH5↑/PHF20↓ | [91] | |
| FTO | Oncogene | Chemo-resistance↑ | FTO↑/YTHDF2/SIVA1↓ | [92] | |
| FTO | miR-96 | Oncogene | Tumorigenesis, progression↑ | AMPKα2↓/FTO↑/m6A↓/MYC↑ | [93] |
| IGF2BP2 | LINC00460 | Oncogene | Proliferation, metastasis↑ | IGF2BP2-DHX9↑/HMGA1↑ | [108] |
| YTHDF1 | Oncogene | Tumorigenesis, metastasis↑ | YTHDF1↑/m6A/ARHGEF2↑ | [109] | |
| YTHDF2 | miR-6125 | Oncogene | Proliferation, growth↓ | YTHDF2↓/m6A/GSK3β↑ | [110] |
| HNRNPA2B1 | MIR100HG | Oncogene | Chemo-resistance, metastasis↑ | hnRNPA2B1↑/m6A/TCF7L2↑ | [111] |
Hypoxia: In addition to the gut microbiota, hypoxia is also closely related to the occurrence, progression, and prognosis of CRC[112]. Hypoxia is the most common component of the TME and affects a series of biological behaviours, such as genetic instability, proliferation, differentiation, metastasis, invasion, metabolism, apoptosis, and other biological behaviours of tumour cells, such as in CRC[113,114]. Moreover, hypoxia can promote neovascularization, i.e., tumour angiogenesis, by activating hypoxia-inducible factor-1 (HIF-1)[115]. In addition, hypoxia can directly or indirectly affect the methylation modification of m6A[116], and m6A modification, in turn, can affect hypoxia and its inducible factors[100]. Further studies have shown that the pathway of hypoxia-regulated CRC progression is closely related to m6A regulatory factors[117], which supports the idea that hypoxia is also one of the upstream mechanisms of m6A regulation in CRC.
In a recent study that utilized controlled hypoxic conditions, activated HIF-1 induced high expression of the lncRNA STEAP3-AS1, and a large amount of STEAP3-AS1 further competed with the m6A reader YTHDF2 for binding to STEAP3 mRNA, which resulted in the protection of STEAP3 mRNA from YTHDF2-mediated degradation. Stabilized expression of STEAP3 in turn activates Wnt/β-catenin, which ultimately promotes CRC cell proliferation and invasion. In summary, hypoxia inhibits m6A-mediated mRNA degradation and plays an important role in the malignant progression and poor prognosis of CRC[118]. Another study demonstrated that hypoxia promotes ubiquitin-mediated degradation of the m6A eraser FTO, which results in lower FTO protein levels, and that low expression of FTO fails to inhibit m6A methylation of metastasis-associated protein 1 transcripts, which stabilizes mRNA expression by binding to IGF2BP2 and thereby contributes to the metastasis of CRC cells[94]. In summary, hypoxia modulates the effect of m6A on CRC and is positively associated with the malignant progression of CRC. In addition, hypoxia has an important effect on the abundance of the gut microbiota, and hypoxia can regulate the type and amount of the gut microbiota[119], which in turn may affect CRC progression through the pathways previously described.
Inflammatory response: Many clinical experiments and epidemiological studies have demonstrated that there is a complex association between inflammation and the development and progression of malignant tumours[120]. Chronic intestinal inflammation is closely associated with the development of IBD and the proliferation and metastasis of CRC[121], and IBD may increase the risk of CRC[122]. Inflammatory cells generated by long-term intestinal inflammation first activate the proinflammatory signalling pathway, which in turn releases cytokines and chemokines, ultimately forming an inflammatory microenvironment[123]. The increased methylation of m6A in the proinflammatory signalling pathway has been supported by studies[124], and it has also been shown that the expression of m6A and m6A regulatory factors may be increased in the inflammatory microenvironment[125]. In addition, some studies have summarized the relevant role of m6A in the development and progression of IBD[10], and other studies have analysed the interaction between m6A regulatory factors and IBD risk genes[26], which suggests a close relationship between m6A and IBD. In summary, intestinal inflammation also plays an important role in m6A regulation during CRC progression.
A study by Wang et al[126] revealed that under normal conditions, the deubiquitinase USP47 reduces the efficiency of YTHDF1-mediated c-Myc translation. Once USP47 is deficient, it leads to high c-Myc protein expression, as well as disruption of Treg cell metabolism, which in turn leads to inflammation and antitumour immunity, including anti-CRC. Wang et al[109] further showed that high expression of YTHDF1 exacerbates inflammation, which promotes the progression and metastasis of inflammatory CRC. In addition, Zhang et al[127] developed two isoform systems, m6AregCluster and m6AsigCluster, for assessing the association of m6A regulators with TMEs such as inflammation in CRC, which helps elucidate the mechanisms and principles involved.
Like hypoxia, intestinal inflammation also causes gut dysbiosis[128], and gut dysbiosis also regulates CRC progression through relevant pathways involving m6A. However, the difference is that gut dysbiosis also promotes intestinal inflammation[129].
Immune response: Immune factors, as important components of the TME, can cause immune tolerance and immunosuppression by regulating immune cells such as dendritic cells and T cells and their signalling pathways, which in turn contribute to the evasion of CRC cells from the host immune system and the proliferation and metastasis of CRC[19]. m6A modification decouples T-cell proliferation from cell survival by controlling the IL-7-mediated JAK-STAT signalling pathway and the TCR-mediated ERK/AKT signalling pathway[130]. Deletion of METTL3 results in decreased infiltration of CD206+ m2-like TAMs and increased infiltration of CD103+ cDC1s and results in tumour suppressor activity, suggesting that METTL3 has a role in driving TME immunosuppression[131]. Chen et al[81] reported that METTL3 drives myeloid-derived suppressor cell (MDSC) accumulation, inhibits the proliferation of immune cells, such as CD4+ T cells and CD8+ T cells, and promotes CRC proliferation, which supports the role of METTL3 in driving TME immunosuppression. In METTL16-overexpressing CRC tissues, a significant decrease in the level of the proliferative biomarker Ki-67 was detected, which was accompanied by an increase in the infiltration levels of CD4+ T cells and CD8+ T cells and increased antitumour activity[132]. Similar conclusions have been reached in studies concerning changes in m6A writers in the breast cancer TME. For example, the expression of METTL14 and ZC3H13 was significantly positively correlated with CD4+ T cells, CD8+ T cells, dendritic cells, macrophages, and neutrophils in breast cancer tissues and exhibited some antitumour activity[133].
With respect to m6A erasers and altered immune responses in the TME, the lack of ALKBH5 increases the m6A modification of interferon-γ (IFN-γ) and C-X-C motif chemokine ligand 2 mRNAs, which decreases the stability of mRNAs in CD4+ T cells and increases their expression[134]. In a humanized mouse model, the knockdown of ALKBH5 was able to reduce the MDSC content, promote increased levels of NK cells, CD4+ T cells, and CD8+ T-cell infiltration, and promote tumour suppressor activity[90]. FTO evades host immune surveillance by regulating glycolytic processes in tumour tissues. Knockdown of FTO impairs tumour cell glycolytic activity, enhances the degree of CD4+ T-cell and CD8+ T-cell infiltration, and restores the immune function of CD8+ T cells (elevated levels of IFN-γ and granzyme B), thereby inhibiting tumour proliferation[135].
To date, findings regarding the associations of m6A readers with changes in immune responses in the TME have not been reported. In knockout experiments, the expression of the vast majority of m6A regulators was positively correlated with the infiltration levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells in CRC tissues. High expression of FMR1, LRPPRC, METTL14, RBMX, YTHDC2, YTHDF2, and YTHDF3 in CRC patients is associated with a poor prognosis (Figure 3).
Currently, various types of m6A regulatory factors used as biomarkers of CRC have demonstrated great clinical value in their diagnosis and prognostic assessment, and great potential exists in their use as therapeutic targets for CRC. This section focuses on summarizing the progress of m6A in the diagnosis and prognostic assessment of CRC and discusses the possibility of m6A regulatory factors as therapeutic targets for CRC using leukaemia and other malignant cancers as examples.
On the basis of the increasing number of studies showing that aberrant m6A regulatory factors are closely associated with CRC progression, m6A regulatory factors are expected to become biomarkers of CRC and play important roles in the diagnosis and prognosis of CRC[136]. For example, many studies have reported high expression of METTL3 in CRC cells or patients and suggested a strong association with poor CRC prognosis[137,138], although a small number of studies reached the opposite conclusion[24]. Some reports have suggested that, considering the heterogeneity of METTL3, more easily detectable downstream target RNAs could be selected as new biomarkers[136], which may require more experimental data support. The results of these limited studies support that METTL16 is elevated in CRC patients[83,87]. In addition, relevant experiments have demonstrated that METTL14 Levels are downregulated in CRC patients and suggest that METTL14 Levels are negatively correlated with CRC progression[88,103]. However, studies on the erasers ALKBH5 and FTO are contradictory, and the levels of ALKBH5 and FTO fluctuate high and low in CRC tissues, making them currently unsuitable as biomarkers. Notably, the existing studies showing how readers such as IGF2BP2 and YTHDF1 affect the progression of CRC are promising. In the foreseeable future, as research clarifies how m6A regulators are expressed in CRC, the next step will be to explore how m6A regulators can be used in the clinical diagnosis of CRC and assessment of prognosis.
