Published online Mar 28, 2024. doi: 10.3748/wjg.v30.i12.1680
Peer-review started: December 4, 2023
First decision: January 23, 2024
Revised: February 1, 2024
Accepted: March 8, 2024
Article in press: March 8, 2024
Published online: March 28, 2024
Processing time: 114 Days and 16.6 Hours
After the study of circulating tumor cells in blood through liquid biopsy (LB), this technique has evolved to encompass the analysis of multiple materials originating from the tumor, such as nucleic acids, extracellular vesicles, tumor-educated platelets, and other metabolites. Additionally, research has extended to include the examination of samples other than blood or plasma, such as saliva, gastric juice, urine, or stool. LB techniques are diverse, intricate, and variable. They must be highly sensitive, and pre-analytical, patient, and tumor-related factors significantly influence the detection threshold, diagnostic method selection, and potential results. Consequently, the implementation of LB in clinical practice still faces several challenges. The potential applications of LB range from early cancer detection to guiding targeted therapy or immunotherapy in both early and advanced cancer cases, monitoring treatment response, early identification of relapses, or assessing patient risk. On the other hand, gastric cancer (GC) is a disease often diagnosed at advanced stages. Despite recent advances in molecular understanding, the currently available treatment options have not substantially improved the prognosis for many of these patients. The application of LB in GC could be highly valuable as a non-invasive method for early diagnosis and for enhancing the management and outcomes of these patients. In this comprehensive review, from a pathologist’s perspective, we provide an overview of the main options available in LB, delve into the fundamental principles of the most studied techniques, explore the potential utility of LB application in the context of GC, and address the obstacles that need to be overcome in the future to make this innovative technique a game-changer in cancer diagnosis and treatment within clinical practice.
Core Tip: Liquid biopsy (LB) has the potential to revolutionize cancer diagnostics, offering reduced invasiveness and improved understanding of tumor heterogeneity. Going beyond examining circulating tumor cells and cell-free DNA in blood, LB studies now explore a variety of structures and non-blood samples. Despite advancements in LB for tumor detection, prognostic assessment and treatment guidance, challenges remain, including complex and expensive techniques, lack of standardization, and suboptimal scientific evidence. In the context of gastric cancer, LB represents a promising approach, especially in advanced stages. This review navigates through LB intricacies, emphasizing its benefits while urging for future improvements to achieve clinical impact.
- Citation: Díaz del Arco C, Fernández Aceñero MJ, Ortega Medina L. Liquid biopsy for gastric cancer: Techniques, applications, and future directions. World J Gastroenterol 2024; 30(12): 1680-1705
- URL: https://www.wjgnet.com/1007-9327/full/v30/i12/1680.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i12.1680
Over the past few decades, remarkable technical and molecular advancements in the field of biological sciences, particularly in cancer research, have given rise to innovative techniques that improve conventional approaches. Liquid biopsy (LB) has emerged as a groundbreaking method for detecting and analyzing specific molecules and cells in bodily fluids. While the majority of LB studies have focused on circulating tumor cells (CTCs) or circulating tumor DNA (ctDNA) in peripheral blood samples, other possibilities, such as the analysis of exosomes or tumor-educated platelets (TEPs), or the utilization of non-blood samples, have come to the forefront. The primary objectives of LB in cancer encompass early tumor diagnosis, treatment monitoring, timely detection of relapses, prognosis determination, and the identification of therapeutic biomarkers in both early and advanced cancer cases. In contrast to conventional biopsy, LB holds the advantages of being a minimally invasive technique that can capture the spatial and temporal tumor heterogeneity, by reflecting various cell clones present in the body at a given moment. However, LB techniques must be highly sensitive, and pre-analytic factors, patient and tumor features significantly impact the detection threshold, diagnostic methodology, and potential outcomes. Additionally, LB techniques are complex, expensive, and currently lack standardization. Consequently, the integration of LB into clinical practice faces several challenges.
On another note, gastric cancer (GC) is a disease with a poor prognosis, particularly in Western countries. Significant differences exist between Western and Asian patients, with GC being more prevalent in Asian countries. Geographic variability has been also observed in clinical, histological, prognostic, molecular and treatment response factors. While some high-incidence countries like Japan and Korea have implemented screening strategies that improve patient prognosis, these techniques are not considered to be cost-effective in Western countries, leading to the detection of most cases at late stages. Furthermore, recent advances in molecular pathology and targeted therapies have not significantly influenced the prognosis of GC patients. Therefore, there is a need to facilitate early diagnosis, improve patient selection for various treatments, and identify new therapeutically relevant biomarkers. In this scenario, the application of LB holds great potential for GC. Previous studies have assessed the utility of analyzing CTCs, ctDNA, and other molecules in peripheral blood samples from GC patients in different contexts. The potential value of using non-blood samples, such as gastric juice or urine, is also under investigation.
In this comprehensive review, from a pathologist’s perspective, we provide an overview of the main options available in LB. We delve into the fundamental principles of the most studied techniques, explore the potential utility of LB in GC, and address the obstacles that need to be overcome in the future to make this innovative technique a game-changer in cancer diagnosis and treatment within clinical practice.
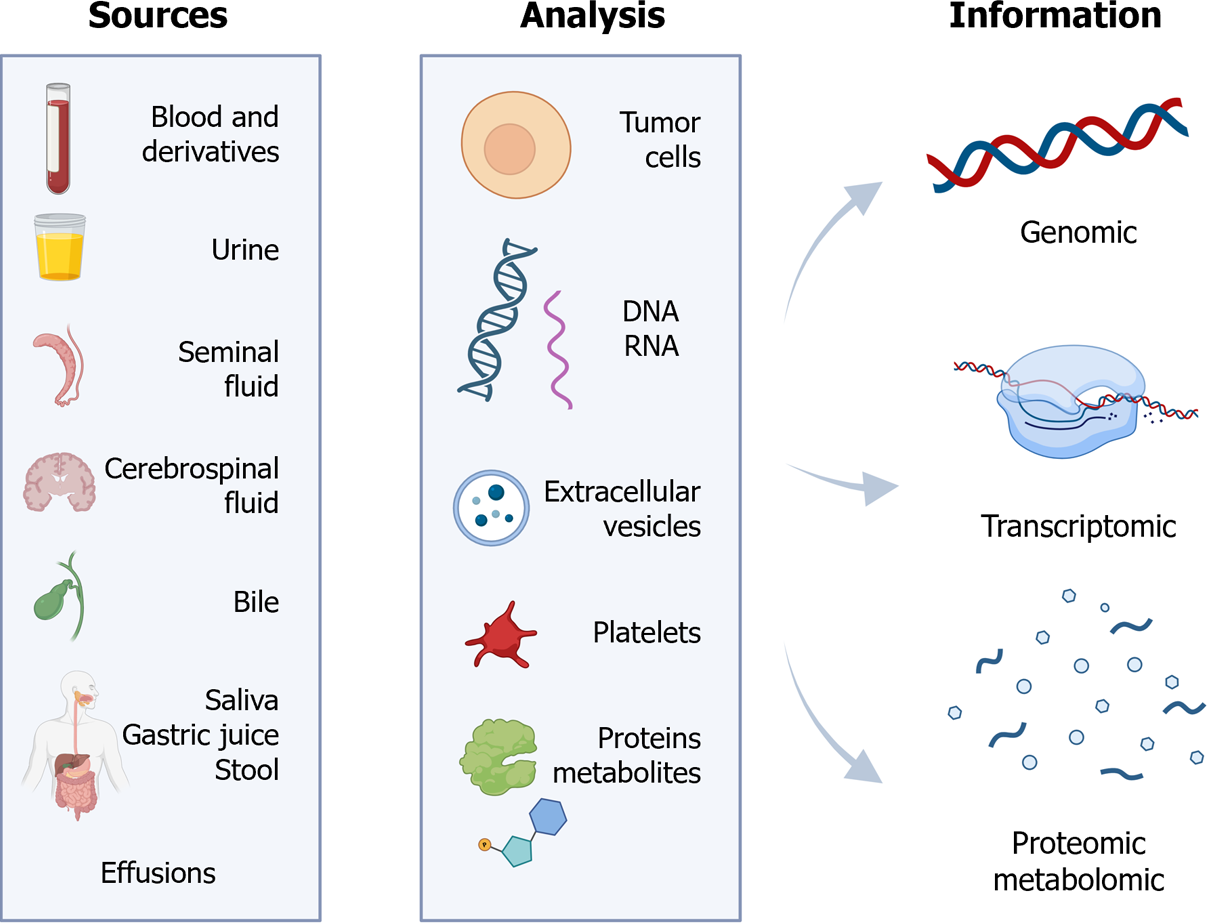
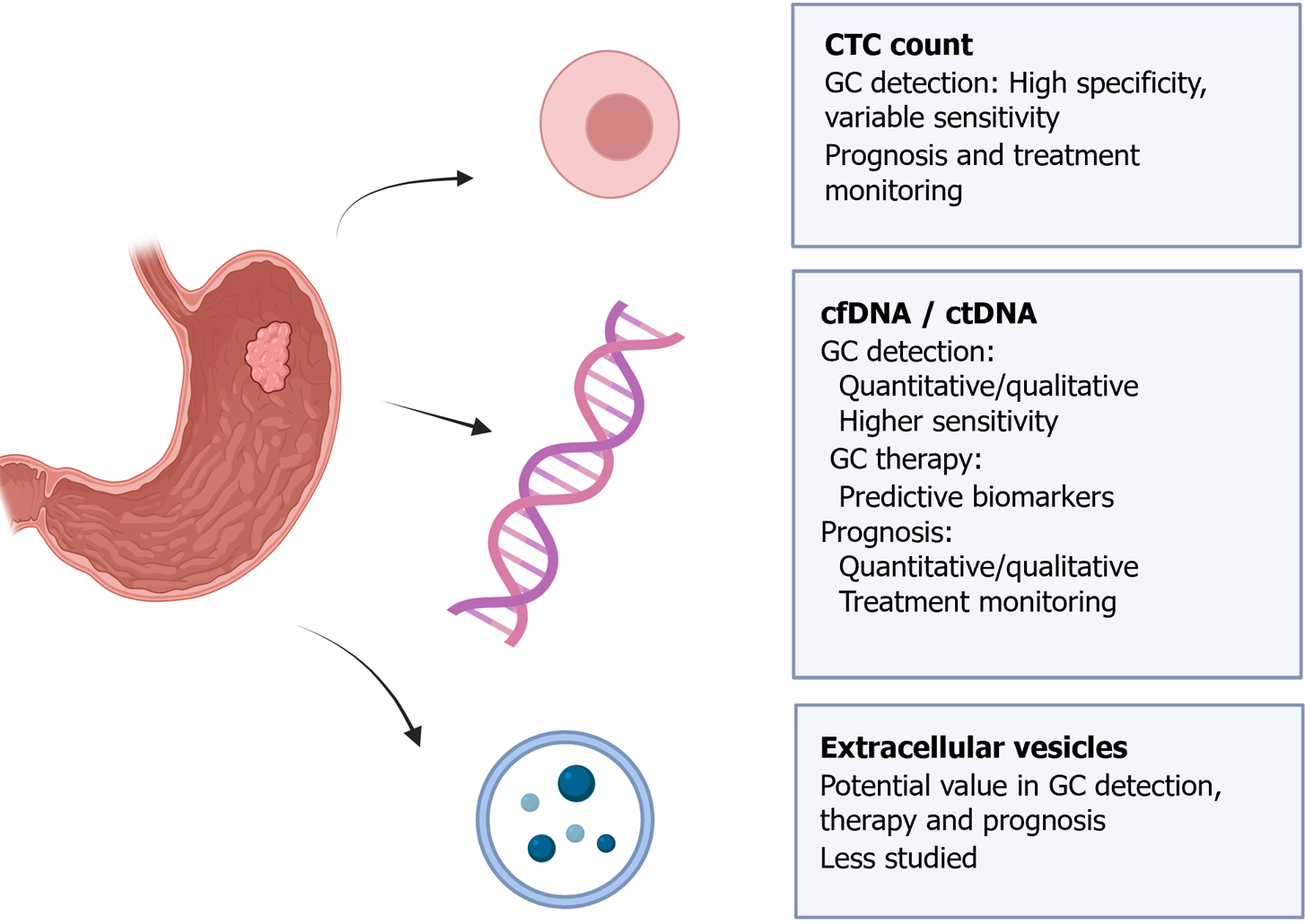
LB is a term commonly used to refer to the detection and analysis of circulating DNA or cells in peripheral blood. However, it encompasses laboratory examinations of various bodily fluids, and includes the detection of diverse tumor-associated structures[1] (Figure 1).
Consequently, LB can be applicable to fluids such as blood, serum, plasma, urine, sputum, or cerebrospinal fluid. In addition, it enables the identification of CTCs, cell-free DNA (cfDNA), ctDNA, cell-free RNA (cfRNA), long non-coding RNA (lncRNA), microRNA (miRNA), extracellular vesicles, TEPs, proteins, or metabolites[2].
Exploring the theoretical foundations of LB reveals its historical roots in ancient Greece, where the belief in the diagnostic potential of bodily fluids was prevalent[3]. Notably, in the 5th century B.C., Hippocrates formulated the humoral theory, linking the onset of disease to imbalances among the four humors of the body[4].
Moving closer to the present, the subspecialty of cytology emerged in the 19th century in the field of pathology. Cytology involves the analysis of fluids for diagnostic purposes, as opposed to the use of “solid” tissue fragments in conventional histology. Exfoliative and aspiration cytology are the two primary branches[5] (Figure 2).
The evolution of cytology in pathology parallels advances in cancer knowledge: originally a tool for morphological diagnosis under an optical microscope, it has become instrumental in the morphological-molecular diagnosis[6]. Despite some overlap between cytology, when employed for current molecular diagnostic purposes, and LB, key distinctions arise. Notably, blood, plasma, or serum samples are not typical subjects of cytological studies. Moreover, cytology enables the correlation of findings from complementary techniques (molecular, immunohistochemical, or histochemical) with the microscopic morphology of the cells present in the sample. Additionally, detection procedures in LB should exhibit heightened sensitivity, owing to the scarcity of target molecules or cells in LB samples.
Narrowing the focus to the field of LB, its foundational contributions can be attributed to Dr. Thomas Ashworth in 1869. In that year, a study discovered the existence of CTCs in the plasma of a patient with metastatic cancer[7]. Sub-sequent observations highlighted the prognostic value of CTCs in oncology, their presence in early-stage tumors, and their potential utility in guiding patient prognosis or treatment[8]. The first commercial approval for CTC utilization in clinical practice materialized in 2004, with the United States Food and Drug Administration (FDA) endorsing the CellSearch® platform for isolating and enumerating CTCs in metastatic breast cancer patients. This approval extended in 2007 and 2008 to encompass patients with colorectal and prostate cancer[9]. While the CellSearch® system remained the sole entity granted for capturing CTCs until 2022, other technologies, such as the Parsortix® PC1 system, have recently garnered approval[10].
The analysis of nucleic acids in blood was addressed later, beginning with cfDNA. The first detection of this molecule in plasma occurred in 1948[11]. Decades later, it was observed that the amount of cfDNA is higher in cancer patients, and it was confirmed that these molecules could originate from tumor cells. In the 1990s, a significant stride was made as mutations began to be discerned using cfDNA extracted from blood and other samples of patients with solid and hematological tumors, followed by the performance of methylation studies[12]. Clinical determination of genetic alterations in cfDNA became possible in 2005, but it was not until 2014 and 2016 that cfDNA-based mutational tests secured approval from two pivotal international regulatory bodies: European Medicines Agency (EMA) and FDA. In 2014, the EMA authorized the identification of EGFR mutations in cfDNA extracted from blood samples of lung carcinoma patients, with the aim of guiding gefitinib treatment when no tissue sample was available[13]. In 2016, the FDA approved the detection of specific EGFR mutations in blood samples to facilitate patient selection for erlotinib treatment[14]. Simultaneously, in 2016, the use of cfDNA was approved to detect the EGFR T790M mutation and guide treatment with osimertinib in lung cancer patients[15].
LB offers notable advantages, including: (1) The non/minimal invasiveness of the technique; (2) the ability to investigate both the spatial and temporal heterogeneity of tumors; and (3) the possibility of conducting serial studies[16]. Considering the molecular divergence between primary tumors and various metastatic sites, it is advisable to study the profile of each of them[17]. This approach proves particularly valuable in patients with already diagnosed primary tumors and metastatic lesions in challenging locations, in order to tailor specific treatments for each cell subclone and enhance the likelihood of eradicating all tumor cells in the body.
However, LB is not without its drawbacks. A significant limitation arises from the substantial methodological variability observed across different studies, hindering the standardization of LB procedures and the comparison of research results. Further exploration of these disadvantages is provided in the section addressing “future challenges”.
As previously mentioned, LB can analyze CTCs, circulating nucleic acids, extracellular vesicles, TEPs, proteins, or metabolites[2]. Among these, the most studied structures are CTCs and cfDNA.
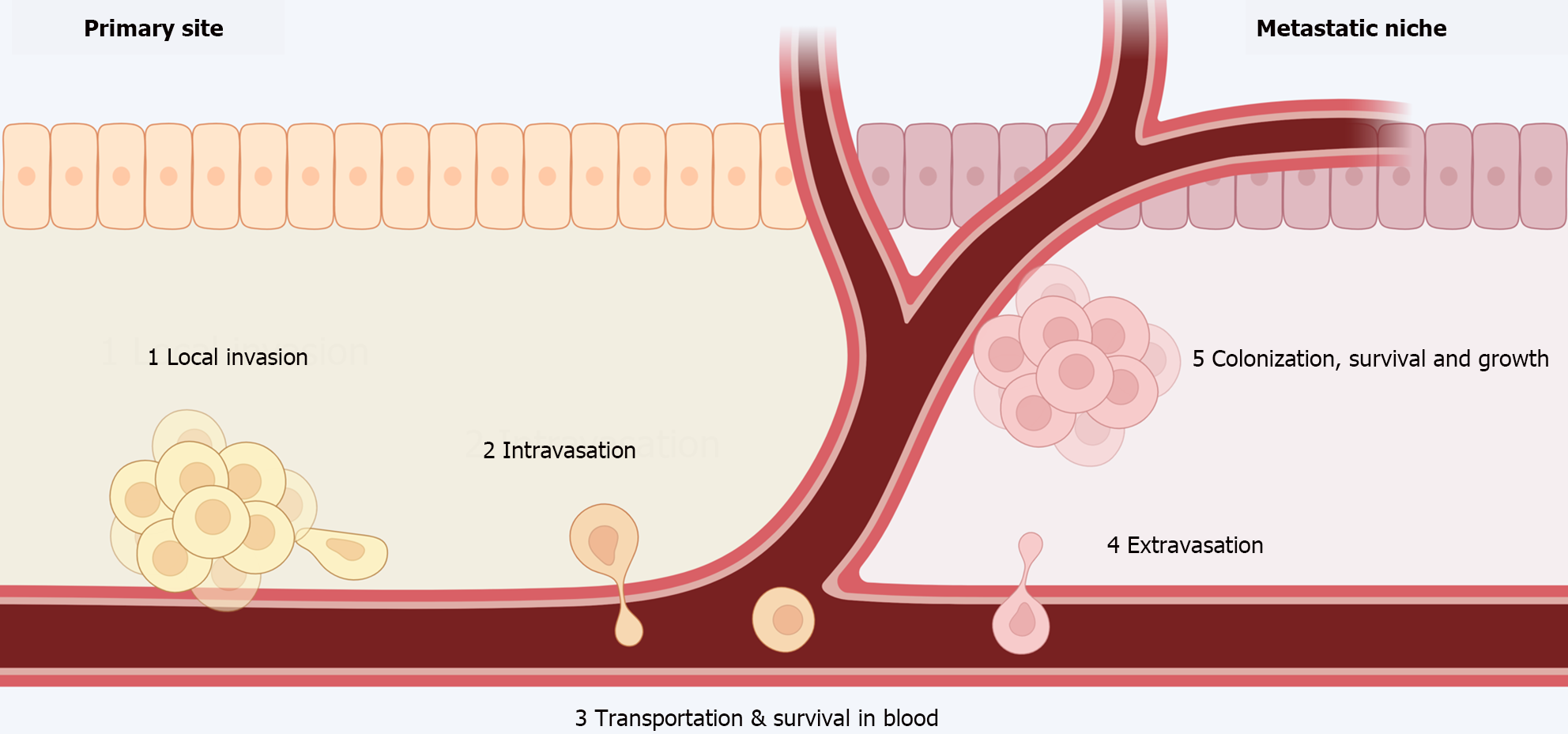
Metastases are the leading cause of therapeutic failure and cancer-related deaths. The metastatic process involves the steps depicted in Figure 3[18,19].
