Published online Mar 21, 2024. doi: 10.3748/wjg.v30.i11.1497
Peer-review started: December 13, 2023
First decision: December 28, 2023
Revised: January 12, 2024
Accepted: March 1, 2024
Article in press: March 1, 2024
Published online: March 21, 2024
Processing time: 99 Days and 0.7 Hours
Esophageal squamous cell carcinoma (ESCC) is a malignant epithelial tumor, characterized by squamous cell differentiation, it is the sixth leading cause of cancer-related deaths globally. The increased mortality rate of ESCC patients is predominantly due to the advanced stage of the disease when discovered, coupled with higher risk of metastasis, which is an exceedingly malignant characteristic of cancer, frequently leading to a high mortality rate. Unfortunately, there is currently no specific and effective marker to predict and treat metastasis in ESCC. MicroRNAs (miRNAs) are a class of small non-coding RNA molecules, approximately 22 nucleotides in length. miRNAs are vital in modulating gene expression and serve pivotal regulatory roles in the occurrence, progression, and prognosis of cancer. Here, we have examined the literature to highlight the intimate correlations between miRNAs and ESCC metastasis, and show that ESCC metastasis is predominantly regulated or regulated by genetic and epigenetic factors. This review proposes a potential role for miRNAs as diagnostic and therapeutic biomarkers for metastasis in ESCC metastasis, with the ultimate aim of reducing the mortality rate among patients with ESCC.
Core Tip: Esophageal squamous cell carcinoma (ESCC) is the sixth leading cause of cancer-related deaths globally predominantly due to metastasis. MicroRNAs (miRNAs), acting either as tumor suppressors or oncogenes, play crucial roles in the development and progression of tumors. We herein discussed the intimate correlations between miRNAs and ESCC metastasis, predominantly associated with genetic and epigenetic regulatory, and aimed to propose the potential role of miRNAs as diagnostic and therapeutic biomarkers for ESCC metastasis.
- Citation: Wei QY, Jin F, Wang ZY, Li BJ, Cao WB, Sun ZY, Mo SJ. MicroRNAs: A novel signature in the metastasis of esophageal squamous cell carcinoma. World J Gastroenterol 2024; 30(11): 1497-1523
- URL: https://www.wjgnet.com/1007-9327/full/v30/i11/1497.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i11.1497
Esophageal cancer, the eighth most prevalent cancer worldwide, ranks as the sixth leading cause of cancer-related deaths[1]. There are two major histological types of esophageal cancer, which include esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC)[2]. Despite significant advancements in the treatment of ESCC treatment, both morbidity and mortality rates continue to rise annually. This is largely due to the advanced stage of the ESCC at diagnosis, combined with an increased propensity for metastasis[3,4]. When ESCC cells become metastatic and acquire the ability to invade surrounding tissues and enter the bloodstream or lymphatic system, they can travel to distant organs such as the liver, lungs, gastrointestinal tract, pancreas, and bones to establish secondary tumors[5].
Cancer metastasis is a complex process that encompasses a series of stages including local invasion (involving epithelial-to-mesenchymal transition, EMT), survival and entry into the bloodstream or lymphatic system (intravasation), exit from the bloodstream or lymphatic systems (extravasation), and proliferation at a new location (colonization). Examining this intricate process can offer valuable insights into the development of novel therapeutic strategies for cancer treatment. Numerous studies have demonstrated that a variety of signaling pathways, primarily receptor tyrosine kinase (RTK), transforming growth factor-β (TGF-β), Wnt/β-catenin, and the interleukin (IL)-6/Stat3 pathways, can partake in the process of cancer metastasis process[6-10]. Furthermore, evidence suggests that abnormally expressed microRNAs (miRNAs) can impact cancer metastasis by modulating these signaling pathways[7-10].
MiRNAs are endogenous short non-coding RNAs (approximately 22 nucleotides) that can regulate gene expression by inhibiting mRNA transcription or by promoting mRNA degradation through sequence- specific binding in the 3'-untranslated region (3' -UTR). They often function as master regulators of gene expression and play crucial roles in the development and progression of tumors by acting as either as tumor suppressors or oncogenes[11]. This review focuses on the role and mechanism of miRNAs in facilitating ESCC metastasis of ESCC, and also aims to highlight the potential use of miRNAs as diagnostic and therapeutic biomarkers in the management of ESCC metastasis.
Tissues from cancer patients, cancer cell lines, and xenograft models are commonly employed to investigate the occurrence and mechanisms of cancer cell metastasis. In the tissues of cancer patients, lymph node metastasis (LNM), vessel invasion (VI), lymph node invasion (LNI), lymphatic invasion (LI), tumor nodes metastasis stage (TNM), and depth of invasion (DI), are often used to evaluate the metastatic characteristics of the cancer. In cancer cells, the cell invasion, migration and EMT often are commonly used to evaluate the ability of tumor cells to metastasize. In xenograft models, the location and quantity of tumor distribution are used to identify the metastatic potential of tumors. Thus, we assess the role and underlying mechanism of miRNA in the metastasis of ESCC by examining its metastatic behavior in tissues, cells, and xenograft models (Tables 1 and 2)[12-15].
| miRNA | Metastasis studies in different sources | Ref. | ||
| Suppressor | Tissue | Cell | Xenograft | |
| miR-1 | LNM, VI, TNM, advanced C stage | I | [148] | |
| miR-10a | LNM, C stage | I, M, EMT | Tumor Me | [29] |
| miR-100 | LNM | I, M | [24] | |
| miR-100-5p | DI, LI, T stage | [149] | ||
| miR-101 | LNM | I, M | Liver Me | [30] |
| miR-10527-5p | LNM, T stage, LI | I, M, EMT, | [9] | |
| LI | ||||
| miR-107 | NS (LNM) | I, M | [152] | |
| miR-122 | I, M, EMT | [116] | ||
| miR-124 | LNM, Me | I, M | [25] | |
| miR-124-3p | LNM, DI, DM, TNM | I, M, EMT | [31,156] | |
| miR-125a-5p | I, M | [157] | ||
| miR-125b | LNM, TNM | I, M, EMT | [26] | |
| miR-1254 | VI, P stage | [159] | ||
| miR-126 | LNM, TNM | I, M | [27] | |
| miR-128-3p | LNM | I, M, EMT | [23] | |
| miR-129 | TNM | I, M | [28,143] | |
| miR-129-2 | LNM, DM, advance clinical TNM | I, M | [164] | |
| MiR-129-2-3p | I, M | [166] | ||
| miR-1294 | LNM, LI, VI | I, M | [84] | |
| miR-130a-5p | I, M | [169] | ||
| miR-133a | LNM, P stage | I, M | [170] | |
| miR-133b | I, M, EMT | [102] | ||
| miR-134 | LNM, TNM | I, M | [32] | |
| miR-136-5p | LNM, TNM | I, M | [168] | |
| miR-137 | I, M, EMT | [174] | ||
| miR-138 | LNM, TNM | [18] | ||
| miR-140-3p | LNM, DM, TNM | I, EMT | [176] | |
| miR-140 | Me | I, M, EMT | [178] | |
| miR-143 | LNM, DI, VI, TNM | I, M | [13] | |
| miR-143-3p | I, M | [180] | ||
| miR-144 | Me, TNM | I, M | Liver Me | [33] |
| miR-145 | LNM, DI, TNM | I, M, EMT | [182, 183] | |
| miR-145-3p | I, M | [185] | ||
| miR-145-5p | I, M, EMT | [187] | ||
| miR-146a | NS (N stage), TNM | I, M, EMT | [189] | |
| miR1469 | LNM, DI | [191] | ||
| miR-148a | I, M, EMT | [193] | ||
| miR-149-5p | I, M | [10] | ||
| miR-150 | LNM, LI, VI, TNM | I, M, EMT | [195] | |
| miR-150-3p | I, M | [197] | ||
| miR-153 | I, M, EMT | [94] | ||
| miR-181a-5p | I, M | DLA Me | [199] | |
| miR-185 | NS (LNM, DM, TNM) | I, M | Lung Me | [15] |
| miR-186-5p | NS (LNM), DM | I, M | [43] | |
| miR-193a-5p | LNM, TNM | [19] | ||
| miR-193b | I, M | [104] | ||
| miR-195 | LNM, VI, TNM | I, M | [202] | |
| miR-198 | I, M | [204] | ||
| miR-202 | LNM | M | [206] | |
| miR203 | LNM, TNM | I, M, EMT | Lung Me | [34,81] |
| miR-203a-3p | I, M, EMT | [209] | ||
| miR-204-5p | LNM, DI | I, M, EMT | [210] | |
| miR-205 | LNM | I, M, EMT | [93,213] | |
| miR206 | LNM, TNM | [20] | ||
| miR-212-3p | I | [215] | ||
| miR-214 | LNM, tumor stage | I, M, EMT | [216,217] | |
| miR-214-3p | I, M | [219] | ||
| miR-216a/b | LNM, TNM | [220] | ||
| miR-217 | I, M | [80] | ||
| miR-218 | LNM, TNM | I, M | [221] | |
| miR-26a | LNM, C stages | I, M | Liver Me | [33,223] |
| miR-27a | LNM | I | [103] | |
| miR-27b-3p | LNM, TNM | I, M, EMT | [225] | |
| miR-29b | LNM | I, M | [88] | |
| miR-29c | MPI | [228] | ||
| miR-30a | LNM | I, M | [148] | |
| miR-30b | NS (N stage) | I, M, EMT | [44] | |
| miR30a-3p | LNM | I, M, EMT | [148] | |
| miR-30c | T, N, M, stage | I, EMT | [95] | |
| miR-30d | LNM, advanced TNM | I, M | [150] | |
| miR-30e-5 | I, M | [126] | ||
| miR-301a | I, M | [151] | ||
| miR-323a-3p | I, M | Lung Me | [153] | |
| miR-326 | I, M | [154] | ||
| miR-328 | I, M | Liver Me | [155] | |
| miR-33a-5p | LNM, TNM | I, M | [111] | |
| miR-335-5p | I, M, EMT | [158] | ||
| miR-338-3p | I, M, EMT | [101] | ||
| miR-338-5p | I, M | Lung Me | [160] | |
| miR-34a | LNM, TNM | I, M, EMT | Liver Me | [36,161] |
| miR-340-5p | LNM, TNM | I, M | [162] | |
| miR-3612 | I, M | [163] | ||
| miR-365 | I, EMT | [165] | ||
| miR-370 | LNM, tumor invasion | [167] | ||
| miR-370-3p | LNM, TNM | I, M | [168] | |
| miR-375 | LNM, LVI, DM, TNM | I, M, EMT | Tumor mobility, I | [37,38] |
| miR-377 | LNM, TNM | I, M | [171] | |
| miR378 | LNM, TNM | I, M, | [172] | |
| miR-378a-3p | I, M | [11] | ||
| miR-422a | I, M | [173] | ||
| miR-429 | LNM, TNM | I, M, EMT | [175] | |
| miR-4324 | LNM, DI | I, M, EMT | [175] | |
| miR-433-3p | I, M | [177] | ||
| miR-4429 | LNM, TNM | I, M, EMT | [114] | |
| miR-449a-5p | LNM, TNM | [179] | ||
| miR-451 | LNM, TNM | [21] | ||
| miR-455-3p | LNM | I | [181] | |
| miR-455-5p | LNM | I, M | [184] | |
| miR-4766-5p | I, M, EMT | [186] | ||
| miR-485-5p | NS (LNM), TNM | I, M, EMT | [188] | |
| miR-486-5p | I | [190] | ||
| miR-490-3p | I, M, EMT | [192] | ||
| miR-493 | I, M | [145] | ||
| miR-497-5p | I, M | [194] | ||
| miR-498 | NS (LNM, DM) | I, M | [196] | |
| miR-508-3p | I, M | [198] | ||