Among the many studies on the effects of m6A regulatory factors on CRC progression, m6A regulatory factors and related pathways are promising therapeutic targets. In addition, several studies have investigated the effects of m6A regulatory factors on chemotherapeutic resistance and sensitivity to radiotherapy, as well as synergistic effects with PD-1/PD-L1 inhibitors for the treatment of CRC. Therefore, the development of corresponding agonists or inhibitors of m6A regulatory factors seems to be a promising therapeutic strategy[139]. For example, METTL3 inhibitors are already in use, and METTL16 inhibitors and METTL14 agonists are envisioned. The roles of ALKBH5 and FTO in CRC progression are not yet clear, and whether they can be used as therapeutic targets still awaits follow-up studies. Therapeutic strategies targeting readers such as IGF2BP2 and YTHDF1, on the other hand, seem to be promising for future consideration.
Dysregulation of m6A modification occurs to varying degrees in patients with various types of cancer. m6A RNA modification plays a role as tumour promoters or tumour suppressors in vivo. On this basis, it is possible to consider targeting different m6A writers, erasers, or readers by designing corresponding inhibitors or agonists and combining them with other therapies to enhance tumour immunity and improve clinical benefits. Although practical reports on the application of relevant drugs in CRC treatment are lacking, many drugs (e.g., FTO inhibitors) have been widely investigated for the treatment of malignant diseases, such as AML, to improve the clinical benefit (Table 4).
| Drug | Role in cancer | Cancer type | Target | IC50 | Mechanism | Ref. |
| STM2457 | Tumour inhibitor | AML | METTL3 | 16.9 nM | m6A↓/HOXA1018↓/MYC19↓ | [142] |
| UZH1a | Tumour inhibitor | AML | METTL3 | 4.6 μM | Inhibits METTL3 catalytic activity and decreases m6A level and mRNA transcription level | [143] |
| CS1 | Tumour inhibitor | AML | FTO | m6A↑/LILRB4↓/MYC↓/CEBPA↓/RARA↑/ASB2↑ | [145] | |
| FB23 | Tumour inhibitor | AML | FTO | 0.8-1.5 μM | m6A↑/MYC↓/CEBPA↓/RARA↑/ASB2↑ | [148] |
| MA | Tumour inhibitor | AML | FTO | 17.4 μM | Binds to FTO and inhibits demethylation | [147] |
| R-2HG | Tumour inhibitor | AML | FTO | m6A↑/FTO↓/MYC↓CEBPA↓ | [146] | |
| Rhein | Tumour inhibitor | Liver cancer | FTO/ALKBH5 | 30 mM | Competitive binding of FTO to substrate binding sites and increased m6A levels | [149] |
| 2-[(1-hydroxy-2-oxo-2-phenylethyl) sulfanyl] acetic acid | Tumour inhibitor | AML | ALKBH5 | 0.84 μM | Binds ALKBH5 and decreases m6A levels | [150] |
| 4-{[(furan-2-yl) methyl]amino}-1,2-diazinane-3,6-dione | Tumour inhibitor | AML | ALKBH5 | 1.79 μM | Binds ALKBH5 and decreases m6A levels | [150] |
The targeted drug STM2457 specifically inhibits METTL3, reduces the m6A levels of METTL3-dependent core leukaemia m6A substrates (including HOXA1018 and MYC19), and decreases the protein translation levels of BRD4 and SP1. Ultimately, STM2457 inhibits AML and has almost negligible toxic effects[140]. Another drug that targets METTL3, UZH1a, exerts inhibitory effects on METTL3 activity by binding to its SAM site to reduce m6A levels and inhibit mRNA transcription[141].
In addition to m6A erasers, several targeted drugs with applications in other types of cancers that could be used in attempts to treat CRC have emerged[142]. Most of these drugs are FTO inhibitors. CS1, a selective FTO inhibitor, has shown potent antileukaemic efficacy by blocking the binding of FTO to MYC, CEBPA, and RARA[143]. R-2-Hydroxyglutaric acid, a tumour suppressor drug that directly inhibits FTO, targets the FTO/m6A/MYC/CEBPA signalling pathway, increases m6A levels and has antitumour effects[144]. Meclofenamic acid is a highly selective inhibitor of FTO that is able to bind FTO and inhibit its demethylation, thus exerting anticancer effects[145]. The mechanism of action of FB23 is similar to that of CS1, which is able to negatively regulate the expression of ASB2, RARA, MYC, and CEBPA and upregulate the level of m6A in AML cells, exerting antitumour effects[146]. In Zhao et al's review[147] of the therapeutic potential for liver disease, Rhein was proposed as a promising drug for the treatment of hepatocellular carcinoma, which competitively binds to the substrate-binding site of FTO and increases the m6A level, thus exerting antitumour effects. To date, few targeted therapeutic agents against ALKBH5 have been developed. 2-[(1-Hydroxy-2-oxo-2-phenylethyl) sulfanyl] acetic acid and 4-{[(furan-2-yl) methyl] amino}-1,2-diazinane-3,6-dione are two unnamed tentative ALKBH5 inhibitors that bind to ALKBH5 and inhibit its activity, reducing m6A levels and exhibiting antiproliferative activity in AML cell lines[148].
There is a lack of research reports on targeted therapeutic agents against m6A readers for cancer treatment, but among them, YTHDF1 and FMR1 are also expected to be new targets in CRC m6A-targeted therapies. In Transwell experiments, after the YTHDF1 gene was knocked down in CRC cells derived from CRC patients, the growth of HCT116 and HT-29 cells in vitro was inhibited, which significantly reduced the ability of CRC cells to migrate and invade in vitro[109]. The upregulation of FMR1 increased EGFR mRNA expression and activated the ERBB signalling pathway in CRC cells, promoting cancer cell proliferation and metastasis[149]. These findings suggest that m6A readers can also be used as targets for CRC therapy and have potential for targeted therapy.
On the other hand, the distribution and metabolic profile of the gut microbiota also affect the effect of tumour immunity in the body. As one of the important upstream mechanisms of m6A, the gut microbiota also has great potential as a biomarker and therapeutic target for CRC. There have been literature reviews on the research progress of gut microbiota biomarkers in the early diagnosis of CRC, especially F. nucleatum[128], and the application of gut microbiota therapies in the prevention and treatment of CRC, especially via faecal microbiota transplantation[150]. In addition, gut microbiota metabolites have a potential role in the diagnosis of CRC[151,152]. A study on the influence of the gut microbiota on m6A modification revealed that the presence of large amounts of butyrate, a metabolite of intestinal microorganisms, can reduce the level of METTL3 and the expression of cyclin E1, which in turn reduces the level of m6A in CRC cells and inhibits their proliferation and metastasis[25]. Some researchers have noted that killing B. fragilis and bacteria associated with mucin degradation, inflammation, and DNA methylation via antibiotics can reverse the dysregulated intestinal microecology in patients and inhibit CRC progression[153]. Notably, if antibiotic therapy disrupts the original intestinal ecological balance, it can instead cause an intestinal inflammatory response and exacerbate the development of intestinal tumours, causing disease progression[154]. In these cases, regulating the gut microbiota profile with drugs and combining it with m6A-targeted therapy may be a novel idea for the treatment of CRC, but antibiotics should also be used carefully to reduce the destruction of the original probiotic flora of the host to avoid disease exacerbation. In addition, the interaction between the host gut microbiota and m6A RNA modification is complex, and the mechanism is still not completely clear. The specific process of regulating the balance of the host gut microbiota to influence the level of host m6A modification and play an anticancer role as a possible therapeutic approach still needs more in-depth research (Table 4).
The morbidity and mortality of CRC in young patients have been gradually increasing in recent decades. m6A modification of RNA-mediated posttranscriptional regulation plays an important role in the development of CRC. In this process, TME factors such as metabolism, hypoxia, inflammation, and immunity are closely related to the progression of CRC, in which the intestinal flora plays an important role.
On the basis of the above findings, we propose that the intestinal flora can participate in the regulation of m6A regulators through the methyl donor pathway and other possible pathways, which in turn affects the progression of CRC through various pathways. In other words, the intestinal flora is one of the important upstream mechanisms by which m6A modification affects CRC progression.
Notably, few studies have directly demonstrated the involvement of the intestinal flora as an upstream mechanism of m6A in the process of CRC. In addition, most existing studies have focused on the regulation of writers, such as METTL3, by the gut flora to affect CRC, and there is a relative gap in studies on the associations between the gut flora and erasers and readers. The former still needs to be supported by more high-quality experimental data, and the latter needs new studies to fill this gap. Moreover, the relationship between METTL3 and CRC progression is still debated, although most experiments support the conclusion that METTL3 is an oncogenic factor. The promotion or inhibition of CRC progression by ALKBH5 and FTO appears to be contradictory. There are relatively few studies on the direct regulation of CRC by readers such as METTL16 and IGF2BP2; however, IGF2BP2 and YTHDF2 are widely involved in the mechanism of CRC regulation by writers and erasers, and METTL14 has essentially been proven to be an oncogenic factor.
