Published online Sep 28, 2024. doi: 10.3748/wjg.v30.i36.4014
Revised: August 25, 2024
Accepted: September 9, 2024
Published online: September 28, 2024
Processing time: 192 Days and 6 Hours
Gastrointestinal disorders encompass a spectrum of conditions affecting various organs within the digestive system, such as the esophagus, stomach, colon, rectum, pancreas, liver, small intestine, and bile ducts. The role of autophagy in the etiology and progression of gastrointestinal diseases has garnered significant attention. This paper seeks to evaluate the impact and mechanisms of autophagy in gastrointestinal disorders by synthesizing recent research findings. Specifically, we delve into inflammation-related gastrointestinal conditions, including ul-cerative colitis, Crohn’s disease, and pancreatitis, as well as gastrointestinal cancers such as esophageal, gastric, and colorectal cancers. Additionally, we provide commentary on a recent publication by Chang et al in the World Journal of Gastroenterology. Our objective is to offer fresh perspectives on the mechanisms and therapeutic approaches for these gastrointestinal ailments. This review aims to offer new perspectives on the mechanisms and therapeutic strategies for gastrointestinal disorders by critically analyzing relevant publications. As discussed, the role of autophagy in gastrointestinal diseases is complex and, at times, contentious. To harness the full therapeutic potential of autophagy in treating these conditions, more in-depth research is imperative.
Core Tip: Extensive research has implicated autophagy in the pathogenesis and advancement of diverse gastrointestinal disorders. Nevertheless, the precise role of autophagy in these ailments remains incompletely understood, and the specific underlying mechanisms remain elusive. Consequently, further investigation is warranted to address these gaps in knowledge.
- Citation: Shao BZ, Zhang WG, Liu ZY, Linghu EQ. Autophagy and its role in gastrointestinal diseases. World J Gastroenterol 2024; 30(36): 4014-4020
- URL: https://www.wjgnet.com/1007-9327/full/v30/i36/4014.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i36.4014
The term “autophagy” was coined in the 1960s[1]. Derived from the Greek roots “auto” (self) and “phagy” (eat), autophagy describes cellular metabolic processes in which cytoplasmic proteins and specific organelles are self-degraded[2]. Since its inception, the mechanisms of autophagy have been extensively explored by the scientific community. Research on autophagy won the Nobel prize in physiology or medicine in 2016[3]. Classic autophagy is classified into three types: Micro-autophagy, chaperone-mediated autophagy, and macro-autophagy[3]. Micro-autophagy represents a nonselective lysosomal process. Chaperone-mediated autophagy