CTCs are tumor cells that detach from the primary tumor and circulate in the blood. They generally constitute a very small percentage of blood-circulating cells, which are predominantly leukocytes[20]. Remarkably, CTCs are considered as rare as one in a billion circulating cells[21]. Furthermore, only a small percentage of these cells will survive and give rise to metastases, underscoring the crucial interaction between the blood and metastatic niche microenvironment and CTCs[22].
Blood circulation and interaction with immune cells: CTCs circulate in the peripheral blood both individually and in clusters, and can interact with various immune cells. This interaction may protect CTCs from the aggressive blood microenvironment and promote their survival, proliferation, migratory capacity, and metastatic potential[23].
Interaction with neutrophils: Neutrophils are the most abundant leukocytes in the blood. The CTC-neutrophil interaction can support tumor progression, either through direct neutrophil-CTC interaction (via adhesion molecules) or indirectly, through the release of certain substances or the formation of structures by neutrophils, including neutrophil extracellular traps (NETs)[24,25].
Interaction with macrophages: In vitro studies have shown that the interaction between CTCs and circulating monocytes in the blood can promote their differentiation into macrophages, the secretion of various mediators for leukocyte recruitment, and the migration and invasiveness of CTCs[26]. Moreover, this interaction may enhance the intravasation of CTCs and epithelial-mesenchymal transition (EMT), boosting tumor heterogeneity and improving the metastatic potential of CTCs[27].
Interaction with platelets: Activated circulating platelets promote the survival of CTCs and their colonization and growth in secondary sites[28]. CTC-platelet aggregates appear to protect CTCs from mechanical stress and NK cells[29]. Additionally, CTC-platelet interactions can promote vascular permeability and CTC adhesion to endothelial cells, facilitating tumor extravasation to metastatic locations[30].
Interaction with myeloid-derived suppressor cells (MDSCs): MDSCs have immunosuppressive and metastasis-promoting capabilities. The MDSC-CTC interaction seems to help CTCs evade T-cell-mediated responses and promote their proliferation[31].
Interaction with cancer-associated fibroblasts (CAFs): Preliminary studies indicate that CTCs can transport CAFs to metastatic locations, and CAFs could protect CTCs from mechanical stress and promote their survival, invasion, and EMT[32,33].
Isolation and detection: As mentioned earlier, CTCs are extremely rare, occurring at a frequency of approximately 1 in a billion peripheral blood cells. This poses a significant technological challenge for their isolation. Advances in detection techniques, as reflected in recent studies, are anticipated to continue progressing[9,23]. Due to their scarcity, enrichment strategies become essential for separating CTCs from blood cells, enabling the acquisition of a CTC-enriched fraction for CTC purification or downstream analyses.
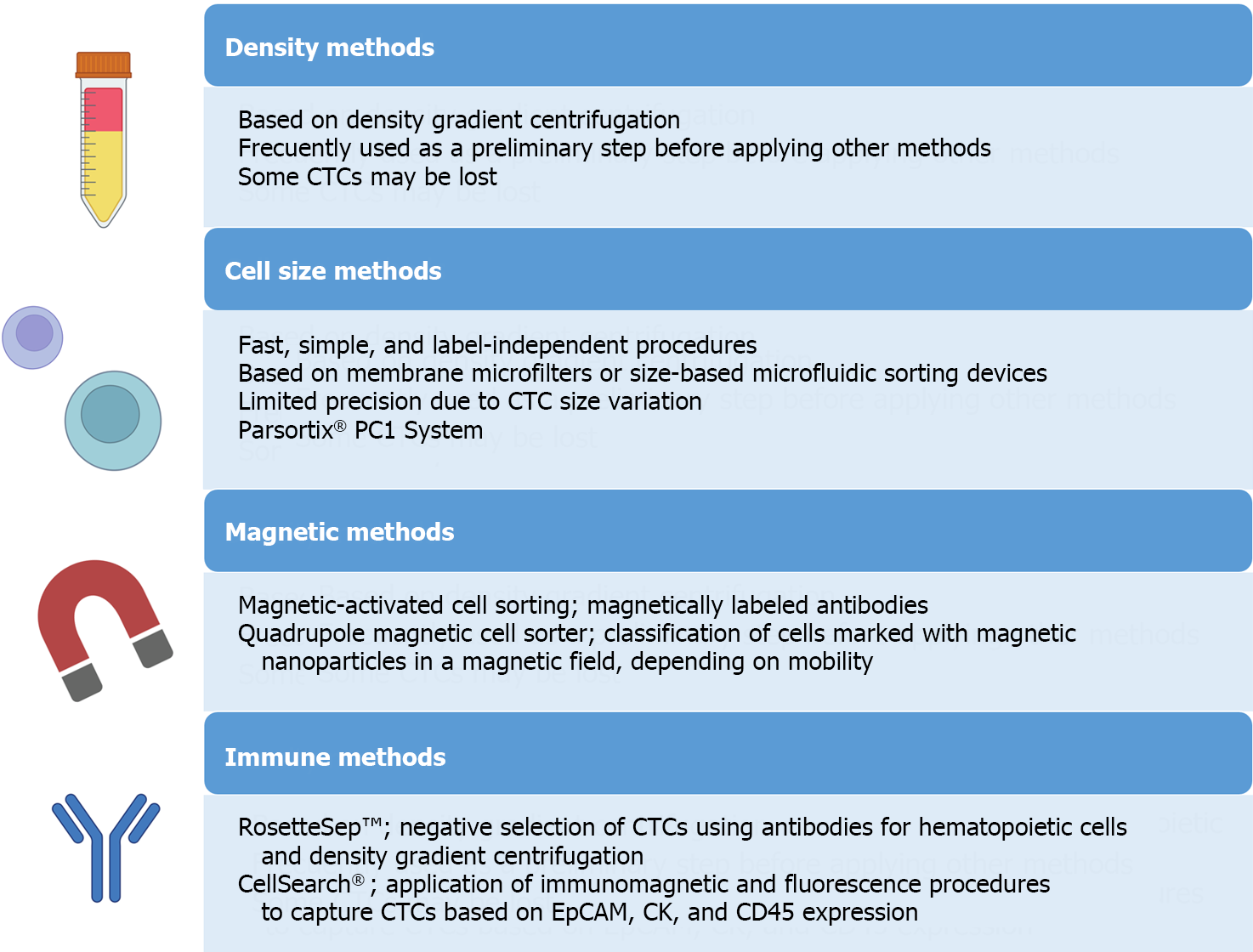
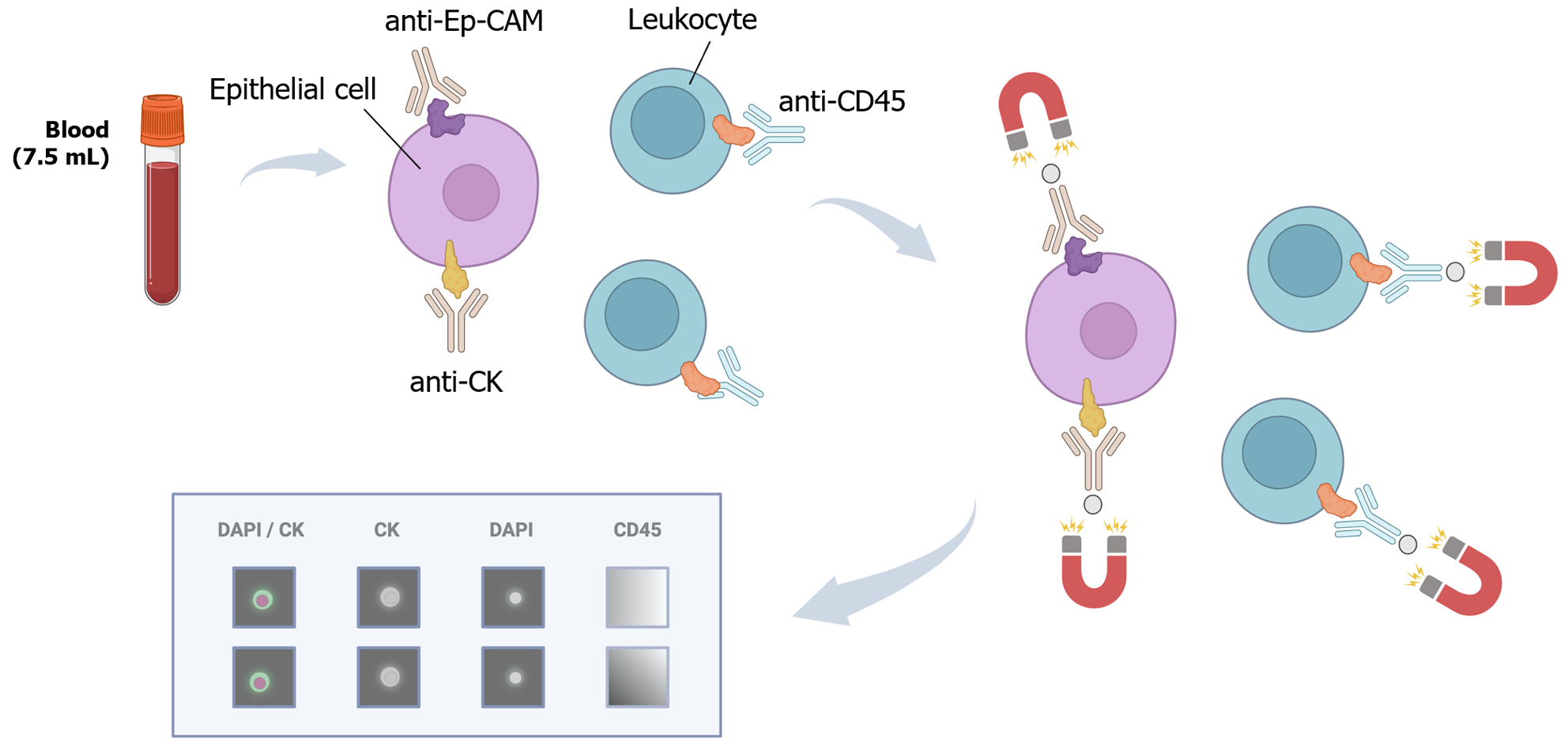
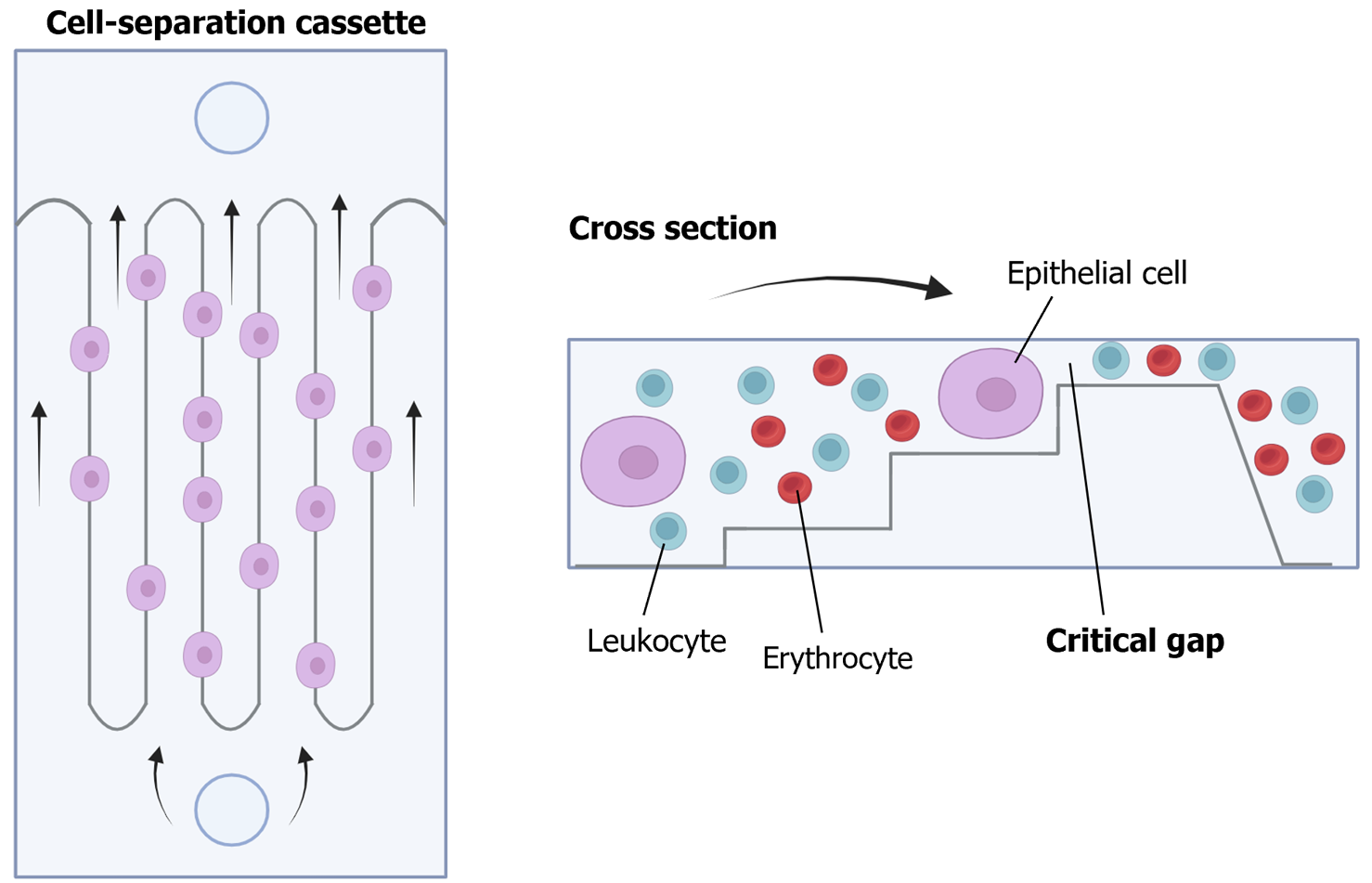
For the isolation and detection of CTCs, various methods have traditionally been employed, including physical methods based on density, deformability, or cell size, magnetic techniques, or immune-based methods[34] (Figure 4). In addition to antibody-based strategies, other techniques such as fluorescence in situ hybridization, cytometric methods, or nucleic acid-based detection (RT-qPCR) can be utilized for CTC detection. Many systems combine these approaches to directly isolate and detect CTCs. These methods can also be categorized as “negative” if they remove immune cells to isolate CTCs or “positive” if they actively select CTCs[35].
It is crucial to note that while the immune positive approach is commonly used, it comes with limitations. The primary challenge lies in the potential loss of CTCs, as a significant number may lose EpCAM expression, due to the EMT phenomenon, the acquisition of stemness features or a histologic type-dependent decrease in expression[36-39]. To overcome this limitation, antigen-based negative methods can be employed, including antibodies to identify blood cells such as CD45. Moreover, novel platforms utilizing microfluidics and/or nanomaterials have emerged[40]. Examples include CTC-iChip, which combines leukocyte marking with antibodies and cell selection based on size, enhancing CTC purity and viability[41]. The main challenges for these new systems lie in their long analysis time and high cost, prompting efforts to automate the process and to reduce costs in subsequent studies[42-45].
Main applications of the study of CTCs: Initially, CTC isolation platforms were used to detect the presence or absence of CTCs in blood, count them, and correlate these counts with patient prognosis or treatment response. For instance, in the years 2004, 2007 and 2008, the CellSearch® system was approved for determining the prognosis of metastatic breast, colorectal and prostate cancer patients, respectively[9] (Figure 5). The identification and enumeration of CTCs in peripheral blood was associated with decreased progression-free and overall survival (OS) in various studies[46,47].
Subsequently, technological advances enabled the study of the genome, transcriptome, and proteome of CTCs. In 2022, the FDA approved the use of the Parsortix® PC1 platform for isolating CTCs in patients with metastatic breast cancer, allowing downstream analysis using validated molecular techniques[10] (Figure 6). This approval specifically pertained to CTC enrichment, and it did not include the identification and enumeration of CTCs with prognostic or patient management purposes. Characterizing and analyzing isolated CTCs can be useful for diagnosis, prognosis determination, treatment response, patient management, and for identifying clinically relevant CTC subtypes, as seen in previous studies including patients with lung, breast, colorectal, hepatocellular, prostate cancer or melanoma[48-57]. Interestingly, differences have been observed between the genomic, transcriptomic, and proteomic profiles of the primary tumor and CTCs[58].
Advantages and disadvantages of CTCs: Studying CTCs within the context of LB presents several advantages over investigating other cells or molecules[23]. Firstly, as CTCs are considered pre-metastatic populations, delving into their study has the potential to significantly enhance our comprehension of the cancer life cycle and the development of metastases. Secondly, CTCs provide an opportunity for cell cultures or can be implanted in mice to establish tumor models, thereby greatly facilitating the personalization of treatment[59].
However, the challenges in analyzing CTCs in blood lie in their scarcity, the absence of entirely sensitive and specific markers for their detection, the difficulty in obtaining viable cells for culture, and the lack of standardization, in addition to high associated costs and large processing times[60,61].
Addressing the first obstacle necessitates the development and enhancement of highly precise technologies for both isolating and detecting CTCs and subsequently analyzing them. As for the second challenge, rescuing the inadequate isolation of CTCs by EpCAM-based technologies is feasible by incorporating both epithelial and mesenchymal cancer markers, or by using marker-independent detection methods[62,63]. The utilization of mesenchymal markers alongside epithelial markers offers the potential to identify distinct CTC populations. Despite the drawback of including circulating mesenchymal cells associated with the tumor as CTCs, some studies suggest that these cells also correlate to patient prognosis and enhance the metastatic potential of CTCs[64]. Concerning the third challenge, most platforms currently in use fail to yield cells suitable for culturing or mouse models due to processes that compromise their viability, primarily during cell labeling and immobilization. Certain technologies, however, enable the retrieval of cells with enhanced viability for subsequent studies[41]. As for the fourth challenge, it is foreseeable that these techniques will evolve, become more automated, and undergo standardization in the coming years. In this process, digital pathology could play an essential role[61].
cfDNA consists of double-stranded DNA fragments found in bodily fluids, originating from both normal and pathological cells[65]. cfDNA typically circulates bound to proteins that shield it from degradation, and it primarily arises from cell death phenomena, including necrosis and apoptosis. Other significant sources of cfDNA include NETosis or extracellular vesicles[66-68]. Elevated levels of cfDNA have been observed in patients with benign lesions, inflammatory diseases, or tissue trauma[69]. cfDNA also holds value in the non-invasive prenatal diagnosis of fetal genetic diseases, as a percentage of fetal DNA is present in the peripheral blood of pregnant women[70]. In cancer patients, cfDNA comprises a mixture of DNA from normal cells and, theoretically, DNA from different tumor subclones and locations, forming ctDNA[71]. Moreover, the presence of cell-free mitochondrial DNA circulating as small fragments has been identified, correlating with the severity of physical or psychological trauma, cancer detection and burden, as well as the occurrence and severity of other diseases[72,73].