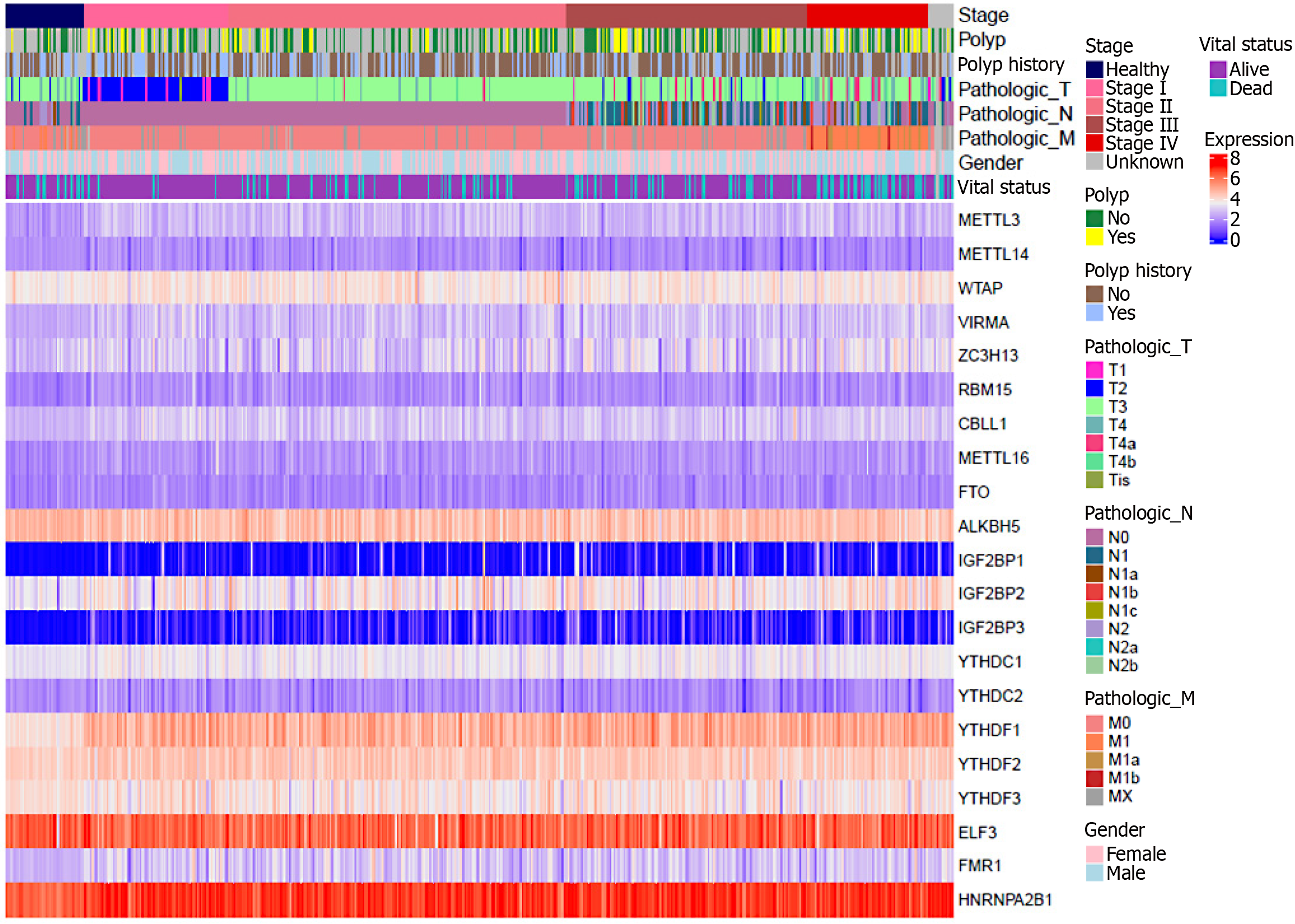
In order to verify the correctness of the above, we grouped and generated a heatmap based on the dataset of two molecular analysis-based articles on CRC[48,155], according to the gender of the patients in the samples, the origin of the tissues, the TNM classification, and the vital status. As can be seen from the heatmap below (Figure 4), compared with normal tissues: M6A regulators such as METTL3, WTAP, VIRMA, ZC3H13, CBLL1, METTL16, IGF2BP1/2/3, YTHDF1/2, FMR1, HNRNPA2B1, etc., showed different degrees of increase in the expression level in the tumor tissues and exhibited oncogenic effects. The expression levels of m6A regulators such as METTL14 and YTHDC1/2 decreased to different degrees in tumor tissues and exhibited cancer-suppressing effects. The relationship between the expression levels of m6A regulators such as ALKBH5, FTO, YTHDF3, ELF3, and RBM15 and the progression of CRC is still unclear, a situation similar to the results of earlier studies by other scholars. It may be related to the sampling method of the data samples, which deserves more experiments to explore and repeat the validation.
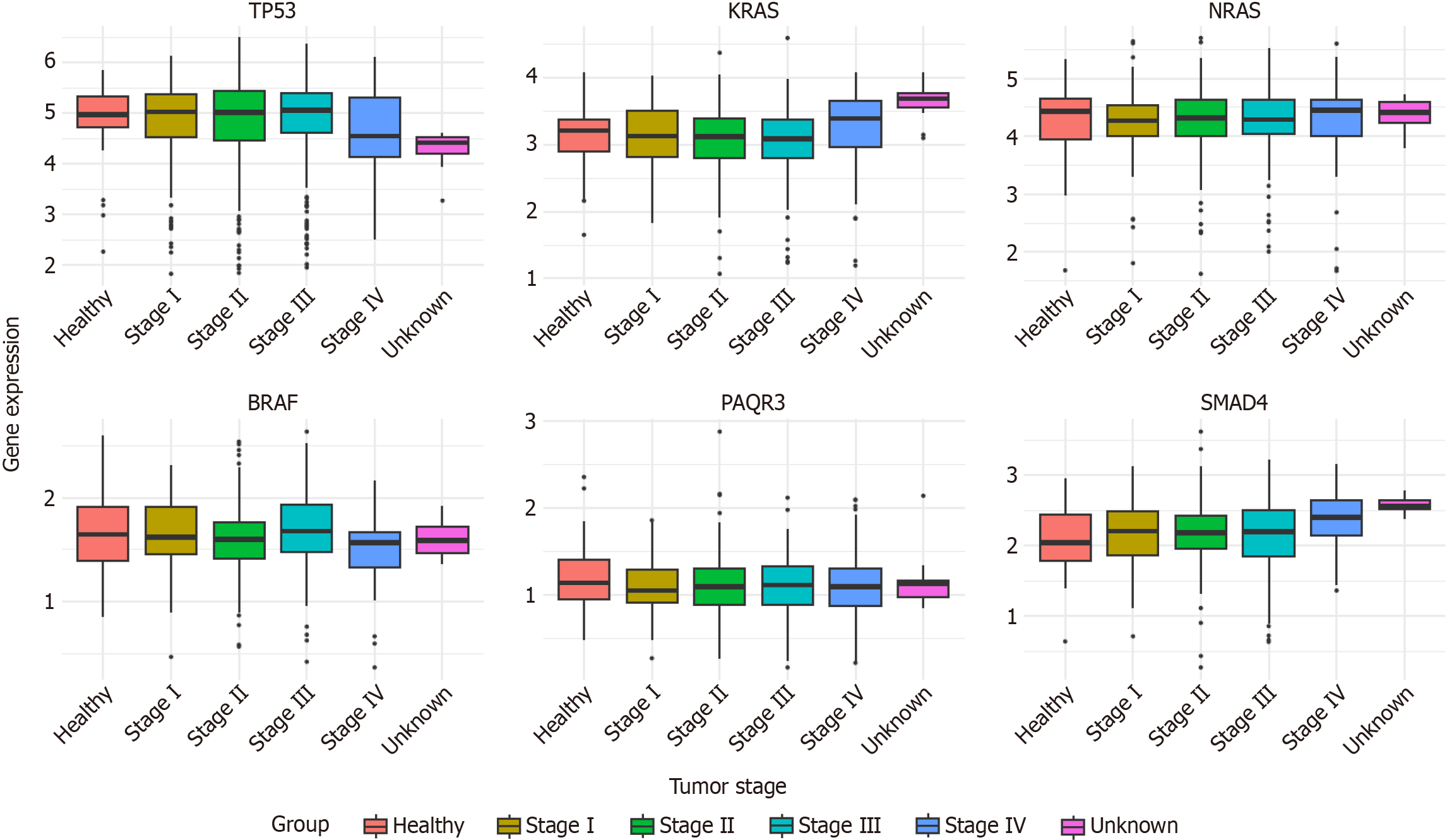
In addition, we selected some representative CRC-associated oncogenes/tumor suppressor genes and generated a box plot based on the RNA-Seq data of COAD and rectum adenocarcinoma from the TCGA database to analyze the differences in their expression levels during CRC progression. From this box plot (Figure 5), it can be seen that compared with normal tissues: The transcript levels of KRAS and SMAD4 increased to different degrees in tumor tissues and showed oncogenic effects; the transcript levels of TP53 and PAQR3 decreased to different degrees in tumor tissues and showed cancer-suppressing effects; the transcript levels of NRAS and BRAF did not increase significantly in tumor tissues, which may be due to the fact that different tissues were sampled at the time of sampling. The reason for this result may be due to a certain degree of mixing of cells from different tissue sources at the time of sampling, resulting in no significant difference in expression in the final data.
