Published online Jun 28, 2024. doi: 10.3748/wjg.v30.i24.3076
Revised: May 21, 2024
Accepted: May 31, 2024
Published online: June 28, 2024
Processing time: 95 Days and 9.2 Hours
Helicobacter pylori (H. pylori) infection is closely associated with gastrointestinal diseases. Our preliminary studies have indicated that H. pylori infection had a significant impact on the mucosal microbiome structure in patients with gastric ulcer (GU) or duodenal ulcer (DU).
To investigate the contributions of H. pylori infection and the mucosal microbiome to the pathogenesis and progression of ulcerative diseases.
Patients with H. pylori infection and either GU or DU, and healthy individuals without H. pylori infection were included. Gastric or duodenal mucosal samples was obtained and subjected to metagenomic sequencing. The compositions of the microbial communities and their metabolic functions in the mucosal tissues were analyzed.
Compared with that in the healthy individuals, the gastric mucosal microbiota in the H. pylori-positive patients with GU was dominated by H. pylori, with signi
H. pylori infection significantly alters the gastric microbiota structure, diversity, and biological functions, which may be important contributing factors for GU.
Core Tip: In this study, we explored the influence and mechanism of action of Helicobacter pylori (H. pylori) on the development of gastric ulcer and duodenal ulcer, which are currently inconclusive. Metagenomic sequencing was performed on mucosal samples from H. pylori-infected patients with gastric ulcer or duodenal ulcer and healthy individuals. Mucosal flora structure, differentially functional genes, and metabolic pathways were analyzed to explore the role that H. pylori plays in the development of gastric ulcer and duodenal ulcer. Ultimately, two key metabolic pathways and related genes contributed from H. pylori were identified.
- Citation: Jin LX, Fang YP, Xia CM, Cai TW, Li QQ, Wang YY, Yan HF, Chen X. Helicobacter pylori infection alters gastric microbiota structure and biological functions in patients with gastric ulcer or duodenal ulcer. World J Gastroenterol 2024; 30(24): 3076-3085
- URL: https://www.wjgnet.com/1007-9327/full/v30/i24/3076.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i24.3076
Upper gastrointestinal ulcers, encompassing gastric ulcer (GU) and duodenal ulcer (DU), are defined as breaches of the mucosal integrity in the stomach or proximal small intestine, resulting in open lesions. Chronic or high-dose use of nonsteroidal anti-inflammatory drugs may impair the ability of the stomach mucosa, increasing the susceptibility of the gastric lining to damage from gastric acid and subsequently leading to ulcer formation[1]. Smoking, excessive alcohol consumption, stress, and genetic predispositions are also risk factors[2]. Another significant risk factor is infection with Helicobacter pylori (H. pylori). H. pylori is bacterium capable of surviving the harsh acidic environment of the stomach. The flagella of H. pylori aid in its adherence to the gastric mucosa, while the secretion of urease helps to create an alkaline environment locally and neutralize the surrounding gastric acid. In addition, an increasing number of studies reveal the presence of variable microbial communities within the stomach[3,4]. A balanced gastrointestinal microbiota sustains health, with the normal microbiota involved in protein metabolism, the synthesis of various vitamins, and the absorption of minerals. Moreover, the microbiota forms a natural biological barrier that enhances immune defenses of the host and prevents the invasion of pathogens. Overproliferation of H. pylori can disrupt the gastric microecological environment, leading to dysbiosis and inflammatory responses that contribute to disease onset, such as chronic gastritis, peptic ulcer, and gastric cancer. The specific underlying mechanisms are not fully understood and remain the focus[5]. Both the duodenum and the stomach can be colonized by H. pylori and develop ulcers, yet whether the pathogenesis of GU as well as DU is consistently related to H. pylori infection remains inconclusive.
This study performed metagenomic sequencing analysis of the gastrointestinal microbiota in H. pylori-infected patients with GU or DU, using healthy individuals as controls. By examining the structure and dominant bacterial groups in the mucosal microbiota across different ulcer types, this research aimed to provide a theoretical basis for investigating the contribution of H. pylori infection and the mucosal microbiota to the pathogenesis and progression of ulcerative diseases.
Patients who were scheduled for gastroscopy examination were enrolled in this study at the First People’s Hospital of Wenling according to the inclusion and exclusion criteria. The inclusion criteria were as follows: (1) 30-55 years of age; (2) consultation for different degrees of GU or DU; (3) no use of antibiotics, bismuth, H2-receptor antagonists, or proton pump inhibitors in the last 2 wk; and (4) positive H. pylori infection detected by a 13C urea breath test. The exclusion criteria were as follows: (1) Severe cardiac, hepatic, or renal impairment; (2) pregnant or lactating women; (3) a history of esophageal or gastrointestinal surgery; (4) inability to correctly express their symptoms, such as psychosis or severe neurosis; and (5) use of nonsteroidal anti-inflammatory drugs or the presence of alcoholism. In addition, healthy volunteers who were examined to ensure that they had no severe gastritis, GU, DU, H. pylori infection, metabolic, cardiovascular, and cerebrovascular diseases, or cancer were selected as a control group. All healthy volunteers also needed to meet the inclusion (except for the 2nd and 4th inclusion criteria) and exclusion criteria. This study was conducted in full compliance with the World Medical Association Declaration of Helsinki. All enrolled patients and healthy individuals were informed about the details of the study and signed an informed consent form.