operates as a discerning form of autophagy that hinges on the recognition of chaperones through specific motifs within the targeted proteins, as well as lysosomal chaperones. Macro-autophagy, on the other hand, is the most extensively investigated variant of autophagy. It is a metabolic process characterized by the formation of double-membraned autophagosomes, which serve as functional units that subsequently merge with lysosomes to facilitate further degradation and recycling. In addition to traditional autophagy, specialized selective autophagy, such as pexophagy and reticulophagy, has been identified. These distinct varieties of selective autophagy embody unique roles and functions within specific organelles and under specific environmental conditions.
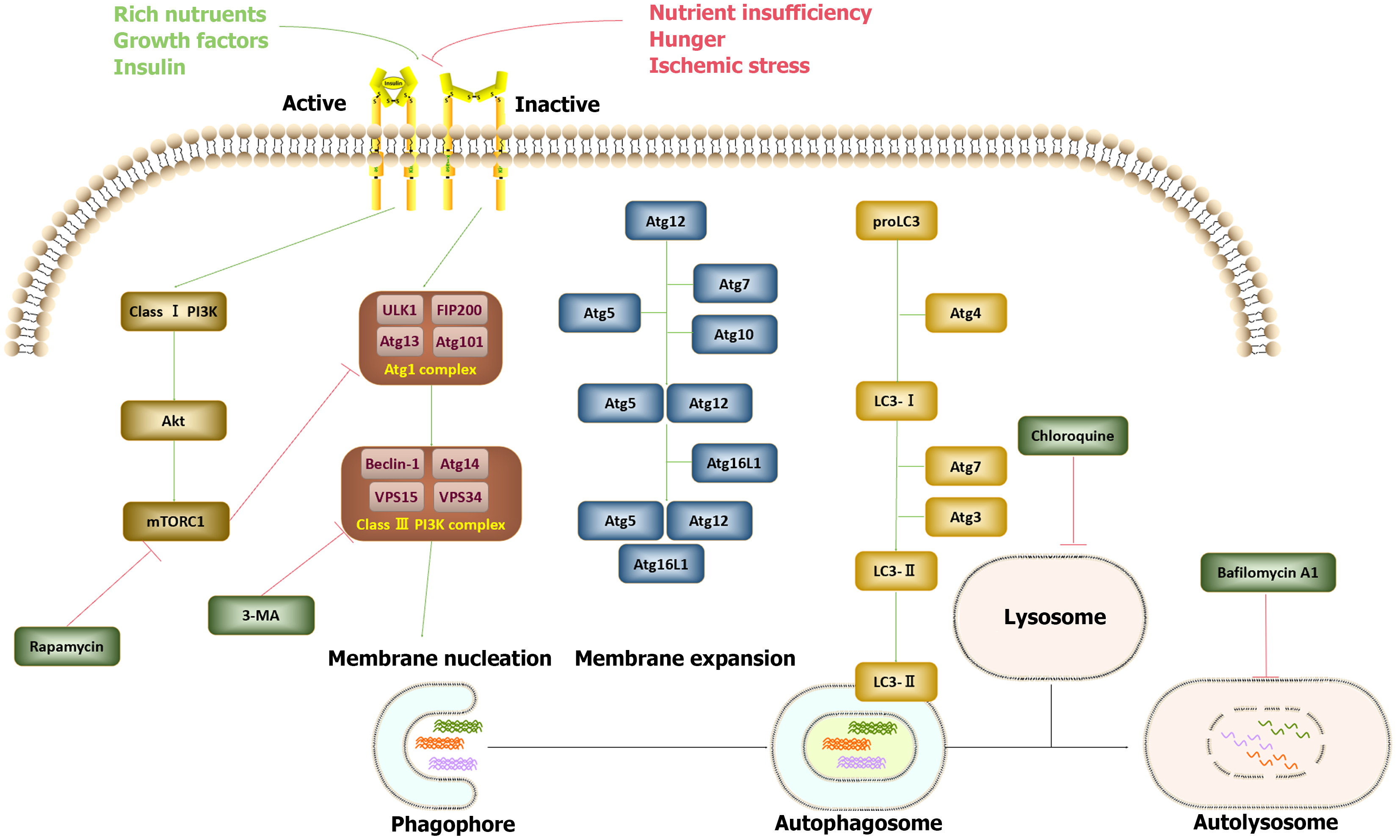
The autophagy-lysosomal system represents one of the classical pathways for protein degradation, alongside the ubiquitin–proteasome system. This intricate process is orchestrated by over 30 autophagy-related genes (Atgs), the majority of which are conserved in mammalian cells. Autophagy induction is a two-step process[4] (illustrated in Figure 1). Initially, under conditions of stress, such as hypoxia, phagophores form to envelop substrates. Phagophore formation necessitates the assembly of the Atg1 complex and the class III phosphatidylinositol 3-kinase (PI3K) complex. The Atg1 complex includes Unc-51-like kinase, focal adhesion kinase family interacting protein of 200 kD, ATG13, and ATG101. Following initiation, the bilayer membrane undergoes a series of processes, including expansion, elongation, and nucleation, leading to the formation of double-membrane, sphere-shaped autophagosomes. This process relies on the assembly of the ATG16L1 complex, which is composed of ATG5, ATG12, and ATG16L1, as well as the participation of two ubiquitin-like proteins, ATG12 and Atg8 (light chain 3). In the second step, autophagosomes subsequently shed their “coat proteins” [light chain 3-II (LC3-II)] and merge with lysosomes to form functional autolysosomes. This process is facilitated by ATG3 and ATG7. The class I PI3K-mammalian target of rapamycin (mTOR) pathway acts as an inhibitory regulator of autophagy, whereas the class III PI3K pathway serves as an autophagy inducer. Various autophagy inducers and inhibitors have been utilized in experimental studies and clinical practice to modulate autophagy levels[5]. For example, rapamycin is commonly employed to increase autophagy levels by inhibiting mTORC1 activation. Conversely, the inhibition of autophagy is achieved through mechanisms such as 3-methyladenine. Additionally, chloroquine disrupts autophagy through its influence on the acidic environment of lysosomes, whereas bafilomycin A1 disturbs the formation of autolysosomes.