Origin and characteristics: According to previous studies, the majority of circulating cfDNA fragments in blood are 150-180 base pairs long, aligning with the DNA content of a nucleosome[74]. The relationship between fragment length and the presence or absence of cancer is contradictory. In various cancer types, both an increase and a decrease in cfDNA integrity have been observed. Generally, it is accepted that ctDNA is more fragmented than cfDNA. Some studies have shown that ctDNA is longer in tumors with a high necrotic rate and shorter in cases with increased apoptosis[75,76].
The percentage of cfDNA is typically low but varies among patients. This percentage is higher in cancer patients, yet, in most cases, it remains below 100 ng/mL. Additionally, in most cancer patients the majority of cfDNA comes from non-tumor cells, including both blood cells and infiltrating non-tumor cells[77]. The reported range of ctDNA relative to cfDNA varies between < 0.5% to 95.0%, being lower in early stages (around 1%) and higher in advanced stages (up to 40%)[78-80]. This percentage varies not only with disease burden but also with features such as proliferation and apoptosis rates, necrosis, inflammation, tumor microenvironment, patient and treatment-related factors.
Finally, cfDNA undergoes rapid turnover, resulting in a very short half-life. Pioneering studies in pregnant women observed that fetal cfDNA in maternal blood has a half-life of approximately 15 min post-partum, becoming undetectable after 2 h[81]. Currently, cfDNA is considered to undergo a two-phase clearance, with a rapid phase within the first 10 min to the first hour and a slow phase with a half-life of 13 h. However, in cancer patients, the half-life of ctDNA seems to be less than 2 h[82].
Main applications of ctDNA analyses: As observed with CTCs, ctDNA has multiple potential clinical applications, including screening, characterizing early disease, detecting molecular residual disease (MRD), predicting relapses, genotyping advanced cancer, early assessment of treatment efficacy, monitoring response, and identifying mechanisms of resistance to therapy.
To date, the FDA has approved several ctDNA-based companion diagnostic assays for the safe and effective use of various targeted therapies, mainly in metastatic tumors[83]. These indications include PCR-based techniques for detecting EGFR or PIK3CA alterations in NSCLC and breast cancer, respectively, as well as the use of next-generation sequencing (NGS) platforms in various cancers, such as metastatic breast, prostate, lung, or ovarian carcinomas. However, a key limitation of currently FDA-approved ctDNA-based assays is that a proportion of patients with mutation-positive tumors in tissue-based testing may not be detected[84].
Ongoing studies aim to validate the analysis of ctDNA in early-stage patients and other clinical contexts, exploring additional genetic alterations and applications beyond specific drug treatments in metastatic patients. For example, in early stages, the detection of MRD can prove valuable in identifying patients at risk of relapse and tailoring treatment accordingly. Across different tumor types, the use of ctDNA has shown high positive predictive value with an acceptable negative predictive value[85,86]. Interestingly, serial ctDNA determinations can enhance the sensitivity of MRD detection. Additionally, relapse can be detected in LB before it becomes visible in other clinical or radiological tests. In the metastatic context, ctDNA determination can be useful for monitoring tumor evolution, assessing treatment resistance, determining patient management, or indicating other treatments, such as immunotherapy[87-89].
Advantages and disadvantages: As advantages over CTCs, ctDNA is more abundant, making its study less technologically complex and potentially more sensitive and specific[90]. CTCs reflect more of the metastasis-initiating cells, while ctDNA represents more of the tumor burden. Thus, ctDNA carries the evolutionary information of both primary and metastatic tumors as cancer cells dynamically evolve in response to intermittent drug treatment. Moreover, in recent years, there has been progress in ctDNA analysis, transitioning from mostly discrepant results to having validated tools that can be implemented in clinical practice.
However, the drawbacks of ctDNA analysis should be considered. Firstly, there is currently no platform that serves all potential applications of ctDNA, as technologies vary depending on whether they are used to detect cancer in early stages, monitor patients in advanced stages, detect genetic alterations for treatment resistance, or other purposes[84]. Pre-analytical variables such as tumor-related factors, patient characteristics, treatment history, collection tube, storage conditions, and processing methods significantly impact results[91]. Secondly, despite the higher abundance of ctDNA compared to CTCs, false negatives may occur due to concentration variations and technique-related issues. Thirdly, false positives have also been detected with cfDNA studies, as normal cells can present somatic alterations and clonal expansion[92]. Finally, the clotting process during serum preparation induces cell lysis, so ctDNA analysis might be hampered by increased levels of high-molecular cfDNA when using serum instead of plasma. Due to this, plasma has been suggested as the better specimen type for ctDNA analysis[93].
While ctDNA and CTCs dominate LB research, several other structures in body fluids hold potential clinical value, albeit with challenges due to limited evidence and procedural standardization.
cfRNA: cfRNA reaches the blood mainly through passive release due to apoptosis and necrosis phenomena, active secretion by microvesicles, and secretion with nucleoproteins or protein-RNA complexes. It is usually found in association with extracellular vesicles and lipoprotein complexes that prevent its degradation and can be secreted by both normal and pathological cells[94]. Free RNA is unstable and degrades within seconds of incubation, requiring these vehicles for stabilization[66].
Beyond serving as a prognostic marker or aiding in early cancer and relapse diagnosis, cfRNA allows the analysis of the expression signature of a tumor. This is particularly valuable for identifying biomarkers, gene expression patterns, understanding intercellular communication, the tumor immune environment, and determining the tissue of origin and/or subtype of a tumor through specific markers[95]. However, challenges include the low quantity of cfRNA in blood, its susceptibility to degradation, and the complexity of its extraction.
Clinical implementation of cfRNA has seen fewer trials compared to ctDNA, with limited available evidence. Notably, the FDA approved one platform in 2012, the PROGENSA PCA3 assay, which detects PCA3 RNA in urine and is useful for determining the risk of prostate cancer[96,97].
Extracellular vesicles: These structures are secreted by all living cells and can be detected in any fluid. They are heterogeneous, represent their cells of origin, and contain associated molecules (proteins, nucleic acids, lipids, and metabolites) relevant for diverse applications in LB[98].
EVs can be classified as exosomes if they come from the intracellular endosomal system, or ectosomes/microvesicles formed directly from the plasma membrane[99]. Additional categories include “apoptotic bodies”, “large oncosomes”, and “migrasomes”[100]. On the other hand, EVs can also be classified as “small EVs” if they are 200 nm or less and “large EVs” if they are larger[101]. In recent years, exomers, small non-membranous particles of 50 nm or less, have also been described, which seem to be related to the regulation of metabolic pathways[102].
The main function of EVs is intercellular communication, with their specific function varying depending on the type and state of the originating cell[100]. In cancer, EVs seem to promote metastasis, angiogenesis, immune evasion, adaptation, and resistance to treatment[103]. Interestingly, they not only serve for communication with nearby cells but also seem to reach distant cells.
For their isolation and detection, techniques similar to those used for CTCs have been developed[104,105]. For their characterization, transmission electron microscopy can be used, or quantification and characterization can be done with methods such as dynamic light scattering technique or nanoparticle tracking analysis[105]. After their isolation, the molecules present in EVs can be studied with lipid, proteomic, transcriptomic, or genomic analysis[98].
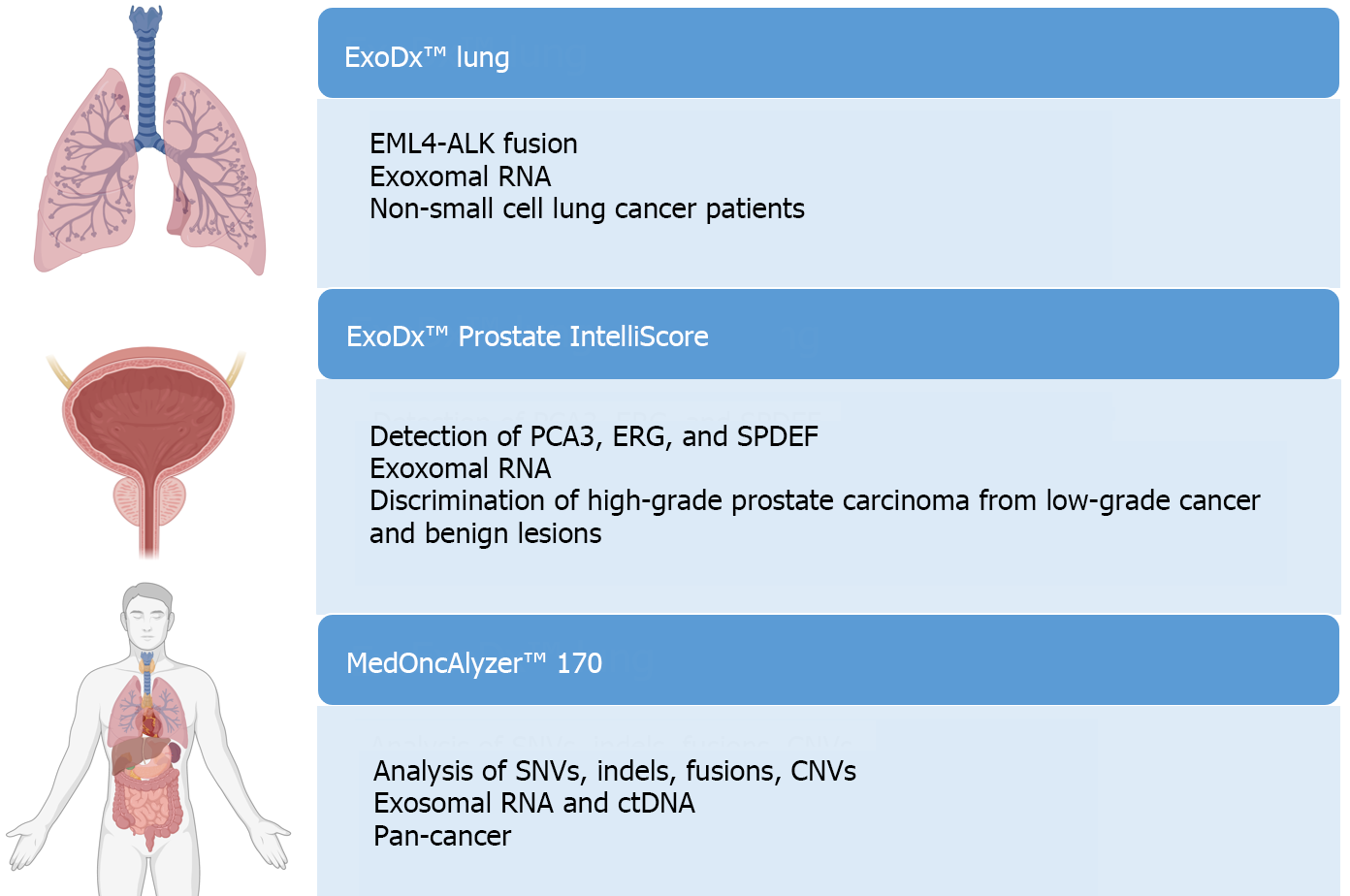
The study of EVs holds clinical potential in detecting biomarkers for cancer diagnostics, treatment monitoring, predicting disease progression, and assessing therapy resistance. However, only three platforms for EV analysis have been commercialized: ExoDx™ Lung, ExoDx™ Prostate IntelliScore, and MedOncAlyzer™ 170[98,104,106] (Figure 7).
The study of EVs has the advantage of encompassing multiple types of molecules, enabling its use in multi-analyte platforms to detect various alterations. In addition, EVs are more abundant than CTCs in all body fluids and more stable than cfDNA due to the protection of their lipid membranes, so EV-based techniques could be more sensitive than these other two approaches. However, EVs are very heterogeneous structures, and there are no detailed characterization techniques available. They are highly variable depending on the physiological or pathological process, and the available evidence is generally scarce.
TEPs: Platelets are anucleated blood cells derived from megakaryocytes that, when activated, form aggregates and secrete a considerable amount of substances into the environment. Traditionally recognized for their role in hemostasis, thrombosis, and wound healing processes, platelets have also been observed to play roles as mediators in cancer development, particularly in the onset of metastasis[107]. TEPs alter their function depending on signals produced by tumor cells, developing a distinctive phenotype and releasing substances that promote cell growth and survival, neoangiogenesis, immune escape, and extracellular matrix remodeling, thereby facilitating tumor progression, migration, and spread[108].
Concerning their potential clinical value, the analysis of TEPs has shown utility in cancer detection, monitoring treatment response, and studying RNA profiles[109].
While blood and its derivatives, such as plasma or serum, are the most commonly used samples, studies on LB can be conducted using various other sample types[110,111]. Depending on the tumor type and/or its anatomical location, these alternative fluids may offer greater sensitivity and specificity compared to blood sources. This is due to their increased contact with the tumor, resulting in a higher concentration of tumor-derived molecules like ctDNA. Additionally, these alternative samples can complement plasma studies, and some can be self-collected by the patient. However, their clinical implementation is complex due to the need to standardize pre-analytical factors, the lack of commercially available kits, and the requirement for invasive procedures to obtain certain samples, such as effusions or cerebrospinal fluid. This complexity may limit their serial use for monitoring purposes. Table 1 summarizes the main applications, advantages, and limitations of non-blood samples, according to the review by Tivey et al[112] and recent research articles[113-115].
| Sample | Tumor | Main features |
| Cerebrospinal fluid | Primary and metastatic CNS | Helpful due to the frequent metastatic seeding, the difficulty in obtaining tissue biopsies from this location, and the obstruction of the blood-brain barrier |
| Urine and seminal fluid | Prostate, urothelial, renal | Renal cancer-derived structures may be limited by glomerular filtration; cfDNA concentration and fragment size in seminal fluid seem to differ between healthy and prostate cancer patients |
| Stool | Colorectal | ctDNA extraction can be complex; ctDNA may be present in low concentrations (mixed with DNA from the bacterial flora) |
| Pleural or peritoneal fluid | Lung, mesothelioma, peritoneal spread | Determination of predictive biomarkers in lung cancer; detection of minimal residual disease or early involvement of the peritoneum |
| Saliva | Oral cancers | Monitoring of minimal residual disease and treatment response; low ctDNA concentration, limited fragment size |
| Bile | Biliary tract | Useful in difficult-to-biopsy cases; invasive procedure |
| Gastric juice | Gastric | Improvement of diagnostic sensitivity; invasive procedure (endoscopy) |
GC ranks as the fifth most common cancer worldwide, with over one million new cases diagnosed in 2020[116]. It exhibits significant geographical variability, being more prevalent in Asian countries and some Eastern European nations, while less common in Western regions[117]. Certain countries, such as Japan and Korea, have implemented screening strategies based on imaging techniques that have improved early diagnosis and prognosis for GC patients[118,119]. However, disparities not only exist between Asian and Western countries regarding GC incidence and early detection. Geographical variations have also been observed in clinical, morphological and molecular features, prognosis, and risk factors for GC[120-122]. In terms of patient outcomes, GC is an aggressive tumor ranking as the third leading cause of cancer death globally. In Western countries, it is often diagnosed at advanced stages, presenting 5-year survival rates of 20%-30%[123,124].
In recent years, significant advances in technical tools have enhanced the understanding of the molecular features and immune environment of tumors, and their potential prognostic and therapeutic roles. This knowledge has led to an expanded therapeutic arsenal, focusing on immunotherapy and targeted therapies, which generally exhibit better efficacy and fewer side effects than conventional treatments. However, these advancements have not significantly improved the prognosis for a substantial portion of GC patients[125].
GC diagnosis relies on the interpretation of tissue biopsies by a pathologist. Microscopically, it can be classified according to the latest World Health Organization (WHO) system from 2019, into four main types: Tubular, poorly cohesive, papillary, and mucinous[126]. According to the Laurén classification, described in 1965 and still used in clinical practice, GC can be divided into intestinal, diffuse, and mixed types[127]. As observed by our research team in a previous study, supporting findings from other investigators, there are differences in clinical, molecular, and prognostic characteristics between intestinal and diffuse GC[128]. Therefore, studying Laurén subtypes separately could enhance patient stratification both in clinical practice and in clinical trials. Other microscopic findings, such as tumor budding or tumor infiltration by inflammatory cells, may also have prognostic implications or aid in treatment selection[129].
After diagnosis, the only curative treatment available for GC is surgery. In early GC cases, which are scarce in Western countries, endoscopic surgery techniques, such as endoscopic mucosal resection or endoscopic submucosal dissection, can be performed[130]. In other resectable cases, total or subtotal gastrectomy, usually associated with D2 lymphadenectomy, is commonly performed. However, in advanced cases, treatment primarily relies on chemotherapy regimens. The only specifically approved targeted therapies for GC are trastuzumab for cases with amplified HER2 and anti-VEGF/VEGFR drugs[131]. Furthermore, GC is included in the FDA-approved indication for pembrolizumab (anti-PD-L1) for all solid tumors in cases with high microsatellite instability (MSI) or deficiency in the DNA mismatch repair system[132].
A broad spectrum of molecular alterations has been identified in GC, owing to the wealth of information generated by new genomic and transcriptomic studies[133]. These alterations can be categorized into isolated changes, disregulation of pathways involved in GC pathogenesis, gene expression profiles, and molecular classifications integrating available molecular information[134-136].
Isolated alterations primarily include mutations and copy number alterations (CNAs), although heterozygosity and gene fusions have also been observed[133]. Mutations in genes such as TP53, BRCA2, cell adhesion genes, or genes encoding proteins in chromatin regulatory complexes or histone regulatory complexes are notable. Affected pathways include the WNT/beta-catenin pathway, alterations in CTNNB1, or different tyrosine kinase receptor pathways (HER, PI3K, etc.). Regarding CNAs, amplifications in genes associated with tyrosine kinase receptor pathways, such as HER2, EGFR, MET, KRAS, or FGFR2, are prominent[137,138]. Amplification of PD-L1 and PD-L2, genes related to the cell cycle, and various transcription factors has also been observed. Epigenetic alterations include the inactivation of various tumor suppressor genes by promoter methylation, such as CDH1, MLH1, CDKN2a, or PTEN.