| miR-515-3p | Me | I, M, EMT | Lung ME | [12] |
| miR-516b | LNM, DI, advanced TNM | I, M, EMT | [200] | |
| miR-518b | LNM | I | [201] | |
| miR-542-3p | NS (metastasis) | I, M | [47] | |
| miR-574-3p | LNM, DI, TNM | I, M | [50] | |
| miR-590 | LNM, TNM | I, M, EMT | [51] | |
| miR-595 | I, M | [203] | ||
| miR-599 | I, M, EMT | [205] | ||
| miR-615-5p | LNM, advanced TNM | I, M | [207] | |
| miR-620 | I, M, EMT | Lung ME | [208] | |
| miR-622 | N stage, TNM | I, M, EMT | [138] | |
| miR-625 | LNM, DM, advanced TNM | I | [211,212] | |
| miR-630 | LNM, P stage | I, M, EMT | [214] | |
| miR-652 | LNM, TNM | I, M | Multiple ME | [39] |
| miR-670-3p | I, M | [139] | ||
| miR-671-5p | I, M, EMT | [218] | ||
| miR-718 | LNM, TNM | [22] | ||
| miR-765 | I, M | [133] | ||
| miR-874-3p | LNM, C stage | I, M | [14] | |
| miR-942-5p | TNM, C stage | I, M, EMT | [222] | |
| miR-98 | LNM, TNM | I, M | [216] | |
| miR-99a | I, M, EMT | [224] | ||
| let-7 | LNM | [226] | ||
| let-7a | TNM | I, M, EMT | LNM | [227] |
| let-7b-5p | I, M | [40] | ||
| miRNA | Metastasis studies in different sources | Ref. | ||
| oncomiR | Tissue | Cell | Xenograft | |
| miR-10b-3p | LNM, C stages | I, M, EMT | Lung, Liver Bone Me | [56,57] |
| miR-10b | NS (LNM) | I, M | [229] | |
| miR-103a-2-5p | I | [68] | ||
| miR-105-5p | I, M | [69] | ||
| miR-106b-5p | TNM | I, M, EMT | Lung Me | [52] |
| miR-1179 | Me TNM | I | [232] | |
| miR-1269 | LNM, TNM | I, M | [59] | |
| miR-1290 | LNM, tumor invasion, TNM | I, M | [113] | |
| miR-130b | I, M | [70] | ||
| miR1323 | I, M | [71] | ||
| miR-135b-5p | LNM | I, M | [236] | |
| MiR-140-5p | I, M, EMT | [92] | ||
| miR-142-3p | LNM | [237] | ||
| miR-1470 | M | [239] | ||
| miR-17-5p | LNM, TNM, DI | I, M | [60,240] | |
| miR-17a | LNM, C stage | [242] | ||
| miR-18a | LNM, TNM | I, M | [61,244] | |
| miR-181b-5p | Advance TNM | M | [107] | |
| miR-181c-3p | DI, TNM | [246] | ||
| miR-19b | LNM, C stage | [242] | ||
| miR-19b-3p | I, M | [72] | ||
| miR-196a | I, M | [74] | ||
| miR-196a-3p | M | [74] | ||
| miR-20b-5p | LNM, C stage | I, M | [249] | |
| miR200a | I, M | [87] | ||
| miR-211 | LNM, LI, VI, TNM | I, M, EMT | [282] | |
| miR-221 | I, M | [251] | ||
| miR-224 | NR (LNM), TNM | I, M | [252] | |
| miR-224-5p | I, M, EMT | [100] | ||
| miR-23a | NS | I, M | Lung Me | [140,254] |
| miR-23b-3p | I, M, EMT | [73] | ||
| MiR-23b-5p | Me | I, M | Lung Me | [257] |
| miR-25 | LNM, TNM | I, M, EMT | Lung Me | [58] |
| miR-25-3p | LNM, TNM | I, M | [62] | |
| miR-30d-5p | LNM, TNM, DI | I, M, EMT | [91,230] | |
| miR-301 | LNM, TNM | I, M | [63] | |
| miR-301a-3p | LNM, tumor invasion, TNM | [231] | ||
| miR-301b | I, M | [75] | ||
| miR-31 | N, M stage, TNM | I, M, EMT | [233] | |
| miR-320b | LNM | I, M, EMT | [53] | |
| miR-330-3p | I, M | [234] | ||
| miR-3656 | I, M | [235] | ||
| miR-373 | LNM, TNM | I, M | [64] | |
| miR-425 | I, M | [119] | ||
| miR-4286 | I, M | [238] | ||
| miR-4443 | TNM | I | [60] | |
| miR-452-5p | LNM, DM | I, M | [241] | |
| miR-483-3p | I, M, EMT | [243] | ||
| miR-483-5p | LNM, TNM | I, M | [65,66] | |
| miR-506 | LNM, TNM | [245] | ||
| miR-5692b | DI, TNM | [246] | ||
| miR-602 | LNM, TNM | I, M | Lung, liver, bone, adrenal gland Me | [54] |
| miR-612 | LNM, metastasis | I, M | [247] | |
| miR-624-3p | I, M, EMT | [76] | ||
| miR-675-3p | I, M, EMT | [248] | ||
| miR-766-3p | LNM, TNM | I, M | [67] | |
| miR-875-5p | I, M | [250] | ||
| miR-9 | LNM | I, M, EMT | Lung Me | [85] |
| miR-92a | LNM, LNI | I, M | [86] | |
| miR-92a-3p | I, M | [77] | ||
| miR-92b-3p | I, M | [253] | ||
| miR-935 | Me | [255] | ||
| miR93 | I, M | [256] | ||
Numerous studies have found that miRNAs, acting either as tumor suppressors or oncogenes, play crucial roles in the development and progression of tumors[13]. As tumor suppressors, some miRNAs can inhibit tumor occurrence and development by targeting genes that have the potential to promote oncogenesis and progression[16]. Conversely, as oncogenes, some miRNAs, often called oncomiRs, can promote tumor malignancy by suppressing the expression of genes that normally prevent cells from becoming cancerous[17]. Multiple studies over the past decade have highlighted the importance of miRNAs also acting as suppressors or oncogenes in ESCC metastasis.
There is a significant reduction in the expression levels of tumor-suppressive miRNAs in both tissues and cells associated with ESCC. These miRNAs substantially influence the malignancy and metastatic potential of ESCC malignancy, including ESCC metastasis (Table 1). For example, miRNAs like miR-138, miR-193a-5p, miR-206, miR-451, and miR-718 are markedly decreased in ESCC-associated tissues, and these decreases are significantly correlated with LNM and the TNM stage[18-22]. Furthermore, the reduced expression of miRNAs, including but not limited to miR-100, miR-124, miR-125b, miR-126, miR-128-3p, and miR-129, are not only positively associated with ESCC metastasis in ESCC patient tissues, but also promotes ESCC cell invasion, migration, and/or EMT processes[23-28]. Several studies have confirmed the inhibitory effects of some miRNAs on ESCC metastasis at the tissue, cell, and animal model levels, including miR-10a, miR-101, miR-124-3p, miR-134, miR-144, miR-203, miR-26a, miR-34a, miR-375, miR-515-3p, miR-652, and let-7b-5p[12,29-40]. Intriguingly, our research reveals that certain miRNAs, including miR-107, miR-146a, miR-185, miR-186-5p, miR-30b, miR-485-5p, miR-498, and miR-542-3p, among others, have minimal effects on ESCC metastasis in tissue samples. However, they can significantly suppress ESCC cell invasion and migration and have a considerable impact on metastasis in xenograft models[15,41-47].
The role of these miRNAs as tumor suppressors heavily relies on the tumor-promoting activities of their target genes. For instance, the significant downregulation of miR-101 in ESCC tissues correlates strongly with LNM. Moreover, miR-101 can suppress tumor metastasis both in vitro (in xenograft models) and in vivo (in ESCC cells). Notably, the inhibitory effect of miR-101 in ESCC migration and invasion is reversed via activation of its target genes, COX-2, MALAT1, or EZH2. These genes are identified as playing crucial roles in promoting tumor development and progression, although not exclusively in ESCC[30,48,49]. Another notable suppressor miRNA is miR-574-3p, whose reduced expression is negatively associated with LNM, TNM stage, and invasion depth. Inhibition of miR-574-3p promotes migration and invasion of ESCC cells in vitro. However, the knockdown of its target genes, FAM3C or MAPK1, can lessen the increase in migration and invasion observed following treatment with the miR-574-3p inhibitor. The majority of studies suggest that both FAM3C and MAPK1 promote malignant tumor development, including proliferation, invasion, and migration[50].
Similar to cancer-suppressive miRNAs, oncomiRs have also been identified as promoters of ESCC tissue infiltration, as well as ESCC cell invasion, migration, and epithelial-mesenchymal transition (EMT) in ESCC (Table 2)[51-77]. The increased expressions of miR-10b-3p, miR-106b-5p, miR-25, miR-320b, miR-602, miR-9, and miR-99b/Let-7e/miR-125a not only show significant correlations with metastasis in ESCC patients, but these miRNAs are also known to increase the invasion, migration, and/or EMT of ESCC cells. Additionally, these miRNAs can promote the metastasis of transplanted tumors, primarily to the lungs[52-58]. Other miRNAs, including miR-1269, miR-17-5p, miR-18a-5p, miR-25-3p, miR-301, miR-373, miR-483-5p, miR-766-3p, and others, display oncogenic roles in metastasis, affecting both ESCC tissues and cells[59-67]. In addition, some miRNAs including but not limited to miR-103a-2-5p, miR-105-5p, miR-130b, miR-1323, miR-19b-3p, miR-196a, miR-23b-3p, and miR-301b display potent pro-metastatic functions, primarily pertaining to cellular activity[68-75].
The oncogenic potential of oncomiRs also hinges on their target genes, which typically act as tumor suppressors by preventing cells from becoming malignant. For instance, PTEN, a well-known tumor suppressor, is downregulated by several oncomiRs, including miR-1323, miR-25-3p, miR-301, miR-624-3p, miR-92a-3p, and others. The expression of these oncomiRs in ESCC tissues and/or cells is significantly upregulated and further research has shown that the effects of PTEN overexpression, including inhibition of cell migration and invasion, effects that can be partially reversed by the aforementioned oncomiRs[62,63,71,76,77]. Programmed cell death 4 (PDCD4), which functions as a tumor suppressor in ESCC, is a direct target of miR-320b. Exosomal miR-320b has been shown to promote LNM and lymphangiogenesis both in vitro and in vivo. Additional in vitro studies have confirmed that re-expression of PDCD4 can not only rescue its downregulation, but can also counteract lymphangiogenesis and LN metastasis mediated by exosomes with high levels of miR-320b[53].
Research evidence indicates that both genetic and epigenetic regulatory mechanisms of miRNAs play a crucial role in tumorigenesis and development, including metastasis in ESCC[54,78].
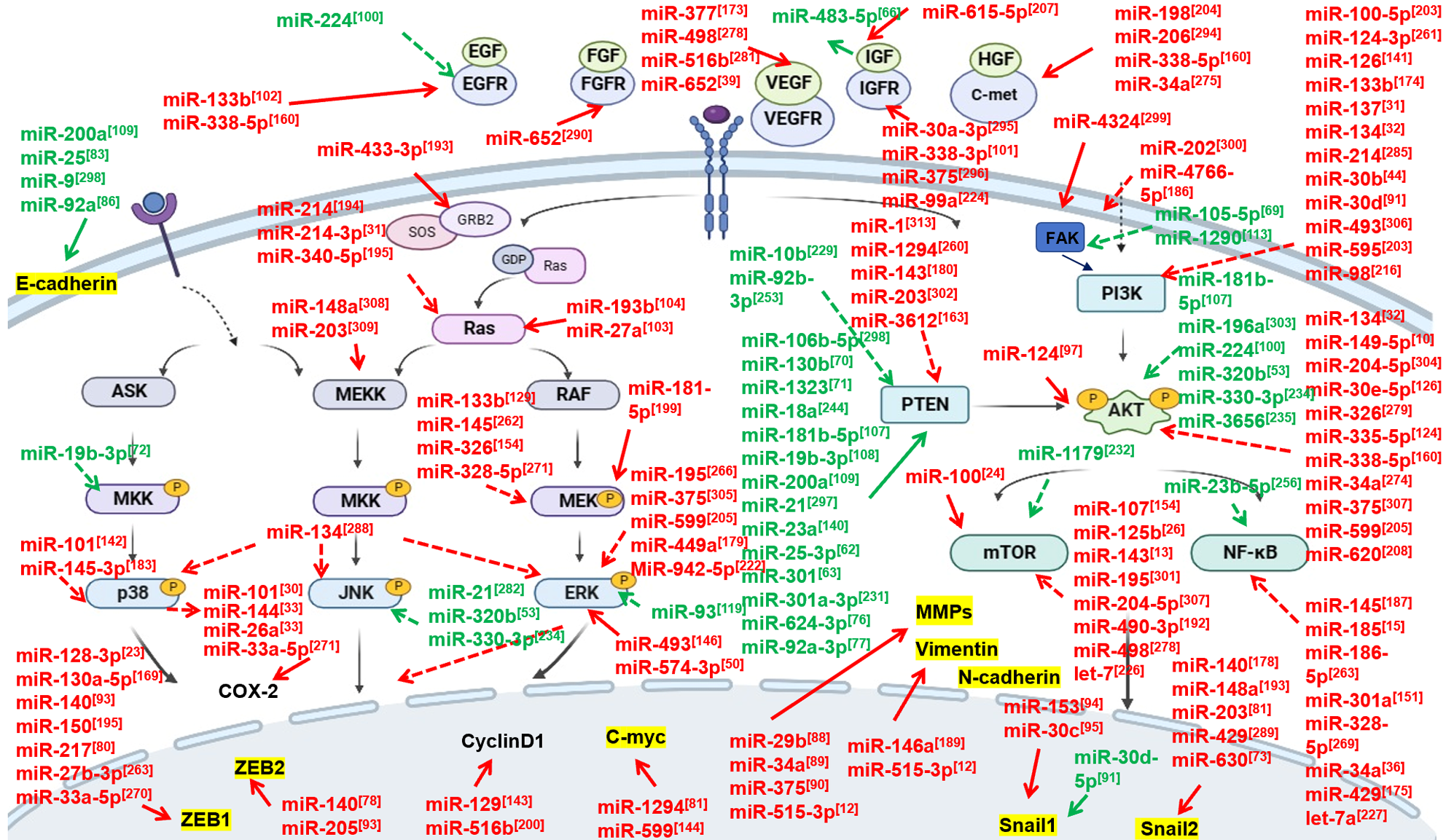
Tumor metastasis-associated genes (TMAGs) represent a category of genes that serve a vital function in the process of metastasis. This group includes metastasis suppressor genes (MSGs), and metastasis promoting genes (MPGs). MSGs and MPGs are primarily involved in various signaling pathways, including the RTK, transforming growth factor-β (TGF-β), Wnt/β-catenin, and interleukin (IL)-6/Stat3 pathways. They also target specific and key factors, including ZEB1/2, Snail1/2, E-cadherin, N-cadherin, C-myc, Vimentin, and MMPs, all of which are recognized as markers of tumor metastasis. Substantial evidence indicates that miRNAs exert their function on ESCC metastasis by targeting tumor suppressor genes or oncogenes, which belonging to the above-mentioned signaling pathways and their downstream signaling molecules[79].