On the basis of previous research both within our laboratory and from other sources[4], autophagy has been demonstrated to actively participate in and regulate numerous diseases across various domains. In the context of gastrointestinal diseases, the role of autophagy in influencing the pathogenesis and progression of these conditions has been extensively investigated. The influence of autophagy is exerted through the regulation of inflammation and immune responses, as well as the modulation of cellular functions and biological status, including gastrointestinal epithelial cells, inflammatory cells, and even cancer cells. A recent publication by Chang et al[6] in the World Journal of Gastroenterology comprehensively reviewed the impact of autophagy on the functions of gastrointestinal cells, including digestive cells, secretory cells, regenerative cells, and physical barriers. The authors illustrate how autophagy plays a pivotal role in influencing several gastrointestinal diseases by modulating inflammatory reactions. As a result, autophagy is a crucial determinant in maintaining homeostasis within the digestive system. In the following sections, we comprehensively examine the role of autophagy in various well-characterized gastrointestinal diseases, including inflammation-associated conditions such as ulcerative colitis (UC), Crohn’s disease (CD), and pancreatitis, as well as gastrointestinal malignancies such as esophageal, gastric, and colorectal cancers.
UC is a type of inflammatory bowel disease (IBD) characterized by chronic inflammation affecting the entire length of the colon and rectum. The pathogenesis of UC involves a complex interplay of factors, including abnormalities in the immune system, environmental influences, disturbances in the gut microbiota, exogenous infections, and specific genetic mutations. However, the precise mechanisms underlying its pathogenesis remain incompletely understood. UC typically manifests as a pattern of inflammation spreading from the rectum to the distal colon, eventually encompassing the entire large intestine. In recent years, considerable attention has been given to exploring the role of autophagy in UC. Studies have revealed a potential association between decreased autophagic activity and the onset of UC[7]. Recent findings have highlighted the potential of activating the intestinal nuclear receptor vitamin D receptor (VDR) via autophagy. This has been demonstrated in several studies[8-10]. Notably, decreased expression of VDR and dysregulated vitamin D/VDR signaling pathways have been observed in patients with UC. Moreover, Zhou et al[11] reported mTOR-dependent deficiency in autophagy flux in human intestinal epithelial cells derived from active UC patients. In dextran sulfate sodium (DSS)-induced UC models, Wu et al[12] reported the contribution of autophagy to gut microbiota homeostasis in UC models. In line with these findings, recent investigations conducted by our research team have demonstrated that inducing autophagy via the adenosine 5’-monophosphate-activated protein kinase-mTOR-p70S6K pathway through the activation of specific receptors alleviates UC symptoms and suppresses intestinal inflammation in DSS-induced mouse models[13]. Collectively, these studies underscore the therapeutic potential of autophagy in ameliorating the onset and progression of UC. Notably, polymorphisms in specific Atgs, including CASP1, SERPINA1, and CCL2, have been linked to an increased risk of UC. These findings identify novel molecular targets for the development of autophagy-modulating therapies for the management of active UC[14].
CD is a distinct entity within the spectrum of IBD. CD manifests as localized or regional enteritis of uncertain etiology and is influenced by a multitude of pathogenetic factors, including compromised immunity, environmental stressors, familial predisposition, and specific genetic mutations. In contrast to UC, CD is more susceptible to genetic influences. Extensive genome-wide association studies conducted in 2007 highlighted the significant association between mutations in certain Atgs, notably ATG16L1 and immunity-related GTPase M (IRGM), and the pathogenesis of CD[15,16]. Moreover, investigations by Rioux et al[17] revealed that polymorphisms in Atgs are related to CD-associated adherent-invasive Escherichia coli replication. Nucleotide binding oligomerization domain-containing protein 2 (NOD2) has emerged as a pivotal inducer of autophagy, orchestrating the recruitment of ATG16L1 to the cell membrane[18]. Notably, mutations in NOD2 observed in CD patients have been linked to disruptions in intestinal microbiota homeostasis and heightened gut inflammatory responses[19,20]. In addition to NOD2, IRGM, ULK-1, and XBP-1, mutations or deletions in other Atgs are highly related to the pathogenesis of IBD, particularly CD, according to recent reviews[21]. Studies utilizing 2,4,6-trinitrobenzenesulfonic acid-induced models have reported that inhibiting autophagy exacerbates CD symptoms and intestinal inflammation, highlighting the protective role of autophagy in CD[22]. Collectively, these findings underscore the specific involvement of autophagy in the onset and progression of CD. On the basis of our insights into the role of autophagy in the pathogenesis and progression of CD, targeting impaired autophagy, particularly that caused by polymorphisms in Atgs, may represent a promising therapeutic approach for CD[23].