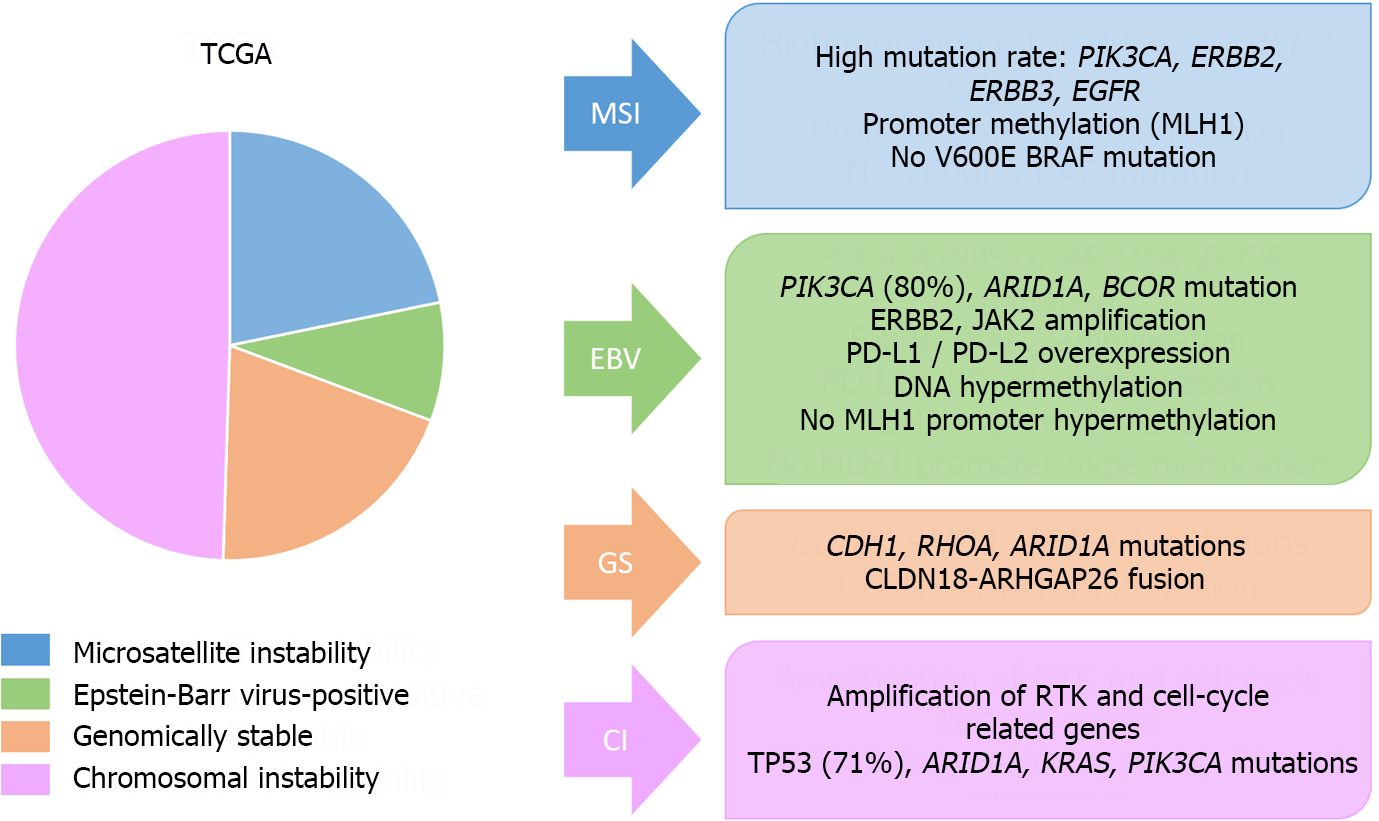
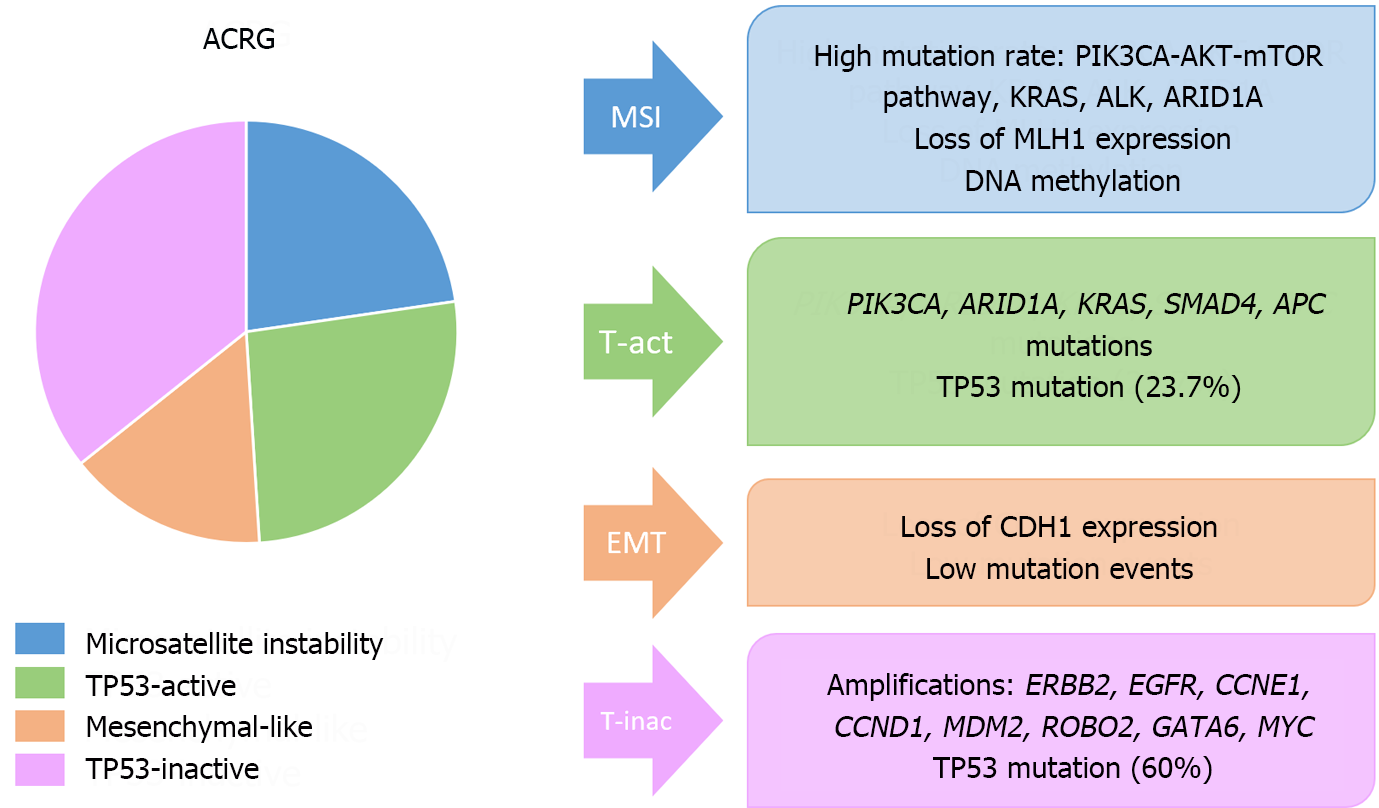
On the other hand, the most important molecular classifications in GC have been published by the Cancer Genome Atlas (TCGA) and the Asian Cancer Research Group (ACRG)[137,138] (Figures 8 and 9). These systems have been associated with clinical, prognostic and predictive factors. Additionally, other molecular classifications have been published by Shah et al[139], Tan et al[140], Lei et al[141], Wang et al[142], Wang et al[143], or Furukawa et al[144].
Unfortunately, these categorizations have not been translated into clinical practice, primarily due to the need for extensive molecular studies and the variety of alterations included in each molecular group. Our research team and other investigators have managed to formulate stratification systems derived from these classifications, primarily those of TCGA and ACRG, using immunohistochemistry as a surrogate marker[145]. However, validation studies are needed to identify the most sensitive and specific markers and cutoff points for their implementation in clinical practice.
Given the previously discussed aspects regarding prognosis, therapeutic options, and the molecular landscape of GC, there is a need to optimize patient classification, identify easily implementable prognostic or therapeutic biomarkers, and enhance the understanding of GC tumorigenesis and progression. These efforts aim to improve patient prognosis and treatment cost-effectiveness. In this context, LB emerges as a non-invasive option to enhance theoretical knowledge with potential immediate clinical impact. However, it should be noted that while research on LB applications is more advanced in other tumors, it is still in early stages for GC. The main applications of LB in GC are outlined in Figure 10. Tables 2 and 3 present the key findings from previous studies regarding the use of CTCs, cfDNA/ctDNA, RNA and proteins in the early detection, prognostic determination, and therapy response of GC. These tables summarize and adapt the data presented in the reviews by Zhang et al[146], Ma et al[147], and Ha et al[148].
| Role | Structure | Approach | Accuracy | Findings/challenges |
| Diagnosis | CTCs | Count | S: 42-85%; E: 90%-99% | Low sensitivity; diverse cutoffs |
| CD44+ | S: 92%; E: 100% | Small sample size | ||
| cfDNA/ctDNA | Methylation | S: 50%-69%; E: 75%-92% | Diverse markers | |
| Quantification | S: 79%-96%; E: 91%-94% | Diverse methodology | ||
| RNA | MiRNA | S: 67%-99%; E: 66%-95% | Diverse markers | |
| CircRNA | S: 78%-89%; E: 45%-84% | Diverse markers | ||
| LncRNA | S: 65%-84%; E: 53%-87% | Diverse markers | ||
| EVs RNA | MiRNA | S: 26%-95%; E: 56%-100% | Up or down expression; diverse markers | |
| CircRNA | S: 41%-87%; E: 68%-98% | Down expression; diverse markers | ||
| LncRNA | S: 70%-88%; E: 60%-98% | Up expression; diverse markers | ||
| Proteins | TTF1-3, CDH17, TFF3, and TXNRD1 | S: 62%-96%; E: 57%-83% | Diverse markers; panels |
| Role | Structure | Approach | Association | Findings/challenges |
| Prognosis | CTCs | Count | ↓OS/D-RFS | Diverse cutoffs |
| EpCAM, CEA, CD44, CD133, TWIST, ploidy, FR, PD-1 | ↓OS/D-RFS | Diverse methodology | ||
| cfDNA/ctDNA | Quantification | ↓OS/D-RFS | Diverse methodology | |
| Amplification: BRAF, FGFR2, MET; mutation: TP53, ARAF | ↓OS | Diverse methodology | ||
| Methylation | ↓OS/DFS | Diverse markers | ||
| EVs RNA | MiRNA | ↓OS/D-RFS | ↑ or ↓ expression; Diverse markers | |
| CircRNA | ↓OS/RFS | ↑ or ↓ expression; diverse markers | ||
| LncRNA | ↓OS/DFS | ↑Expression; diverse markers | ||
| Predictive | CTCs | Count | Response to ST | |
| HER2, PD-L1 status | Trastuzumab resistance/IT | |||
| cfDNA/ctDNA | Quantification | Response to ST-CT/anti PD-1 | ||
| Panels Amplification: HER2; mutation: PIK3CA, NF1, HER2, EGFR, TP53, BRCA MSI | Targeted therapy; response to ST; trastuzumab resistance; treatment monitoring; IT | |||
GC detection: Previous studies demonstrated an elevated number of CTCs in GC patients compared to healthy controls, and this count tends to increase with the progression of the disease[149]. Concerning the use of CTCs for GC detection, there is generally a high specificity (> 95%), while sensitivity varies depending on the method, patient factors, and tumor characteristics, mainly GC stage. A 2013 meta-analysis showed a pooled sensitivity and specificity of CTC detection for GC diagnosis of 42% and 99%, respectively[150]. It is worth noting that most studies included in this analysis employed LB techniques based on RT-PCR for detecting CK or carcinoembryonic antigen (CEA) expression[151]. The cutoff point for considering a result as positive has not been standardized, although a previous study found that the limit of 2 CTCs per 7.5 mL of blood presented the highest sensitivity and specificity at 85.3% and 90.3%, respectively[152]. Interestingly, this study included resectable patients, mostly in stage pT1 (54%), and without nodal metastasis (59%). These researchers detected CTCs using a centrifugal microfluidic system with a fluid-assisted separation technique.
Unfortunately, due to the scarcity of CTCs detected with currently available methods, the application of LB for early detection of GC is limited.
Characterization of GC: CTCs in GC also hold potential for assessing prognostic or predictive biomarkers. Biomarkers associated with approved targeted therapies are particularly valuable, as LB enables their non-invasive analysis in GC patients. In cases with metastatic disease, where accessing metastases can be challenging, these biomarkers become especially crucial. CTCs may better represent systemic tumor heterogeneity, and LB techniques serve as a valuable complement to the study of tissue samples from the primary tumor. Additionally, LB allows for serial determinations during the disease course, which can be useful for monitoring genotypic and phenotypic changes in the tumor cell burden of the body. Previous studies have highlighted the utility of evaluating PD-L1 expression or HER2 status in CTCs from GC patients[153,154].
Correlation with clinicopathological data and prognosis: Previous research has established correlations between the number of CTCs and various clinicopathological factors, including Laurén type, histological grade, perineural infiltration, lymphovascular invasion, TNM stage, and cellular proliferation (Ki-67 index). Diffuse cases according to the Laurén classification, tumors with high histological grade, perineural and vascular infiltration, higher proliferation rate, and at more advanced stages are associated with a higher likelihood of detectable CTCs[153,155].
According to some authors, the CTC count correlates better with GC prognosis than other currently available markers in clinical practice[152]. CTC detection has been variably correlated with the OS and recurrence-free/disease-free survival (RFS/DFS) of GC patients, with cutoff points generally between 1-5 CTCs[156-158]. A recent meta-analysis including 14 retrospective studies and over 1000 patients showed that CTC-positive patients, with a cutoff of 2.8, had significantly lower OS. The relationship between OS and CTCs depended on factors such as the chosen cutoff, the sample size of the study, the type of treatment (mainly resected vs. unresected GC), and the method employed for CTC detection. Furthermore, CTC-positive patients showed higher TNM stage, high histological grade, and more frequent distant metastases[159].
Treatment monitoring: The serial determination of CTCs proves valuable in assessing treatment response in various tumors. In GC, patients with a higher CTC count tend to exhibit a poorer response to chemotherapy and a worse prognosis compared to those with a lower count[160]. Additionally, the rapid decline of CTCs following treatment initiation is associated with a better prognosis, and the conversion to a higher number of CTCs may be an early indicator of treatment failure.
Main approved tests: ctDNA analysis stands out as the predominant LB approach for gastrointestinal tumors, with approved indications in GC, particularly for biomarker determination. For instance, ctDNA can be employed to assess HER2 status in GC cases where tissue testing is impractical or urgent treatment decisions are required[84].
GC detection: For the detection of GC, quantitative analysis of ctDNA (total copy number analysis in blood or serum) or qualitative detection, involving the identification of tumor-specific genetic changes, can be utilized (Table 2)[161]. Similar to CTCs, the cfDNA/ctDNA load is found to be higher in GC patients compared to healthy controls or those with preneoplastic lesions, escalating with tumor stage[162].
Overall, ctDNA analysis has demonstrated higher sensitivity than CTCs for GC detection[163]. Notably, the study by Kim et al[163] achieved a sensitivity and specificity exceeding 90%. Similarly, Qian et al[164] observed that cfDNA concentration was more sensitive than conventional markers like CEA, carbohydrate antigen (CA) 19-9, CA72-4, or CA50 in distinguishing between patients with benign gastric disease or healthy individuals and those with early GC. However, other studies have shown contradictory results. Cabel et al[165] observed limited sensitivity of ctDNA testing in resectable patients. A meta-analysis published in 2017 calculated a pooled sensitivity and specificity of the presence of certain ctDNA markers of 62% and 95%, respectively[166]. It should be noted that the sensitivity of these determinations, as seen with CTCs, is influenced by factors related to the detection technique and the tumor, particularly its size and extension[167]. In early-stage tumors, the sensitivities observed in previous studies are not yet sufficient to recommend the implementation of these tests in clinical practice[161].
In addition to assessing ctDNA levels, as previously mentioned, a qualitative study of ctDNA can also be conducted. For instance, the analysis of DNA methylation patterns for GC screening, such as the Galleri multicancer detection test, has been explored[168]. However, the sensitivity of this technique is notably influenced by the disease stage, ranging from 16.7% in stage I GC to 100% in stage IV tumors[162].
Characterization of GC: The study of ctDNA proves particularly valuable for determining prognostic or therapeutic biomarkers. ctDNA sequencing has been useful in gastrointestinal tumors, revealing various genetic and epigenetic alterations, such as alterations in EGFR, HER2, TP53, PIK3CA, MSI, or germline mutations of BRCA[169]. Similar to CTCs, ctDNA analysis offers the advantages of non/minimal invasiveness and representation of tumor heterogeneity compared to tissue biopsy, with higher sensitivity than CTCs, although it should be noted that the sensitivity of detecting mutations in ctDNA may also be influenced by the tumor stage.
Regarding the study of HER2 amplification by LB, of clinical importance in GC, a previous study published by Gao et al[170] analyzed 70 patients and found a 91.4% correlation between tumor tissues and ctDNA, indicating that LB is a good surrogate for HER2 analysis in GC.
As seen with CTCs, the presence of certain ctDNA markers has been variably associated with various clinicopathological factors, including tumor size, level of infiltration, number of lymph node metastases, presence of distant metastases, or Helicobacter pylori infection[166].
Correlation with prognosis: Previous studies have demonstrated that the detection of ctDNA correlates with a worse prognosis, linking it to OS, DFS/RFS, and/or the presence of recurrences or distant metastases[171,172]. The 2017 meta-analysis conducted by Gao, encompassing 16 studies and over 1000 GC patients, further confirmed the relationship between a high level of ctDNA and significantly worse OS and DFS in GC[167]. Subsequently, the meta-analysis by Min et al[173] included 34 articles with more than 5000 subjects, revealing a significant association between ctDNA detection in blood and OS, DFS, and PFS, with hazard ratios of 2.74, 3.13, and 3.04, respectively.
Therefore, the study of ctDNA holds promise for detecting MRD and predicting the likelihood of recurrences after treatment. In the study by Yang et al[167], it was noted that all patients with detectable ctDNA after surgery experienced recurrences, with ctDNA detected in blood an average of 6 months before recurrence detection by imaging methods. Additionally, the investigation of various genetic or epigenetic alterations in ctDNA can also correlate with patient prognosis. For example, a previous study analyzed CBLB mutations in ctDNA, revealing that patients with these alterations presented significantly shorter OS and DFS[171].
Treatment monitoring: In the realm of treatment monitoring, Zhou et al[171] observed that combining the analysis of ctDNA levels with traditional tumor markers and radiological control improved the accuracy of the latter. Kim et al[174] studied cancer-specific rearrangements in ctDNA and observed positive ctDNA detection 4 months before detecting a clinical recurrence. Moreover, previous studies have shown that postoperative ctDNA detection, the persistence of high ctDNA levels, or their increase can be an early indicator of recurrence in patients with resected GC[175,176]. Lastly, ctDNA analysis shows potential for monitoring the response to immunotherapy with anti-PD-1 agents or resistance to trastuzumab in patients with metastatic GC[177,178] (Table 3).
As mentioned earlier, exosomes hold significant interest because they are composed of multiple structures derived from tumors, enabling studies beyond genomic analysis. Notably, they are more abundant and stable than ctDNA and can be found in non-blood fluids, making them particularly valuable when analyzing non-blood samples[179].
In GC, exosomes seem to play an important role in tumorigenesis and tumor progression, influencing angiogenesis, immune evasion, cell proliferation, invasiveness, and therapy resistance[180-182]. Additionally, exosomes are implicated in the peritoneal dissemination of GC[183].