Research indicates that miRNAs can regulate the metastasis of ESCC by directly targeting markers associated with tumor metastasis (Figures 1 and 2), including ZEB1/2, Snail1/2, E-cadherin, N-cadherin, C-myc, Vimentin, and members of the Matrix Metalloproteinase family (MMPs), etc.[12,26,78,80-84].
E-cadherin, predominantly found in epithelial tissues, is crucial for maintaining cell adhesion, polarity, and tissue architecture. Within the context of cancer metastasis, E-cadherin acts as a tumor suppressor. It serves as a critical biomarker and regulator of tumor metastasis, and its altered expression may indicate cancer progression. miR-9, miR-25, miR-92a, and miR-200a have been found to either directly or partially mediate tumor metastasis (including LNM, EMT, invasion, and migration) through E-cadherin[83,85-87].
The MMP family consists of enzymes mainly responsible for the degradation of extracellular matrix (ECM) components. In cancer, MMPs promote tumor growth, invasion, and metastasis by breaking down the ECM barriers, thereby facilitating cancer cell migration. Furthermore, they are involved in angiogenesis, a process of new blood vessel formation critical for tumor growth, survival, and metastasis. MMP2, MMP3, MMP9, and MMP13 are targets for miR-29b, miR-515-3p, miR-34a,and miR-375, respectively. These miRNAs are wholly or partly involved in ESCC metastasis[12,88-90].
Additionally, several transcription factors (TFs), particularly ZEB1/2 and Snail1/2, play critical roles in tumor metastasis. Their primary function is to downregulate the expression of epithelial markers (like E-cadherin) and upregulate the expression of mesenchymal markers (like N-cadherin and Vimentin), thereby driving the process of EMT process[81,82,91,92]. ZEB2, is a direct target of miR-140 and miR-205, and it reverses the effects of these miRNAs on EMT, migration, and invasion of ESCC cells[78,93]. miR-30c, miR-30d-5p, miR-203, and miR-153 are known to directly target Snail1, a critical regulator of metastasis. Overexpression of these miRNAs, either individually or in combination, can counteract metastatic behaviors in whole or in part, thereby presenting a potential therapeutic strategy for mitigating cancer metastasis[82,91,94,95].
The RTKs signaling pathway serves as a central conduit for communication within and between cells, regulating a multitude of cellular functions including cell growth, differentiation, invasion, and migration[7]. This pathway comes into effect when specific signals, typically growth factors or hormones such as epidermal growth factor (EGF), fibroblast growth factor (FGF), vascular endothelial growth factor (VEGF), hepatocyte growth factor (HGF), and insulin-like growth factor (IGF), bind to their corresponding RTK on the cell surface. Upon activation, the RTK acts as a platform for various intracellular signaling pathways, but primarily, the mitogen-activated protein kinases (MAPKs) and phosphatidylinositol 3-kinase (PI3K) pathways. MAPK signaling, a common and highly conserved pathway, primarily includes extracellular signaling-associated kinases (ERK1/2), Jun amino-terminal kinases (JNK), and p38-MAPK. The PI3K/Akt pathway can be irregularly activated via several mechanisms, including various genomic alterations such as mutations in PTEN, Akt, and mTOR mechanism[7,71,96,97]. Dysregulation of the RTK signaling pathway is implicated in the malignant progress
Numerous miRNAs have been identified as crucial regulators in ESCC metastasis by targeting RTK signaling, suggesting a critical crosstalk between miRNAs and the RTK signaling is necessary for ESCC metastasis (Figure 1)[100,101]. Certain miRNAs exert control over ESCC metastasis by directly interacting with growth factors or growth factor receptors, such as RTKs, subsequently regulating their downstream signaling pathways and molecules. For instance, miR-133b can repress ESCC cell invasion and metastasis by targeting the EGFR, which inhibits the PI3K/AKT signaling pathway and curbs the expression of downstream molecules such as N-cadherin, MMP-2, MMP-9, and E-cadherin[102]. Similarly, the downregulation of miR-338-3p, whose target is the IGF1R, activates the Raf/MEK/ERK pathway and affects the expression levels of E-cadherin, N-cadherin, and vimentin, and thus promoting ESCC cell invasion, migration, and EMT in ESCC[101]. Certain miRNAs are known to directly target the downstream signaling of the RTK pathway to influence ESCC metastasis. For example, miR-27a and miR-193b, functioning as tumor suppressors, are downregulated in ESCC and suppress cell migration and invasion via the direct regulation of KRAS, which is linked to the MAPK/ERK signaling pathway[103,104]. The tumor suppressor protein PTEN is able to dephosphorylate Akt to lessen its activation, thus blocking all downstream signaling events controlled by Akt and acting as a negative regulator of PI3K. Numerous miRNAs, including but not limited to miR-106b-5p, miR-130b, miR-1323, miR-18a, miR-181b-5p, miR-19b-3p, and miR-200a, exhibit a conspicuous overexpression and a negative association with LNM and/or TNM classification in ESCC. This overexpression fosters the progression of ESCC by facilitating cell invasion, metastasis, and/or EMT[52,71,105-109]. Furthermore, most of miRNAs whether indirectly or directly targeting PTEN can decrease the anticancer effect of PTEN, and promote the invasion and metastasis of ESCC[70,71,108].
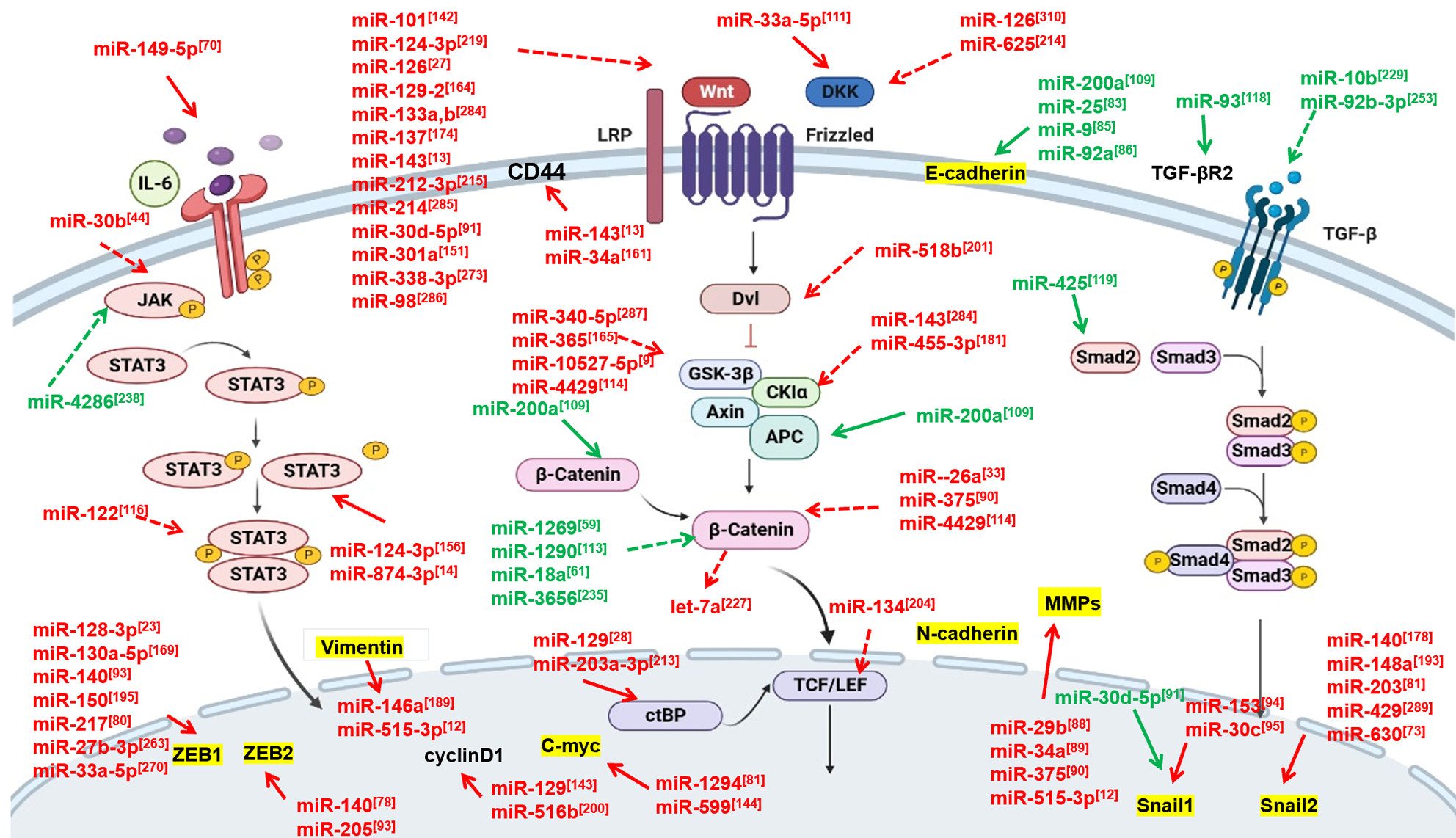
The Wnt/β-catenin signaling pathway assumes a pivotal role in various physiological processes, such as cell proliferation, differentiation, migration, and apoptosis. This signaling pathway consists of several components, including Wnts, receptors (such as Frizzled, FZD and low-density lipoprotein receptor-related proteins, LRP), Dishevelled (Dsh/Dvl), β-catenin, glycogen synthase kinase 3β (GSK-3β), Axin, and APC (adenomatous polyposis coli). Research has shown that the Wnt/β-catenin signaling pathway is intricately linked with tumor development, partly as a significant contributor to the onset and metastasis of ESCC, including metastasis[98,110].
Studies suggest that miRNAs can directly or indirectly modulate various components of the Wnt/β-catenin pathway, thereby either activating or inhibiting ESCC metastasis (Figure 2). For instance, miR-33a-5p, which exhibits diminished expression in ESCC tissues and correlates with higher TNM staging and LNM, can directly target Dickkopf-related protein 1 (DKK1) thereby influencing the metastasis of ESCC via the Wnt/β-catenin pathway[111]. Additionally, a majority of miRNAs associated with ESCC were found to indirectly target molecules associated with the Wnt/β-catenin pathway. One such example is miR-340, which is significantly downregulated in ESCC tissues and is highly correlated with LNM and TNM stages. phosphoserine aminotransferase 1 (PSAT1), via the GSK3β/β-catenin/Cyclin D1 pathway, was identified as a direct target of miR-340, although it can also partially counteracts the miR-340 mediated inhibition of viability, invasion, and EMT in ESCC cells[112]. Beyond miR-200a, Numerous miRNAs, including suppressor miRNAs and oncomiRs, have been found to indirectly regulate β-catenin, playing a critical role in ESCC metastasis. For example, both of miR-1269 and miR-1290 act as oncomiRs, and are upregulated in ESCC tissues and cell lines. They are associated with positive LNM and/or advanced TNM stage, and their overexpression promotes the malignant transformation of ESCC cells, including migration and invasion, by directly targeting SRY-box transcription factor 6, interactive with β-catenin (SOX6)[59,113]. Furthermore, the downregulation of miR-4429 has been shown to foster ESCC cell invasion and migration by partially regulating β-catenin through the direct targeting of SRXN1[114].