In addition to UC and CD, pancreatitis has emerged as another prevalent inflammation-related gastrointestinal disorder. Pancreatitis, characterized by pancreatic self-digestion primarily mediated by trypsin, manifests as damage to and necrosis of pancreatic tissue due to the overactivation of self-protective inflammatory and immune responses. Extensive evidence underscores the close association between autophagy and pancreatitis. As previously documented, the onset of acute pancreatitis triggers a significant increase in autophagosome formation within pancreatic acinar cells[24]. However, recent findings have revealed elevated levels of both LC3-II and sequestosome 1 in mouse models of pancreatitis[25], suggesting impaired autophagy. Furthermore, multiple investigations have demonstrated a decrease in lysosome formation in cerulein-treated mouse pancreatic acinar cells, as evidenced by the downregulation of lysosome-associated membrane proteins (LAMP1 and LAMP2), which play crucial roles in stabilizing lysosomal membranes and safeguarding the cytoplasm from acid hydrolases[26,27]. These studies underscore the importance of restoring autophagic function in the management of pancreatitis.
Gastrointestinal cancers encompass a spectrum of malignancies affecting tissues within the gastrointestinal tract, such as the esophagus, stomach, colon, and rectum[28]. Esophageal cancer has been extensively linked with autophagy. A recent study revealed that 22 autophagic long-chain noncoding ribonucleic acids are strongly associated with the survival rate of patients with esophageal adenocarcinoma[29]. Research by Fang et al[30] revealed that the induction of autophagy protected esophageal cancer cells exposed to diketopyrrolopyrrole treatment. Moreover, autophagy is related to microRNA-193b-related chemoresistance to 5-fluorouracil[31]. Furthermore, microRNA-498 has been identified as a suppressor of esophageal cancer through the inhibition of autophagy[32]. These findings underscore the intricate interplay between autophagy and esophageal cancer progression, suggesting potential avenues for therapeutic intervention in this malignancy. In contrast, several studies have presented conflicting findings. A recent investigation demonstrated that silencing autophagy exacerbates radiotherapy resistance in esophageal squamous cell carcinoma[33]. Moreover, inducing autophagy can elicit autophagy-mediated cell cycle arrest[34,35]. We hypothesize that the conflicting outcomes of autophagy in esophageal cancer may stem from variations in the induction methods used and the extent of autophagy activation. Therefore, further research is urgently needed to clarify the exact function of autophagy in esophageal cancer.
Gastric cancer, also known as stomach cancer, is one of the most formidable human malignancies globally and poses a significant health burden [36,37]. Recent investigations have extensively probed the role of autophagy in gastric cancer. Numerous studies suggest that modulating autophagy levels could be a promising therapeutic strategy for gastric cancer treatment. Notably, the interplay between autophagy and programmed cell death (PD)-1 has garnered considerable attention. Wang et al[38] demonstrated that inhibiting autophagy could augment PD-L1 expression, thereby increasing sensitivity to PD-L1-targeted immunotherapy. Moreover, studies have revealed the involvement of unfolded protein response-induced autophagy activation mediated by SEC62, an endoplasmic reticulum membrane protein implicated in protein transport, in promoting gastric cancer metastasis[39]. However, the role of autophagy in the context of Helicobacter pylori (H. pylori) infection is complex. Autophagy has been shown to confer protection against H. pylori infection[40], thereby influencing the incidence of gastric cancer[40,41]. Prolonged exposure to H. pylori has been demonstrated to suppress autophagy in gastric epithelial cells[42]. Furthermore, the degradative functions of autophagy induced by vitamin D3 are effective in protecting gastric epithelial cells against H. pylori infection[43]. These findings underscore the need to carefully consider the multifaceted and occasionally contradictory effects of autophagy in the treatment of gastric cancer, particularly its influence on H. pylori infection.
Colorectal cancer is a prevalent malignant tumor affecting the digestive tract[21]. It is the fourth most prevalent malignancy globally and accounts for approximately 11% of all diagnosed malignant tumors worldwide[44]. Extensive research, including investigations conducted by our team and other researchers, has highlighted the role of autophagy in regulating inflammation in diseases[45-47]. Similarly, in colorectal cancer, autophagy has emerged as a significant player in modulating inflammatory and immune processes. Notably, autophagy has metabolic homeostatic roles centered on preserving regulatory Treg cells[48]. Additionally, induction of mitophagy has been shown to stimulate antitumor adaptive immunity[49]. Conversely, a deficiency in autophagy has been associated with tumor progression in colorectal cancer[50,51]. Intriguingly, however, a separate study revealed that the Thr300Ala variation of ATG16L1, a pivotal Atg, was correlated with improved overall survival in human patients with colorectal cancer[52]. These data suggest that the inflammatory and immunoregulatory functions of autophagy may play crucial roles in mitigating tumorigenesis. Additionally, specific mutations in Atgs could offer therapeutic advantages in colorectal cancer.