Duodenal mucosal biopsy specimens from patients with DU and gastric mucosal biopsy specimens from patients with GU and healthy individuals were collected. The samples were immediately stored at -80 °C and centralized for metagenome sequencing.
Genomic DNA was extracted from all mucosal samples using the Invitrogen PureLink genomic DNA kit (Life Technologies, United States), according to the manufacturer’s instructions. Nucleic acids were characterized by 1% agarose gel electrophoresis, quantified using a Qubit 2.0 fluorometer, and stored at -20 °C.
DNA libraries were prepared by using the Illumina TruSeq DNA PCR-free sample prep kits (low-throughput protocol) following the manufacturer’s instructions. One microgram of each genomic DNA sample was sheared using a Covaris S2 sonication device (Covaris, United States) to obtain 350 bp inserts. After sonication, the DNA fragments were end repaired, and the appropriate library size was selected using different ratios of sample purification beads. Then the samples were A-tailed and ligated to adapters. Finally, the size and concentration of the DNA fragments were assessed using the KAPA library quantification kit for the Illumina platforms (Kapa Biosystems, United States). Approximately 8 pmol of each 350-bp library was paired-end sequenced (2 × 100 bp) on an Illumina PE150 instrument (Illumina, United States).
The raw data were preprocessed to remove low-quality data. Fastp was used to remove the splice sequences automatically, and low-quality sequences (average quality score < 15) and short sequences (final length < 15 bp) were removed. Bowtie2 (parameter: --very-sensitive) was used to remove the host sequences. Finally, valid sequences (clean data) were obtained.
All valid sequences from all samples were annotated and categorized using Kraken2 to investigate the species composition and diversity of the samples. The classifier matched each k-mer in the query sequence to the lowest common ancestor of all genomes containing the given k-mer for classification. The alpha diversity indices, including Chao1, Shannon, and Simpson indices, were calculated with QIIME, and statistical significance of differences was determined using the Kruskal-Wallis test. To explore beta diversity, principal coordinate analysis (PCoA) on weighted UniFrac distances was used. Permutational multivariate analysis of variance (Adonis function) and analysis of similarities (Anosim function) by Bray-Curtis distances were carried out with the PRIMER 6 software package to compare microbial communities among the groups. A linear discriminant analysis (LDA) effective size algorithm was introduced using the LDA effective size (LEfSe) tool to identify the features that were most likely to explain differences among the groups by an LDA score greater than 2.
HUMAnN3 was used to compare the clean reads with the information in a protein database (UniRef90; based on DIAMOND). After filtering out the reads that failed to be compared using the HUMAnN3 default comparison parameters (translated query coverage threshold = 90.0; prescreen threshold = 0.01; evalue threshold = 1.0; and translated subject coverage threshold = 50.0), the relative abundance of each protein in UniRef90 and the relative abundance of the corresponding function in each functional database were determined. The MetaCyc database (https://metacyc.org/) and KEGG ORTHOLOGY database (https://www.genome.jp/kegg/ko.html) were used for further metabolic pathway and protein function annotation. Based on the gene annotation information, combined with the functional abundance information, a comparative analysis of metabolic pathways between samples was performed, and the information was subsequently entered into iPath 3 for visualization (https://pathways.embl.de/).
Continuous variables with a normal distribution are presented as the mean ± SD, and nonnormally distributed variables are reported as the median (interquartile range). Categorical variables are presented as percentages. The Kruskal-Wallis test or Wilcoxon test was used to assess differences between the groups. Statistical analyses were performed using SPSS 19, and a two-sided P value < 0.05 was considered to indicate statistical significance.
A total of 12 participants, including 4 patients with GU (group A, H. pylori-positive, 2 females and 2 males), 4 with DU (group B, H. pylori-positive, 2 females and 2 males), and 4 healthy individuals (group C, H. pylori-negative, 2 females and 2 males), were recruited for this study. No significant differences in the gender ratios (50% vs 50% vs 50%) or age [48 (5.00) vs 44 (5.25) vs 47.5 (5.75) years; P = 0.607] were detected among the groups. All participants did not use probiotics for 2 mo and ate a normal diet as usual.