IL-6, a pleiotropic cytokine, is implicated in a diverse array of pathological processes including chronic inflammation, autoimmune diseases, and tumors. and other diseases. IL-6 is known to activate the downstream JAK/STAT3 signaling pathway, a hallmark of many malignant tumors, thereby contributing to tumor onset and, progression, and metastasis[115]. In ESCC, miRNAs serve as crucial effectors to the IL-6/STAT3 pathway, which is consistently involved in cancer metastasis (Figure 2). One such instance is miR-149-5p, a direct target of IL-6. The overexpression of miR-149-5p suppresses IL-6 expression at both the mRNA and protein levels, thereby inhibiting the invasion and migration of ESCC cells[10]. Similarly, miR-874-3p, is found downregulated in ESCC tissues and cell lines and exhibits a statistically significant association with LNM and clinical stage. Its overexpression markedly suppresses migration and invasion in ESCC cells by directly targeting STAT3[14]. Intriguingly, several miRNAs, such as miR-30b and miR-122, have been found to modulate ESCC cell invasion, migration, and EMT by indirectly regulating JAK and STAT3, respectively[44,116].
The transforming growth factor-β (TGF-β) signaling pathway is a classical signal transduction pathway, encompassing both the conventional Smad-dependent pathway and the non-Smad-dependent pathways. The canonical pathway is mediated through the transcription factors SMAD2, SMAD3, and SMAD4, while the non-canonical pathways include MAPK, nuclear factor (NF)-κB, and PI3K/AKT/mTOR signaling, etc[8]. TGF-β signaling has been identified as a pivotal regulator in the modulation of the progression and metastasis of various types of cancer, including in ESCC. In the context of ESCC, our discussion primarily focuses on the role of the canonical pathway. TGF-β, induces EMT activation via the TGF-β/Smad signaling pathway[117].
In recent years, there has been a surge in investigations demonstrating that miRNAs play a pivotal role in the metastasis of ESCC through the modulation of the TGF-β pathway (Figure 2). For instance, miR-93-5p is highly expressed in ESCC cells and is known to enhance cell proliferation, migration, and invasion, while simultaneously suppressing apoptosis in ESCC cells. It achieves these effects by targeting and downregulating the expression of TGFβR2[118]. Similarly, miR-425 is overexpressed in both ESCC tissues and plasma, and positively influences cell migration and invasion. It functions as an oncogene by specifically targeting the 3'-untranslated region (3'-UTR) of SMAD2, thereby modulating its expression and subsequent cellular behavior[119].
Epigenetics refers to the study of heritable changes in gene expression that occur without a change in DNA sequence. Epigenetic changes occur in extreme cases and contain several regulatory mechanisms, including noncoding RNAs (ncRNAs), DNA methylation, heterochromatin, polycomb and trithorax proteins and 3D genome architecture[120]. In recent years, accumulating evidences suggest that different expression patterns of ncRNA play increasingly important roles in cancer. ncRNAs are made of distinct classes, including noncoding RNA (lncRNA), miRNA, circRNA, rRNA, tRNA, and so on[121-123]. Among them, more exciting progress has been made in the study of the role and mechanism of lncRNA, circRNA and the impact of DNA methylation of miRNAs in tumor processes (Table 3).
| miRNA (Suppressor) | Epigenetic regulation | Ref. |
| miR-101 | ~/lncMALAT1 | [48,49,142] |
| lncXIST/~/EZH2 | ||
| PSMA3-AS1/~/EZH2 | ||
| miR-122 | LINC01410/~/ PKM2 | [259] |
| miR-129 | lncNEAT1/~/CtBP2; | [28,145] |
| lnc XIST/~/CCND1 | ||
| miR-130a-5p | CCL18-induced lncHOTAIR/~/ZEB1 | [169] |
| miR-133b | TTN-AS1/~/FSCN1 | [102,128,129,261] |
| NCK1-AS1/~/ENPEP | ||
| lncKCNQ1OT1/~/EGFRTHAP9-AS1/~/SOX4 | ||
| miR-145 | lncROR/~/FSCN1 | [262] |
| miR-140-5p | lncSNHG16/~/ZEB1 | [92] |
| miR-186-5p | lncSNHG6/~/HIF-1α | [264] |
| miR-195 | lncSNHG1/~/Cdc42 | [265,266] |
| miR203 | circ_0008717/~/Slug | [35,267] |
| circPRMT5/~ | ||
| miR206 | circ_0008726/~ | [20] |
| miR-214 | FAM83A-AS1/~/CDC25B | [130,268] |
| LINC00963/~/RAB14 | ||
| miR-30a | OIP5AS1/~/VOPP1 | [131] |
| miR-30d-5p | DDX11-AS1/~/SNAI1 | [91] |
| miR-30e-5p | lncDLEU2/~/ E2F7 | [126] |
| miR-301a | lncGAS5/~/CXCR4 | [151] |
| miR-328 | lncLOC146880/~/FSCN1 | [269] |
| miR-33a-5p | lncDANCR/~/ZEB1 | [270,271] |
| lncCASC15/~/PTGS2 | ||
| miR-335-5p | lncTHAP9-AS1/~/SGMS2 | [124] |
| miR-338-3p | lncBRAF/~/IGF1R | [101,273] |
| lncSNHG17/~/SOX4 | ||
| miR-34a | lncMIR31HG/~/c-Met | [274] |
| miR-340-5p | LINC00662/~/HOXB2 | [162] |
| miR-378a-3p | SLC2A1-AS1/~/Glut1 | [132] |
| miR-498 | lncTUG1/~/Cdc42 | [46,294,278] |
| AFAP1-AS1/~/VEGFA | ||
| lncTUG1/~/XBP1 | ||
| miR-508-3p | lncPCAT-1/~/ANXA10 | [198] |
| miR-516b | lncJPX/~/VEGFA | [281] |
| miR-765 | MAFG-AS1/~/PDX1 | [135] |
| oncomiR | ||
| miR-18a-5p | lncPART1/~/SOX6 | [61] |
| miR-21 | LncRNA-IUR/~/PTEN | [134,282] |
| HAND2-AS1/~ | ||
| miR-1290 | circ_0086414/~/SPARCL1circ_0001946/~/SOX6 | [113,283] |
| miR1323 | circLARP4/~/PTEN | [71] |
| miR-224 | circ_0007624/~/CPEB3 | [100] |
| miR-23a | circFoxo3/miR-23a | [140] |
| miR-125a-5p | lncHOTAIR/~/HK2 | [258] |
| miR-124 | circHIPK3/~/AKT3 | [25,136] |
| circ2646/~/PLP2 | ||
| miR-1294 | circ 0004370/~/LASP1 | [260] |
| miR-140-3p | circNTRK2/~/NRIP1 | [176,222] |
| circ0087378/~/E2F3 | ||
| miR-198 | circLPAR3/~/C-met | [204] |
| miR-217 | circZDHHC5/~/ZEB1 | [80] |
| miR-27b-3p | circLONP2/~/ZEB1 | [263] |
| miR-3612 | circ_0006948/~/LASP1 | [163] |
| miR-422a | circUBAP2/~/Rab10 | [173] |
| miR-433-3p | circ_0023984/~/REV3L | [177] |
| miR-4766-5p | circPDE3B/~/LAMA1 | [186] |
| miR-490-3p | circ_0006948/ ~/HMGA2 | [192] |
| miR-497-5p | circ-AGFG1/~/SLC1A5 | [194] |
| miR-595 | circNRIP1/~/SEMA4D | [203] |
| miR-599 | circHIPK3/~/c-myc | [144,205] |
| circ_0030018/~/ENAH | ||
| miR-615-5p | circOGDH/~/PDX1 | [139] |
| miR-622 | circ_0001273/~/SLC1A5 | [138] |
| miR-670-3p | circABCB10/~ | [139] |
| miR-874-3p | circ0000977/~ | [272] |
| miR-126 | DNMT1/~/ADAM9 | [141] |
| miR-137 | CTCF/(DNA methylation) /~/EZH2+PXN | [174] |
| miR-107 | circSFMBT2/~/SLC1A5 | [275,276] |
| LINC00152/~/Rab10 | ||
| miR-149-5p | lnc GACAT3/~/FOXM1 | [10,277] |
| circ_0000654/~/IL-6 | ||
| miR-326 | LINC01711/~/FSCN1 | [154,279] |
| circATIC/~/ID1 | ||
| miR-377 | LncNEAT1/~/E2F3 | [171,280] |
| circ_0072088/~/VEGF | ||
| miR-493 | LINC00324/~/MAPK1 | [145,146] |
| circFIG4/~/E2F3 | ||
| miR-124-3p | lncZFAS1/~/STAT3 | [31,156] |
| DNMT1/~/BCAT1 ~/EZH2/H3K27me3 | ||
| miR-10b-3p | DNA methylation; Hypoxia | [56,57] |
| miR-320b | DGCR8+METTL3- N6 -methyladenosine (m6A)/~ | [53] |
| miR-483-5p | Igf2 methylationgene promoter/Igf2/~/ARHGDIA | [66] |
| miR-602 | CpG hypomethylation/~ | [54] |
| miR-515-3p | Promoter hypomethylation | [12] |
LncRNA are ncRNAs with lengths > 200 nucleotides. They represent diverse types of RNA molecules with limited or no protein-coding capability, and different biological functions depending on their subcellular location. Cytoplasmic lncRNAs exhibit vital roles in cancer, often acting as tumor suppressors or oncogenes through the modulation of miRNAs[124]. Recently, accumulated evidences has demonstrated that lncRNAs are involved in ESCC metastasis by acting as competing endogenous RNAs (ceRNAs) that “sponge” miRNAs to block their function, and then, up-regulate the downstream genes[125]. For example, lncRNA DLEU2, is up-regulated in EC tissues and associated with poor prognosis. The overexpression of this lncRNA DLEU2 increased the proliferation, migration and invasion abilities of ESCC by suppressing miR-30e-5p and then directly targeting E2F7. Another lncRNA, LncRNA-IUR up-regulates PTEN by “sponging” miR-21 and acting as a tumor suppressor in ESCC metastasis[126]. Antisense lncRNAs, a special type of lncRNA, are antisense RNAs with partial exon overlap with forward genes. They can also regulate gene expression by competitively binding to certain miRNA in ESCC metastasis, such as PSMA3-AS1, ZFAS1, TTN-AS1, NCK1-AS1, THAP9-AS1, FAM83A-AS1, OIP5AS1, DDX11-AS1, SLC2A1-AS1, and MAFG-AS1[48,91,124,127-133]. MAFG-AS1 is significantly up-regulated in ESCC tissues and cell lines, and accelerates ESCC cell proliferation, migration, invasion and aerobic glycolysis by competitively adsorbing miR-765, a negative modulator of PDX1 expression of PDX1. Overexpression of lncRNA HAND2-AS1 its overexpression inhibited cell proliferation, migration, and invasion by downregulation of miRNA-21 in ESCC cells[133,134].
CircRNAs also act as miRNA “sponges” and comprise a large class of endogenous non-coding RNA with covalently closed loops. CircRNAs have independent functions from the linear transcripts that are transcribed from identical genes[135]. Recently, it was demonstrated that numerous circRNAs are differentially expressed in ESCC, and their dysfunction is linked to ESCC metastasis. The typical circRNAs contain circHIPK3, circ2646, circOGDH, circ_0001273, circABCB10, circ_0001946, circLARP4, circ_0007624, circFoxo3[25,71,100,113,136-140]. circ-OGDH was upregulated in ESCC cells and its inhibition repressed cell proliferation, migration, and invasion, and reduced miR-615-5p-induced cellular glutamine metabolism to regulate PDX1 expression[137]. circLARP4 expression was observably downregulated in ESCC, and its overexpression circLARP4 restrained cell proliferation and migration in ESCC cell. And circLARP4 was also able to act as a sponge for miR-1323 and negatively modulated miR-1323 via the PTEN/PI3K/AKT pathway in ESCC[71].