This paper provides an in-depth exploration of the role of autophagy in several prominent gastrointestinal disorders, including inflammation-related conditions such as UC, CD, and pancreatitis, as well as gastrointestinal cancers, including esophageal, gastric, and colorectal cancers. Our discussion underscores the intricate and occasionally competitive effects of autophagy on gastrointestinal ailments. The complex and sometimes contradictory roles of autophagy in gastrointestinal diseases may be attributed to its varying effects at different stages of disease development, the differential induction of autophagy, or the intricate nature of gastrointestinal pathogenesis and progression. Despite extensive research in this area, the precise mechanisms through which autophagy operates in gastrointestinal diseases remain elusive. The complexity of the role of autophagy in gastrointestinal diseases has hindered the successful clinical application of autophagy-modulating therapies. Consequently, further studies are essential to fully elucidate the role of autophagy in these diseases and to develop appropriate modulation strategies tailored to different stages of disease progression.
The authors would like to thank Professor Li X for his help in language editing.
| 1. | Sakai Y, Oku M. ATG and ESCRT control multiple modes of microautophagy. FEBS Lett. 2024;598:48-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 2. | Kumar V, Jurkunas UV. Mitochondrial Dysfunction and Mitophagy in Fuchs Endothelial Corneal Dystrophy. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 47] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 3. | Zhu Y, Liu F, Jian F, Rong Y. Recent progresses in the late stages of autophagy. Cell Insight. 2024;3:100152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 4. | Shao BZ, Wang P, Bai Y. Editorial: Autophagy in Inflammation Related Diseases. Front Pharmacol. 2022;13:912487. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 5. | Wang P, Shao BZ, Deng Z, Chen S, Yue Z, Miao CY. Autophagy in ischemic stroke. Prog Neurobiol. 2018;163-164:98-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 328] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 6. | Chang YF, Li JJ, Liu T, Wei CQ, Ma LW, Nikolenko VN, Chang WL. Morphological and biochemical characteristics associated with autophagy in gastrointestinal diseases. World J Gastroenterol. 2024;30:1524-1532. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 3] [Cited by in RCA: 8] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 7. | Hu X, Deng J, Yu T, Chen S, Ge Y, Zhou Z, Guo Y, Ying H, Zhai Q, Chen Y, Yuan F, Niu Y, Shu W, Chen H, Ma C, Liu Z, Guo F. ATF4 Deficiency Promotes Intestinal Inflammation in Mice by Reducing Uptake of Glutamine and Expression of Antimicrobial Peptides. Gastroenterology. 2019;156:1098-1111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 104] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 8. | Bakke D, Sun J. Ancient Nuclear Receptor VDR With New Functions: Microbiome and Inflammation. Inflamm Bowel Dis. 2018;24:1149-1154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 106] [Article Influence: 13.3] [Reference Citation Analysis (1)] |
| 9. | Law AD, Dutta U, Kochhar R, Vaishnavi C, Kumar S, Noor T, Bhadada S, Singh K. Vitamin D deficiency in adult patients with ulcerative colitis: Prevalence and relationship with disease severity, extent, and duration. Indian J Gastroenterol. 2019;38:6-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 10. | Karimi S, Tabataba-Vakili S, Yari Z, Alborzi F, Hedayati M, Ebrahimi-Daryani N, Hekmatdoost A. The effects of two vitamin D regimens on ulcerative colitis activity index, quality of life and oxidant/anti-oxidant status. Nutr J. 2019;18:16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 11. | Zhou M, Xu W, Wang J, Yan J, Shi Y, Zhang C, Ge W, Wu J, Du P, Chen Y. Boosting mTOR-dependent autophagy via upstream TLR4-MyD88-MAPK signalling and downstream NF-κB pathway quenches intestinal inflammation and oxidative stress injury. EBioMedicine. 2018;35:345-360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 260] [Cited by in RCA: 297] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 12. | Wu MY, Liu L, Wang EJ, Xiao HT, Cai CZ, Wang J, Su H, Wang Y, Tan J, Zhang Z, Wang J, Yao M, Ouyang DF, Yue Z, Li M, Chen Y, Bian ZX, Lu JH. PI3KC3 complex subunit NRBF2 is required for apoptotic cell clearance to restrict intestinal inflammation. Autophagy. 2021;17:1096-1111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (1)] |