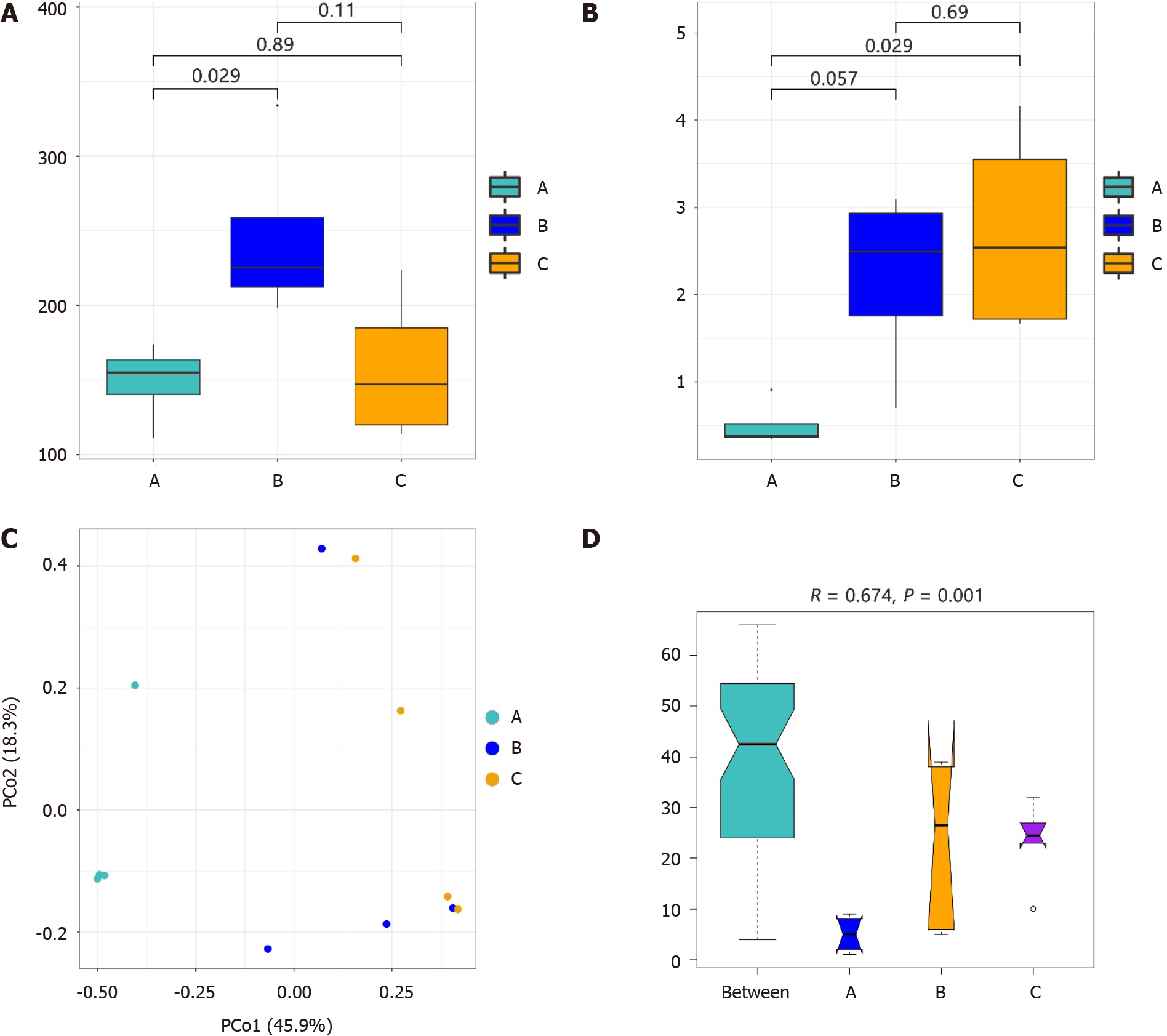
An average of 3135070 high-quality sequence reads per sample were obtained after the quality-filtering steps. After annotating and categorizing the valid sequences, 149, 246, and 158 species were identified in groups A, B, and C, respectively. The results of the alpha diversity of the microbiota are shown in Figure 1A and B. The Chao1 index of the duodenal mucosal microbiota (group B) was greater than that of the gastric mucosa microbiota (groups A and C), especially compared with that of group A (Wilcoxon rank-sum test, P = 0.029). However, alpha diversity in groups B and C was greater than that in group A based on the Shannon index (P = 0.029 between groups A and C).
The similarities in the bacterial community structure are shown in Figure 1C, and significant differences were detected among the groups (Anosim, R = 0.674, P = 0.001; Figure 1D). The microbial structure in the gastric mucosal tissue was different from that in the duodenum of patients with H. pylori infection. We also found that H. pylori infection in the gastric mucosa influenced the structure of the duodenal mucosal microbiota. The gastric mucosal microbiota structure differed between groups A and C.