Concerning their clinical application, several studies have analyzed exosomes and exosomal-derived RNAs and proteins[184-187]. The detection of these molecules has shown diagnostic, prognostic, predictive, or therapeutic potential. In the diagnostic domain, miRNAs, lncRNAs, circRNAs, and proteins such as TRIM 3 have been studied[188]. Generally, miRNAs have been studied in combination (as profiles), while lncRNAs and circRNAs have been investigated individually[179]. For prognostic value, various miRNAs, lncRNAs, circRNAs, and proteins such as TGF-β1, apolipoprotein E, or MET have been analyzed[187,189,190]. Interestingly, the exosomal PD-L1 content has been found to be an independent predictor of OS and negatively correlated with the CD4+ and CD8+ T cell count in a cohort of metastatic GC patients[191].
An intriguing aspect of exosomes, apart from the previously described functions, is their potential role as a drug delivery method in GC[192].
A comprehensive systematic review performed by Lopes et al[115] compiled over 80 articles related to the use of LB for GC detection utilizing non-blood samples. The majority of articles included Asian patients, and they mainly focused on samples of gastric juice and urine, and to a lesser extent, salivary or stool samples. It should be noted that studies on peritoneal LB for purposes beyond diagnosis were excluded from the review.
Regarding gastric juice, Yamamoto et al[193] analyzed the BARHL2 methylation in ctDNA and exosomes, observing differences between GC samples and controls and concluding that this alteration could assist in the early diagnosis of GC, with high sensitivity and specificity. Other researchers have analyzed different types of RNA, proteins, amino acids, microbiota, or metabolites in gastric juice, primarily aiming to detect GC and improve the sensitivity of upper gastrointestinal endoscopy. Proteins such as pepsin A and gastricsin, along with miR-21 and miR-133a, have shown promising discriminatory value[115].
In urine samples, according to the review by Lopes et al[115], the markers with the best diagnostic value in isolation were the metabolite glycerol and endothelial lipase. Profiles including combinations of metabolites or amino acids have also been published, showing good diagnostic accuracy in these samples. Studies using saliva have focused on detecting lectins and RNA molecules, while stool analysis has explored several proteins, TERT promoter methylation (DNA), or microbiota profiles.
In patients with peritoneal involvement, an increase in certain exosome-derived miRNAs in peritoneal fluid, such as miR-21-5p, miR-92a-3p, miR-223-3p, and miR-342-3p, has been detected[179]. Furthermore, previous studies have observed that the levels of various miRNAs correlate with OS and RFS.
It is worth noting that despite the scarcity of studies on non-blood samples, molecules with diagnostic value have been detected, which could also be used in disease monitoring. These samples present potential value as a source of molecules for determining prognosis, treatment, or patient management[194]. As mentioned earlier, analyzing these fluids can offer heightened sensitivity as they are in direct proximity to tumor cells, potentially yielding distinct information from blood samples. Additionally, certain samples like urine can be self-collected by patients, leading to an improvement in their quality of life and simplifying the process of conducting serial LBs for follow-up purposes.
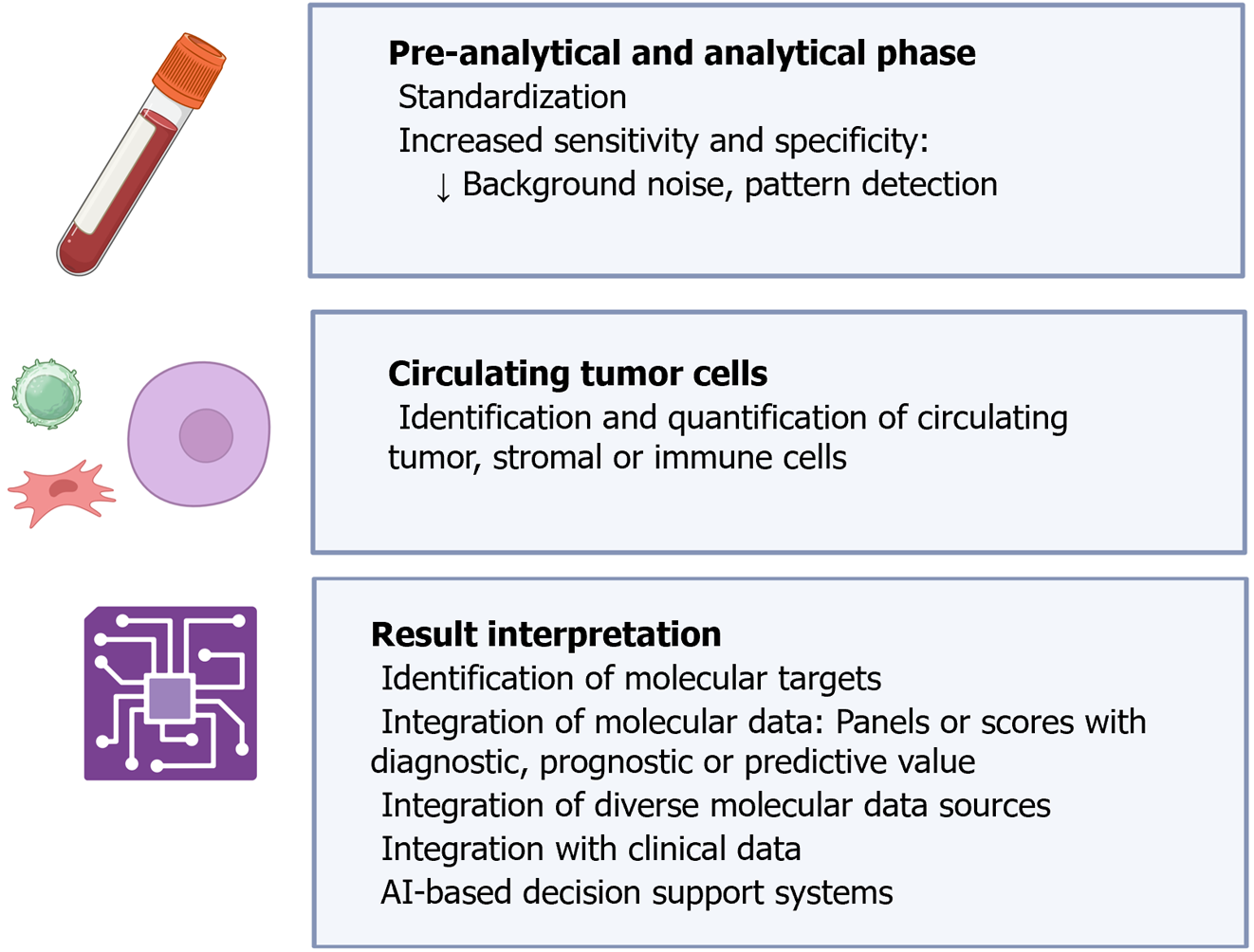
In some clinical specialties, such as radiology or pathology, digitization and artificial intelligence (AI) tools have substantially improved the workflow and diagnostic accuracy of various techniques where they have been integrated[195,196]. Specifically in pathology, the digitization of slides and the utilization of algorithms have opened an almost infinite world of possibilities, facilitating not only the morphological analysis of samples but also the management of large amounts of data. For instance, it is now feasible to identify and quantify cancer cells, intratumoral and peritumoral stromal cells, or inflammatory cells separately[197]. Additionally, algorithms have been developed for standardizing the interpretation of complementary techniques such as PD-L1 expression[198]. In the field of molecular pathology, AI is useful for combining genomic, transcriptomic, proteomic and metabolomic information, and due to its ability to integrate various types of data, it can also serve as a bridge between clinical and molecular features, advancing personalized cancer management[199]. Many of these advancements are potentially applicable to LB. Although published studies on this topic are not abundant, some authors have highlighted the potential of AI-assisted LB, mainly regarding the interpretation of results from cfDNA/ctDNA analyses[200-204]. The application of AI in the study of CTCs would be more similar to current digital pathology, allowing the identification of tumor cells and accompanying stromal or immune cells both by morphology and by the expression of various markers, decreasing inter- and intra-observer variability[205]. Combining our experience as pathologists and the findings of previous studies, we have summarized the potential applications of AI in LB in Figure 11.
The genomic, transcriptomic, proteomic, and metabolomic information that LB can provide, coupled with the development of high-throughput techniques such as NGS or mass spectrometry, and the ability to integrate vast amounts of data through AI tools, facilitates the integration of LB into the multi-omics era. Consequently, cfDNA/ctDNA emerges as a significant source of genomic data, cfRNA as a source of transcriptomic data, and CTCs or EVs as reservoirs of multiple sources of molecular information. Multi-omics studies of GC tissues have been pivotal in integrating molecular information and elucidating the molecular landscape of GC, as observed in the ACRG or TCGA classifications. In LB, novel biomarkers have been identified that could be useful in the early diagnosis or treatment of GC. These biomarkers encompass a range of elements such as plasma proteins, somatic mutations, genomic or proteomic signatures. Additionally, ongoing multi-omics clinical trials are investigating diverse blood-based biomarkers for diagnosing GC or detecting peritoneal involvement in GC patients[206,207].
Several clinical trials are currently underway to assess the efficacy of LB in diagnosing, determining patient prognosis, or assessing treatment response in GC[146,208]. The majority of these studies have been designed in China, with a smaller proportion occurring in European countries and the United States. While most of these trials focus on analyzing ctDNA using blood specimens, some also evaluate CTCs and samples from peritoneal lavage or gastric fluid. The primary objectives of these trials include predicting prognosis, with a smaller subset aimed at assessing response to chemotherapy, immunotherapy, and/or trastuzumab, primarily in the adjuvant or advanced settings.
In the following section, we outline the primary challenges related to LB, with a focus on GC. It is important to note that while the authors concentrate on GC, many of the challenges discussed are also applicable to other tumor types.
Due to recent technical advances, the sensitivity of LB studies has increased progressively. However, in GC, it is still not sufficient for clinical application, except for limited approvals, such as the use of blood specimens as a rescue sample when a traditional sample is unavailable for specific indications. The strong dependence of LB sensitivity on the disease stage makes it nearly impossible to use it as a screening tool without improving the detection capacity of available techniques.
Deep analyses and broad-spectrum molecular tools, such as whole-genome sequencing, have been more frequently used in LB than in conventional tissue sample analysis in clinical practice. This may lead to the detection of variants of undetermined significance or even false positives. In this regard, some researchers suggest that the use of algorithms or machine learning tools could be useful to improve the quality of the data obtained in LB studies[209].
A major limitation of applying LB in clinical practice for GC is the poor quality of the available evidence, largely due to the small sample size of the majority of conducted studies. Another added problem is the heterogeneity of the patients included, especially concerning the stage at diagnosis, a factor that, as previously noted, substantially influences technique sensitivity.
Western and Asian GC patients exhibit differences in multiple aspects, including clinical, histological, prognostic, or molecular features. While most studies have been conducted in Asian patients, LB studies in low-prevalence populations, such as Western populations, are necessary to analyze the accuracy and validate different techniques. This requires inter-institutional collaboration and the establishment of shared, high-quality databases to collect a significant number of cases.
Standardization of sample collection, maintenance, processing procedures, and detection and characterization methods for different molecules is essential in LB. However, there is considerable variability in LB technologies, making it almost impossible to compare results between most of the published studies. The development of protocols by scientific societies and responsible entities has the potential to improve the comparability between studies and the quality of the results obtained.
Although LB is assumed to represent the total tumor burden of the body better than targeted biopsies, it is unclear whether LB may overrepresent a specific cell clone, limiting its value as a broadly representative test. Therefore, the combination of LB with traditional biopsies, imaging techniques, and analytical values, along with the performance of serial LB studies, may be useful to improve the capture of tumor heterogeneity.
Most LB studies have focused on the analysis of CTCs or ctDNA. However, exosomes and other cells and molecules present differences and certain advantages compared to CTCs and ctDNA, with great diagnostic, therapeutic and management potential. Therefore, technical improvement, standardization of detection methods, and biomarker studies using these samples are necessary to exploit their full potential.
In GC and other tumor types, peripheral blood is the most commonly used sample in LB. Despite some advantages associated with non-blood samples, their exploration has been limited. Fewer studies have been published, often with small sample sizes. Additionally, the methods for collecting, maintaining, and processing non-blood samples, as well as the technologies for detecting molecules of interest, exhibit wide variations across studies. It is crucial to acknowledge that the conditions of non-blood samples differ from those of peripheral blood (e.g., pH or density), so procedures commonly used in blood might need to be modified for other sample types. Furthermore, in GC, most studies have been conducted for diagnostic purposes, and the analysis of non-blood samples to detect molecules with prognostic or therapeutic value would also be interesting.
LB can provide a vast amount of information at various molecular levels. In this context, the utilization of AI methodologies offers numerous advantages in both pre-analytical and analytical stages, as well as in result interpretation.
LB has emerged as a non- or minimally invasive technique for studying various molecules and cells in bodily fluids, offering advantages in capturing spatial and temporal tumor heterogeneity. It is particularly interesting in patients with advanced tumors and multiple metastatic locations.
Despite its advantages, LB faces challenges, including complex and expensive techniques, lack of standardization and scientific evidence of suboptimal quality due to small sample sizes and procedure variability.
LB presents numerous analytic approaches, serving multiple potential purposes such as early tumor diagnosis, prognostic stratification, treatment monitoring, detection of therapeutic resistance, or analysis of predictive biomarkers.
Various components, including CTCs, cfDNA/ctDNA, RNA, extracellular vesicles, or TEPs can be analyzed in LB, using peripheral blood or other fluids like urine, seminal fluid, saliva, effusions, cerebrospinal fluid, bile, or gastric juice.
CTC analysis offers insights into pre-metastatic cell populations and the genome, transcriptome, proteome, and metabolome of tumor cells. Challenges include enhancing sensitivity, improving capture techniques for better cell viability, identifying more sensitive and specific markers for cell isolation, or developing antigen-independent platforms.
cfDNA/ctDNA is more abundant than CTCs, requires less technological complexity and generally provides greater sensitivity and specificity. Challenges include the use of different platforms depending on the analytic context, the occurrence of false negatives and positives and the inability to conduct transcriptomic, proteomic or metabolomic analyses.
The use of non-blood samples has potential advantages, including greater sensitivity and specificity than peripheral blood samples, depending on the circumstances. Furthermore, some of these samples can be collected by the patients themselves. However, the available evidence on the use of these samples in LB is very limited.
GC is an aggressive tumor that presents clinical, histological, and molecular differences between Western and Asian countries. It is generally diagnosed at advanced stages and has a poor prognosis.
The only curative option for GC is surgery. In advanced tumors, the main therapeutic approach is chemotherapy, and in recent years, a few targeted therapies (anti-HER2 and anti-VEGF/VEGFR) and immunotherapy (anti-PD-L1) have been added to the therapeutic arsenal.
Molecular classifications of GC have been developed and have shown potential prognostic and therapeutic value. Unfortunately, these classifications have not yet been translated into clinical practice.
In GC, there is a need to develop cost-effective screening tools for countries with lower prevalence, improve patient stratification for treatment and clinical trials, increase knowledge about tumor progression mechanisms, and identify new biomarkers with therapeutic value.
CTC studies in GC have shown that both their count and characterization can be useful for detecting GC, determining patient prognosis, monitoring the disease, and identifying therapeutic biomarkers, generally with high specificity.
cfDNA/ctDNA analysis is the predominant LB approach in gastrointestinal tumors. In GC, HER2 status can be assessed using ctDNA in cases where a tissue sample is not available or treatment initiation is urgent.
cfDNA/ctDNA analysis in GC can be useful for determining prognosis and treatment monitoring. Still, for its implementation as a screening tool, the sensitivity of detection techniques needs improvement since its concentration is highly dependent on the tumor stage.
Future advancements in LB should prioritize enhancing technique sensitivity, minimizing false positives, deciphering the pathogenicity of variants of uncertain significance, increasing sample sizes and follow-up times, accounting for geographical disparities, standardizing detection methodologies, developing specific AI techniques, and exploring alternative structures beyond CTCs and cfDNA/ctDNA in both blood and non-blood specimens.
In conclusion, LB emerges as a promising tool in GC research, offering significant advantages in terms of reduced invasiveness and enhanced capture of tumor heterogeneity. Despite challenges such as technical complexity and lack of standardization, the diverse analytical approaches in LB open a broad range of possibilities for early diagnosis, prognostic stratification, treatment monitoring, and identification of therapeutic biomarkers. The analysis of CTCs, cfDNA/ctDNA and other structures in blood and non-blood samples provides unique insights into GC and other tumors.
Despite recent advances in the molecular understanding of GC and the application of LB, fundamental challenges persist. These include the standardization of methods and platforms, improvement of sensitivity, reduction of false positives, and the need to increase the number of patients included in research studies to enhance the available evidence. International collaboration and the establishment of high-quality shared databases are essential to comprehensively address these challenges and propel the field forward.