| 13. | Ke P, Shao BZ, Xu ZQ, Wei W, Han BZ, Chen XW, Su DF, Liu C. Activation of Cannabinoid Receptor 2 Ameliorates DSS-Induced Colitis through Inhibiting NLRP3 Inflammasome in Macrophages. PLoS One. 2016;11:e0155076. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 91] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 14. | Qiu P, Liu L, Fang J, Zhang M, Wang H, Peng Y, Chen M, Liu J, Wang F, Zhao Q. Identification of Pharmacological Autophagy Regulators of Active Ulcerative Colitis. Front Pharmacol. 2021;12:769718. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 15. | Tysk C, Lindberg E, Järnerot G, Flodérus-Myrhed B. Ulcerative colitis and Crohn's disease in an unselected population of monozygotic and dizygotic twins. A study of heritability and the influence of smoking. Gut. 1988;29:990-996. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 543] [Cited by in RCA: 524] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 16. | Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, Günther S, Prescott NJ, Onnie CM, Häsler R, Sipos B, Fölsch UR, Lengauer T, Platzer M, Mathew CG, Krawczak M, Schreiber S. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1519] [Cited by in RCA: 1479] [Article Influence: 77.8] [Reference Citation Analysis (0)] |
| 17. | Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, Shugart YY, Griffiths AM, Targan SR, Ippoliti AF, Bernard EJ, Mei L, Nicolae DL, Regueiro M, Schumm LP, Steinhart AH, Rotter JI, Duerr RH, Cho JH, Daly MJ, Brant SR. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596-604. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1435] [Cited by in RCA: 1402] [Article Influence: 73.8] [Reference Citation Analysis (0)] |
| 18. | Park SC, Jeen YT. Genetic Studies of Inflammatory Bowel Disease-Focusing on Asian Patients. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 68] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 19. | Belyayev L, Hawksworth J, Khan K, Kaufman S, Subramanian S, Kroemer A, Loh K, Girlanda R, Fishbein TM, Matsumoto CS. Immunologic Complications and Graft Survival in Crohn's Disease and NOD2 Mutant Non-Crohn's Disease Adult Recipients Following Intestine Transplantation. Transplant Direct. 2020;6:e556. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 20. | Frade-Proud'Hon-Clerc S, Smol T, Frenois F, Sand O, Vaillant E, Dhennin V, Bonnefond A, Froguel P, Fumery M, Guillon-Dellac N, Gower-Rousseau C, Vasseur F. A Novel Rare Missense Variation of the NOD2 Gene: Evidencesof Implication in Crohn's Disease. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 21. | Wang SL, Shao BZ, Zhao SB, Fang J, Gu L, Miao CY, Li ZS, Bai Y. Impact of Paneth Cell Autophagy on Inflammatory Bowel Disease. Front Immunol. 2018;9:693. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 22. | Macias-Ceja DC, Cosín-Roger J, Ortiz-Masiá D, Salvador P, Hernández C, Esplugues JV, Calatayud S, Barrachina MD. Stimulation of autophagy prevents intestinal mucosal inflammation and ameliorates murine colitis. Br J Pharmacol. 2017;174:2501-2511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 70] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 23. | Alula KM, Theiss AL. Autophagy in Crohn's Disease: Converging on Dysfunctional Innate Immunity. Cells. 2023;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 25] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 24. | Gukovskaya AS, Gukovsky I, Algül H, Habtezion A. Autophagy, Inflammation, and Immune Dysfunction in the Pathogenesis of Pancreatitis. Gastroenterology. 2017;153:1212-1226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 284] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 25. | Wang S, Chao X, Jiang X, Wang T, Rodriguez Y, Yang L, Pacher P, Ni HM, Ding WX. Loss of acinar cell VMP1 triggers spontaneous pancreatitis in mice. Autophagy. 2022;18:1572-1582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 26. | Wang S, Ni HM, Chao X, Wang H, Bridges B, Kumer S, Schmitt T, Mareninova O, Gukovskaya A, De Lisle RC, Ballabio A, Pacher P, Ding WX. Impaired TFEB-mediated lysosomal biogenesis promotes the development of pancreatitis in mice and is associated with human pancreatitis. Autophagy. 2019;15:1954-1969. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 91] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 27. | Zhang T, Gan Y, Zhu S. Association between autophagy and acute pancreatitis. Front Genet. 2023;14:998035. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 28. | Griffin-Sobel JP. Gastrointestinal Cancers: Screening and Early Detection. Semin Oncol Nurs. 2017;33:165-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 21] [Article Influence: 2.3] [Reference Citation Analysis (1)] |