On the basis of these findings above, future research can first consider more experimental studies that focus on the influence of the intestinal flora on CRC progression by altering the expression levels of different m6A regulators. Second, research can continue to elucidate the relationship between other individual m6A regulators and CRC progression, resolve controversial issues and fill in gaps in related research, which may help to explain the mechanism by which multiple regulators act together in CRC. m6A regulators and the gut microbiota have shown great potential as diagnostic biomarkers and therapeutic targets for CRC, and we highlight some of the therapeutic advances in the former. For example, although m6A-targeted therapies have not yet been applied to the treatment of CRC, METTL3 inhibitors, FTO inhibitors, and ALKBH5 inhibitors have already shown good efficacy in the treatment of other malignant tumours, especially AML. However, many challenges remain. For example, the efficacy and risk of the existing m6A regulator inhibitors used in the clinic for CRC therapy are not yet known, and other inhibitors or agonists that target m6A regulators still need to be developed. In addition, the combination of m6A-targeted therapies with gut microbiota therapies, as well as influencing the level of m6A modifications in the host by regulating the balance of the host gut microbiota and exerting anticancer effects, is promising and may help to improve the treatment of CRC.
| 1. | GBD 2019 Colorectal Cancer Collaborators. Global, regional, and national burden of colorectal cancer and its risk factors, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Gastroenterol Hepatol. 2022;7:627-647. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 327] [Cited by in RCA: 451] [Article Influence: 112.8] [Reference Citation Analysis (1)] |
| 2. | Man J, Zhang T, Yin X, Chen H, Zhang Y, Zhang X, Chen J, Yang X, Lu M. Spatiotemporal Trends of Colorectal Cancer Mortality Due to Low Physical Activity and High Body Mass Index From 1990 to 2019: A Global, Regional and National Analysis. Front Med (Lausanne). 2021;8:800426. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 3. | Hardikar S, Newcomb PA, Campbell PT, Win AK, Lindor NM, Buchanan DD, Makar KW, Jenkins MA, Potter JD, Phipps AI. Prediagnostic Physical Activity and Colorectal Cancer Survival: Overall and Stratified by Tumor Characteristics. Cancer Epidemiol Biomarkers Prev. 2015;24:1130-1137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 4. | Li Y, Zhou D, Liu Q, Zhu W, Ye Z, He C. Gene Polymorphisms of m6A Erasers FTO and ALKBH1 Associated with Susceptibility to Gastric Cancer. Pharmgenomics Pers Med. 2022;15:547-559. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 5. | Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 520] [Cited by in RCA: 966] [Article Influence: 107.3] [Reference Citation Analysis (2)] |
| 6. | Silva DAS, Tremblay MS, Souza MFM, Mooney M, Naghavi M, Malta DC. Mortality and years of life lost by colorectal cancer attributable to physical inactivity in Brazil (1990-2015): Findings from the Global Burden of Disease Study. PLoS One. 2018;13:e0190943. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 7. | Wang X, Lu X, Wang P, Chen Q, Xiong L, Tang M, Hong C, Lin X, Shi K, Liang L, Lin J. SRSF9 promotes colorectal cancer progression via stabilizing DSN1 mRNA in an m6A-related manner. J Transl Med. 2022;20:198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 8. | Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother. 2019;112:108613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 631] [Article Influence: 90.1] [Reference Citation Analysis (0)] |
| 9. | Haruehanroengra P, Zheng YY, Zhou Y, Huang Y, Sheng J. RNA modifications and cancer. RNA Biol. 2020;17:1560-1575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 131] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 10. | Zhang J, Song B, Zeng Y, Xu C, Gao L, Guo Y, Liu J. m6A modification in inflammatory bowel disease provides new insights into clinical applications. Biomed Pharmacother. 2023;159:114298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 26] [Reference Citation Analysis (0)] |
| 11. | Liu Z, Gao L, Cheng L, Lv G, Sun B, Wang G, Tang Q. The roles of N6-methyladenosine and its target regulatory noncoding RNAs in tumors: classification, mechanisms, and potential therapeutic implications. Exp Mol Med. 2023;55:487-501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 65] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 12. | Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, Jin KX, Wang X, Huang CM, Fu Y, Ge XM, Song SH, Jeong HS, Yanagisawa H, Niu Y, Jia GF, Wu W, Tong WM, Okamoto A, He C, Rendtlew Danielsen JM, Wang XJ, Yang YG. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403-1419. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 656] [Cited by in RCA: 963] [Article Influence: 80.3] [Reference Citation Analysis (6)] |
| 13. | Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 5' UTR m(6)A Promotes Cap-Independent Translation. Cell. 2015;163:999-1010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1005] [Cited by in RCA: 1513] [Article Influence: 137.5] [Reference Citation Analysis (0)] |
| 14. | Lee Y, Choe J, Park OH, Kim YK. Molecular Mechanisms Driving mRNA Degradation by m(6)A Modification. Trends Genet. 2020;36:177-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 316] [Article Influence: 52.7] [Reference Citation Analysis (0)] |
| 15. | Qu F, Tsegay PS, Liu Y. N(6)-Methyladenosine, DNA Repair, and Genome Stability. Front Mol Biosci. 2021;8:645823. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 16. | Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, Carter AC, Flynn RA, Zhou C, Lim KS, Dedon P, Wernig M, Mullen AC, Xing Y, Giallourakis CC, Chang HY. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707-719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 758] [Cited by in RCA: 1029] [Article Influence: 85.8] [Reference Citation Analysis (0)] |
| 17. | Huang P, Liu M, Zhang J, Zhong X, Zhong C. The Potential Role of m(6)A in the Regulation of TBI-Induced BGA Dysfunction. Antioxidants (Basel). 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 18. | Ito-Kureha T, Leoni C, Borland K, Cantini G, Bataclan M, Metzger RN, Ammann G, Krug AB, Marsico A, Kaiser S, Canzar S, Feske S, Monticelli S, König J, Heissmeyer V. The function of Wtap in N(6)-adenosine methylation of mRNAs controls T cell receptor signaling and survival of T cells. Nat Immunol. 2022;23:1208-1221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 64] [Reference Citation Analysis (0)] |
| 19. | Xu X, Huang J, Ocansey DKW, Xia Y, Zhao Z, Xu Z, Yan Y, Zhang X, Mao F. The Emerging Clinical Application of m6A RNA Modification in Inflammatory Bowel Disease and Its Associated Colorectal Cancer. J Inflamm Res. 2021;14:3289-3306. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 20. | He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18:176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 359] [Cited by in RCA: 1020] [Article Influence: 145.7] [Reference Citation Analysis (0)] |
| 21. | Chen Y, Zheng X, Wu C. The Role of the Tumor Microenvironment and Treatment Strategies in Colorectal Cancer. Front Immunol. 2021;12:792691. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 76] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 22. | Kasprzak A. The Role of Tumor Microenvironment Cells in Colorectal Cancer (CRC) Cachexia. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 128] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 23. | Xia C, Cai Y, Ren S, Xia C. Role of microbes in colorectal cancer therapy: Cross-talk between the microbiome and tumor microenvironment. Front Pharmacol. 2022;13:1051330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 24. | Chen S, Zhang L, Li M, Zhang Y, Sun M, Wang L, Lin J, Cui Y, Chen Q, Jin C, Li X, Wang B, Chen H, Zhou T, Wang L, Hsu CH, Zhuo W. Fusobacterium nucleatum reduces METTL3-mediated m(6)A modification and contributes to colorectal cancer metastasis. Nat Commun. 2022;13:1248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 178] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 25. | Zhu W, Si Y, Xu J, Lin Y, Wang JZ, Cao M, Sun S, Ding Q, Zhu L, Wei JF. Methyltransferase like 3 promotes colorectal cancer proliferation by stabilizing CCNE1 mRNA in an m6A-dependent manner. J Cell Mol Med. 2020;24:3521-3533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 91] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 26. | Nie K, Yi J, Yang Y, Deng M, Yang Y, Wang T, Chen X, Zhang Z, Wang X. A Broad m6A Modification Landscape in Inflammatory Bowel Disease. Front Cell Dev Biol. 2021;9:782636. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (1)] |
| 27. | Cuevas-Sierra A, Ramos-Lopez O, Riezu-Boj JI, Milagro FI, Martinez JA. Diet, Gut Microbiota, and Obesity: Links with Host Genetics and Epigenetics and Potential Applications. Adv Nutr. 2019;10:S17-S30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 380] [Cited by in RCA: 340] [Article Influence: 48.6] [Reference Citation Analysis (0)] |
| 28. | Luo J, Yu J, Peng X. Could partial nonstarch polysaccharides ameliorate cancer by altering m(6)A RNA methylation in hosts through intestinal microbiota? Crit Rev Food Sci Nutr. 2022;62:8319-8334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 29. | Stiemsma LT, Nakamura RE, Nguyen JG, Michels KB. Does Consumption of Fermented Foods Modify the Human Gut Microbiota? J Nutr. 2020;150:1680-1692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 95] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 30. | Liu WW, Zhang ZY, Wang F, Wang H. Emerging roles of m6A RNA modification in cancer therapeutic resistance. Exp Hematol Oncol. 2023;12:21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 31] [Reference Citation Analysis (0)] |