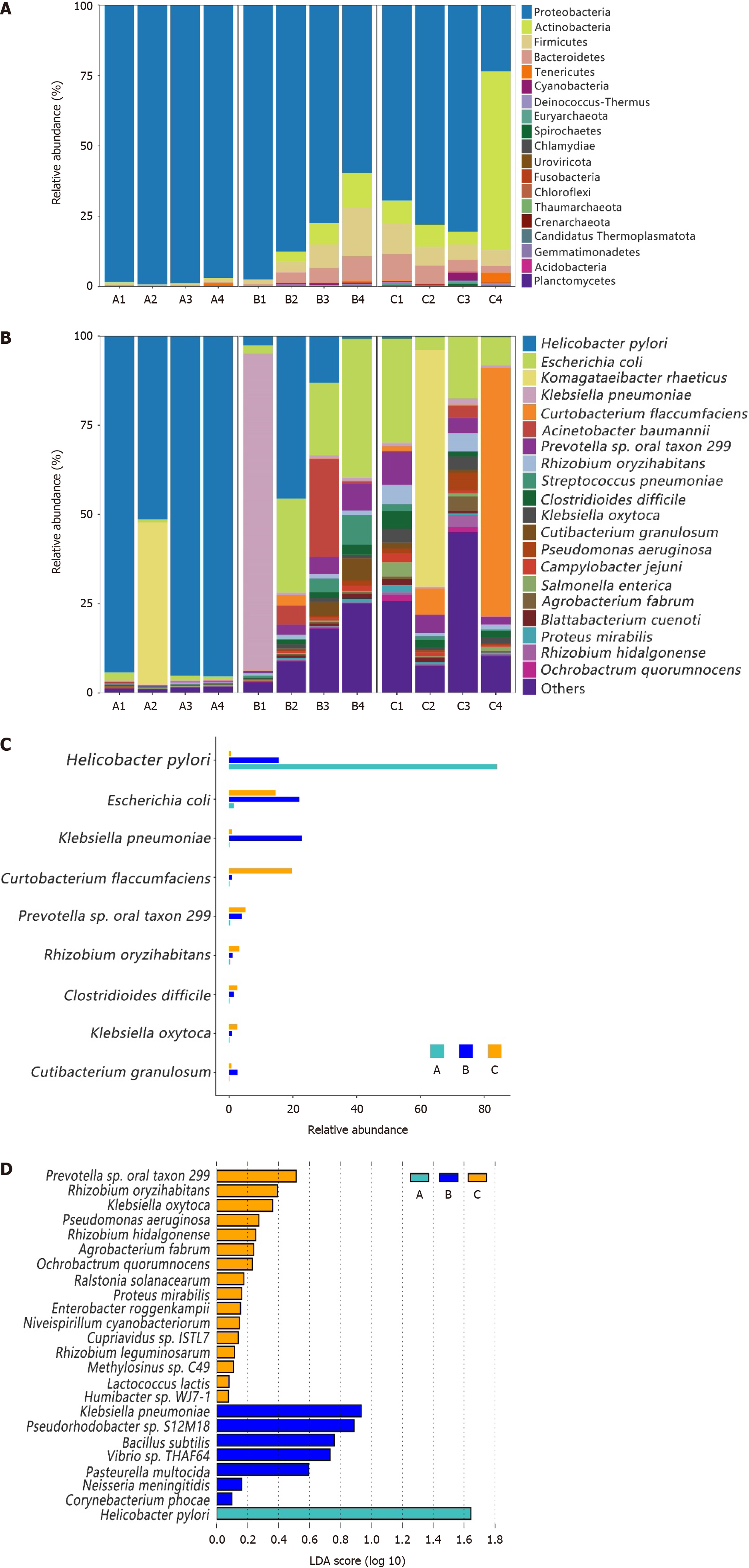
Most of the gastric and duodenal mucosal bacteria detected in this study belonged to the following four phyla: Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes (Figure 2A). Except for two unique highly abundant species, the main 11 species (with average percentages above 1%) in the gastric microbiota from group C comprised up to 58.48% of the total microbiota and included the following species: Escherichia coli (E. coli), Prevotella sp. oral taxon 299, Rhizobium oryzihabitans, Curtobacterium flaccumfaciens, Klebsiella oxytoca, Clostridioides difficile, Pseudomonas aeruginosa, Salmonella enterica, Agrobacterium fabrum, Campylobacter jejuni, and Rhizobium hidalgonense (Figure 2B). However, the main species in the gastric microbiota from group A (H. pylori-positive) was H. pylori, which comprised up to 94% of the total microbiota, except for the A2 sample, in which Komagataeibacter rhaeticus and H. pylori accounted for 45.49% and 51.45%, respectively. In the duodenal mucosal microbiota, the species E. coli, H. pylori, Acinetobacter baumannii, Prevotella sp. oral taxon 299, Streptococcus pneumoniae, Cutibacterium granulosum, Clostridioides difficile, and Rhizobium oryzihabitans were found to be enriched in patients with DU and H. pylori infection (group B) (Figure 2B). We compared the proportions of dominant species among the groups and found that most of them significantly differed (P < 0.05), as shown in Figure 2C.