DNA methylation is a form of DNA chemical modification that can alter genetic expression without altering the DNA sequence. It is a common occurrence in malignancies and is implicated in tumor initiation and progress[31]. Methylation of the CPG island, usually located in the promoter region of genes, its methylation also plays an important role in ESCC metastasis by up-regulating expression of various miRNAs, includingmiR-483-5p, miR-602[54,66]. miR-483-5p was overexpressed in preoperative serum and cancer tissues and is significantly correlated with the TNM stage and LNM. Low methylation of the Igf2 gene promoter region may promote the expression of Igf2, which directly affects the expression of miR-483-5p and its target genes, including Rho GDP dissociation inhibitor α, activated leukocyte cell adhesion molecule, and suppressor of cytokine signaling 3[66]. miR-602 expression was also increased in human ESCC and significantly correlated with LNM and TNM stage, and showed positive effects on ESCC cell invasion and migration by targeting Fork head box (FOX)K2 (FOXK2). Additionally, direct evidence has shown that the overexpression of miR-602 in the ESCC tissues was correlated with promoter hypomethylation and that demethylation of the promoter genes could upregulate the expression of miR-602[54]. DNA methyltransferases (DNMTs) often are involved in DNA methylation, thereby often modifying gene function through the regulation of gene expression. In ESCC, miR-124-3p shows a high correlation with TNM stage and can directly targets the mRNA 3’UTR region of BCAT1. The expression of miR-124-3p itself is regulated by hypermethylation-silencing regulation mediated by DNA methyltransferase 1 (DNMT1)[31]. The DNMT1/miR-126 epigenetic circuit contributes to ESCC proliferation and migration via ADAM9/EGFR/AKT signaling: ADAM9 has been identified as a key target of miR-126 and ectopic expression of miR-126 or silencing of ADAM9 reduced ESCC cell proliferation and migration by inhibiting EGFR-AKT signaling; Downregulation of miR-126 was due to promoter hypermethylation of its host gene Egfl7, and DNMT1 was aberrantly upregulated in ESCC and is responsible for the hypermethylation of Egfl7; Intriguingly, overexpression of miR-126 suppressed DNMT1,was suppressed by overexpression of miR-126, indicating the existence of a regulatory feedback circuit[141].
A more complex and dynamic epigenetic regulatory network of miRNA has been shown to be positively involved in ESCC metastasis. For example, one lncRNA, circRNA can “spong” several miRNAs. lncXIST, serves as oncogene and is significantly upregulated in ESCC tissues and cells, and is significantly associated with a poor prognosis in ESCC patients. Downregulation of lncXIST can inhibit ESCC cell proliferation, migration and invasion by elevating expression of miR-101 and decreasing the expression of EZH2. Downregulation of lncXIST also functions by inhibiting CCND1 expression via “sponging” of miR-129-5p[142,143]. circHIPK3 is highly expressed in ESCC tissues and cell lines and is associated with advanced TNM stage, LNM and tumor size. circHIPK3 can promote ESCC cell proliferation, invasion, and migration by modulating the miR-599/c-MYC axis, as well as via the regulation of ESCC cell proliferation, migration, and EMT by absorbing miR-124 via target AKT3[25,144]. Not more than that, the same miRNA can be regulated by different epigenetic mechanisms. For example, miR-493-5p, is dramatically downregulated in ESCC tissues and cells due to “sponging” from both circFIG4 and linc00324, both of which modulated ESCC progression including ESCC cell invasion and migration via targeting the miR-493-5p/E2F3 and miR-493-5p/MAPK1 axis, respectively[145-147]. Another example, miR-124-3p, was found to have positive correlations with ESCC invasion and migration, as well as EMT via the regulation of lncZFAS1 and through DNA methylation[31,127].
ESCC is a cancer that originates from the squamous epithelial cells lining the esophagus. It is notorious for its aggressive progression and high propensity for metastasis or dispersion to other regions of the body. In ESCC, metastasis contributes to approximately 90% of cancer-associated fatalities, primarily due to the resulting disruption of digestive function, complications arising from metastasis, an overall deterioration of health, and poor prognostic outcomes. The treatment of metastatic ESCC is intricate, possibly necessitating a combination of therapeutic modalities including surgery, radiation therapy, chemotherapy, and targeted treatments. However, these interventions frequently carry their own array of side effects and complications, and their efficacy can be restricted in the face of metastatic progression. As such, deciphering the mechanisms that underpin metastasis and pioneering effective measures for its prevention and treatment are pivotal objectives in ESCC research.
The metastatic process of ESCC is influenced by a variety of factors, including tumor characteristics, host factors, microenvironmental elements, as well as genetic and epigenetic variations. Growing evidence suggests that miRNA signatures represent an epigenetic mechanism that can profoundly impact various stages of metastasis by regulating gene expression. This regulation occurs through multiple signaling pathways, primarily including the RTK, TGF-β, Wnt/β-catenin, and IL6/Stat3 pathways. Given their regulatory capabilities, miRNAs have surfaced as potential novel diagnostic, prognostic, and therapeutic markers for ESCC metastasis. In their capacity as diagnostic and prognostic markers, certain miRNAs are consistently overexpressed or underexpressed in ESCC tissues. These miRNAs show negative or positive associations with characteristics of metastasis, such as tumor invasive depth, lymphatic/vascular invasion, lymph node metastasis, and distant metastasis. Intriguingly, specific miRNAs have even been linked to metastatic potential, being associated with the propensity of a tumor to metastasize, the severity of metastasis, and the likely sites of metastatic spread. As therapeutic targets, miRNAs that suppress ESCC metastasis can be restored through the application of miRNA mimics, while those promoting ESCC metastasis can be suppressed using anti-miRNA molecules. The regulation of miRNAs is also influenced by other epigenetic factors, including lncRNAs, circRNAs, and DNA methylation. Additionally, miRNAs play a crucial role in molding the tumor microenvironment, affecting angiogenesis, and modulating the immune response to tumors. Given their central roles, miRNAs hold significant potential as metastatic biomarkers for both the diagnosis and therapy of ESCC.
Recent advancements in miRNA-based therapy revolve around two principal tactics: the suppression of oncogenic miRNAs to reinstate the activity of their targeted tumor suppressor genes, and the augmentation of tumor suppressor miRNAs to dampen the expression of the oncogenes they regulate. Although they are typically downregulated in cancers, enhancing the expression of tumor suppressor miRNAs can correct the aberrant overexpression of oncogenes they normally control. Additionally, the artificial introduction of tumor suppressor miRNAs can replenish deficient miRNA levels, thereby targeting and disrupting cellular pathways that contribute to tumor development and metastasis. At present, a variety of therapeutic strategies capitalizing on miRNA mimics or inhibitors are being intensively investigated. One such example is MRX34, a liposome-encapsulated miR-34 mimic that has been introduced into clinical trials for the treatment of patients with primary liver cancer or other malignancies involving the liver. Despite the significant therapeutic potential of miRNAs, several challenges loom must be addressed before their comprehensive integration into clinical practice. One of the foremost challenges in the field of miRNA therapy lies in the potential for off-target effects. These occur when the treatment unintentionally affects genes that have no direct connection to the disease being addressed, leading to unintended and potentially detrimental side effects. Such inadvertent gene regulation can be attributed to the inherent sequence similarities among miRNAs. Moreover, the expansive binding capacity of miRNAs exacerbates this issue; a single miRNA molecule may attach to multiple, disparate mRNAs, thereby simultaneously regulating a host of unrelated genes. This promiscuity in target selection underscores the complexity of achieving precise therapeutic outcomes with miRNA-based interventions. Another substantial challenge in miRNA therapy is achieving effective delivery and ensuring stability. The targeted transport of miRNAs or their inhibitors to designated tissues or cells without loss of function is an ongoing obstacle. Additionally, miRNAs are inherently unstable in the bloodstream due to rapid degradation by nucleases, which calls for the development of sophisticated systems capable of safeguarding these fragile therapeutic agents during delivery. Furthermore, the administration of exogenous miRNA or miRNA mimics may elicit an immune response that could undermine the safety and efficacy of the treatment. Determining the optimal dosage of miRNAs is equally critical, as incorrect dosing could compromise the therapeutic balance, impacting both treatment outcomes and patient well-being. Finally, there are still limitations in accurately predicting miRNA targets, understanding miRNA-mRNA interactions, and quantifying miRNA expression levels, all of which can impede the development of miRNA-based therapies.
As research advances and technology evolves, scientists and healthcare professionals are pioneering methods to navigate the complexities of miRNA therapy. For instance, the integration of large-scale functional screenings, sophisticated bioinformatics analyses, and strategic chemical modifications are being employed to attenuate off-target effects and bolster specificity. Moreover, an array of innovative delivery mechanisms, including nanocarriers, viral vectors, and exosome-based systems, are being utilized to shield miRNAs from enzymatic degradation and to ensure precise tissue-specific targeting with elevated efficiency. Enhancements in the structural design of miRNAs to assist in evasion of immune detection stand as a testament to the proactive measures being taken to preclude unsolicited immune responses. Additionally, gaining a nuanced understanding of the complex miRNA-mRNA interactions, as well as the epigenetic mechanisms at play, is necessary in order to illuminate new potential applications for miRNA-based interventions.
In summary, persistent explorations in the field of miRNA research are unlocking significant opportunities for improving diagnostic accuracy, refining prognostic predictions, and advancing the therapeutic strategies employed in the battle against cancer. With steadfast commitment to research, the future of miRNAs in medicine appears to be both promising and profound.