| 31. | Flamand MN, Tegowski M, Meyer KD. The Proteins of mRNA Modification: Writers, Readers, and Erasers. Annu Rev Biochem. 2023;92:145-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 120] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 32. | Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, Yang C, Chen Y. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6:74. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 265] [Cited by in RCA: 1586] [Article Influence: 317.2] [Reference Citation Analysis (0)] |
| 33. | Wang T, Kong S, Tao M, Ju S. The potential role of RNA N6-methyladenosine in Cancer progression. Mol Cancer. 2020;19:88. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 218] [Cited by in RCA: 793] [Article Influence: 132.2] [Reference Citation Analysis (0)] |
| 34. | Song N, Cui K, Zhang K, Yang J, Liu J, Miao Z, Zhao F, Meng H, Chen L, Chen C, Li Y, Shao M, Zhang J, Wang H. The Role of m6A RNA Methylation in Cancer: Implication for Nature Products Anti-Cancer Research. Front Pharmacol. 2022;13:933332. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 35. | Sendinc E, Shi Y. RNA m6A methylation across the transcriptome. Mol Cell. 2023;83:428-441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 284] [Reference Citation Analysis (0)] |
| 36. | Shen D, Wang B, Gao Y, Zhao L, Bi Y, Zhang J, Wang N, Kang H, Pang J, Liu Y, Pang L, Chen ZS, Zheng YC, Liu HM. Detailed resume of RNA m(6)A demethylases. Acta Pharm Sin B. 2022;12:2193-2205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 60] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 37. | Zhu TY, Hong LL, Ling ZQ. Oncofetal protein IGF2BPs in human cancer: functions, mechanisms and therapeutic potential. Biomark Res. 2023;11:62. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 73] [Reference Citation Analysis (0)] |
| 38. | Ge X, Xue G, Ding Y, Li R, Hu K, Xu T, Sun M, Liao W, Zhao B, Wen C, Du J. The Loss of YTHDC1 in Gut Macrophages Exacerbates Inflammatory Bowel Disease. Adv Sci (Weinh). 2023;10:e2205620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 39. | Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, Qi M, Lu Z, Shi H, Wang J, Cheng Y, Luo G, Dai Q, Liu M, Guo X, Sha J, Shen B, He C. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115-1127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 657] [Cited by in RCA: 837] [Article Influence: 93.0] [Reference Citation Analysis (0)] |
| 40. | Sikorski V, Selberg S, Lalowski M, Karelson M, Kankuri E. The structure and function of YTHDF epitranscriptomic m(6)A readers. Trends Pharmacol Sci. 2023;44:335-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 67] [Reference Citation Analysis (0)] |
| 41. | Orouji E, Peitsch WK, Orouji A, Houben R, Utikal J. Oncogenic Role of an Epigenetic Reader of m(6)A RNA Modification: YTHDF1 in Merkel Cell Carcinoma. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 42. | Zhang G, Xu Y, Wang X, Zhu Y, Wang L, Zhang W, Wang Y, Gao Y, Wu X, Cheng Y, Sun Q, Chen D. Dynamic FMR1 granule phase switch instructed by m6A modification contributes to maternal RNA decay. Nat Commun. 2022;13:859. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 60] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 43. | Chen X, Yang HT, Zhang B, Phillips JW, Cheng D, Rigo F, Witte ON, Xing Y, Black DL. The RNA-binding proteins hnRNP H and F regulate splicing of a MYC-dependent HRAS exon in prostate cancer cells. Proc Natl Acad Sci U S A. 2023;120:e2220190120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 44. | Xu P, Yang J, Chen Z, Zhang X, Xia Y, Wang S, Wang W, Xu Z. N6-methyladenosine modification of CENPF mRNA facilitates gastric cancer metastasis via regulating FAK nuclear export. Cancer Commun (Lond). 2023;43:685-705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 31] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 45. | Li M, Yang L, Chan AKN, Pokharel SP, Liu Q, Mattson N, Xu X, Chang WH, Miyashita K, Singh P, Zhang L, Li M, Wu J, Wang J, Chen B, Chan LN, Lee J, Zhang XH, Rosen ST, Müschen M, Qi J, Chen J, Hiom K, Bishop AJR, Chen CW. Epigenetic Control of Translation Checkpoint and Tumor Progression via RUVBL1-EEF1A1 Axis. Adv Sci (Weinh). 2023;10:e2206584. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 46. | Zhang Q, Zhu Y, Cao X, Tan W, Yu J, Lu Y, Kang R, Wang X, Li E. The epigenetic regulatory mechanism of PIWI/piRNAs in human cancers. Mol Cancer. 2023;22:45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 50] [Reference Citation Analysis (0)] |
| 47. | Zhang N, Zuo Y, Peng Y, Zuo L. Function of N6-Methyladenosine Modification in Tumors. J Oncol. 2021;2021:6461552. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 62] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 48. | Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330-337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6773] [Cited by in RCA: 6858] [Article Influence: 489.9] [Reference Citation Analysis (10)] |
| 49. | Wang Y, Wang Y, Patel H, Chen J, Wang J, Chen ZS, Wang H. Epigenetic modification of m(6)A regulator proteins in cancer. Mol Cancer. 2023;22:102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 122] [Article Influence: 40.7] [Reference Citation Analysis (0)] |
| 50. | Yang W, Zou J, Li Y, Liu R, Yan Z, Chen S, Zhao X, Guo W, Huang M, Li W, Zhu X, Chen Z. Corrigendum: Longitudinal Circulating Tumor DNA Profiling in Metastatic Colorectal Cancer During Anti-EGFR Therapy. Front Oncol. 2022;12:903586. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 51. | Huang J, Xu P, Chen Y. A Retrospective Study from a Single Center to Identify Hematological Factors that Distinguish Between Patients with Colorectal Carcinoma and Colorectal Adenoma. Med Sci Monit. 2022;28:e936745. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 52. | Godlewski J, Kmiec Z. Colorectal Cancer Invasion and Atrophy of the Enteric Nervous System: Potential Feedback and Impact on Cancer Progression. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 53. | Herszényi L, Sipos F, Galamb O, Solymosi N, Hritz I, Miheller P, Berczi L, Molnár B, Tulassay Z. Matrix metalloproteinase-9 expression in the normal mucosa-adenoma-dysplasia-adenocarcinoma sequence of the colon. Pathol Oncol Res. 2008;14:31-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 54. | Zheng W, Wang M, Chai X, Pan F, Xu M, Wang Y, Lan L, Hu F, Zhang Z, Chen Z. Targeted metabolomics analysis of nucleosides and the identification of biomarkers for colorectal adenomas and colorectal cancer. Front Mol Biosci. 2023;10:1163089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 55. | Pan J, Liu F, Xiao X, Xu R, Dai L, Zhu M, Xu H, Xu Y, Zhao A, Zhou W, Dang Y, Ji G. METTL3 promotes colorectal carcinoma progression by regulating the m6A-CRB3-Hippo axis. J Exp Clin Cancer Res. 2022;41:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 77] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 56. | Yang P, Wang Q, Liu A, Zhu J, Feng J. ALKBH5 Holds Prognostic Values and Inhibits the Metastasis of Colon Cancer. Pathol Oncol Res. 2020;26:1615-1623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 69] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 57. | Yan G, An Y, Xu B, Wang N, Sun X, Sun M. Potential Impact of ALKBH5 and YTHDF1 on Tumor Immunity in Colon Adenocarcinoma. Front Oncol. 2021;11:670490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 58. | Ji J, Liu S, Liang Y, Zheng G. Comprehensive analysis of m6A regulators and relationship with tumor microenvironment, immunotherapy strategies in colorectal adenocarcinoma. BMC Genom Data. 2023;24:44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 59. | Kuai D, Zhu S, Shi H, Yang R, Liu T, Liu H, Min L, Zhang S. Aberrant expression of m(6)A mRNA methylation regulators in colorectal adenoma and adenocarcinoma. Life Sci. 2021;273:119258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 60. | Liu X, Liu L, Dong Z, Li J, Yu Y, Chen X, Ren F, Cui G, Sun R. Expression patterns and prognostic value of m(6)A-related genes in colorectal cancer. Am J Transl Res. 2019;11:3972-3991. [PubMed] |
| 61. | Fang M, Yao J, Zhang H, Sun J, Yin Y, Shi H, Jiang G, Shi X. Specific deletion of Mettl3 in IECs triggers the development of spontaneous colitis and dysbiosis of T lymphocytes in mice. Clin Exp Immunol. 2024;217:57-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 62. | Lu TX, Zheng Z, Zhang L, Sun HL, Bissonnette M, Huang H, He C. A New Model of Spontaneous Colitis in Mice Induced by Deletion of an RNA m(6)A Methyltransferase Component METTL14 in T Cells. Cell Mol Gastroenterol Hepatol. 2020;10:747-761. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 110] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 63. | Zhang T, Ding C, Chen H, Zhao J, Chen Z, Chen B, Mao K, Hao Y, Roulis M, Xu H, Kluger Y, Zou Q, Ye Y, Zhan M, Flavell RA, Li HB. m(6)A mRNA modification maintains colonic epithelial cell homeostasis via NF-κB-mediated antiapoptotic pathway. Sci Adv. 2022;8:eabl5723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 74] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 64. | Motawi TK, Shaker OG, Amr G, Senousy MA. RNA methylation machinery and m(6)A target genes as circulating biomarkers of ulcerative colitis and Crohn's disease: Correlation with disease activity, location, and inflammatory cytokines. Clin Chim Acta. 2024;561:119831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 65. | Chen Y, Lei J, He S. m(6)A Modification Mediates Mucosal Immune Microenvironment and Therapeutic Response in Inflammatory Bowel Disease. Front Cell Dev Biol. 2021;9:692160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 66. | Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 771] [Cited by in RCA: 918] [Article Influence: 65.6] [Reference Citation Analysis (0)] |