The specific taxa that were most likely to contribute to the differences among the groups are shown in Figure 2D. Sixteen species, including Rhizobium oryzihabitans, Klebsiella oxytoca, Pseudomonas aeruginosa, Rhizobium hidalgonenses, and Agrobacterium fabrumI, were enriched in the gastric mucosal microbiota of healthy individuals (group C), while 7 species, including Bacillus subtilis, Pasteurella multocida, and Neisseria meningitides, were enriched in the duodenal mucosal microbiota of H. pylori-positive patients with DU. Only H. pylori was enriched in the gastric mucosal microbiota of patients who were positive for H. pylori. We observed specific taxa within each group. However, they were not suffi
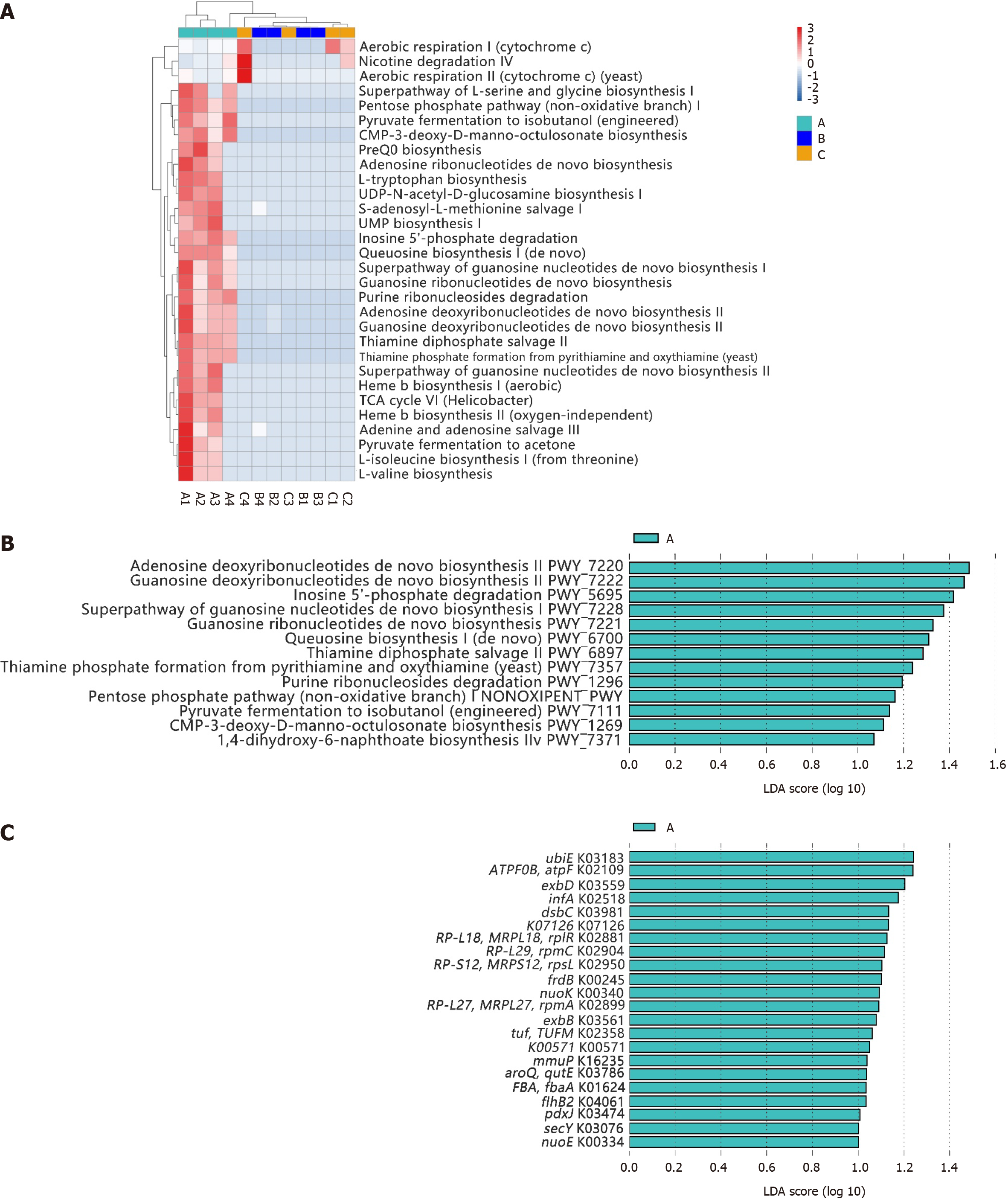
The top 30 identified pathways are shown in Figure 3A. Similar to the microbial compositions, differences in the major predicted functional profiles between gastric microbiota in participants with or without H. pylori infection were significant (group A vs group B). The duodenal microbiota in participants with H. pylori infection (group B) was similar to the gastric microbiota in healthy individuals (group C). The LEfSe algorithm revealed 13 significantly different pathways in group A, mainly related to the synthesis, conversion, and degradation of nucleotides (PWY-5695, PWY-7220, PWY-7221, PWY-7222, PWY-7228, and PWY0-1296), carbohydrate metabolism (PWY-1269), and vitamin metabolism (PWY-6897, PWY-7357, and PWY-7371). Additionally, a specific biosynthetic pathway was identified [PWY-1269, queuosine (Q) biosynthesis I (de novo)], which is involved in the biosynthesis of Q and focused on the modification of transfer RNA (tRNA) (Figure 3B). The synthesis and degradation of inosine (PWY-5695), adenosine (PWY-7220), and guanosine (PWY-7221 and PWY-7222) are directly related to the composition of DNA and RNA, while the synthesis of thiamine phosphate (PWY-6897 and PWY-7357) and menaquinone (MK; PWY-7371) is associated with the activity of certain cofactors and enzymes. The source of the species that was most likely to contribute to the different functions was H. pylori (Supplementary Figure 1).