| 1. | Sheikh M, Roshandel G, McCormack V, Malekzadeh R. Current Status and Future Prospects for Esophageal Cancer. Cancers (Basel). 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 163] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 2. | Yang J, Liu X, Cao S, Dong X, Rao S, Cai K. Understanding Esophageal Cancer: The Challenges and Opportunities for the Next Decade. Front Oncol. 2020;10:1727. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 128] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 3. | He S, Xu J, Liu X, Zhen Y. Advances and challenges in the treatment of esophageal cancer. Acta Pharm Sin B. 2021;11:3379-3392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 202] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 4. | Zheng S, Liu B, Guan X. The Role of Tumor Microenvironment in Invasion and Metastasis of Esophageal Squamous Cell Carcinoma. Front Oncol. 2022;12:911285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 5. | Li Q, Deng M, Xi M, Zhu Y, Hu Y. Characteristics and Treatment of Brain Metastases from Esophageal Squamous Cell Carcinoma. J Cancer. 2018;9:901-905. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 6. | Suhail Y, Cain MP, Vanaja K, Kurywchak PA, Levchenko A, Kalluri R, Kshitiz. Systems Biology of Cancer Metastasis. Cell Syst. 2019;9:109-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 306] [Article Influence: 43.7] [Reference Citation Analysis (0)] |
| 7. | Regad T. Targeting RTK Signaling Pathways in Cancer. Cancers (Basel). 2015;7:1758-1784. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 223] [Cited by in RCA: 297] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 8. | Xie F, Ling L, van Dam H, Zhou F, Zhang L. TGF-β signaling in cancer metastasis. Acta Biochim Biophys Sin (Shanghai). 2018;50:121-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 190] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 9. | Xiao Z, Feng X, Zhou Y, Li P, Luo J, Zhang W, Zhou J, Zhao J, Wang D, Wang Y, Tian Z, Zhao X. Exosomal miR-10527-5p Inhibits Migration, Invasion, Lymphangiogenesis and Lymphatic Metastasis by Affecting Wnt/β-Catenin Signaling via Rab10 in Esophageal Squamous Cell Carcinoma. Int J Nanomedicine. 2023;18:95-114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 33] [Reference Citation Analysis (0)] |
| 10. | Xu Z, Tie X, Li N, Yi Z, Shen F, Zhang Y. Circular RNA hsa_circ_0000654 promotes esophageal squamous cell carcinoma progression by regulating the miR-149-5p/IL-6/STAT3 pathway. IUBMB Life. 2020;72:426-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 11. | Ding N, Sun X, Wang T, Huang L, Wen J, Zhou Y. miR378a3p exerts tumor suppressive function on the tumorigenesis of esophageal squamous cell carcinoma by targeting Rab10. Int J Mol Med. 2018;42:381-391. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 12. | Hu HF, Xu WW, Zhang WX, Yan X, Li YJ, Li B, He QY. Identification of miR-515-3p and its targets, vimentin and MMP3, as a key regulatory mechanism in esophageal cancer metastasis: functional and clinical significance. Signal Transduct Target Ther. 2020;5:271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 13. | Ni Y, Meng L, Wang L, Dong W, Shen H, Wang G, Liu Q, Du J. MicroRNA-143 functions as a tumor suppressor in human esophageal squamous cell carcinoma. Gene. 2013;517:197-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 56] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 14. | Yuan RB, Zhang SH, He Y, Zhang XY, Zhang YB. MiR-874-3p is an independent prognostic factor and functions as an anti-oncomir in esophageal squamous cell carcinoma via targeting STAT3. Eur Rev Med Pharmacol Sci. 2018;22:7265-7273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 15. | Jing R, Chen W, Wang H, Ju S, Cong H, Sun B, Jin Q, Chu S, Xu L, Cui M. Plasma miR-185 is decreased in patients with esophageal squamous cell carcinoma and might suppress tumor migration and invasion by targeting RAGE. Am J Physiol Gastrointest Liver Physiol. 2015;309:G719-G729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 16. | Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1765] [Cited by in RCA: 1996] [Article Influence: 99.8] [Reference Citation Analysis (0)] |
| 17. | Hill M, Tran N. miRNA interplay: mechanisms and consequences in cancer. Dis Model Mech. 2021;14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 476] [Article Influence: 95.2] [Reference Citation Analysis (0)] |
| 18. | Zheng S, Zhang X, Wang X, Li J. Downregulation of miR-138 predicts poor prognosis in patients with esophageal squamous cell carcinoma. Cancer Biomark. 2017;20:49-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 19. | Guo Y, Jin J, Li L, Zhang X, Dong X, He Y. Expression and role of microRNA-193a-5p in esophageal cancer. Zhongliu. 2019;39:784-794. |
| 20. | Han T, Shi M, Chen G, Hao J. Circ_0008726 promotes malignant progression of ESCC cells through miR-206/HOXA13 pathway. Gen Thorac Cardiovasc Surg. 2023;71:33-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 21. | Zang W, Wang Y, Du Y, Xuan X, Wang T, Li M, Ma Y, Li P, Chen X, Dong Z, Zhao G. Differential expression profiling of microRNAs and their potential involvement in esophageal squamous cell carcinoma. Tumour Biol. 2014;35:3295-3304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 22. | Sun L, Dong S, Dong C, Sun K, Meng W, Lv P, Yin H, Ming L, He F. Predictive value of plasma miRNA-718 for esophageal squamous cell carcinoma. Cancer Biomark. 2016;16:265-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 23. | Zhao L, Li R, Xu S, Li Y, Zhao P, Dong W, Liu Z, Zhao Q, Tan B. Tumor suppressor miR-128-3p inhibits metastasis and epithelial-mesenchymal transition by targeting ZEB1 in esophageal squamous-cell cancer. Acta Biochim Biophys Sin (Shanghai). 2018;50:171-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 57] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 24. | Zhang N, Fu H, Song L, Ding Y, Wang X, Zhao C, Zhao Y, Jiao F. MicroRNA-100 promotes migration and invasion through mammalian target of rapamycin in esophageal squamous cell carcinoma. Oncol Rep. 2014;32:1409-1418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 25. | Yao D, Lin S, Chen S, Wang Z. circHIPK3 regulates cell proliferation and migration by sponging microRNA-124 and regulating serine/threonine kinase 3 expression in esophageal squamous cell carcinoma. Bioengineered. 2022;13:9767-9780. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 26. | Mei LL, Wang WJ, Qiu YT, Xie XF, Bai J, Shi ZZ. miR-125b-5p functions as a tumor suppressor gene partially by regulating HMGA2 in esophageal squamous cell carcinoma. PLoS One. 2017;12:e0185636. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 58] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 27. | Li H, Meng F, Ma J, Yu Y, Hua X, Qin J, Li Y. Insulin receptor substrate-1 and Golgi phosphoprotein 3 are downstream targets of miR126 in esophageal squamous cell carcinoma. Oncol Rep. 2014;32:1225-1233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 28. | Li Y, Chen D, Gao X, Li X, Shi G. LncRNA NEAT1 Regulates Cell Viability and Invasion in Esophageal Squamous Cell Carcinoma through the miR-129/CTBP2 Axis. Dis Markers. 2017;2017:5314649. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 69] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 29. | Liu Y, Wang X, Jiang X, Yan P, Zhan L, Zhu H, Wang T, Wen J. Tumor-suppressive microRNA-10a inhibits cell proliferation and metastasis by targeting Tiam1 in esophageal squamous cell carcinoma. J Cell Biochem. 2019;120:7845-7857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 30. | Shao Y, Li P, Zhu ST, Yue JP, Ji XJ, He Z, Ma D, Wang L, Wang YJ, Zong Y, Wu YD, Zhang ST. Cyclooxygenase-2, a Potential Therapeutic Target, Is Regulated by miR-101 in Esophageal Squamous Cell Carcinoma. PLoS One. 2015;10:e0140642. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 31. | Zeng B, Zhang X, Zhao J, Wei Z, Zhu H, Fu M, Zou D, Feng Y, Luo H, Lei Y. The role of DNMT1/hsa-miR-124-3p/BCAT1 pathway in regulating growth and invasion of esophageal squamous cell carcinoma. BMC Cancer. 2019;19:609. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 32. | Yuan Y, Wang Q, Cao F, Han B, Xu L. MiRNA-134 suppresses esophageal squamous cell carcinoma progression by targeting FOXM1. Int J Clin Exp Pathol. 2019;12:2130-2138. [PubMed] |
| 33. | Shao Y, Li P, Zhu ST, Yue JP, Ji XJ, Ma D, Wang L, Wang YJ, Zong Y, Wu YD, Zhang ST. MiR-26a and miR-144 inhibit proliferation and metastasis of esophageal squamous cell cancer by inhibiting cyclooxygenase-2. Oncotarget. 2016;7:15173-15186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 46] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 34. | Liu Z, Huang Y, Han Z, Shen Z, Yu S, Wang T, Dong Z, Kang M. Exosome-mediated miR-25/miR-203 as a potential biomarker for esophageal squamous cell carcinoma: improving early diagnosis and revealing malignancy. Transl Cancer Res. 2021;10:5174-5182. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 35. | Wang T, Wang J, Ren W, Chen S, Cheng YF, Zhang XM. CircRNA-0008717 promotes cell proliferation, migration, and invasion by regulating miR-203/Slug in esophageal cancer cells. Ann Transl Med. 2020;8:999. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 36. | Cui XB, Peng H, Li RR, Mu JQ, Yang L, Li N, Liu CX, Hu JM, Li SG, Wei Y, Laibo-Yin, Zhou H, Li F, Chen YZ. MicroRNA-34a functions as a tumor suppressor by directly targeting oncogenic PLCE1 in Kazakh esophageal squamous cell carcinoma. Oncotarget. 2017;8:92454-92469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 37. | Hu C, Lv L, Peng J, Liu D, Wang X, Zhou Y, Huo J. MicroRNA-375 suppresses esophageal cancer cell growth and invasion by repressing metadherin expression. Oncol Lett. 2017;13:4769-4775. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 38. | Li J, Li X, Li Y, Yang H, Wang L, Qin Y, Liu H, Fu L, Guan XY. Cell-specific detection of miR-375 downregulation for predicting the prognosis of esophageal squamous cell carcinoma by miRNA in situ hybridization. PLoS One. 2013;8:e53582. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 39. | Gao P, Wang D, Liu M, Chen S, Yang Z, Zhang J, Wang H, Niu Y, Wang W, Yang J, Sun G. DNA methylation-mediated repression of exosomal miR-652-5p expression promotes oesophageal squamous cell carcinoma aggressiveness by targeting PARG and VEGF pathways. PLoS Genet. 2020;16:e1008592. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 40. | Zheng S, Liu Q, Ma R, Tan D, Shen T, Zhang X, Lu X. Let-7b-5p inhibits proliferation and motility in squamous cell carcinoma cells through negative modulation of KIAA1377. Cell Biol Int. 2019;43:634-641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 41. | Zhang P, Zhang W, Jiang J, Shen Z, Chen S, Yu S, Kang M. MiR-107 inhibits the malignant biological behavior of esophageal squamous cell carcinoma by targeting TPM3. J Gastrointest Oncol. 2022;13:1541-1555. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 42. | Liu H, Ren G, Zhu L, Liu X, He X. The upregulation of miRNA-146a inhibited biological behaviors of ESCC through inhibition of IRS2. Tumour Biol. 2016;37:4641-4647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 43. | Xu C, Li B, Zhao S, Jin B, Jia R, Ge J, Xu H. MicroRNA-186-5p Inhibits Proliferation And Metastasis Of Esophageal Cancer By Mediating HOXA9. Onco Targets Ther. 2019;12:8905-8914. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 44. | Meng L, Wang F, Sun S, Zheng Y, Ding Z, Sun Y, Li B, Meng Q, Xu M. MicroRNA-30b targets CBX3 and regulates cell proliferation, apoptosis, and migration in esophageal squamous cell carcinoma via the JAK2/STAT3 signaling pathway. Int J Clin Exp Pathol. 2017;10:11828-11837. [PubMed] |