| 29. | Wu L, Zheng Y, Ruan X, Wu D, Xu P, Liu J, Wu D, Li X. Long-chain noncoding ribonucleic acids affect the survival and prognosis of patients with esophageal adenocarcinoma through the autophagy pathway: construction of a prognostic model. Anticancer Drugs. 2022;33:e590-e603. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 86] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 30. | Fang F, Li Y, Chang L. Mechanism of autophagy regulating chemoresistance in esophageal cancer cells. Exp Mol Pathol. 2020;117:104564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 31. | Nyhan MJ, O'Donovan TR, Boersma AW, Wiemer EA, McKenna SL. MiR-193b promotes autophagy and non-apoptotic cell death in oesophageal cancer cells. BMC Cancer. 2016;16:101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 32. | Li D, Yan M, Sun F, Song J, Hu X, Yu S, Tang L, Deng S. miR-498 inhibits autophagy and M2-like polarization of tumor-associated macrophages in esophageal cancer via MDM2/ATF3. Epigenomics. 2021;13:1013-1030. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 33. | Yao X, Chen H, Xu B, Lu J, Gu J, Chen F, Ju M, Sun X. The ATPase subunit of ATP6V1C1 inhibits autophagy and enhances radiotherapy resistance in esophageal squamous cell carcinoma. Gene. 2021;768:145261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 34. | Ma Q, Liao H, Xu L, Li Q, Zou J, Sun R, Xiao D, Liu C, Pu W, Cheng J, Zhou X, Huang G, Yao L, Zhong X, Guo X. Autophagy-dependent cell cycle arrest in esophageal cancer cells exposed to dihydroartemisinin. Chin Med. 2020;15:37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 35. | Chen X, He LY, Lai S, He Y. Dihydroartemisinin inhibits the migration of esophageal cancer cells by inducing autophagy. Oncol Lett. 2020;20:94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 36. | Yeoh KG, Tan P. Mapping the genomic diaspora of gastric cancer. Nat Rev Cancer. 2022;22:71-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 124] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 37. | Ajani JA, D'Amico TA, Bentrem DJ, Chao J, Cooke D, Corvera C, Das P, Enzinger PC, Enzler T, Fanta P, Farjah F, Gerdes H, Gibson MK, Hochwald S, Hofstetter WL, Ilson DH, Keswani RN, Kim S, Kleinberg LR, Klempner SJ, Lacy J, Ly QP, Matkowskyj KA, McNamara M, Mulcahy MF, Outlaw D, Park H, Perry KA, Pimiento J, Poultsides GA, Reznik S, Roses RE, Strong VE, Su S, Wang HL, Wiesner G, Willett CG, Yakoub D, Yoon H, McMillian N, Pluchino LA. Gastric Cancer, Version 2.2022, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2022;20:167-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 1179] [Article Influence: 294.8] [Reference Citation Analysis (0)] |
| 38. | Wang X, Wu WKK, Gao J, Li Z, Dong B, Lin X, Li Y, Li Y, Gong J, Qi C, Peng Z, Yu J, Shen L. Autophagy inhibition enhances PD-L1 expression in gastric cancer. J Exp Clin Cancer Res. 2019;38:140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 134] [Article Influence: 19.1] [Reference Citation Analysis (1)] |
| 39. | Su S, Shi YT, Chu Y, Jiang MZ, Wu N, Xu B, Zhou H, Lin JC, Jin YR, Li XF, Liang J. Sec62 promotes gastric cancer metastasis through mediating UPR-induced autophagy activation. Cell Mol Life Sci. 2022;79:133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 40. | Raju D, Hussey S, Ang M, Terebiznik MR, Sibony M, Galindo-Mata E, Gupta V, Blanke SR, Delgado A, Romero-Gallo J, Ramjeet MS, Mascarenhas H, Peek RM, Correa P, Streutker C, Hold G, Kunstmann E, Yoshimori T, Silverberg MS, Girardin SE, Philpott DJ, El Omar E, Jones NL. Vacuolating cytotoxin and variants in Atg16L1 that disrupt autophagy promote Helicobacter pylori infection in humans. Gastroenterology. 