Additionally, the LEfSe algorithm revealed 22 significantly different orthologous groups in group A, including K03183 (ubiE), K02109 (TPF0B, atpF), K03559 (exbD), K02518 (infA), and K03981 (dsbC) (Figure 3C). These groups play roles across several critical biological nodes, such as nucleotide and carbohydrate metabolism, quinone substance metabolism, flagella synthesis, and synthesis of transport proteins. Visual display diagrams of the differences in metabolic pathways (Supplementary Figure 2) and microbial metabolic pathways (Supplementary Figure 3) were generated.
The primary objective of this study was to explore the gastric microbiota composition in H. pylori-infected patients with GU and its correlation with the pathogenesis and development of GU based on metagenomic sequencing. The secondary objective of this research was to investigate the duodenal microbiota composition in H. pylori-infected patients with DU and its relationship to ulcer formation.
Compared with those in healthy individuals, the gastric microbial communities in the H. pylori-infected patients with GUs were dominated by a single bacterial group, with H. pylori as the predominant species and other bacteria present at considerably lower relative abundances. These findings suggest that the stomach, with its acidic environment, may harbor a lower absolute quantity of colonizing bacteria. Hence, a high proportion of H. pylori could reduce the relative abundances of other bacterial species. Additionally, after colonization, H. pylori may suppress the survival of other members of the gastric microbiota through mechanisms such as releasing virulence factors, altering gastric acidity, inducing host immune responses, and competing with resident microbial populations[6-8]. Zheng et al[9] analyzed the gastric microbiome and observed significant decreases in the relative abundances of Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, Gemmatimonadetes, and Verrucomicrobia compared with those in the H. pylori-negative group. Moreover, at the genus level, Achromobacter, Devosia, Halomonas, Mycobacterium, Pseudomonas, Serratia, Sphingopyxis, and Stenotrophomonas were more abundant in the H. pylori-negative group, while only Helicobacter was more abundant in the H. pylori-positive group, similar to our results. In this study, the gastric microbiota of healthy individuals was found to be diverse, with 16 species including Rhizobium oryzihabitans, Klebsiella oxytoca, Pseudomonas aeruginosa, Rhizobium hidalgonenses, and Agrobacterium fabrum being enriched in the gastric mucosal microbiota. In contrast, in the stomachs of H. pylori-infected patients with GU, H. pylori accounted for as much as 94% of the microbial population, thus representing a superdominant species. The Chao1 index indicated that the microbial diversity in the stomach was low. However, there was no significant difference in the gastric microbial diversity between those infected and not infected with H. pylori. Nevertheless, the microbial evenness in the H. pylori-infected GU group was significantly lower than that in the healthy control group (group A vs group C, P = 0.029).
Furthermore, we found that the differential functions identified in group A were almost exclusively derived from H. pylori. The Q biosynthesis I pathway involves the conversion of preQ0 to preQ1, followed by the attachment of preQ1 to tRNA through the action of tRNA guanine transglycosylase and, finally, by the transformation into Q by the QueA and QueG/QueH enzymes. This process is part of the process of tRNA Q-modification. tRNAs with Q-modifications can regulate the translation of genes containing NAU codons (termed Q-genes), such as queH in H. pylori, within the cell[10,11]. These Q-genes are primarily enriched in functions associated with cellular adhesion, biofilm formation, and virulence. Jorge Díaz-Rullo[12] studied three model organisms – E. coli, Bacillus subtilis, and yeast – and discovered Q enhanced biofilm formation in both Gram-negative and -positive bacteria. Q-tRNA also specifically induced the expression of virulence factors (such as lipopolysaccharide) in pathogenic E. coli ST131 strains, as well as the expression of toxins involved in interbacterial competition in Pseudomonas putida. Experimental evidence suggests that Q-tRNA can regulate bacterial adhesion and biofilm formation by modulating the translation of Q-genes, thereby enhancing the virulence of most human pathogenic bacteria. The release of adhesion and virulence factors, and biofilm formation by H. pylori are closely associated with the development and progression of GU. Upon infecting the host, H. pylori produces surface adhesins that attach to the gastric mucosa[13]. These adhesins contain toxins such as neutrophil-activating proteins, which stimulate the production of free radicals, leading to local tissue damage, increased synthesis of inflammatory cytokines such as IL-8 and tumor necrosis factor α, and inhibition of prostaglandin synthesis, resulting in inflammation and ulcer formation[14]. Damage to the inner wall of the gastric mucosa in the main body region of the stomach leads to acid sipping into epithelial cells, further inflaming the area and causing ulcers or exacerbating existing ulcers. H. pylori also interferes with normal mechanisms for repairing damaged gastric tissue, thus delaying the healing of existing ulcers or promoting the development of new ulcers[15]. Research by Díaz-Rullo et al[12] did not use H. pylori as a model organism to study the role of Q in regulating bacterial adhesion, virulence, and biofilm formation, which warrants further validation.