| 45. | Han DL, Wang LL, Zhang GF, Yang WF, Chai J, Lin HM, Fu Z, Yu JM. MiRNA-485-5p, inhibits esophageal cancer cells proliferation and invasion by down-regulating O-linked N-acetylglucosamine transferase. Eur Rev Med Pharmacol Sci. 2019;23:2809-2816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 46. | Wang Z, Liu J, Wang R, Wang Q, Liang R, Tang J. Long Non-Coding RNA Taurine Upregulated Gene 1 (TUG1) Downregulation Constrains Cell Proliferation and Invasion through Regulating Cell Division Cycle 42 (CDC42) Expression Via MiR-498 in Esophageal Squamous Cell Carcinoma Cells. Med Sci Monit. 2020;26:e919714. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 47. | Sun J, Deng Y, Shi J, Yang W. MicroRNA5423p represses OTUB1 expression to inhibit migration and invasion of esophageal cancer cells. Mol Med Rep. 2020;21:35-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 48. | Qiu BQ, Lin XH, Ye XD, Huang W, Pei X, Xiong D, Long X, Zhu SQ, Lu F, Lin K, Zhang XQ, Xu JJ, Sheng LL, Zhang XM, Zhang PF, Wu YB. Long non-coding RNA PSMA3-AS1 promotes malignant phenotypes of esophageal cancer by modulating the miR-101/EZH2 axis as a ceRNA. Aging (Albany NY). 2020;12:1843-1856. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 49. | Wang X, Li M, Wang Z, Han S, Tang X, Ge Y, Zhou L, Zhou C, Yuan Q, Yang M. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 2015;290:3925-3935. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 262] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 50. | Jin LL, Zhang SJ, Lu GX, Lv F, Shang R, Yang J. miR-574-3p inhibits proliferation and invasion in esophageal cancer by targeting FAM3C and MAPK1. Kaohsiung J Med Sci. 2020;36:318-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 51. | Guan H, Liu J, Lv P, Zhou L, Zhang J, Cao W. MicroRNA590 inhibits migration, invasion and epithelialtomesenchymal transition of esophageal squamous cell carcinoma by targeting lowdensity lipoprotein receptorrelated protein 6. Oncol Rep. 2020;44:1385-1392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 52. | Yang F, Sun Z, Wang D, Du T. MiR-106b-5p regulates esophageal squamous cell carcinoma progression by binding to HPGD. BMC Cancer. 2022;22:308. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 53. | Liu T, Li P, Li J, Qi Q, Sun Z, Shi S, Xie Y, Liu S, Wang Y, Du L, Wang C. Exosomal and intracellular miR-320b promotes lymphatic metastasis in esophageal squamous cell carcinoma. Mol Ther Oncolytics. 2021;23:163-180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 54. | Liu M, Yu J, Wang D, Niu Y, Chen S, Gao P, Yang Z, Wang H, Zhang J, Zhang C, Zhao Y, Hu W, Sun G. Epigenetically Upregulated MicroRNA-602 Is Involved in a Negative Feedback Loop with FOXK2 in Esophageal Squamous Cell Carcinoma. Mol Ther. 2019;27:1796-1809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 55. | Ma J, Zhan Y, Xu Z, Li Y, Luo A, Ding F, Cao X, Chen H, Liu Z. ZEB1 induced miR-99b/let-7e/miR-125a cluster promotes invasion and metastasis in esophageal squamous cell carcinoma. Cancer Lett. 2017;398:37-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 62] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 56. | Zhang Q, Zhang J, Fu Z, Dong L, Tang Y, Xu C, Wang H, Zhang T, Wu Y, Dong C, Shao S, Wang G. Hypoxia-induced microRNA-10b-3p promotes esophageal squamous cell carcinoma growth and metastasis by targeting TSGA10. Aging (Albany NY). 2019;11:10374-10384. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 57. | Lu YF, Yu JR, Yang Z, Zhu GX, Gao P, Wang H, Chen SY, Zhang J, Liu MY, Niu Y, Wei XM, Wang W, Ye FJ, Zhang LX, Zhao Y, Sun GG. Promoter hypomethylation mediated upregulation of MicroRNA-10b-3p targets FOXO3 to promote the progression of esophageal squamous cell carcinoma (ESCC). J Exp Clin Cancer Res. 2018;37:301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 58. | Xu X, Chen Z, Zhao X, Wang J, Ding D, Wang Z, Tan F, Tan X, Zhou F, Sun J, Sun N, Gao Y, Shao K, Li N, Qiu B, He J. MicroRNA-25 promotes cell migration and invasion in esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 2012;421:640-645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 96] [Article Influence: 6.9] [Reference Citation Analysis (1)] |
| 59. | Bai X, Wang Q, Rui X, Li X, Wang X. Upregulation of miR-1269 Contributes to the Progression of Esophageal Squamous Cell Cancer Cells and Is Associated With Poor Prognosis. Technol Cancer Res Treat. 2021;20:1533033820985858. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 60. | Wang X, Han J, Liu Y, Hu J, Li M, Chen X, Xu L. miR-17-5p and miR-4443 Promote Esophageal Squamous Cell Carcinoma Development by Targeting TIMP2. Front Oncol. 2021;11:605894. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 61. | Zhao Y, Zhang Q, Liu H, Wang N, Zhang X, Yang S. lncRNA PART1, manipulated by transcriptional factor FOXP2, suppresses proliferation and invasion in ESCC by regulating the miR18a5p/SOX6 signaling axis. Oncol Rep. 2021;45:1118-1132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 62. | Zhang L, Tong Z, Sun Z, Zhu G, Shen E, Huang Y. MiR-25-3p targets PTEN to regulate the migration, invasion, and apoptosis of esophageal cancer cells via the PI3K/AKT pathway. Biosci Rep. 2020;40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 63. | Wang B, Hua P, Wang R, Li J, Zhang G, Jin C, Zhang Y. Inhibited MicroRNA-301 Restrains Angiogenesis and Cell Growth in Esophageal Squamous Cell Carcinoma by Elevating PTEN. Nanoscale Res Lett. 2021;16:3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 64. | Liu W, Li M, Chen X, Zhang D, Wei L, Zhang Z, Wang S, Meng L, Zhu S, Li B. MicroRNA-373 promotes migration and invasion in human esophageal squamous cell carcinoma by inhibiting TIMP3 expression. Am J Cancer Res. 2016;6:1-14. [PubMed] |
| 65. | Chen Y, Wang H, Zhu S, Lan X. miR-483-5p promotes esophageal cancer progression by targeting KCNQ1. Biochem Biophys Res Commun. 2020;531:615-621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 66. | Zhang H, Shi X, Chang W, Li Y, Wang L. Epigenetic alterations of the Igf2 promoter and the effect of miR4835p on its target gene expression in esophageal squamous cell carcinoma. Mol Med Rep. 2018;17:2251-2256. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 67. | Liu S, Lin Z, Zheng Z, Rao W, Lin Y, Chen H, Xie Q, Chen Y, Hu Z. Serum exosomal microRNA-766-3p expression is associated with poor prognosis of esophageal squamous cell carcinoma. Cancer Sci. 2020;111:3881-3892. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 68. | Gao DC, Hou B, Zhou D, Liu QX, Zhang K, Lu X, Zhang J, Zheng H, Dai JG. Tumor-derived exosomal miR-103a-2-5p facilitates esophageal squamous cell carcinoma cell proliferation and migration. Eur Rev Med Pharmacol Sci. 2020;24:6097-6110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 69. | He B, Zhang K, Han X, Su C, Zhao J, Wang G, Zhang L, Hu W. Extracellular Vesicle-Derived miR-105-5p Promotes Malignant Phenotypes of Esophageal Squamous Cell Carcinoma by Targeting SPARCL1 via FAK/AKT Signaling Pathway. Front Genet. 2022;13:819699. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 70. | Yu T, Cao R, Li S, Fu M, Ren L, Chen W, Zhu H, Zhan Q, Shi R. MiR-130b plays an oncogenic role by repressing PTEN expression in esophageal squamous cell carcinoma cells. BMC Cancer. 2015;15:29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 79] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 71. | Chen Z, Yao N, Gu H, Song Y, Ye Z, Li L, Lu P, Shao Q. Circular RNA_LARP4 Sponges miR-1323 and Hampers Progression of Esophageal Squamous Cell Carcinoma Through Modulating PTEN/PI3K/AKT Pathway. Dig Dis Sci. 2020;65:2272-2283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 72. | Zhang Y, Lu W, Chen Y, Lin Y, Yang X, Wang H, Liu Z. The miR-19b-3p-MAP2K3-STAT3 feedback loop regulates cell proliferation and invasion in esophageal squamous cell carcinoma. Mol Oncol. 2021;15:1566-1583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 73. | Zhang J, Zhang Y, Tan X, Zhang Q, Liu C. MiR-23b-3p induces the proliferation and metastasis of esophageal squamous cell carcinomas cells through the inhibition of EBF3. Acta Biochim Biophys Sin (Shanghai). 2018;50:605-614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 74. | Hu CM, Peng J, Lv L, Wang XH, Huo JR, Liu DL. MiR-196a promotes the proliferation and migration of esophageal cancer via the UHRF2/TET2 axis. Mol Cell Biochem. 2022;477:537-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 75. | Fu G, Pei Z, Song N. Oncogenic microRNA-301b regulates tumor repressor dystrobrevin alpha to facilitate cell growth, invasion and migration in esophageal cancer. Esophagus. 2021;18:315-325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 76. | He Y, Mingyan E, Wang C, Liu G, Shi M, Liu S. CircVRK1 regulates tumor progression and radioresistance in esophageal squamous cell carcinoma by regulating miR-624-3p/PTEN/PI3K/AKT signaling pathway. Int J Biol Macromol. 2019;125:116-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 66] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 77. | Li X, Guo S, Min L, Guo Q, Zhang S. miR-92a-3p promotes the proliferation, migration and invasion of esophageal squamous cell cancer by regulating PTEN. Int J Mol Med. 2019;44:973-981. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 78. | Yang S, Li X, Shen W, Hu H, Li C, Han G. MicroRNA-140 Represses Esophageal Cancer Progression via Targeting ZEB2 to Regulate Wnt/β-Catenin Pathway. J Surg Res. 2021;257:267-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 79. | Iiizumi M, Liu W, Pai SK, Furuta E, Watabe K. Drug development against metastasis-related genes and their pathways: a rationale for cancer therapy. Biochim Biophys Acta. 2008;1786:87-104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 55] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 80. | Wang Q, Yang L, Fan Y, Tang W, Sun H, Xu Z, Zhou J, Zhang Y, Zhu B, Cao X. Circ-ZDHHC5 Accelerates Esophageal Squamous Cell Carcinoma Progression in vitro via miR-217/ZEB1 Axis. Front Cell Dev Biol. 2020;8:570305. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 81. | Xiao JN, Yan TH, Yu RM, Gao Y, Zeng WL, Lu SW, Que HX, Liu ZP, Jiang JH. Long non-coding RNA UCA1 regulates the expression of Snail2 by miR-203 to promote hepatocellular carcinoma progression. J Cancer Res Clin Oncol. 2017;143:981-990. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 78] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 82. | Li G, Qi HW, Dong HG, Bai P, Sun M, Liu HY. Targeting deubiquitinating enzyme USP26 by microRNA-203 regulates Snail1's pro-metastatic functions in esophageal cancer. Cancer Cell Int. 2020;20:355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 83. | Liu B, Li X, Li C, Xu R, Sun X. miR-25 mediates metastasis and epithelial-mesenchymal-transition in human esophageal squamous cell carcinoma via regulation of E-cadherin signaling. Bioengineered. 2019;10:679-688. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 84. | Liu K, Li L, Rusidanmu A, Wang Y, Lv X. Down-Regulation of MiR-1294 is Related to Dismal Prognosis of Patients with Esophageal Squamous Cell Carcinoma through Elevating C-MYC Expression. Cell Physiol Biochem. 2015;36:100-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 85. | Song Y, Li J, Zhu Y, Dai Y, Zeng T, Liu L, Wang H, Qin Y, Zeng M, Guan XY, Li Y. MicroRNA-9 promotes tumor metastasis via repressing E-cadherin in esophageal squamous cell carcinoma. Oncotarget. 2014;5:11669-11680. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 95] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 86. | Chen ZL, Zhao XH, Wang JW, Li BZ, Wang Z, Sun J, Tan FW, Ding DP, Xu XH, Zhou F, Tan XG, Hang J, Shi SS, Feng XL, He J. microRNA-92a promotes lymph node metastasis of human esophageal squamous cell carcinoma via E-cadherin. J Biol Chem. 2011;286:10725-10734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 126] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 87. | Li P, Liu X, Xing W, Qiu H, Li R, Liu S, Sun H. Exosome-derived miR-200a promotes esophageal cancer cell proliferation and migration via the mediating Keap1 expression. Mol Cell Biochem. 2022;477:1295-1308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 88. | Qi Y, Li X, Zhao S. miR-29b inhibits the progression of esophageal squamous cell carcinoma by targeting MMP-2. Neoplasma. 2015;62:384-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 89. | Yang L, Song X, Zhu J, Li M, Ji Y, Wu F, Chen Y, Cui X, Hu J, Wang L, Cao Y, Wei Y, Zhang W, Li F. Tumor suppressor microRNA-34a inhibits cell migration and invasion by targeting MMP-2/MMP-9/FNDC3B in esophageal squamous cell carcinoma. Int J Oncol. 2017;51:378-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 90. | Osako Y, Seki N, Kita Y, Yonemori K, Koshizuka K, Kurozumi A, Omoto I, Sasaki K, Uchikado Y, Kurahara H, Maemura K, Natsugoe S. Regulation of MMP13 by antitumor microRNA-375 markedly inhibits cancer cell migration and invasion in esophageal squamous cell carcinoma. Int J Oncol. 2016;49:2255-2264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 91. | Guo Y, Sun P, Guo W, Yin Q, Han J, Sheng S, Liang J, Dong Z. LncRNA DDX11 antisense RNA 1 promotes EMT process of esophageal squamous cell carcinoma by sponging miR-30d-5p to regulate SNAI1/ZEB2 expression and Wnt/β-catenin pathway. Bioengineered. 2021;12:11425-11440. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 92. | Zhang K, Chen J, Song H, Chen LB. SNHG16/miR-140-5p axis promotes esophagus cancer cell proliferation, migration and EMT formation through regulating ZEB1. Oncotarget. 2018;9:1028-1040. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 70] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 93. | Matsushima K, Isomoto H, Yamaguchi N, Inoue N, Machida H, Nakayama T, Hayashi T, Kunizaki M, Hidaka S, Nagayasu T, Nakashima M, Ujifuku K, Mitsutake N, Ohtsuru A, Yamashita S, Korpal M, Kang Y, Gregory PA, Goodall GJ, Kohno S, Nakao K. MiRNA-205 modulates cellular invasion and migration via regulating zinc finger E-box binding homeobox 2 expression in esophageal squamous cell carcinoma cells. J Transl Med. 2011;9:30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 118] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 94. | Zuo J, Wang D, Shen H, Liu F, Han J, Zhang X. MicroRNA-153 inhibits tumor progression in esophageal squamous cell carcinoma by targeting SNAI1. Tumour Biol. 2016;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 95. | Ma T, Zhao Y, Lu Q, Lu Y, Liu Z, Xue T, Shao Y. MicroRNA-30c functions as a tumor suppressor via targeting SNAI1 in esophageal squamous cell carcinoma. Biomed Pharmacother. 2018;98:680-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 96. | Huang Z, Tian Z, Zhao Y, Zhu F, Liu W, Wang X. MAPK Signaling Pathway Is Essential for Female Reproductive Regulation in the Cabbage Beetle, Colaphellus bowringi. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 97. | Guanen Q, Junjie S, Baolin W, Chaoyang W, Yajuan Y, Jing L, Junpeng L, Gaili N, Zhongping W, Jun W. MiR-214 promotes cell meastasis and inhibites apoptosis of esophageal squamous cell carcinoma via PI3K/AKT/mTOR signaling pathway. Biomed Pharmacother. 2018;105:350-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 98. | An L, Li M, Jia Q. Mechanisms of radiotherapy resistance and radiosensitization strategies for esophageal squamous cell carcinoma. Mol Cancer. 2023;22:140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 117] [Reference Citation Analysis (0)] |