The authors would like to express their gratitude to Tony Punk (contact: contacto@tonypunk.es) for his contribution in creating Figure 2. Figures 1, 3-7, 10, and 11 were generated by C. Díaz del Arco using the Biorender tool under an individual license (https://www.biorender.com/).
| 1. | Connal S, Cameron JM, Sala A, Brennan PM, Palmer DS, Palmer JD, Perlow H, Baker MJ. Liquid biopsies: the future of cancer early detection. J Transl Med. 2023;21:118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 198] [Article Influence: 66.0] [Reference Citation Analysis (1)] |
| 2. | Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic - implementation issues and future challenges. Nat Rev Clin Oncol. 2021;18:297-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 820] [Article Influence: 164.0] [Reference Citation Analysis (0)] |
| 3. | Di Zazzo E, Intrieri M, Davinelli S. Liquid Biopsy and Cancer: An Ongoing Story. J Clin Med. 2023;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 4. | Bujalkova M, Straka S, Jureckova A. Hippocrates’ humoral pathology in nowaday’s reflections. Bratisl Lek Listy. 2001;102:489-492. [PubMed] |
| 5. | Jain D, Ramachandrappa VS, Singh V, Malik PS, Madan K, Faruq M, Guleria R. Use of Exfoliative Specimens and Fine-Needle Aspiration Smears for Mutation Testing in Lung Adenocarcinoma. Acta Cytol. 2017;61:455-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 6. | Díaz Del Arco C, Fernández Aceñero MJ. Molecular diagnostic techniques in cytopathology: From the bench to the patient’s bedside. Diagn Cytopathol. 2018;46:620-623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 7. | Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3360] [Cited by in RCA: 3447] [Article Influence: 156.7] [Reference Citation Analysis (0)] |
| 8. | Zhou H, Zhu L, Song J, Wang G, Li P, Li W, Luo P, Sun X, Wu J, Liu Y, Zhu S, Zhang Y. Liquid biopsy at the frontier of detection, prognosis and progression monitoring in colorectal cancer. Mol Cancer. 2022;21:86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 184] [Article Influence: 46.0] [Reference Citation Analysis (1)] |
| 9. | Millner LM, Linder MW, Valdes R Jr. Circulating tumor cells: a review of present methods and the need to identify heterogeneous phenotypes. Ann Clin Lab Sci. 2013;43:295-304. [PubMed] |
| 10. | Templeman A, Miller MC, Cooke MJ, O’Shannessy DJ, Gurung Y, Pereira T, Peters SG, Piano M, Teo M, Khazan N, Kim K, Cohen E, Lopez HB, Alvarez F, Ciccioli M, Pailhes-Jimenez AS. Analytical performance of the FDA-cleared Parsortix(®) PC1 system. J Circ Biomark. 2023;12:26-33. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 11. | Mandel P, METAIS P. [Nuclear Acids In Human Blood Plasma]. C R Seances Soc Biol Fil. 1948;142:241-243. [PubMed] |
| 12. | Caldas C, Hahn SA, Hruban RH, Redston MS, Yeo CJ, Kern SE. Detection of K-ras mutations in the stool of patients with pancreatic adenocarcinoma and pancreatic ductal hyperplasia. Cancer Res. 1994;54:3568-3573. [PubMed] |
| 13. | Herbreteau G, Vallée A, Charpentier S, Normanno N, Hofman P, Denis MG. Circulating free tumor DNA in non-small cell lung cancer (NSCLC): clinical application and future perspectives. J Thorac Dis. 2019;11:S113-S126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 14. | Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14:531-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1017] [Cited by in RCA: 1459] [Article Influence: 162.1] [Reference Citation Analysis (0)] |
| 15. | Karachaliou N, Sosa AE, Molina MA, Centelles Ruiz M, Rosell R. Possible application of circulating free tumor DNA in non-small cell lung cancer patients. J Thorac Dis. 2017;9:S1364-S1372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 16. | Gilson P, Merlin JL, Harlé A. Deciphering Tumour Heterogeneity: From Tissue to Liquid Biopsy. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 78] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 17. | Harbst K, Lauss M, Cirenajwis H, Winter C, Howlin J, Törngren T, Kvist A, Nodin B, Olsson E, Häkkinen J, Jirström K, Staaf J, Lundgren L, Olsson H, Ingvar C, Gruvberger-Saal SK, Saal LH, Jönsson G. Molecular and genetic diversity in the metastatic process of melanoma. J Pathol. 2014;233:39-50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 50] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 18. | van Zijl F, Krupitza G, Mikulits W. Initial steps of metastasis: cell invasion and endothelial transmigration. Mutat Res. 2011;728:23-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 469] [Cited by in RCA: 633] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 19. | Ruscitto F, Roda N, Priami C, Migliaccio E, Pelicci PG. Beyond Genetics: Metastasis as an Adaptive Response in Breast Cancer. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 20. | Akpe V, Kim TH, Brown CL, Cock IE. Circulating tumour cells: a broad perspective. J R Soc Interface. 2020;17:20200065. [RCA] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 21. | Woo D, Yu M. Circulating tumor cells as "liquid biopsies" to understand cancer metastasis. Transl Res. 2018;201:128-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 22. | Lin D, Shen L, Luo M, Zhang K, Li J, Yang Q, Zhu F, Zhou D, Zheng S, Chen Y, Zhou J. Circulating tumor cells: biology and clinical significance. Signal Transduct Target Ther. 2021;6:404. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 298] [Cited by in RCA: 618] [Article Influence: 123.6] [Reference Citation Analysis (0)] |
| 23. | Liu J, Lian J, Chen Y, Zhao X, Du C, Xu Y, Hu H, Rao H, Hong X. Circulating Tumor Cells (CTCs): A Unique Model of Cancer Metastases and Non-invasive Biomarkers of Therapeutic Response. Front Genet. 2021;12:734595. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 24. | Guo B, Oliver TG. Partners in Crime: Neutrophil-CTC Collusion in Metastasis. Trends Immunol. 2019;40:556-559. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 25. | Saini M, Szczerba BM, Aceto N. Circulating Tumor Cell-Neutrophil Tango along the Metastatic Process. Cancer Res. 2019;79:6067-6073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 26. | Hamilton G, Rath B. Circulating tumor cell interactions with macrophages: implications for biology and treatment. Transl Lung Cancer Res. 2017;6:418-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 27. | Osmulski PA, Cunsolo A, Chen M, Qian Y, Lin CL, Hung CN, Mahalingam D, Kirma NB, Chen CL, Taverna JA, Liss MA, Thompson IM, Huang TH, Gaczynska ME. Contacts with Macrophages Promote an Aggressive Nanomechanical Phenotype of Circulating Tumor Cells in Prostate Cancer. Cancer Res. 2021;81:4110-4123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 28. | Lou XL, Sun J, Gong SQ, Yu XF, Gong R, Deng H. Interaction between circulating cancer cells and platelets: clinical implication. Chin J Cancer Res. 2015;27:450-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 61] [Reference Citation Analysis (0)] |
| 29. | Hayes B, Brady L, Sheill G, Baird AM, Guinan E, Stanfill B, Dunne J, Holden D, Vlajnic T, Casey O, Murphy V, Greene J, Allott EH, Hussey J, Cahill F, Van Hemelrijck M, Peat N, Mucci LA, Cunningham M, Grogan L, Lynch T, Manecksha RP, McCaffrey J, O’Donnell DM, Sheils O, O’Leary JJ, Rudman S, McDermott R, Finn S. Circulating Tumour Cell Numbers Correlate with Platelet Count and Circulating Lymphocyte Subsets in Men with Advanced Prostate Cancer: Data from the ExPeCT Clinical Trial (CTRIAL-IE 15-21). Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 30. | Liu Y, Zhang Y, Ding Y, Zhuang R. Platelet-mediated tumor metastasis mechanism and the role of cell adhesion molecules. Crit Rev Oncol Hematol. 2021;167:103502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 31. | Liu Q, Liao Q, Zhao Y. Myeloid-derived suppressor cells (MDSC) facilitate distant metastasis of malignancies by shielding circulating tumor cells (CTC) from immune surveillance. Med Hypotheses. 2016;87:34-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 32. | Hurtado P, Martínez-Pena I, Piñeiro R. Dangerous Liaisons: Circulating Tumor Cells (CTCs) and Cancer-Associated Fibroblasts (CAFs). Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 66] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 33. | Ortiz-Otero N, Clinch AB, Hope J, Wang W, Reinhart-King CA, King MR. Cancer associated fibroblasts confer shear resistance to circulating tumor cells during prostate cancer metastatic progression. Oncotarget. 2020;11:1037-1050. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 34. | Gribko A, Künzel J, Wünsch D, Lu Q, Nagel SM, Knauer SK, Stauber RH, Ding GB. Is small smarter? Nanomaterial-based detection and elimination of circulating tumor cells: current knowledge and perspectives. Int J Nanomedicine. 2019;14:4187-4209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 35. | Kang H, Kim J, Cho H, Han KH. Evaluation of Positive and Negative Methods for Isolation of Circulating Tumor Cells by Lateral Magnetophoresis. Micromachines (Basel). 2019;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 36. | Brown TC, Sankpal NV, Gillanders WE. Functional Implications of the Dynamic Regulation of EpCAM during Epithelial-to-Mesenchymal Transition. Biomolecules. 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 37. | Wendt MK, Taylor MA, Schiemann BJ, Schiemann WP. Down-regulation of epithelial cadherin is required to initiate metastatic outgrowth of breast cancer. Mol Biol Cell. 2011;22:2423-2435. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 146] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 38. | Jinesh GG, Brohl AS. Classical epithelial-mesenchymal transition (EMT) and alternative cell death process-driven blebbishield metastatic-witch (BMW) pathways to cancer metastasis. Signal Transduct Target Ther. 2022;7:296. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 65] [Reference Citation Analysis (3)] |
| 39. | Punnoose EA, Atwal SK, Spoerke JM, Savage H, Pandita A, Yeh RF, Pirzkall A, Fine BM, Amler LC, Chen DS, Lackner MR. Molecular biomarker analyses using circulating tumor cells. PLoS One. 2010;5:e12517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 228] [Cited by in RCA: 230] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 40. | Descamps L, Le Roy D, Deman AL. Microfluidic-Based Technologies for CTC Isolation: A Review of 10 Years of Intense Efforts towards Liquid Biopsy. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 79] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 41. | Karabacak NM, Spuhler PS, Fachin F, Lim EJ, Pai V, Ozkumur E, Martel JM, Kojic N, Smith K, Chen PI, Yang J, Hwang H, Morgan B, Trautwein J, Barber TA, Stott SL, Maheswaran S, Kapur R, Haber DA, Toner M. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat Protoc. 2014;9:694-710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 525] [Cited by in RCA: 540] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 42. | Zhang X, Zhu Z, Xiang N, Long F, Ni Z. Automated Microfluidic Instrument for Label-Free and High-Throughput Cell Separation. Anal Chem. 2018;90:4212-4220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 43. | Jia Z, Yuan H, Zhao X, Yin J, Cong H, Gao W, Jin Q, Jia C, Zhao J. Single-cell genetic analysis of lung tumor cells based on self-driving micro-cavity array chip. Talanta. 2021;226:122172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 44. | Wang J, Li Y, Wang R, Han C, Xu S, You T, Xia J, Xu X, Wang D, Tang H, Yang C, Chen X, Peng Z. A Fully Automated and Integrated Microfluidic System for Efficient CTC Detection and Its Application in Hepatocellular Carcinoma Screening and Prognosis. ACS Appl Mater Interfaces. 2021;13:30174-30186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 45. | Lee J, Kwak B. Simultaneous on-chip isolation and characterization of circulating tumor cell sub-populations. Biosens Bioelectron. 2020;168:112564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 46. | Müller V, Riethdorf S, Rack B, Janni W, Fasching PA, Solomayer E, Aktas B, Kasimir-Bauer S, Pantel K, Fehm T; DETECT study group. Prognostic impact of circulating tumor cells assessed with the CellSearch System™ and AdnaTest Breast™ in metastatic breast cancer patients: the DETECT study. Breast Cancer Res. 2012;14:R118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 148] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 47. | Lawrence R, Watters M, Davies CR, Pantel K, Lu YJ. Circulating tumour cells for early detection of clinically relevant cancer. Nat Rev Clin Oncol. 2023;20:487-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 213] [Reference Citation Analysis (0)] |
| 48. | Carter L, Rothwell DG, Mesquita B, Smowton C, Leong HS, Fernandez-Gutierrez F, Li Y, Burt DJ, Antonello J, Morrow CJ, Hodgkinson CL, Morris K, Priest L, Carter M, Miller C, Hughes A, Blackhall F, Dive C, Brady G. Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer. Nat Med. 2017;23:114-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 244] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 49. | Chiu CG, Nakamura Y, Chong KK, Huang SK, Kawas NP, Triche T, Elashoff D, Kiyohara E, Irie RF, Morton DL, Hoon DS. Genome-wide characterization of circulating tumor cells identifies novel prognostic genomic alterations in systemic melanoma metastasis. Clin Chem. 2014;60:873-885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 50. | Li SH, Wu MH, Wang HM, Hsu PC, Fang YF, Wang CL, Chu HC, Lin HC, Lee LY, Wu CY, Yang CT, Chen JS, Hsieh JC. Circulating EGFR Mutations in Patients with Lung Adenocarcinoma by Circulating Tumor Cell Isolation Systems: A Concordance Study. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 51. | Jiang JH, Gao J, Chen CY, Ao YQ, Li J, Lu Y, Fang W, Wang HK, de Castro DG, Santarpia M, Hashimoto M, Yuan YF, Ding JY. Circulating tumor cell methylation profiles reveal the classification and evolution of non-small cell lung cancer. Transl Lung Cancer Res. 2022;11:224-237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 52. | Pailler E, Faugeroux V, Oulhen M, Catelain C, Farace F. Routine clinical use of circulating tumor cells for diagnosis of mutations and chromosomal rearrangements in non-small cell lung cancer-ready for prime-time? Transl Lung Cancer Res. 2017;6:444-453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 53. | Negishi R, Yamakawa H, Kobayashi T, Horikawa M, Shimoyama T, Koizumi F, Sawada T, Oboki K, Omuro Y, Funasaka C, Kageyama A, Kanemasa Y, Tanaka T, Matsunaga T, Yoshino T. Transcriptomic profiling of single circulating tumor cells provides insight into human metastatic gastric cancer. Commun Biol. 2022;5:20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 39] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 54. | Dong L, Du X, Lu C, Zhang Z, Huang CY, Yang L, Warren S, Kuczler MD, Reyes DK, Luo J, Amend SR, Xue W, Pienta KJ. RNA profiling of circulating tumor cells systemically captured from diagnostic leukapheresis products in prostate cancer patients. Mater Today Bio. 2022;17:100474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 55. | Mostert B, Sieuwerts AM, Bolt-de Vries J, Kraan J, Lalmahomed Z, van Galen A, van der Spoel P, de Weerd V, Ramírez-Moreno R, Smid M, Verhoef C, IJzermans JN, Gratama JW, Sleijfer S, Foekens JA, Martens JW. mRNA expression profiles in circulating tumor cells of metastatic colorectal cancer patients. Mol Oncol. 2015;9:920-932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 56. | Lee YT, Sun N, Kim M, Wang JJ, Tran BV, Zhang RY, Qi D, Zhang C, Chen PJ, Sadeghi S, Finn RS, Saab S, Han SB, Busuttil RW, Pei R, Zhu Y, Tseng HR, You S, Yang JD, Agopian VG. Circulating Tumor Cell-Based Messenger RNA Scoring System for Prognostication of Hepatocellular Carcinoma: Translating Tissue-Based Messenger RNA Profiling Into a Noninvasive Setting. Liver Transpl. 2022;28:200-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 57. | Lin SY, Chang SC, Lam S, Irene Ramos R, Tran K, Ohe S, Salomon MP, Bhagat AAS, Teck Lim C, Fischer TD, Foshag LJ, Boley CL, O’Day SJ, Hoon DSB. Prospective Molecular Profiling of Circulating Tumor Cells from Patients with Melanoma Receiving Combinatorial Immunotherapy. Clin Chem. 2020;66:169-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 58. | Jiang M, Jin S, Han J, Li T, Shi J, Zhong Q, Li W, Tang W, Huang Q, Zong H. Detection and clinical significance of circulating tumor cells in colorectal cancer. Biomark Res. 2021;9:85. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 59. | Fernández-Santiago C, López-López R, Piñeiro R. Models to study CTCs and CTC culture methods. Int Rev Cell Mol Biol. 2023;381:57-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 60. | van der Toom EE, Verdone JE, Gorin MA, Pienta KJ. Technical challenges in the isolation and analysis of circulating tumor cells. Oncotarget. 2016;7:62754-62766. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 92] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 61. | Bankó P, Lee SY, Nagygyörgy V, Zrínyi M, Chae CH, Cho DH, Telekes A. Technologies for circulating tumor cell separation from whole blood. J Hematol Oncol. 2019;12:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 238] [Cited by in RCA: 235] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 62. | Zhao XH, Wang ZR, Chen CL, Di L, Bi ZF, Li ZH, Liu YM. Molecular detection of epithelial-mesenchymal transition markers in circulating tumor cells from pancreatic cancer patients: Potential role in clinical practice. World J Gastroenterol. 2019;25:138-150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 42] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 63. | Deng Z, Wu S, Wang Y, Shi D. Circulating tumor cell isolation for cancer diagnosis and prognosis. EBioMedicine. 2022;83:104237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 178] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 64. | Götze J, Nitschke C, Uzunoglu FG, Pantel K, Sinn M, Wikman H. Tumor-Stroma Interaction in PDAC as a New Approach for Liquid Biopsy and its Potential Clinical Implications. Front Cell Dev Biol. 2022;10:918795. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 65. | de Miranda FS, Barauna VG, Dos Santos L, Costa G, Vassallo PF, Campos LCG. Properties and Application of Cell-Free DNA as a Clinical Biomarker. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 57] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 66. | Hassan S, Shehzad A, Khan SA, Miran W, Khan S, Lee YS. Diagnostic and Therapeutic Potential of Circulating-Free DNA and Cell-Free RNA in Cancer Management. Biomedicines. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 67. | Cahilog Z, Zhao H, Wu L, Alam A, Eguchi S, Weng H, Ma D. The Role of Neutrophil NETosis in Organ Injury: Novel Inflammatory Cell Death Mechanisms. Inflammation. 2020;43:2021-2032. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 94] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 68. | Vorobjeva NV, Chernyak BV. NETosis: Molecular Mechanisms, Role in Physiology and Pathology. Biochemistry (Mosc). 2020;85:1178-1190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 336] [Article Influence: 56.0] [Reference Citation Analysis (0)] |
| 69. | Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer--a survey. Biochim Biophys Acta. 2007;1775:181-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 442] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 70. | Bianchi DW, Chiu RWK. Sequencing of Circulating Cell-free DNA during Pregnancy. N Engl J Med. 2018;379:464-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 231] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 71. | Schou JV, Larsen FO, Sørensen BS, Abrantes R, Boysen AK, Johansen JS, Jensen BV, Nielsen DL, Spindler KL. Circulating cell-free DNA as predictor of treatment failure after neoadjuvant chemo-radiotherapy before surgery in patients with locally advanced rectal cancer. Ann Oncol. 2018;29:610-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 72. | van der Pol Y, Moldovan N, Ramaker J, Bootsma S, Lenos KJ, Vermeulen L, Sandhu S, Bahce I, Pegtel DM, Wong SQ, Dawson SJ, Chandrananda D, Mouliere F. The landscape of cell-free mitochondrial DNA in liquid biopsy for cancer detection. Genome Biol. 2023;24:229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 41] [Reference Citation Analysis (0)] |