The 1,4-dihydroxy-6-naphthoate biosynthesis II pathway is involved in the synthesis of demethylmenaquinones (DMKs) by H. pylori via a unique futalosine pathway. This pathway was enriched in H. pylori-infected patients with GU (group A). Additionally, in group A, we detected an enrichment of the differentially expressed gene uibE, which plays a significant role in the biosynthesis of MK. MK and DMK are low-molecular-weight lipophilic components found within the cytoplasmic membrane of many bacterial species. The majority of anaerobic bacteria, both Gram-negative and -positive, contain MK or DMK as their primary quinones. MK possesses certain cytotoxic properties. It may promote oxidative stress-induced cellular damage by generating reactive oxygen species and depleting glutathione, which leads to damage to cell membranes, DNA, and cell organelles[16]. MK can also activate the mitochondrial pathway and caspase-8- and Bid-dependent pathways to mediate apoptosis[17]. MK and H2O2 were used to induce oxidative stress in a model Enterococcus faecium strain (MTCC 3031). Oxidative stress resulted in a significant reduction in the redox ratio (NADPH/NADP) by 55% (with MK) and 28% (with H2O2), leading to decreased bacterial folate synthesis. Reduced levels of folate can induce the development of colorectal cancer[18]. Additionally, oxidative stress significantly decreased the density of bacterial growth by 61% (with MK) and the biomass yield by 21% (with H2O2)[19]. These findings suggest that the oxidative stress induced by MK could contribute to damage to gastric mucosal tissue and may be one of the factors promoting the development and progression of GU.
Compared to patients with GU, those with DU exhibited a more diverse microbial structure in the duodenal mucosa, which were consistent with our earlier investigation using 16S ribosomal RNA sequencing[20]. Interestingly, compared to H. pylori-negative patients with DU, H. pylori was not the absolutely predominant bacteria, and had less effect on the diversity of duodenal mucosal flora in H. pylori-positive patients with DU in our another study on duodenal mucosal flora[21]. Additionally, H. pylori infection in the gastric mucosa also did not influence the structure of the duodenal mucosal microbiota[22]. We speculate that this might be due to the higher diversity of flora in the duodenal mucosa and the more intricate and tight-knit inter-flora interactions between the flora, which enable it to preserve a more stable duodenal flora. In our study, all four duodenal mucosal samples showed a certain proportion of H. pylori (ranging from 0.89% to 45.53%). H. pylori infection has been shown to reduce bicarbonate secretion in the duodenal mucosa, which can contribute to ulcer formation. Furthermore, H. pylori infection can affect the expression and function of CFTR and SLC26A6 in the duodenal mucosa, further impairing the ability of the duodenum to secrete bicarbonate[23]. However, owing to the greater diversity of the duodenal mucosal microbiota, H. pylori may be one of several factors contributing to the development of DU. Clinically, GU typically has a longer disease course than DU and is more likely to undergo malignant transformation, suggesting that H. pylori infection causes greater damage to gastric mucosal tissues than to the duodenum. The mechanisms underlying the development of DU require further investigation.
In summary, this study revealed that in H. pylori-positive patients with GU, the gastric mucosal microbiota was dominated by H. pylori, with a significant reduction in biodiversity, and intergroup differential functions were all derived from H. pylori. Pathways and related genes involved in tRNA Q-modification as well as the synthesis of MK and DMK were enriched in H. pylori-positive patients with GU, suggesting a potential association with the development and progression of GU. The microbial diversity in H. pylori-positive DU patients was not significantly different from that in healthy controls, indicating that H. pylori infection may be only one of several factors contributing to the occurrence of DU. The mechanisms underlying the development of DU require further investigation.