2012;142:1160-1171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 184] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 41. | Muhammad JS, Nanjo S, Ando T, Yamashita S, Maekita T, Ushijima T, Tabuchi Y, Sugiyama T. Autophagy impairment by Helicobacter pylori-induced methylation silencing of MAP1LC3Av1 promotes gastric carcinogenesis. Int J Cancer. 2017;140:2272-2283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 65] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 42. | He Y, Wang C, Zhang X, Lu X, Xing J, Lv J, Guo M, Huo X, Liu X, Lu J, Du X, Li C, Chen Z. Sustained Exposure to Helicobacter pylori Lysate Inhibits Apoptosis and Autophagy of Gastric Epithelial Cells. Front Oncol. 2020;10:581364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 43. | Hu W, Zhang L, Li MX, Shen J, Liu XD, Xiao ZG, Wu DL, Ho IHT, Wu JCY, Cheung CKY, Zhang YC, Lau AHY, Ashktorab H, Smoot DT, Fang EF, Chan MTV, Gin T, Gong W, Wu WKK, Cho CH. Vitamin D3 activates the autolysosomal degradation function against Helicobacter pylori through the PDIA3 receptor in gastric epithelial cells. Autophagy. 2019;15:707-725. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 117] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 44. | Zhang B, Wang HE, Bai YM, Tsai SJ, Su TP, Chen TJ, Wang YP, Chen MH. Inflammatory bowel disease is associated with higher dementia risk: a nationwide longitudinal study. Gut. 2021;70:85-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 218] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 45. | Shao BZ, Wang SL, Fang J, Li ZS, Bai Y, Wu K. Alpha7 Nicotinic Acetylcholine Receptor Alleviates Inflammatory Bowel Disease Through Induction of AMPK-mTOR-p70S6K-Mediated Autophagy. Inflammation. 2019;42:1666-1679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 46. | Xu W, Hua Z, Wang Y, Tang W, Ou W, Liu F, Yang Y, Ding W, Wang Z, Cui L, Ge W, Gu Y, Wang X, Chen Y, Liu CY, Du P. AMBRA1 promotes intestinal inflammation by antagonizing PP4R1/PP4c mediated IKK dephosphorylation in an autophagy-independent manner. Cell Death Differ. 2024;31:618-634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 47. | Wu MY, Wang EJ, Ye RD, Lu JH. Enhancement of LC3-associated efferocytosis for the alleviation of intestinal inflammation. Autophagy. 2024;20:1442-1443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 48. | Wei J, Long L, Yang K, Guy C, Shrestha S, Chen Z, Wu C, Vogel P, Neale G, Green DR, Chi H. Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat Immunol. 2016;17:277-285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 265] [Cited by in RCA: 374] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 49. | Ziegler PK, Bollrath J, Pallangyo CK, Matsutani T, Canli Ö, De Oliveira T, Diamanti MA, Müller N, Gamrekelashvili J, Putoczki T, Horst D, Mankan AK, Öner MG, Müller S, Müller-Höcker J, Kirchner T, Slotta-Huspenina J, Taketo MM, Reinheckel T, Dröse S, Larner AC, Wels WS, Ernst M, Greten TF, Arkan MC, Korn T, Wirth D, Greten FR. Mitophagy in Intestinal Epithelial Cells Triggers Adaptive Immunity during Tumorigenesis. Cell. 2018;174:88-101.e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 114] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 50. | Lucas C, Salesse L, Hoang MHT, Bonnet M, Sauvanet P, Larabi A, Godfraind C, Gagnière J, Pezet D, Rosenstiel P, Barnich N, Bonnet R, Dalmasso G, Nguyen HTT. Autophagy of Intestinal Epithelial Cells Inhibits Colorectal Carcinogenesis Induced by Colibactin-Producing Escherichia coli in Apc(Min/+) Mice. Gastroenterology. 2020;158:1373-1388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 69] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 51. | Liu X, Chen J, Long X, Lan J, Liu X, Zhou M, Zhang S, Zhou J. RSL1D1 promotes the progression of colorectal cancer through RAN-mediated autophagy suppression. Cell Death Dis. 2022;13:43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 52. | Grimm WA, Messer JS, Murphy SF, Nero T, Lodolce JP, Weber CR, Logsdon MF, Bartulis S, Sylvester BE, Springer A, Dougherty U, Niewold TB, Kupfer SS, Ellis N, Huo D, Bissonnette M, Boone DL. The Thr300Ala variant in ATG16L1 is associated with improved survival in human colorectal cancer and enhanced production of type I interferon. Gut. 2016;65:456-464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 77] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/