| 73. | Marcatti M, Saada J, Okereke I, Wade CE, Bossmann SH, Motamedi M, Szczesny B. Quantification of Circulating Cell Free Mitochondrial DNA in Extracellular Vesicles with PicoGreen™ in Liquid Biopsies: Fast Assessment of Disease/Trauma Severity. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 74. | Calabuig-Fariñas S, Jantus-Lewintre E, Herreros-Pomares A, Camps C. Circulating tumor cells vs circulating tumor DNA in lung cancer-which one will win? Transl Lung Cancer Res 2016; 5: 466-482. [RCA] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 75] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 75. | Madhavan D, Wallwiener M, Bents K, Zucknick M, Nees J, Schott S, Cuk K, Riethdorf S, Trumpp A, Pantel K, Sohn C, Schneeweiss A, Surowy H, Burwinkel B. Plasma DNA integrity as a biomarker for primary and metastatic breast cancer and potential marker for early diagnosis. Breast Cancer Res Treat. 2014;146:163-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 130] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 76. | Papadopoulos N. Pathophysiology of ctDNA Release into the Circulation and Its Characteristics: What Is Important for Clinical Applications. Recent Results Cancer Res. 2020;215:163-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 77. | Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, Rajan S, Humphray S, Becq J, Halsall D, Wallis M, Bentley D, Caldas C, Rosenfeld N. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199-1209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1713] [Cited by in RCA: 1759] [Article Influence: 135.3] [Reference Citation Analysis (0)] |
| 78. | Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016;35:347-376. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 418] [Cited by in RCA: 625] [Article Influence: 69.4] [Reference Citation Analysis (0)] |
| 79. | Mouliere F, Chandrananda D, Piskorz AM, Moore EK, Morris J, Ahlborn LB, Mair R, Goranova T, Marass F, Heider K, Wan JCM, Supernat A, Hudecova I, Gounaris I, Ros S, Jimenez-Linan M, Garcia-Corbacho J, Patel K, Østrup O, Murphy S, Eldridge MD, Gale D, Stewart GD, Burge J, Cooper WN, van der Heijden MS, Massie CE, Watts C, Corrie P, Pacey S, Brindle KM, Baird RD, Mau-Sørensen M, Parkinson CA, Smith CG, Brenton JD, Rosenfeld N. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med. 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 622] [Cited by in RCA: 760] [Article Influence: 95.0] [Reference Citation Analysis (0)] |
| 80. | Neumann MHD, Bender S, Krahn T, Schlange T. ctDNA and CTCs in Liquid Biopsy - Current Status and Where We Need to Progress. Comput Struct Biotechnol J. 2018;16:190-195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 165] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 81. | Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet. 1999;64:218-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 812] [Cited by in RCA: 820] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 82. | Cheng F, Su L, Qian C. Circulating tumor DNA: a promising biomarker in the liquid biopsy of cancer. Oncotarget. 2016;7:48832-48841. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 279] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 83. | Vellanki PJ, Ghosh S, Pathak A, Fusco MJ, Bloomquist EW, Tang S, Singh H, Philip R, Pazdur R, Beaver JA. Regulatory implications of ctDNA in immuno-oncology for solid tumors. J Immunother Cancer. 2023;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 44] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 84. | Pascual J, Attard G, Bidard FC, Curigliano G, De Mattos-Arruda L, Diehn M, Italiano A, Lindberg J, Merker JD, Montagut C, Normanno N, Pantel K, Pentheroudakis G, Popat S, Reis-Filho JS, Tie J, Seoane J, Tarazona N, Yoshino T, Turner NC. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2022;33:750-768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 461] [Article Influence: 115.3] [Reference Citation Analysis (0)] |
| 85. | Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, Silliman N, Tacey M, Wong HL, Christie M, Kosmider S, Skinner I, Wong R, Steel M, Tran B, Desai J, Jones I, Haydon A, Hayes T, Price TJ, Strausberg RL, Diaz LA Jr, Papadopoulos N, Kinzler KW, Vogelstein B, Gibbs P. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8:346ra92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 729] [Cited by in RCA: 1097] [Article Influence: 121.9] [Reference Citation Analysis (0)] |
| 86. | Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, Le Quesne J, Moore DA, Veeriah S, Rosenthal R, Marafioti T, Kirkizlar E, Watkins TBK, McGranahan N, Ward S, Martinson L, Riley J, Fraioli F, Al Bakir M, Grönroos E, Zambrana F, Endozo R, Bi WL, Fennessy FM, Sponer N, Johnson D, Laycock J, Shafi S, Czyzewska-Khan J, Rowan A, Chambers T, Matthews N, Turajlic S, Hiley C, Lee SM, Forster MD, Ahmad T, Falzon M, Borg E, Lawrence D, Hayward M, Kolvekar S, Panagiotopoulos N, Janes SM, Thakrar R, Ahmed A, Blackhall F, Summers Y, Hafez D, Naik A, Ganguly A, Kareht S, Shah R, Joseph L, Marie Quinn A, Crosbie PA, Naidu B, Middleton G, Langman G, Trotter S, Nicolson M, Remmen H, Kerr K, Chetty M, Gomersall L, Fennell DA, Nakas A, Rathinam S, Anand G, Khan S, Russell P, Ezhil V, Ismail B, Irvin-Sellers M, Prakash V, Lester JF, Kornaszewska M, Attanoos R, Adams H, Davies H, Oukrif D, Akarca AU, Hartley JA, Lowe HL, Lock S, Iles N, Bell H, Ngai Y, Elgar G, Szallasi Z, Schwarz RF, Herrero J, Stewart A, Quezada SA, Peggs KS, Van Loo P, Dive C, Lin CJ, Rabinowitz M, Aerts HJWL, Hackshaw A, Shaw JA, Zimmermann BG; TRACERx consortium; PEACE consortium, Swanton C. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545:446-451. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1195] [Cited by in RCA: 1390] [Article Influence: 154.4] [Reference Citation Analysis (0)] |
| 87. | Stadler JC, Belloum Y, Deitert B, Sementsov M, Heidrich I, Gebhardt C, Keller L, Pantel K. Current and Future Clinical Applications of ctDNA in Immuno-Oncology. Cancer Res. 2022;82:349-358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 121] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 88. | Ma C, Yang X, Xing W, Yu H, Si T, Guo Z. Detection of circulating tumor DNA from non-small cell lung cancer brain metastasis in cerebrospinal fluid samples. Thorac Cancer. 2020;11:588-593. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 71] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 89. | Miller AM, Karajannis MA. Current Role and Future Potential of CSF ctDNA for the Diagnosis and Clinical Management of Pediatric Central Nervous System Tumors. J Natl Compr Canc Netw. 2022;20:1363-1369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 90. | Tan CR, Zhou L, El-Deiry WS. Circulating Tumor Cells Versus Circulating Tumor DNA in Colorectal Cancer: Pros and Cons. Curr Colorectal Cancer Rep. 2016;12:151-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 56] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 91. | Grölz D, Hauch S, Schlumpberger M, Guenther K, Voss T, Sprenger-Haussels M, Oelmüller U. Liquid Biopsy Preservation Solutions for Standardized Pre-Analytical Workflows-Venous Whole Blood and Plasma. Curr Pathobiol Rep. 2018;6:275-286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 69] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 92. | Chan HT, Chin YM, Nakamura Y, Low SK. Clonal Hematopoiesis in Liquid Biopsy: From Biological Noise to Valuable Clinical Implications. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 129] [Article Influence: 21.5] [Reference Citation Analysis (1)] |
| 93. | Merker JD, Oxnard GR, Compton C, Diehn M, Hurley P, Lazar AJ, Lindeman N, Lockwood CM, Rai AJ, Schilsky RL, Tsimberidou AM, Vasalos P, Billman BL, Oliver TK, Bruinooge SS, Hayes DF, Turner NC. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol. 2018;36:1631-1641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 494] [Cited by in RCA: 675] [Article Influence: 84.4] [Reference Citation Analysis (0)] |
| 94. | Gold B, Cankovic M, Furtado LV, Meier F, Gocke CD. Do circulating tumor cells, exosomes, and circulating tumor nucleic acids have clinical utility? A report of the association for molecular pathology. J Mol Diagn. 2015;17:209-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 160] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 95. | Kan CM, Pei XM, Yeung MHY, Jin N, Ng SSM, Tsang HF, Cho WCS, Yim AK, Yu AC, Wong SCC. Exploring the Role of Circulating Cell-Free RNA in the Development of Colorectal Cancer. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 25] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 96. | Gittelman MC, Hertzman B, Bailen J, Williams T, Koziol I, Henderson RJ, Efros M, Bidair M, Ward JF. PCA3 molecular urine test as a predictor of repeat prostate biopsy outcome in men with previous negative biopsies: a prospective multicenter clinical study. J Urol. 2013;190:64-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 100] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 97. | Mugoni V, Ciani Y, Nardella C, Demichelis F. Circulating RNAs in prostate cancer patients. Cancer Lett. 2022;524:57-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 63] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 98. | Liu J, Chen Y, Pei F, Zeng C, Yao Y, Liao W, Zhao Z. Extracellular Vesicles in Liquid Biopsies: Potential for Disease Diagnosis. Biomed Res Int. 2021;2021:6611244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 99. | Meldolesi J. Extracellular vesicles (exosomes and ectosomes) play key roles in the pathology of brain diseases. Mol Biomed. 2021;2:18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 100. | Zhang Y, Liu Y, Liu H, Tang WH. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci. 2019;9:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1505] [Cited by in RCA: 1541] [Article Influence: 220.1] [Reference Citation Analysis (1)] |
| 101. | Manickam DS. Delivery of mitochondria via extracellular vesicles - A new horizon in drug delivery. J Control Release. 2022;343:400-407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 102. | Anand S, Samuel M, Mathivanan S. Exomeres: A New Member of Extracellular Vesicles Family. Subcell Biochem. 2021;97:89-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 78] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 103. | O’Neill CP, Gilligan KE, Dwyer RM. Role of Extracellular Vesicles (EVs) in Cell Stress Response and Resistance to Cancer Therapy. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 81] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 104. | Irmer B, Chandrabalan S, Maas L, Bleckmann A, Menck K. Extracellular Vesicles in Liquid Biopsies as Biomarkers for Solid Tumors. Cancers (Basel). 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 85] [Reference Citation Analysis (0)] |
| 105. | Yu D, Li Y, Wang M, Gu J, Xu W, Cai H, Fang X, Zhang X. Exosomes as a new frontier of cancer liquid biopsy. Mol Cancer. 2022;21:56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 335] [Cited by in RCA: 576] [Article Influence: 144.0] [Reference Citation Analysis (0)] |
| 106. | Wu K, Xing F, Wu SY, Watabe K. Extracellular vesicles as emerging targets in cancer: Recent development from bench to bedside. Biochim Biophys Acta Rev Cancer. 2017;1868:538-563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 124] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 107. | Roweth HG, Battinelli EM. Lessons to learn from tumor-educated platelets. Blood. 2021;137:3174-3180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 110] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 108. | Best MG, Wesseling P, Wurdinger T. Tumor-Educated Platelets as a Noninvasive Biomarker Source for Cancer Detection and Progression Monitoring. Cancer Res. 2018;78:3407-3412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 191] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 109. | Antunes-Ferreira M, Koppers-Lalic D, Würdinger T. Circulating platelets as liquid biopsy sources for cancer detection. Mol Oncol. 2021;15:1727-1743. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 110. | Yan X, Liu C. Application of Non-Blood-Derived Fluid Biopsy in Monitoring Minimal Residual Diseases of Lung Cancer. Front Surg. 2022;9:865040. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 111. | Ponti G, Manfredini M, Tomasi A. Non-blood sources of cell-free DNA for cancer molecular profiling in clinical pathology and oncology. Crit Rev Oncol Hematol. 2019;141:36-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 61] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 112. | Tivey A, Church M, Rothwell D, Dive C, Cook N. Circulating tumour DNA - looking beyond the blood. Nat Rev Clin Oncol. 2022;19:600-612. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 219] [Article Influence: 54.8] [Reference Citation Analysis (0)] |
| 113. | Ponti G, Maccaferri M, Manfredini M, Micali S, Torricelli F, Milandri R, Del Prete C, Ciarrocchi A, Ruini C, Benassi L, Bettelli S, Kaleci S, Ozben T, Tomasi A. Quick assessment of cell-free DNA in seminal fluid and fragment size for early non-invasive prostate cancer diagnosis. Clin Chim Acta. 2019;497:76-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 114. | Abraham JE, Maranian MJ, Spiteri I, Russell R, Ingle S, Luccarini C, Earl HM, Pharoah PP, Dunning AM, Caldas C. Saliva samples are a viable alternative to blood samples as a source of DNA for high throughput genotyping. BMC Med Genomics. 2012;5:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 113] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 115. | Lopes C, Chaves J, Ortigão R, Dinis-Ribeiro M, Pereira C. Gastric cancer detection by non-blood-based liquid biopsies: A systematic review looking into the last decade of research. United European Gastroenterol J. 2023;11:114-130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 116. | Alagesan P, Goodwin JC, Garman KS, Epplein M. Cancer Progress and Priorities: Gastric Cancer. Cancer Epidemiol Biomarkers Prev. 2023;32:473-486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 117. | Yamaoka Y, Kato M, Asaka M. Geographic differences in gastric cancer incidence can be explained by differences between Helicobacter pylori strains. Intern Med. 2008;47:1077-1083. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 180] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 118. | Hamashima C, Kim Y, Choi KS. Comparison of guidelines and management for Gastric Cancer Screening between Korea and Japan. Value Heal. 2015;18:A272. [DOI] [Full Text] |
| 119. | Jun JK, Choi KS, Lee HY, Suh M, Park B, Song SH, Jung KW, Lee CW, Choi IJ, Park EC, Lee D. Effectiveness of the Korean National Cancer Screening Program in Reducing Gastric Cancer Mortality. Gastroenterology. 2017;152:1319-1328.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 405] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 120. | Russo AE, Strong VE. Gastric Cancer Etiology and Management in Asia and the West. Annu Rev Med. 2019;70:353-367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 124] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 121. | Ryun Park S. Management of gastric cancer: East vs West. Curr Probl Cancer. 2015;39:315-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 122. | Lin SJ, Gagnon-Bartsch JA, Tan IB, Earle S, Ruff L, Pettinger K, Ylstra B, van Grieken N, Rha SY, Chung HC, Lee JS, Cheong JH, Noh SH, Aoyama T, Miyagi Y, Tsuburaya A, Yoshikawa T, Ajani JA, Boussioutas A, Yeoh KG, Yong WP, So J, Lee J, Kang WK, Kim S, Kameda Y, Arai T, Zur Hausen A, Speed TP, Grabsch HI, Tan P. Signatures of tumour immunity distinguish Asian and non-Asian gastric adenocarcinomas. Gut. 2015;64:1721-1731. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 189] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 123. | Mukkamalla SKR, Recio-Boiles A, Babiker HM. Gastric Cancer. 2023 Jul 4. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2024 Jan-. [PubMed] |
| 124. | Ilic M, Ilic I. Epidemiology of stomach cancer. World J Gastroenterol. 2022;28:1187-1203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 239] [Cited by in RCA: 292] [Article Influence: 73.0] [Reference Citation Analysis (23)] |
| 125. | Alshehri A, Alanezi H, Kim BS. Prognosis factors of advanced gastric cancer according to sex and age. World J Clin Cases. 2020;8:1608-1619. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 10] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 126. | Conti CB, Agnesi S, Scaravaglio M, Masseria P, Dinelli ME, Oldani M, Uggeri F. Early Gastric Cancer: Update on Prevention, Diagnosis and Treatment. Int J Environ Res Public Health. 2023;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 111] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 127. | Koemans WJ, Luijten JCHBM, van der Kaaij RT, Grootscholten C, Snaebjornsson P, Verhoeven RHA, van Sandick JW. The metastatic pattern of intestinal and diffuse type gastric carcinoma - A Dutch national cohort study. Cancer Epidemiol. 2020;69:101846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 128. | Díaz Del Arco C, Estrada Muñoz L, Ortega Medina L, Molina Roldán E, Cerón Nieto MÁ, García Gómez de Las Heras S, Fernández Aceñero MJ. Clinicopathological differences, risk factors and prognostic scores for western patients with intestinal and diffuse-type gastric cancer. World J Gastrointest Oncol. 2022;14:1162-1174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 9] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 129. | Díaz Del Arco C, Ortega Medina L, Estrada Muñoz L, García Gómez de Las Heras S, Fernández Aceñero MJ. Is there still a place for conventional histopathology in the age of molecular medicine? Laurén classification, inflammatory infiltration and other current topics in gastric cancer diagnosis and prognosis. Histol Histopathol. 2021;36:587-613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 130. | Min YW, Min BH, Lee JH, Kim JJ. Endoscopic treatment for early gastric cancer. World J Gastroenterol. 2014;20:4566-4573. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 39] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (1)] |
| 131. | Nevo Y, Ferri L. Current management of gastric adenocarcinoma: a narrative review. J Gastrointest Oncol. 2023;14:1933-1948. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 132. | Tateo V, Marchese PV, Mollica V, Massari F, Kurzrock R, Adashek JJ. Agnostic Approvals in Oncology: Getting the Right Drug to the Right Patient with the Right Genomics. Pharmaceuticals (Basel). 2023;16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 59] [Reference Citation Analysis (0)] |
| 133. | Chia NY, Tan P. Molecular classification of gastric cancer. Ann Oncol. 2016;27:763-769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 284] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 134. | Molaei F, Forghanifard MM, Fahim Y, Abbaszadegan MR. Molecular Signaling in Tumorigenesis of Gastric Cancer. Iran Biomed J. 2018;22:217-230. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 31] [Reference Citation Analysis (0)] |
| 135. | Jiang HB, Yang TJ, Lu P, Ma YJ. Gene expression profiling of gastric cancer. Eur Rev Med Pharmacol Sci. 2014;18:2109-2115. [PubMed] |
| 136. | Wang Q, Liu G, Hu C. Molecular Classification of Gastric Adenocarcinoma. Gastroenterology Res. 2019;12:275-282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 137. | Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5015] [Cited by in RCA: 5087] [Article Influence: 423.9] [Reference Citation Analysis (4)] |
| 138. | Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, Ye XS, Do IG, Liu S, Gong L, Fu J, Jin JG, Choi MG, Sohn TS, Lee JH, Bae JM, Kim ST, Park SH, Sohn I, Jung SH, Tan P, Chen R, Hardwick J, Kang WK, Ayers M, Hongyue D, Reinhard C, Loboda A, Kim S, Aggarwal A. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1071] [Cited by in RCA: 1660] [Article Influence: 150.9] [Reference Citation Analysis (0)] |
| 139. | Shah MA, Khanin R, Tang L, Janjigian YY, Klimstra DS, Gerdes H, Kelsen DP. Molecular classification of gastric cancer: a new paradigm. Clin Cancer Res. 2011;17:2693-2701. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 243] [Cited by in RCA: 246] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 140. | Tan IB, Ivanova T, Lim KH, Ong CW, Deng N, Lee J, Tan SH, Wu J, Lee MH, Ooi CH, Rha SY, Wong WK, Boussioutas A, Yeoh KG, So J, Yong WP, Tsuburaya A, Grabsch H, Toh HC, Rozen S, Cheong JH, Noh SH, Wan WK, Ajani JA, Lee JS, Tellez MS, Tan P. Intrinsic subtypes of gastric cancer, based on gene expression pattern, predict survival and respond differently to chemotherapy. Gastroenterology. 2011;141:476-485, 485.e1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 273] [Cited by in RCA: 280] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 141. | Lei Z, Tan IB, Das K, Deng N, Zouridis H, Pattison S, Chua C, Feng Z, Guan YK, Ooi CH, Ivanova T, Zhang S, Lee M, Wu J, Ngo A, Manesh S, Tan E, Teh BT, So JB, Goh LK, Boussioutas A, Lim TK, Flotow H, Tan P, Rozen SG. Identification of molecular subtypes of gastric cancer with different responses to PI3-kinase inhibitors and 5-fluorouracil. Gastroenterology. 2013;145:554-565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 352] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 142. | Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, Siu HC, Deng S, Chu KM, Law S, Chan KH, Chan AS, Tsui WY, Ho SL, Chan AK, Man JL, Foglizzo V, Ng MK, Ching YP, Cheng GH, Xie T, Fernandez J, Li VS, Clevers H, Rejto PA, Mao M, Leung SY. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 694] [Cited by in RCA: 861] [Article Influence: 71.8] [Reference Citation Analysis (0)] |