| 1. | Li R, Wang W, Ma Y, Chen H. Analysis of risk factors for ulcer recurrence and upper gastrointestinal bleeding in children with peptic ulcer treated with Helicobacter pylori eradication therapy. Transl Pediatr. 2023;12:618-630. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (33)] |
| 2. | Rosa IMS, Evangelista AP, Cabral LFF, Gripp MSDM, Machado MC. Úlcera Péptica - uma revisão abrangente sobre a etiologia, epidemiologia, diagnóstico, tratamento, complicações e prevenção. Braz J Hea Rev. 2023;6:24086-24095. [DOI] [Full Text] |
| 3. | Schroeder BO. Outlook on the gut microbiota. Gastroenterol Rep (Oxf). 2022;10:goac024. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (33)] |
| 4. | Ye J, Feng T, Su L, Li J, Gong Y, Ma X. Interactions between Helicobacter pylori infection and host metabolic homeostasis: A comprehensive review. Helicobacter. 2023;28:e13030. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (33)] |
| 5. | Wang Z, Shao SL, Xu XH, Zhao X, Wang MY, Chen A, Cong HY. Helicobacter pylori and gastric microbiota homeostasis: progress and prospects. Future Microbiol. 2023;18:137-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (34)] |
| 6. | Ren S, Cai P, Liu Y, Wang T, Zhang Y, Li Q, Gu Y, Wei L, Yan C, Jin G. Prevalence of Helicobacter pylori infection in China: A systematic review and meta-analysis. J Gastroenterol Hepatol. 2022;37:464-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 161] [Article Influence: 40.3] [Reference Citation Analysis (36)] |
| 7. | Hooi JKY, Lai WY, Ng WK, Suen MMY, Underwood FE, Tanyingoh D, Malfertheiner P, Graham DY, Wong VWS, Wu JCY, Chan FKL, Sung JJY, Kaplan GG, Ng SC. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology. 2017;153:420-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1361] [Cited by in RCA: 2207] [Article Influence: 245.2] [Reference Citation Analysis (3)] |
| 8. | Iino C, Shimoyama T. Impact of Helicobacter pylori infection on gut microbiota. World J Gastroenterol. 2021;27:6224-6230. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 39] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (3)] |
| 9. | Zheng W, Miao J, Luo L, Long G, Chen B, Shu X, Gu W, Peng K, Li F, Zhao H, Botchway BOA, Fang M, Jiang M. The Effects of Helicobacter pylori Infection on Microbiota Associated With Gastric Mucosa and Immune Factors in Children. Front Immunol. 2021;12:625586. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 10. | Tuorto F, Legrand C, Cirzi C, Federico G, Liebers R, Müller M, Ehrenhofer-Murray AE, Dittmar G, Gröne HJ, Lyko F. Queuosine-modified tRNAs confer nutritional control of protein translation. EMBO J. 2018;37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 153] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 11. | Kulkarni S, Rubio MAT, Hegedűsová E, Ross RL, Limbach PA, Alfonzo JD, Paris Z. Preferential import of queuosine-modified tRNAs into Trypanosoma brucei mitochondrion is critical for organellar protein synthesis. Nucleic Acids Res. 2021;49:8247-8260. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 12. | Díaz-Rullo J, González-Pastor JE. tRNA queuosine modification is involved in biofilm formation and virulence in bacteria. Nucleic Acids Res. 2023;51:9821-9837. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 34] [Reference Citation Analysis (0)] |
| 13. | Chmiela M, Kupcinskas J. Review: Pathogenesis of Helicobacter pylori infection. Helicobacter. 2019;24 Suppl 1:e12638. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 77] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 14. | Kao CY, Sheu BS, Wu JJ. Helicobacter pylori infection: An overview of bacterial virulence factors and pathogenesis. Biomed J. 2016;39:14-23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 199] [Cited by in RCA: 317] [Article Influence: 31.7] [Reference Citation Analysis (3)] |
| 15. | Sgouras DN, Trang TT, Yamaoka Y. Pathogenesis of Helicobacter pylori Infection. Helicobacter. 2015;20 Suppl 1:8-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 70] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 16. | Loor G, Kondapalli J, Schriewer JM, Chandel NS, Vanden Hoek TL, Schumacker PT. Menadione triggers cell death through ROS-dependent mechanisms involving PARP activation without requiring apoptosis. Free Radic Biol Med. 2010;49:1925-1936. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 247] [Cited by in RCA: 243] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 17. | Kim YJ, Shin YK, Sohn DS, Lee CS. Menadione induces the formation of reactive oxygen species and depletion of GSH-mediated apoptosis and inhibits the FAK-mediated cell invasion. Naunyn Schmiedebergs Arch Pharmacol. 2014;387:799-809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 18. | Keum N, Giovannucci EL. Folic acid fortification and colorectal cancer risk. Am J Prev Med. 2014;46:S65-S72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 19. | Jose S, Bhalla P, Suraishkumar GK. Oxidative stress decreases the redox ratio and folate content in the gut microbe, Enterococcus durans (MTCC 3031). Sci Rep. 2018;8:12138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 20. | Chen X, Xia C, Li Q, Jin L, Zheng L, Wu Z. Comparisons Between Bacterial Communities in Mucosa in Patients With Gastric Antrum Ulcer and a Duodenal Ulcer. Front Cell Infect Microbiol. 2018;8:126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 21. | Chen X, Xia CM, Dai ZY, Li QQ, Jin LX, Lin PL, Zhang ZC, Zhong HB, Yan HF, Wang GP, Dai L, Zhong YF, Wu ZB. The Relationship Between Helicobacter pylori Colonization and Duodenal flora Structure in Duodenal Ulcer. Curr Biotechnol. 2019;5:536-542. [DOI] [Full Text] |
| 22. | Gong J, Li L, Zuo X, Li Y. Change of the duodenal mucosa-associated microbiota is related to intestinal metaplasia. BMC Microbiol. 2019;19:275. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 23. | Wen G, Jin H, Deng S, Xu J, Liu X, Xie R, Tuo B. Effects of Helicobacter pylori Infection on the Expressions and Functional Activities of Human Duodenal Mucosal Bicarbonate Transport Proteins. Helicobacter. 2016;21:536-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: Https://creativecommons.org/Licenses/by-nc/4.0/