| 67. | Su H, Cheung H, Lau HC, Chen H, Zhang X, Qin N, Wang Y, Chan MTV, Wu WKK, Chen H. Crosstalk between gut microbiota and RNA N6-methyladenosine modification in cancer. FEMS Microbiol Rev. 2023;47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 68. | Tang Q, Huang H, Xu H, Xia H, Zhang C, Ye D, Bi F. Endogenous Coriobacteriaceae enriched by a high-fat diet promotes colorectal tumorigenesis through the CPT1A-ERK axis. NPJ Biofilms Microbiomes. 2024;10:5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 28] [Reference Citation Analysis (0)] |
| 69. | Zhu X, Xu P, Zhu R, Gao W, Yin W, Lan P, Zhu L, Jiao N. Multi-kingdom microbial signatures in excess body weight colorectal cancer based on global metagenomic analysis. Commun Biol. 2024;7:24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 70. | Tan X, Wang Y, Gong T. The interplay between oral microbiota, gut microbiota and systematic diseases. J Oral Microbiol. 2023;15:2213112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 70] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 71. | Rye MS, Garrett KL, Holt RA, Platell CF, McCoy MJ. Fusobacterium nucleatum and Bacteroides fragilis detection in colorectal tumours: Optimal target site and correlation with total bacterial load. PLoS One. 2022;17:e0262416. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 72. | Qi X, Liu Y, Hussein S, Choi G, Kimchi ET, Staveley-O'Carroll KF, Li G. The Species of Gut Bacteria Associated with Antitumor Immunity in Cancer Therapy. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 73. | Deng J, Yuan W, Tan Q, Wei X, Ma J. Non-absorbable antibiotic treatment inhibits colorectal cancer liver metastasis by modulating deoxycholic acid metabolism by intestinal microbes. J Cancer. 2022;13:764-774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 74. | Zhang L, Liu J, Deng M, Chen X, Jiang L, Zhang J, Tao L, Yu W, Qiu Y. Enterococcus faecalis promotes the progression of colorectal cancer via its metabolite: biliverdin. J Transl Med. 2023;21:72. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 43] [Reference Citation Analysis (0)] |
| 75. | Lawrence GW, Begley M, Cotter PD, Guinane CM. Potential Use of Biotherapeutic Bacteria to Target Colorectal Cancer-Associated Taxa. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 76. | Diakité MT, Diakité B, Koné A, Balam S, Fofana D, Diallo D, Kassogué Y, Traoré CB, Kamaté B, Ba D, Ly M, Ba M, Koné B, Maiga AI, Achenbach C, Holl J, Murphy R, Hou L, Maiga M. Relationships between gut microbiota, red meat consumption and colorectal cancer. J Carcinog Mutagen. 2022;13. [PubMed] |
| 77. | Zhang X, Yu D, Wu D, Gao X, Shao F, Zhao M, Wang J, Ma J, Wang W, Qin X, Chen Y, Xia P, Wang S. Tissue-resident Lachnospiraceae family bacteria protect against colorectal carcinogenesis by promoting tumor immune surveillance. Cell Host Microbe. 2023;31:418-432.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 176] [Reference Citation Analysis (0)] |
| 78. | Deng X, Yang J, Zhang Y, Chen X, Wang C, Suo H, Song J. An Update on the Pivotal Roles of Probiotics, Their Components, and Metabolites in Preventing Colon Cancer. Foods. 2023;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 79. | Kang X, Liu C, Ding Y, Ni Y, Ji F, Lau HCH, Jiang L, Sung JJ, Wong SH, Yu J. Roseburia intestinalis generated butyrate boosts anti-PD-1 efficacy in colorectal cancer by activating cytotoxic CD8(+) T cells. Gut. 2023;72:2112-2122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 226] [Article Influence: 75.3] [Reference Citation Analysis (0)] |
| 80. | Shao L, Guo YP, Wang L, Chen MY, Zhang W, Deng S, Huang WH. Effects of ginsenoside compound K on colitis-associated colorectal cancer and gut microbiota profiles in mice. Ann Transl Med. 2022;10:408. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 81. | Chen H, Pan Y, Zhou Q, Liang C, Wong CC, Zhou Y, Huang D, Liu W, Zhai J, Gou H, Su H, Zhang X, Xu H, Wang Y, Kang W, Kei Wu WK, Yu J. METTL3 Inhibits Antitumor Immunity by Targeting m(6)A-BHLHE41-CXCL1/CXCR2 Axis to Promote Colorectal Cancer. Gastroenterology. 2022;163:891-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 185] [Article Influence: 46.3] [Reference Citation Analysis (4)] |
| 82. | Shen C, Xuan B, Yan T, Ma Y, Xu P, Tian X, Zhang X, Cao Y, Ma D, Zhu X, Zhang Y, Fang JY, Chen H, Hong J. m(6)A-dependent glycolysis enhances colorectal cancer progression. Mol Cancer. 2020;19:72. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 316] [Article Influence: 52.7] [Reference Citation Analysis (0)] |
| 83. | Wei W, Zhang ZY, Shi B, Cai Y, Zhang HS, Sun CL, Fei YF, Zhong W, Zhang S, Wang C, He B, Jiang GM, Wang H. METTL16 promotes glycolytic metabolism reprogramming and colorectal cancer progression. J Exp Clin Cancer Res. 2023;42:151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 52] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 84. | Chen X, Xu M, Xu X, Zeng K, Liu X, Sun L, Pan B, He B, Pan Y, Sun H, Xia X, Wang S. RETRACTED: METTL14 Suppresses CRC Progression via Regulating N6-Methyladenosine-Dependent Primary miR-375 Processing. Mol Ther. 2020;28:599-612. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 140] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 85. | Guo S, Chen F, Li L, Dou S, Li Q, Huang Y, Li Z, Liu W, Zhang G. Intracellular Fusobacterium nucleatum infection increases METTL3-mediated m6A methylation to promote the metastasis of esophageal squamous cell carcinoma. J Adv Res. 2024;61:165-178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 46] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 86. | Jabs S, Biton A, Bécavin C, Nahori MA, Ghozlane A, Pagliuso A, Spanò G, Guérineau V, Touboul D, Giai Gianetto Q, Chaze T, Matondo M, Dillies MA, Cossart P. Impact of the gut microbiota on the m(6)A epitranscriptome of mouse cecum and liver. Nat Commun. 2020;11:1344. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 91] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 87. | Wang A, Sun Y, Wang X, Yan Z, Wang D, Zeng L, Lu Q. m(6)A methyltransferase METTL16 mediates immune evasion of colorectal cancer cells via epigenetically regulating PD-L1 expression. Aging (Albany NY). 2023;15:8444-8457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 88. | Cao Y, Wang Z, Yan Y, Ji L, He J, Xuan B, Shen C, Ma Y, Jiang S, Ma D, Tong T, Zhang X, Gao Z, Zhu X, Fang JY, Chen H, Hong J. Enterotoxigenic Bacteroidesfragilis Promotes Intestinal Inflammation and Malignancy by Inhibiting Exosome-Packaged miR-149-3p. Gastroenterology. 2021;161:1552-1566.e12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 215] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 89. | Sun L, Ma L, Zhang H, Cao Y, Wang C, Hou N, Huang N, von Deneen KM, Zhao C, Shi Y, Pan Y, Wang M, Ji G, Nie Y. Fto Deficiency Reduces Anxiety- and Depression-Like Behaviors in Mice via Alterations in Gut Microbiota. Theranostics. 2019;9:721-733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 106] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 90. | Zhai J, Chen H, Wong CC, Peng Y, Gou H, Zhang J, Pan Y, Chen D, Lin Y, Wang S, Kang W, To KF, Chen Z, Nie Y, He HH, Sung JJ, Yu J. ALKBH5 Drives Immune Suppression Via Targeting AXIN2 to Promote Colorectal Cancer and Is a Target for Boosting Immunotherapy. Gastroenterology. 2023;165:445-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 112] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 91. | Zhang Z, Wang L, Zhao L, Wang Q, Yang C, Zhang M, Wang B, Jiang K, Ye Y, Wang S, Shen Z. N6-methyladenosine demethylase ALKBH5 suppresses colorectal cancer progression potentially by decreasing PHF20 mRNA methylation. Clin Transl Med. 2022;12:e940. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 39] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 92. | Lin Z, Wan AH, Sun L, Liang H, Niu Y, Deng Y, Yan S, Wang QP, Bu X, Zhang X, Hu K, Wan G, He W. N6-methyladenosine demethylase FTO enhances chemo-resistance in colorectal cancer through SIVA1-mediated apoptosis. Mol Ther. 2023;31:517-534. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 61] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 93. | Yue C, Chen J, Li Z, Li L, Chen J, Guo Y. microRNA-96 promotes occurrence and progression of colorectal cancer via regulation of the AMPKα2-FTO-m6A/MYC axis. J Exp Clin Cancer Res. 2020;39:240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 112] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 94. | Ruan DY, Li T, Wang YN, Meng Q, Li Y, Yu K, Wang M, Lin JF, Luo LZ, Wang DS, Lin JZ, Bai L, Liu ZX, Zhao Q, Wu XY, Ju HQ, Xu RH. FTO downregulation mediated by hypoxia facilitates colorectal cancer metastasis. Oncogene. 2021;40:5168-5181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 111] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 95. | Ye M, Chen J, Lu F, Zhao M, Wu S, Hu C, Yu P, Kan J, Bai J, Tian Y, Tang Q. Down-regulated FTO and ALKBH5 co-operatively activates FOXO signaling through m6A methylation modification in HK2 mRNA mediated by IGF2BP2 to enhance glycolysis in colorectal cancer. Cell Biosci. 2023;13:148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 44] [Reference Citation Analysis (0)] |