| 143. | Wang H, Ding Y, Chen Y, Jiang J, Lu J, Kong M, Mo F, Huang Y, Zhao W, Fang P, Chen X, Teng X, Xu N, Lu Y, Yu X, Li Z, Zhang J, Wang H, Bao X, Zhou D, Chi Y, Zhou T, Zhou Z, Chen S, Teng L. A novel genomic classification system of gastric cancer via integrating multidimensional genomic characteristics. Gastric Cancer. 2021;24:1227-1241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 144. | Furukawa K, Hatakeyama K, Terashima M, Nagashima T, Urakami K, Ohshima K, Notsu A, Sugino T, Yagi T, Fujiya K, Kamiya S, Hikage M, Tanizawa Y, Bando E, Kanai Y, Akiyama Y, Yamaguchi K. Molecular classification of gastric cancer predicts survival in patients undergoing radical gastrectomy based on project HOPE. Gastric Cancer. 2022;25:138-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 145. | Díaz Del Arco C, Estrada Muñoz L, Molina Roldán E, Cerón Nieto MÁ, Ortega Medina L, García Gómez de Las Heras S, Fernández Aceñero MJ. Immunohistochemical classification of gastric cancer based on new molecular biomarkers: a potential predictor of survival. Virchows Arch. 2018;473:687-695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 146. | Zhang Z, Wu H, Chong W, Shang L, Jing C, Li L. Liquid biopsy in gastric cancer: predictive and prognostic biomarkers. Cell Death Dis. 2022;13:903. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 82] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 147. | Ma S, Zhou M, Xu Y, Gu X, Zou M, Abudushalamu G, Yao Y, Fan X, Wu G. Clinical application and detection techniques of liquid biopsy in gastric cancer. Mol Cancer. 2023;22:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 126] [Reference Citation Analysis (0)] |
| 148. | Han HS, Lee KW. Liquid Biopsy: An Emerging Diagnostic, Prognostic, and Predictive Tool in Gastric Cancer. J Gastric Cancer. 2024;24:4-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 21] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 149. | Thanh Huong P, Gurshaney S, Thanh Binh N, Gia Pham A, Hoang Nguyen H, Thanh Nguyen X, Pham-The H, Tran PT, Truong Vu K, Xuan Duong N, Pelucchi C, La Vecchia C, Boffetta P, Nguyen HD, Luu HN. Emerging Role of Circulating Tumor Cells in Gastric Cancer. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 150. | Tang L, Zhao S, Liu W, Parchim NF, Huang J, Tang Y, Gan P, Zhong M. Diagnostic accuracy of circulating tumor cells detection in gastric cancer: systematic review and meta-analysis. BMC Cancer. 2013;13:314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 151. | Lee MW, Kim GH, Jeon HK, Park SJ. Clinical Application of Circulating Tumor Cells in Gastric Cancer. Gut Liver. 2019;13:394-401. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 152. | Kang HM, Kim GH, Jeon HK, Kim DH, Jeon TY, Park DY, Jeong H, Chun WJ, Kim MH, Park J, Lim M, Kim TH, Cho YK. Circulating tumor cells detected by lab-on-a-disc: Role in early diagnosis of gastric cancer. PLoS One. 2017;12:e0180251. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 61] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 153. | Cheng B, Tong G, Wu X, Cai W, Li Z, Tong Z, He L, Yu S, Wang S. Enumeration And Characterization Of Circulating Tumor Cells And Its Application In Advanced Gastric Cancer. Onco Targets Ther. 2019;12:7887-7896. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 154. | Iwatsuki M, Toyoshima K, Watanabe M, Hayashi N, Ishimoto T, Eto K, Iwagami S, Baba Y, Yoshida N, Hayashi A, Ohta Y, Baba H. Frequency of HER2 expression of circulating tumour cells in patients with metastatic or recurrent gastrointestinal cancer. Br J Cancer. 2013;109:2829-2832. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 155. | Huang X, Gao P, Sun J, Chen X, Song Y, Zhao J, Xu H, Wang Z. Clinicopathological and prognostic significance of circulating tumor cells in patients with gastric cancer: a meta-analysis. Int J Cancer. 2015;136:21-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 156. | Hiraiwa K, Takeuchi H, Hasegawa H, Saikawa Y, Suda K, Ando T, Kumagai K, Irino T, Yoshikawa T, Matsuda S, Kitajima M, Kitagawa Y. Clinical significance of circulating tumor cells in blood from patients with gastrointestinal cancers. Ann Surg Oncol. 2008;15:3092-3100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 151] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 157. | Zhou J, Ma X, Bi F, Liu M. Clinical significance of circulating tumor cells in gastric cancer patients. Oncotarget. 2017;8:25713-25720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 158. | Zheng X, Fan L, Zhou P, Ma H, Huang S, Yu D, Zhao L, Yang S, Liu J, Huang A, Cai C, Dai X, Zhang T. Detection of Circulating Tumor Cells and Circulating Tumor Microemboli in Gastric Cancer. Transl Oncol. 2017;10:431-441. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 159. | Li Z, Song M, Han S, Jin C, Yang J. The prognostic role of circulating tumor cells in gastric cancer: A meta-analysis. Front Oncol. 2022;12:963091. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 160. | Li Y, Gong J, Zhang Q, Lu Z, Gao J, Li Y, Cao Y, Shen L. Dynamic monitoring of circulating tumour cells to evaluate therapeutic efficacy in advanced gastric cancer. Br J Cancer. 2016;114:138-145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 79] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 161. | Chen W, Yan H, Li X, Ge K, Wu J. Circulating tumor DNA detection and its application status in gastric cancer: a narrative review. Transl Cancer Res. 2021;10:529-536. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 162. | Mencel J, Slater S, Cartwright E, Starling N. The Role of ctDNA in Gastric Cancer. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 163. | Kim K, Shin DG, Park MK, Baik SH, Kim TH, Kim S, Lee S. Circulating cell-free DNA as a promising biomarker in patients with gastric cancer: diagnostic validity and significant reduction of cfDNA after surgical resection. Ann Surg Treat Res. 2014;86:136-142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 117] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 164. | Qian C, Ju S, Qi J, Zhao J, Shen X, Jing R, Yu J, Li L, Shi Y, Zhang L, Wang Z, Cong H. Alu-based cell-free DNA: a novel biomarker for screening of gastric cancer. Oncotarget. 2017;8:54037-54045. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 165. | Cabel L, Decraene C, Bieche I, Pierga JY, Bennamoun M, Fuks D, Ferraz JM, Lefevre M, Baulande S, Bernard V, Vacher S, Mariani P, Proudhon C, Bidard FC, Louvet C. Limited Sensitivity of Circulating Tumor DNA Detection by Droplet Digital PCR in Non-Metastatic Operable Gastric Cancer Patients. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 166. | Gao Y, Zhang K, Xi H, Cai A, Wu X, Cui J, Li J, Qiao Z, Wei B, Chen L. Diagnostic and prognostic value of circulating tumor DNA in gastric cancer: a meta-analysis. Oncotarget. 2017;8:6330-6340. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 167. | Yang J, Gong Y, Lam VK, Shi Y, Guan Y, Zhang Y, Ji L, Chen Y, Zhao Y, Qian F, Chen J, Li P, Zhang F, Wang J, Zhang X, Yang L, Kopetz S, Futreal PA, Zhang J, Yi X, Xia X, Yu P. Deep sequencing of circulating tumor DNA detects molecular residual disease and predicts recurrence in gastric cancer. Cell Death Dis. 2020;11:346. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 140] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 168. | Klein EA, Richards D, Cohn A, Tummala M, Lapham R, Cosgrove D, Chung G, Clement J, Gao J, Hunkapiller N, Jamshidi A, Kurtzman KN, Seiden MV, Swanton C, Liu MC. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann Oncol. 2021;32:1167-1177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 630] [Article Influence: 126.0] [Reference Citation Analysis (0)] |
| 169. | Nakamura Y, Taniguchi H, Ikeda M, Bando H, Kato K, Morizane C, Esaki T, Komatsu Y, Kawamoto Y, Takahashi N, Ueno M, Kagawa Y, Nishina T, Kato T, Yamamoto Y, Furuse J, Denda T, Kawakami H, Oki E, Nakajima T, Nishida N, Yamaguchi K, Yasui H, Goto M, Matsuhashi N, Ohtsubo K, Yamazaki K, Tsuji A, Okamoto W, Tsuchihara K, Yamanaka T, Miki I, Sakamoto Y, Ichiki H, Hata M, Yamashita R, Ohtsu A, Odegaard JI, Yoshino T. Clinical utility of circulating tumor DNA sequencing in advanced gastrointestinal cancer: SCRUM-Japan GI-SCREEN and GOZILA studies. Nat Med. 2020;26:1859-1864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 269] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 170. | Gao J, Wang H, Zang W, Li B, Rao G, Li L, Yu Y, Li Z, Dong B, Lu Z, Jiang Z, Shen L. Circulating tumor DNA functions as an alternative for tissue to overcome tumor heterogeneity in advanced gastric cancer. Cancer Sci. 2017;108:1881-1887. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 171. | Zhou H, Liu H, Li J, Wang J, Fu X, Li Y, Mao S, Du J. Postoperative circulating tumor DNA detection and CBLB mutations are prognostic biomarkers for gastric cancer. Genes Genomics. 2023;45:1037-1046. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 172. | Fang WL, Lan YT, Huang KH, Liu CA, Hung YP, Lin CH, Jhang FY, Chang SC, Chen MH, Chao Y, Lin WC, Lo SS, Fen-Yau Li A, Wu CW, Chiou SH, Shyr YM. Clinical significance of circulating plasma DNA in gastric cancer. Int J Cancer. 2016;138:2974-2983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 65] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 173. | Min L, Chen J, Yu M, Yang K, Liu D. Utilizing circulating tumour DNA as a prognostic predictor of gastric cancer: a meta-analysis. Biomarkers. 2023;28:427-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 174. | Kim YW, Kim YH, Song Y, Kim HS, Sim HW, Poojan S, Eom BW, Kook MC, Joo J, Hong KM. Monitoring circulating tumor DNA by analyzing personalized cancer-specific rearrangements to detect recurrence in gastric cancer. Exp Mol Med. 2019;51:1-10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 175. | Pu WY, Zhang R, Xiao L, Wu YY, Gong W, Lv XD, Zhong FY, Zhuang ZX, Bai XM, Li K, Xing CG. Prediction of cancer progression in a group of 73 gastric cancer patients by circulating cell-free DNA. BMC Cancer. 2016;16:943. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 176. | Lan YT, Chen MH, Fang WL, Hsieh CC, Lin CH, Jhang FY, Yang SH, Lin JK, Chen WS, Jiang JK, Lin PC, Chang SC. Clinical relevance of cell-free DNA in gastrointestinal tract malignancy. Oncotarget. 2017;8:3009-3017. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 177. | Kim ST, Cristescu R, Bass AJ, Kim KM, Odegaard JI, Kim K, Liu XQ, Sher X, Jung H, Lee M, Lee S, Park SH, Park JO, Park YS, Lim HY, Lee H, Choi M, Talasaz A, Kang PS, Cheng J, Loboda A, Lee J, Kang WK. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med. 2018;24:1449-1458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 643] [Cited by in RCA: 1235] [Article Influence: 154.4] [Reference Citation Analysis (0)] |
| 178. | Wang DS, Liu ZX, Lu YX, Bao H, Wu X, Zeng ZL, Liu Z, Zhao Q, He CY, Lu JH, Wang ZQ, Qiu MZ, Wang F, Wang FH, Li YH, Wang XN, Xie D, Jia WH, Shao YW, Xu RH. Liquid biopsies to track trastuzumab resistance in metastatic HER2-positive gastric cancer. Gut. 2019;68:1152-1161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 145] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 179. | Yu J, Ostowari A, Gonda A, Mashayekhi K, Dayyani F, Hughes CCW, Senthil M. Exosomes as a Source of Biomarkers for Gastrointestinal Cancers. Cancers (Basel). 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 180. | Li C, Liu DR, Li GG, Wang HH, Li XW, Zhang W, Wu YL, Chen L. CD97 promotes gastric cancer cell proliferation and invasion through exosome-mediated MAPK signaling pathway. World J Gastroenterol. 2015;21:6215-6228. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 82] [Cited by in RCA: 100] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 181. | Stec M, Szatanek R, Baj-Krzyworzeka M, Baran J, Zembala M, Barbasz J, Waligórska A, Dobrucki JW, Mytar B, Szczepanik A, Siedlar M, Drabik G, Urbanowicz B. Interactions of tumour-derived micro(nano)vesicles with human gastric cancer cells. J Transl Med. 2015;13:376. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 182. | Qu JL, Qu XJ, Zhao MF, Teng YE, Zhang Y, Hou KZ, Jiang YH, Yang XH, Liu YP. Gastric cancer exosomes promote tumour cell proliferation through PI3K/Akt and MAPK/ERK activation. Dig Liver Dis. 2009;41:875-880. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 255] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 183. | Chen X, Wang H, Huang Y, Chen Y, Chen C, Zhuo W, Teng L. Comprehensive Roles and Future Perspectives of Exosomes in Peritoneal Metastasis of Gastric Cancer. Front Oncol. 2021;11:684871. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 184. | Ren J, Zhou Q, Li H, Li J, Pang L, Su L, Gu Q, Zhu Z, Liu B. Characterization of exosomal RNAs derived from human gastric cancer cells by deep sequencing. Tumour Biol. 2017;39:1010428317695012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 185. | Zhang H, Deng T, Liu R, Bai M, Zhou L, Wang X, Li S, Yang H, Li J, Ning T, Huang D, Li H, Zhang L, Ying G, Ba Y. Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat Commun. 2017;8:15016. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 271] [Cited by in RCA: 428] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 186. | Gu J, Qian H, Shen L, Zhang X, Zhu W, Huang L, Yan Y, Mao F, Zhao C, Shi Y, Xu W. Gastric cancer exosomes trigger differentiation of umbilical cord derived mesenchymal stem cells to carcinoma-associated fibroblasts through TGF-β/Smad pathway. PLoS One. 2012;7:e52465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 221] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 187. | Yen EY, Miaw SC, Yu JS, Lai IR. Exosomal TGF-β1 is correlated with lymphatic metastasis of gastric cancers. Am J Cancer Res. 2017;7:2199-2208. [PubMed] |
| 188. | Fu H, Yang H, Zhang X, Wang B, Mao J, Li X, Wang M, Zhang B, Sun Z, Qian H, Xu W. Exosomal TRIM3 is a novel marker and therapy target for gastric cancer. J Exp Clin Cancer Res. 2018;37:162. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 103] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 189. | Zheng P, Luo Q, Wang W, Li J, Wang T, Wang P, Chen L, Zhang P, Chen H, Liu Y, Dong P, Xie G, Ma Y, Jiang L, Yuan X, Shen L. Tumor-associated macrophages-derived exosomes promote the migration of gastric cancer cells by transfer of functional Apolipoprotein E. Cell Death Dis. 2018;9:434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 302] [Article Influence: 37.8] [Reference Citation Analysis (1)] |
| 190. | Che Y, Geng B, Xu Y, Miao X, Chen L, Mu X, Pan J, Zhang C, Zhao T, Wang C, Li X, Wen H, Liu Z, You Q. Helicobacter pylori-induced exosomal MET educates tumour-associated macrophages to promote gastric cancer progression. J Cell Mol Med. 2018;22:5708-5719. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 57] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 191. | Fan Y, Che X, Qu J, Hou K, Wen T, Li Z, Li C, Wang S, Xu L, Liu Y, Qu X. Exosomal PD-L1 Retains Immunosuppressive Activity and is Associated with Gastric Cancer Prognosis. Ann Surg Oncol. 2019;26:3745-3755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 152] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 192. | Wang X, Zhang H, Bai M, Ning T, Ge S, Deng T, Liu R, Zhang L, Ying G, Ba Y. Exosomes Serve as Nanoparticles to Deliver Anti-miR-214 to Reverse Chemoresistance to Cisplatin in Gastric Cancer. Mol Ther. 2018;26:774-783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 178] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 193. | Yamamoto H, Watanabe Y, Oikawa R, Morita R, Yoshida Y, Maehata T, Yasuda H, Itoh F. BARHL2 Methylation Using Gastric Wash DNA or Gastric Juice Exosomal DNA is a Useful Marker For Early Detection of Gastric Cancer in an H. pylori-Independent Manner. Clin Transl Gastroenterol. 2016;7:e184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 69] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 194. | Gareev I, Ahmad A, Wang J, Beilerli A, Ilyasova T, Sufianov A, Beylerli O. Gastric juice non-coding RNAs as potential biomarkers for gastric cancer. Front Physiol. 2023;14:1179582. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 195. | Holzinger A, Haibe-Kains B, Jurisica I. Why imaging data alone is not enough: AI-based integration of imaging, omics, and clinical data. Eur J Nucl Med Mol Imaging. 2019;46:2722-2730. [RCA] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 64] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 196. | Shafi S, Parwani AV. Artificial intelligence in diagnostic pathology. Diagn Pathol. 2023;18:109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 136] [Reference Citation Analysis (0)] |
| 197. | Kim I, Kang K, Song Y, Kim TJ. Application of Artificial Intelligence in Pathology: Trends and Challenges. Diagnostics (Basel). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 54] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 198. | Pan B, Kang Y, Jin Y, Yang L, Zheng Y, Cui L, Sun J, Feng J, Li Y, Guo L, Liang Z. Automated tumor proportion scoring for PD-L1 expression based on multistage ensemble strategy in non-small cell lung cancer. J Transl Med. 2021;19:249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 199. | Cirillo D, Núñez-Carpintero I, Valencia A. Artificial intelligence in cancer research: learning at different levels of data granularity. Mol Oncol. 2021;15:817-829. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 200. | Aquino IMC, Pascut D. Liquid biopsy: New opportunities for precision medicine in hepatocellular carcinoma care. Ann Hepatol. 2023;29:101176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 201. | Visser E, Genet SAAM, de Kock RPPA, van den Borne BEEM, Youssef-El Soud M, Belderbos HNA, Stege G, de Saegher MEA, van ‘t Westeinde SC, Brunsveld L, Broeren MAC, van de Kerkhof D, Deiman BALM, Eduati F, Scharnhorst V. Liquid biopsy-based decision support algorithms for diagnosis and subtyping of lung cancer. Lung Cancer. 2023;178:28-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 25] [Reference Citation Analysis (0)] |
| 202. | Chen L, Abou-Alfa GK, Zheng B, Liu JF, Bai J, Du LT, Qian YS, Fan R, Liu XL, Wu L, Hou JL, Wang HY; PreCar Team. Genome-scale profiling of circulating cell-free DNA signatures for early detection of hepatocellular carcinoma in cirrhotic patients. Cell Res. 2021;31:589-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 81] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 203. | Liu S, Wu J, Xia Q, Liu H, Li W, Xia X, Wang J. Finding new cancer epigenetic and genetic biomarkers from cell-free DNA by combining SALP-seq and machine learning. Comput Struct Biotechnol J. 2020;18:1891-1903. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 204. | Roth P, Wischhusen J, Happold C, Chandran PA, Hofer S, Eisele G, Weller M, Keller A. A specific miRNA signature in the peripheral blood of glioblastoma patients. J Neurochem. 2011;118:449-457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 168] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 205. | Ma J, Yang J, Jin Y, Cheng S, Huang S, Zhang N, Wang Y. Artificial Intelligence Based on Blood Biomarkers Including CTCs Predicts Outcomes in Epithelial Ovarian Cancer: A Prospective Study. Onco Targets Ther. 2021;14:3267-3280. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 206. | Shi XJ, Wei Y, Ji B. Systems Biology of Gastric Cancer: Perspectives on the Omics-Based Diagnosis and Treatment. Front Mol Biosci. 2020;7:203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 207. | Matsuoka T, Yashiro M. Novel biomarkers for early detection of gastric cancer. World J Gastroenterol. 2023;29:2515-2533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 53] [Reference Citation Analysis (1)] |
| 208. | Lengyel CG, Hussain S, Trapani D, El Bairi K, Altuna SC, Seeber A, Odhiambo A, Habeeb BS, Seid F. The Emerging Role of Liquid Biopsy in Gastric Cancer. J Clin Med. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 209. | Grizzi G, Salati M, Bonomi M, Ratti M, Holladay L, De Grandis MC, Spada D, Baiocchi GL, Ghidini M. Circulating Tumor DNA in Gastric Adenocarcinoma: Future Clinical Applications and Perspectives. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 210. | BioRender. [cited 2 February 2024]. Available from: https://www.biorender.com/. |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author’s Membership in Professional Societies: Spanish Society of Pathology, No. 3147; Spanish Society for Scientific Documentation and Information, No. 3982.
Specialty type: Oncology
Country/Territory of origin: Spain
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Jiang J, China; Long X, China; Peng XC, China S-Editor: Chen YL L-Editor: A P-Editor: Cai YX