| 96. | Yao B, Zhang Q, Yang Z, An F, Nie H, Wang H, Yang C, Sun J, Chen K, Zhou J, Bai B, Gu S, Zhao W, Zhan Q. CircEZH2/miR-133b/IGF2BP2 aggravates colorectal cancer progression via enhancing the stability of m(6)A-modified CREB1 mRNA. Mol Cancer. 2022;21:140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 108] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 97. | Bao Y, Zhai J, Chen H, Wong CC, Liang C, Ding Y, Huang D, Gou H, Chen D, Pan Y, Kang W, To KF, Yu J. Targeting m(6)A reader YTHDF1 augments antitumour immunity and boosts anti-PD-1 efficacy in colorectal cancer. Gut. 2023;72:1497-1509. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 178] [Article Influence: 59.3] [Reference Citation Analysis (0)] |
| 98. | Liang W, Liu H, Zeng Z, Liang Z, Xie H, Li W, Xiong L, Liu Z, Chen M, Jie H, Zheng X, Huang L, Kang L. KRT17 Promotes T-lymphocyte Infiltration Through the YTHDF2-CXCL10 Axis in Colorectal Cancer. Cancer Immunol Res. 2023;11:875-894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 56] [Reference Citation Analysis (0)] |
| 99. | Zhang Y, Huang W, Yuan Y, Li J, Wu J, Yu J, He Y, Wei Z, Zhang C. Long non-coding RNA H19 promotes colorectal cancer metastasis via binding to hnRNPA2B1. J Exp Clin Cancer Res. 2020;39:141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 107] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 100. | Zhang K, Zhang T, Yang Y, Tu W, Huang H, Wang Y, Chen Y, Pan K, Chen Z. N(6)-methyladenosine-mediated LDHA induction potentiates chemoresistance of colorectal cancer cells through metabolic reprogramming. Theranostics. 2022;12:4802-4817. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 83] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 101. | Chen H, Gao S, Liu W, Wong CC, Wu J, Wu J, Liu D, Gou H, Kang W, Zhai J, Li C, Su H, Wang S, Soares F, Han J, He HH, Yu J. RNA N(6)-Methyladenosine Methyltransferase METTL3 Facilitates Colorectal Cancer by Activating the m(6)A-GLUT1-mTORC1 Axis and Is a Therapeutic Target. Gastroenterology. 2021;160:1284-1300.e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 225] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 102. | Chen C, Yuan W, Zhou Q, Shao B, Guo Y, Wang W, Yang S, Guo Y, Zhao L, Dang Q, Yang X, Wang G, Kang Q, Ji Z, Liu J, Sun Z. N6-methyladenosine-induced circ1662 promotes metastasis of colorectal cancer by accelerating YAP1 nuclear localization. Theranostics. 2021;11:4298-4315. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 130] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 103. | Yang X, Zhang S, He C, Xue P, Zhang L, He Z, Zang L, Feng B, Sun J, Zheng M. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020;19:46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 168] [Cited by in RCA: 416] [Article Influence: 69.3] [Reference Citation Analysis (0)] |
| 104. | Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B, Li C, Sun L, Qin J, Xu T, He B, Pan Y, Sun H, Wang S. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol Cancer. 2020;19:106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 299] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 105. | Wu S, Yun J, Tang W, Familiari G, Relucenti M, Wu J, Li X, Chen H, Chen R. Therapeutic m(6)A Eraser ALKBH5 mRNA-Loaded Exosome-Liposome Hybrid Nanoparticles Inhibit Progression of Colorectal Cancer in Preclinical Tumor Models. ACS Nano. 2023;17:11838-11854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 74] [Reference Citation Analysis (0)] |
| 106. | Shen D, Lin J, Xie Y, Zhuang Z, Xu G, Peng S, Tang G, Bai L, Zhu M, Zhang Y, Huang Z, Wang P, Liu X, Huang M, Luo Y, Wang X, Yu H. RNA demethylase ALKBH5 promotes colorectal cancer progression by posttranscriptional activation of RAB5A in an m6A-YTHDF2-dependent manner. Clin Transl Med. 2023;13:e1279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 32] [Reference Citation Analysis (0)] |
| 107. | Shao Y, Liu Z, Song X, Sun R, Zhou Y, Zhang D, Sun H, Huang J, Wu C, Gu W, Zheng X, Jiang J. ALKBH5/YTHDF2-mediated m6A modification of circAFF2 enhances radiosensitivity of colorectal cancer by inhibiting Cullin neddylation. Clin Transl Med. 2023;13:e1318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 58] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 108. | Hou P, Meng S, Li M, Lin T, Chu S, Li Z, Zheng J, Gu Y, Bai J. LINC00460/DHX9/IGF2BP2 complex promotes colorectal cancer proliferation and metastasis by mediating HMGA1 mRNA stability depending on m6A modification. J Exp Clin Cancer Res. 2021;40:52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 161] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 109. | Wang S, Gao S, Zeng Y, Zhu L, Mo Y, Wong CC, Bao Y, Su P, Zhai J, Wang L, Soares F, Xu X, Chen H, Hezaveh K, Ci X, He A, McGaha T, O'Brien C, Rottapel R, Kang W, Wu J, Zheng G, Cai Z, Yu J, He HH. N6-Methyladenosine Reader YTHDF1 Promotes ARHGEF2 Translation and RhoA Signaling in Colorectal Cancer. Gastroenterology. 2022;162:1183-1196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 137] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 110. | Li H, Zhang N, Jiao X, Wang C, Sun W, He Y, Ren G, Huang S, Li M, Chang Y, Jin Z, Xie Q, Zhang X, Huang H, Jin H. Downregulation of microRNA-6125 promotes colorectal cancer growth through YTHDF2-dependent recognition of N6-methyladenosine-modified GSK3β. Clin Transl Med. 2021;11:e602. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 111. | Liu H, Li D, Sun L, Qin H, Fan A, Meng L, Graves-Deal R, Glass SE, Franklin JL, Liu Q, Wang J, Yeatman TJ, Guo H, Zong H, Jin S, Chen Z, Deng T, Fang Y, Li C, Karijolich J, Patton JG, Wang X, Nie Y, Fan D, Coffey RJ, Zhao X, Lu Y. Interaction of lncRNA MIR100HG with hnRNPA2B1 facilitates m(6)A-dependent stabilization of TCF7L2 mRNA and colorectal cancer progression. Mol Cancer. 2022;21:74. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 147] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 112. | Zichittella C, Barreca MM, Cordaro A, Corrado C, Alessandro R, Conigliaro A. Mir-675-5p supports hypoxia-induced drug resistance in colorectal cancer cells. BMC Cancer. 2022;22:567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 113. | Yao J, Song Y, Yu X, Lin Z. Interaction between N(6)-methyladenosine modification and the tumor microenvironment in colorectal cancer. Mol Med. 2023;29:129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 114. | Lai Q, Li W, Wang H, Xu S, Deng Z. Emerging role of circRNAs in cancer under hypoxia. Oncol Lett. 2022;24:372. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 115. | Manuelli V, Pecorari C, Filomeni G, Zito E. Regulation of redox signaling in HIF-1-dependent tumor angiogenesis. FEBS J. 2022;289:5413-5425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 116. | Sun H, Li K, Zhang X, Liu J, Zhang M, Meng H, Yi C. m(6)Am-seq reveals the dynamic m(6)Am methylation in the human transcriptome. Nat Commun. 2021;12:4778. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 117. | Zhang F, Liu H, Duan M, Wang G, Zhang Z, Wang Y, Qian Y, Yang Z, Jiang X. Crosstalk among m(6)A RNA methylation, hypoxia and metabolic reprogramming in TME: from immunosuppressive microenvironment to clinical application. J Hematol Oncol. 2022;15:84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 65] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 118. | Zhou L, Jiang J, Huang Z, Jin P, Peng L, Luo M, Zhang Z, Chen Y, Xie N, Gao W, Nice EC, Li JQ, Chen HN, Huang C. Hypoxia-induced lncRNA STEAP3-AS1 activates Wnt/β-catenin signaling to promote colorectal cancer progression by preventing m(6)A-mediated degradation of STEAP3 mRNA. Mol Cancer. 2022;21:168. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 114] [Reference Citation Analysis (0)] |
| 119. | Zheng Z, Bian C, Wang H, Su J, Meng L, Xin Y, Jiang X. Prediction of immunotherapy efficacy and immunomodulatory role of hypoxia in colorectal cancer. Ther Adv Med Oncol. 2022;14:17588359221138383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 120. | Zhao N, Ruan M, Koestler DC, Lu J, Salas LA, Kelsey KT, Platz EA, Michaud DS. Methylation-derived inflammatory measures and lung cancer risk and survival. Clin Epigenetics. 2021;13:222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 121. | Bersuder E, Terciolo C, Lechevrel M, Martin E, Quesnelle C, Freund JN, Reimund JM, Gross I. Mesalazine initiates an anti-oncogenic β-catenin / MUCDHL negative feed-back loop in colon cancer cells by cell-specific mechanisms. Biomed Pharmacother. 2022;146:112543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 122. | Hibino S, Kawazoe T, Kasahara H, Itoh S, Ishimoto T, Sakata-Yanagimoto M, Taniguchi K. Inflammation-Induced Tumorigenesis and Metastasis. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 207] [Article Influence: 41.4] [Reference Citation Analysis (0)] |
| 123. | Lee HM, Lee HJ, Chang JE. Inflammatory Cytokine: An Attractive Target for Cancer Treatment. Biomedicines. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 45] [Reference Citation Analysis (0)] |
| 124. | Sun J, Cheng B, Su Y, Li M, Ma S, Zhang Y, Zhang A, Cai S, Bao Q, Wang S, Zhu P. The Potential Role of m6A RNA Methylation in the Aging Process and Aging-Associated Diseases. Front Genet. 2022;13:869950. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 125. | Sebastian-delaCruz M, Olazagoitia-Garmendia A, Gonzalez-Moro I, Santin I, Garcia-Etxebarria K, Castellanos-Rubio A. Implication of m6A mRNA Methylation in Susceptibility to Inflammatory Bowel Disease. Epigenomes. 2020;4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 126. | Wang A, Huang H, Shi JH, Yu X, Ding R, Zhang Y, Han Q, Ni ZY, Li X, Zhao R, Zou Q. USP47 inhibits m6A-dependent c-Myc translation to maintain regulatory T cell metabolic and functional homeostasis. J Clin Invest. 2023;133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 36] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 127. | Zhang Y, Zhang K, Gong H, Li Q, Man L, Jin Q, Zhang L, Li S. Links Between N (6)-Methyladenosine and Tumor Microenvironments in Colorectal Cancer. Front Cell Dev Biol. 2022;10:807129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 128. | Wang Z, Dan W, Zhang N, Fang J, Yang Y. Colorectal cancer and gut microbiota studies in China. Gut Microbes. 2023;15:2236364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 83] [Reference Citation Analysis (0)] |
| 129. | Sánchez-Alcoholado L, Ramos-Molina B, Otero A, Laborda-Illanes A, Ordóñez R, Medina JA, Gómez-Millán J, Queipo-Ortuño MI. The Role of the Gut Microbiome in Colorectal Cancer Development and Therapy Response. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 234] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 130. | Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, Bailis W, Cao G, Kroehling L, Chen Y, Wang G, Broughton JP, Chen YG, Kluger Y, Simon MD, Chang HY, Yin Z, Flavell RA. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548:338-342. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 421] [Cited by in RCA: 762] [Article Influence: 84.7] [Reference Citation Analysis (0)] |
| 131. | Xiong J, He J, Zhu J, Pan J, Liao W, Ye H, Wang H, Song Y, Du Y, Cui B, Xue M, Zheng W, Kong X, Jiang K, Ding K, Lai L, Wang Q. Lactylation-driven METTL3-mediated RNA m(6)A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol Cell. 2022;82:1660-1677.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 538] [Article Influence: 134.5] [Reference Citation Analysis (0)] |
| 132. | Zong X, Zhao J, Wang H, Lu Z, Wang F, Du H, Wang Y. Mettl3 Deficiency Sustains Long-Chain Fatty Acid Absorption through Suppressing Traf6-Dependent Inflammation Response. J Immunol. 2019;202:567-578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 97] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 133. | Gong PJ, Shao YC, Yang Y, Song WJ, He X, Zeng YF, Huang SR, Wei L, Zhang JW. Analysis of N6-Methyladenosine Methyltransferase Reveals METTL14 and ZC3H13 as Tumor Suppressor Genes in Breast Cancer. Front Oncol. 2020;10:578963. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 90] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 134. | Zhou J, Zhang X, Hu J, Qu R, Yu Z, Xu H, Chen H, Yan L, Ding C, Zou Q, Ye Y, Wang Z, Flavell RA, Li HB. m(6)A demethylase ALKBH5 controls CD4(+) T cell pathogenicity and promotes autoimmunity. Sci Adv. 2021;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 149] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 135. | Liu Y, Liang G, Xu H, Dong W, Dong Z, Qiu Z, Zhang Z, Li F, Huang Y, Li Y, Wu J, Yin S, Zhang Y, Guo P, Liu J, Xi JJ, Jiang P, Han D, Yang CG, Xu MM. Tumors exploit FTO-mediated regulation of glycolytic metabolism to evade immune surveillance. Cell Metab. 2021;33:1221-1233.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 269] [Article Influence: 53.8] [Reference Citation Analysis (0)] |
| 136. | Li J, Liang L, Yang Y, Li X, Ma Y. N(6)-methyladenosine as a biological and clinical determinant in colorectal cancer: progression and future direction. Theranostics. 2021;11:2581-2593. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 137. | Lu Y, Wang XM, Li ZS, Wu AJ, Cheng WX. Hsa_circ_0001658 accelerates the progression of colorectal cancer through miR-590-5p/METTL3 regulatory axis. World J Gastrointest Oncol. 2023;15:76-89. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 1] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 138. | Jiang X, Jin Z, Yang Y, Zheng X, Chen S, Wang S, Zhang X, Qu N. m6A modification on the fate of colorectal cancer: functions and mechanisms of cell proliferation and tumorigenesis. Front Oncol. 2023;13:1162300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 139. | Fang Z, Hu Y, Hu J, Huang Y, Zheng S, Guo C. The crucial roles of N(6)-methyladenosine (m(6)A) modification in the carcinogenesis and progression of colorectal cancer. Cell Biosci. 2021;11:72. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 140. | Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G, Pilka ES, Aspris D, Leggate D, Hendrick AG, Webster NA, Andrews B, Fosbeary R, Guest P, Irigoyen N, Eleftheriou M, Gozdecka M, Dias JML, Bannister AJ, Vick B, Jeremias I, Vassiliou GS, Rausch O, Tzelepis K, Kouzarides T. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. 2021;593:597-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 497] [Cited by in RCA: 973] [Article Influence: 194.6] [Reference Citation Analysis (6)] |
| 141. | Jiang D, Hou J, Qian Y, Gao Y, Gao X, Wei S. YTHDF1-regulated expression of TEAD1 contributes to the maintenance of intestinal stem cells. Biochem Biophys Res Commun. 2021;557:85-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 142. | Qiu FS, He JQ, Zhong YS, Guo MY, Yu CH. Implications of m6A methylation and microbiota interaction in non-small cell lung cancer: From basics to therapeutics. Front Cell Infect Microbiol. 2022;12:972655. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 143. | Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M, Deng X, Li H, Huang Y, Gao L, Li C, Zhao Z, Robinson S, Tan B, Qing Y, Qin X, Prince E, Xie J, Qin H, Li W, Shen C, Sun J, Kulkarni P, Weng H, Huang H, Chen Z, Zhang B, Wu X, Olsen MJ, Müschen M, Marcucci G, Salgia R, Li L, Fathi AT, Li Z, Mulloy JC, Wei M, Horne D, Chen J. Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell. 2020;38:79-96.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 258] [Cited by in RCA: 586] [Article Influence: 97.7] [Reference Citation Analysis (0)] |
| 144. | Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, Deng X, Wang Y, Weng X, Hu C, Yu M, Skibbe J, Dai Q, Zou D, Wu T, Yu K, Weng H, Huang H, Ferchen K, Qin X, Zhang B, Qi J, Sasaki AT, Plas DR, Bradner JE, Wei M, Marcucci G, Jiang X, Mulloy JC, Jin J, He C, Chen J. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell. 2018;172:90-105.e23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 495] [Cited by in RCA: 883] [Article Influence: 98.1] [Reference Citation Analysis (0)] |
| 145. | Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, Gan J, Jiang H, Jia GF, Luo C, Yang CG. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43:373-384. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 424] [Cited by in RCA: 555] [Article Influence: 50.5] [Reference Citation Analysis (6)] |
| 146. | Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, Ni T, Zhang ZS, Zhang T, Li C, Han L, Zhu Z, Lian F, Wei J, Deng Q, Wang Y, Wunderlich M, Gao Z, Pan G, Zhong D, Zhou H, Zhang N, Gan J, Jiang H, Mulloy JC, Qian Z, Chen J, Yang CG. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell. 2019;35:677-691.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 499] [Cited by in RCA: 693] [Article Influence: 99.0] [Reference Citation Analysis (0)] |
| 147. | Zhao Z, Meng J, Su R, Zhang J, Chen J, Ma X, Xia Q. Epitranscriptomics in liver disease: Basic concepts and therapeutic potential. J Hepatol. 2020;73:664-679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 113] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 148. | Selberg S, Seli N, Kankuri E, Karelson M. Rational Design of Novel Anticancer Small-Molecule RNA m6A Demethylase ALKBH5 Inhibitors. ACS Omega. 2021;6:13310-13320. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 98] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 149. | Hu Y, Gao Q, Ma S, Yu P, Ding S, Yao X, Zhang Z, Lu S, Lu M, Zhang J, Wang Y, Qian X, Zhong J. FMR1 promotes the progression of colorectal cancer cell by stabilizing EGFR mRNA in an m(6)A-dependent manner. Cell Death Dis. 2022;13:941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 34] [Reference Citation Analysis (0)] |
| 150. | Saeed M, Shoaib A, Kandimalla R, Javed S, Almatroudi A, Gupta R, Aqil F. Microbe-based therapies for colorectal cancer: Advantages and limitations. Semin Cancer Biol. 2022;86:652-665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 151. | Qu R, Zhang Y, Ma Y, Zhou X, Sun L, Jiang C, Zhang Z, Fu W. Role of the Gut Microbiota and Its Metabolites in Tumorigenesis or Development of Colorectal Cancer. Adv Sci (Weinh). 2023;10:e2205563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 146] [Article Influence: 48.7] [Reference Citation Analysis (0)] |
| 152. | Liu Y, Lau HC, Yu J. Microbial metabolites in colorectal tumorigenesis and cancer therapy. Gut Microbes. 2023;15:2203968. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 47] [Reference Citation Analysis (0)] |
| 153. | Zackular JP, Baxter NT, Iverson KD, Sadler WD, Petrosino JF, Chen GY, Schloss PD. The gut microbiome modulates colon tumorigenesis. mBio. 2013;4:e00692-e00613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 433] [Cited by in RCA: 544] [Article Influence: 41.8] [Reference Citation Analysis (1)] |
| 154. | Cao H, Xu M, Dong W, Deng B, Wang S, Zhang Y, Wang S, Luo S, Wang W, Qi Y, Gao J, Cao X, Yan F, Wang B. Secondary bile acid-induced dysbiosis promotes intestinal carcinogenesis. Int J Cancer. 2017;140:2545-2556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 186] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 155. | Liu Y, Sethi NS, Hinoue T, Schneider BG, Cherniack AD, Sanchez-Vega F, Seoane JA, Farshidfar F, Bowlby R, Islam M, Kim J, Chatila W, Akbani R, Kanchi RS, Rabkin CS, Willis JE, Wang KK, McCall SJ, Mishra L, Ojesina AI, Bullman S, Pedamallu CS, Lazar AJ, Sakai R; Cancer Genome Atlas Research Network, Thorsson V, Bass AJ, Laird PW. Comparative Molecular Analysis of Gastrointestinal Adenocarcinomas. Cancer Cell. 2018;33:721-735.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 434] [Cited by in RCA: 403] [Article Influence: 50.4] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/licenses/by-nc/4.0/