| 99. | Kashyap MK, Abdel-Rahman O. Expression, regulation and targeting of receptor tyrosine kinases in esophageal squamous cell carcinoma. Mol Cancer. 2018;17:54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 100. | Cheng J, Ma H, Yan M, Zhang Z, Xing W. Circ_0007624 suppresses the development of esophageal squamous cell carcinoma via targeting miR-224-5p/CPEB3 to inactivate the EGFR/PI3K/AKT signaling. Cell Signal. 2022;99:110448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 101. | Song W, Wang K, Yang X, Dai W, Fan Z. Long noncoding RNA BANCR mediates esophageal squamous cell carcinoma progression by regulating the IGF1R/Raf/MEK/ERK pathway via miR3383p. Int J Mol Med. 2020;46:1377-1388. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 102. | Xu H, Miao J, Liu S, Liu H, Zhang L, Zhang Q. Long non-coding RNA KCNQ1 overlapping transcript 1 promotes the progression of esophageal squamous cell carcinoma by adsorbing microRNA-133b. Clinics (Sao Paulo). 2021;76:e2175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 103. | Zhu L, Wang Z, Fan Q, Wang R, Sun Y. microRNA-27a functions as a tumor suppressor in esophageal squamous cell carcinoma by targeting KRAS. Oncol Rep. 2014;31:280-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 104. | Kang M, Li Y, Zhu S, Zhang S, Guo S, Li P. MicroRNA-193b acts as a tumor suppressor gene in human esophageal squamous cell carcinoma via target regulation of KRAS. Oncol Lett. 2019;17:3965-3973. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 105. | Zhu Y, Ma Y, Peng H, Gong L, Xiao M, Xiang L, He D, Cao K. MiR-130b promotes the progression of oesophageal squamous cell carcinoma by targeting SASH1. J Cell Mol Med. 2019;23:93-103. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 106. | Noor MT, Seehra N, Rajput J, Sharma R, Thakur BS. Evaluation of Roles of MicroRNA-21 and MicroRNA-18a in Esophageal Squamous Cell Carcinoma and Comparison of Their Changes in Expression Post-Chemoradiotherapy. Gastroenterology Res. 2020;13:107-113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 107. | Wang Y, Lu J, Chen L, Bian H, Hu J, Li D, Xia C, Xu H. Tumor-Derived EV-Encapsulated miR-181b-5p Induces Angiogenesis to Foster Tumorigenesis and Metastasis of ESCC. Mol Ther Nucleic Acids. 2020;20:421-437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 108. | Zeng Q, Zhu Z, Song L, He Z. Transferred by exosomes-derived MiR-19b-3p targets PTEN to regulate esophageal cancer cell apoptosis, migration and invasion. Biosci Rep. 2020;40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 109. | Yang B, Liu Y, Li L, Deng H, Xian L. MicroRNA200a promotes esophageal squamous cell carcinoma cell proliferation, migration and invasion through extensive target genes. Mol Med Rep. 2020;21:2073-2084. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 110. | Cui D, Cheung AL. Roles of microRNAs in tumorigenesis and metastasis of esophageal squamous cell carcinoma. World J Clin Oncol. 2021;12:609-622. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 111. | Song Q, Liu H, Li C, Liang H. miR-33a-5p inhibits the progression of esophageal cancer through the DKK1-mediated Wnt/β-catenin pathway. Aging (Albany NY). 2021;13:20481-20494. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 112. | Yan S, Jiang H, Fang S, Yin F, Wang Z, Jia Y, Sun X, Wu S, Jiang T, Mao A. MicroRNA-340 Inhibits Esophageal Cancer Cell Growth and Invasion by Targeting Phosphoserine Aminotransferase 1. Cell Physiol Biochem. 2015;37:375-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 113. | Wang J, Yao W, Li J, Zhang Q, Wei L. Identification of a novel circ_0001946/miR-1290/SOX6 ceRNA network in esophageal squamous cell cancer. Thorac Cancer. 2022;13:1299-1310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 114. | Luo J, Tian Z, Zhou Y, Xiao Z, Park SY, Sun H, Zhuang T, Wang Y, Li P, Zhao X. CircABCA13 acts as a miR-4429 sponge to facilitate esophageal squamous cell carcinoma development by stabilizing SRXN1. Cancer Sci. 2023;114:2835-2847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 115. | Johnson DE, O'Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15:234-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1929] [Cited by in RCA: 2318] [Article Influence: 289.8] [Reference Citation Analysis (0)] |
| 116. | Wang J, Yu PY, Yu JP, Luo JD, Sun ZQ, Sun F, Kong Z, Wang JL. KIF22 promotes progress of esophageal squamous cell carcinoma cells and is negatively regulated by miR-122. Am J Transl Res. 2021;13:4152-4166. [PubMed] |
| 117. | Hao Y, Baker D, Ten Dijke P. TGF-β-Mediated Epithelial-Mesenchymal Transition and Cancer Metastasis. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 363] [Cited by in RCA: 879] [Article Influence: 125.6] [Reference Citation Analysis (0)] |
| 118. | Cai Y, Ruan W, Ding J, Wei N, Wang J, Zhang H, Ma N, Weng G, Su WK, Lin Y, Zhu K. miR935p regulates the occurrence and development of esophageal carcinoma epithelial cells by targeting TGFβR2. Int J Mol Med. 2021;47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 119. | Liu L, Zhao Z, Zhou W, Fan X, Zhan Q, Song Y. Enhanced Expression of miR-425 Promotes Esophageal Squamous Cell Carcinoma Tumorigenesis by Targeting SMAD2. J Genet Genomics. 2015;42:601-611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 120. | Recillas-Targa F. Cancer Epigenetics: An Overview. Arch Med Res. 2022;53:732-740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 79] [Reference Citation Analysis (0)] |
| 121. | Kazimierczyk M, Wrzesinski J. Long Non-Coding RNA Epigenetics. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 122. | Ali Syeda Z, Langden SSS, Munkhzul C, Lee M, Song SJ. Regulatory Mechanism of MicroRNA Expression in Cancer. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 502] [Cited by in RCA: 725] [Article Influence: 120.8] [Reference Citation Analysis (0)] |
| 123. | Denisenko O. Epigenetics of Ribosomal RNA Genes. Biochemistry (Mosc). 2022;87:S103-SS31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 124. | Pan Q, Li B, Zhang J, Du X, Gu D. LncRNA THAP9-AS1 accelerates cell growth of esophageal squamous cell carcinoma through sponging miR-335-5p to regulate SGMS2. Pathol Res Pract. 2021;224:153526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 125. | Yang F, Wen S, Zhang Y, Xu Y, Lv H, Zhu Y, Wang M, Su P, Huang C, Tian Z. Identifying potential metastasis-related long non-coding RNAs, microRNAs, and message RNAs in the esophageal squamous cell carcinoma. J Cell Biochem. 2019;120:13202-13215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 126. | Lu T, Wang R, Cai H, Cui Y. Long non-coding RNA DLEU2 promotes the progression of esophageal cancer through miR-30e-5p/E2F7 axis. Biomed Pharmacother. 2020;123:109650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 127. | Li Z, Qin X, Bian W, Li Y, Shan B, Yao Z, Li S. Exosomal lncRNA ZFAS1 regulates esophageal squamous cell carcinoma cell proliferation, invasion, migration and apoptosis via microRNA-124/STAT3 axis. J Exp Clin Cancer Res. 2019;38:477. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 143] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 128. | Lin C, Zhang S, Wang Y, Nice E, Guo C, Zhang E, Yu L, Li M, Liu C, Hu L, Hao J, Qi W, Xu H. Functional Role of a Novel Long Noncoding RNA TTN-AS1 in Esophageal Squamous Cell Carcinoma Progression and Metastasis. Clin Cancer Res. 2018;24:486-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 116] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 129. | He XY, Wang XQ, Xiao QL, Liu D, Xu QR, Liu S. Long non-coding RNA NCK1-AS1 functions as a ceRNA to regulate cell viability and invasion in esophageal squamous cell carcinoma via microRNA-133b/ENPEP axis. Cell Cycle. 2023;22:596-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 130. | Jia J, Li H, Chu J, Sheng J, Wang C, Jia Z, Meng W, Yin H, Wan J, He F. LncRNA FAM83A-AS1 promotes ESCC progression by regulating miR-214/CDC25B axis. J Cancer. 2021;12:1200-1211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 131. | Xu J, Chen Z, Fang Z, Chen S, Guo Y, Liu X, Chen K. Long non-coding RNA OIP5-AS1 promotes the progression of esophageal cancer by regulating miR-30a/VOPP1 expression. Oncol Lett. 2021;22:651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 132. | Liu H, Zhang Q, Song Y, Hao Y, Cui Y, Zhang X, Qin Y, Zhu G, Wang F, Dang J, Ma S, Zhang Y, Guo W, Li S, Guan F, Fan T. Long non-coding RNA SLC2A1-AS1 induced by GLI3 promotes aerobic glycolysis and progression in esophageal squamous cell carcinoma by sponging miR-378a-3p to enhance Glut1 expression. J Exp Clin Cancer Res. 2021;40:287. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 133. | Qian CJ, Xu ZR, Chen LY, Wang YC, Yao J. LncRNA MAFG-AS1 Accelerates Cell Migration, Invasion and Aerobic Glycolysis of Esophageal Squamous Cell Carcinoma Cells via miR-765/PDX1 Axis. Cancer Manag Res. 2020;12:6895-6908. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 134. | Yan Y, Li S, Wang S, Rubegni P, Tognetti L, Zhang J, Yan L. Long noncoding RNA HAND2-AS1 inhibits cancer cell proliferation, migration, and invasion in esophagus squamous cell carcinoma by regulating microRNA-21. J Cell Biochem. 2019;120:9564-9571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 135. | Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555-565. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 796] [Cited by in RCA: 1114] [Article Influence: 123.8] [Reference Citation Analysis (0)] |
| 136. | Zeng B, Liu Z, Zhu H, Zhang X, Yang W, Li X, Cheng C. CircRNA_2646 functions as a ceRNA to promote progression of esophageal squamous cell carcinoma via inhibiting miR-124/PLP2 signaling pathway. Cell Death Discov. 2021;7:99. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 137. | Liang Z, Zhao B, Hou J, Zheng J, Xin G. CircRNA circ-OGDH (hsa_circ_0003340) Acts as a ceRNA to Regulate Glutamine Metabolism and Esophageal Squamous Cell Carcinoma Progression by the miR-615-5p/PDX1 Axis. Cancer Manag Res. 2021;13:3041-3053. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 138. | Wu B, Chen Y, Xie X, Liang H, Peng F, Che W. Circ_0001273 downregulation inhibits the growth, migration and glutamine metabolism of esophageal cancer cells via targeting the miR-622/SLC1A5 signaling axis. Thorac Cancer. 2022;13:1795-1805. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 139. | Zhang WQ, Liu KQ, Pei YX, Tan J, Ma JB, Zhao J. Circ-ABCB10 promotes proliferation and invasion of esophageal squamous cell carcinoma cells by modulating microRNA-670-3p. Eur Rev Med Pharmacol Sci. 2020;24:6088-6096. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 140. | Xing Y, Zha WJ, Li XM, Li H, Gao F, Ye T, Du WQ, Liu YC. Circular RNA circ-Foxo3 inhibits esophageal squamous cell cancer progression via the miR-23a/PTEN axis. J Cell Biochem. 2020;121:2595-2605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 73] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 141. | Liu R, Gu J, Jiang P, Zheng Y, Liu X, Jiang X, Huang E, Xiong S, Xu F, Liu G, Ge D, Chu Y. DNMT1-microRNA126 epigenetic circuit contributes to esophageal squamous cell carcinoma growth via ADAM9-EGFR-AKT signaling. Clin Cancer Res. 2015;21:854-863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 95] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 142. | Wu X, Dinglin X, Wang X, Luo W, Shen Q, Li Y, Gu L, Zhou Q, Zhu H, Tan C, Yang X, Zhang Z. Long noncoding RNA XIST promotes malignancies of esophageal squamous cell carcinoma via regulation of miR-101/EZH2. Oncotarget. 2017;8:76015-76028. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 143. | Wang H, Li H, Yu Y, Jiang Q, Zhang R, Sun H, Xing W, Li Y. Long non-coding RNA XIST promotes the progression of esophageal squamous cell carcinoma through sponging miR-129-5p and upregulating CCND1 expression. Cell Cycle. 2021;20:39-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 144. | Ba Y, Liu Y, Li C, Zhu Y, Xing W. HIPK3 Promotes Growth and Metastasis of Esophageal Squamous Cell Carcinoma via Regulation of miR-599/c-MYC Axis. Onco Targets Ther. 2020;13:1967-1978. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 145. | Huang Z, Wang C, Zhao X. circFIG 4 drives the carcinogenesis and metastasis of esophagus cancer via the miR-493-5p/E2F3 axis. Thorac Cancer. 2022;13:783-794. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 146. | Sharma U, Kaur Rana M, Singh K, Jain A. LINC00324 promotes cell proliferation and metastasis of esophageal squamous cell carcinoma through sponging miR-493-5p via MAPK signaling pathway. Biochem Pharmacol. 2023;207:115372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 147. | Liu W, Li M, Chen X, Zhu S, Shi H, Zhang D, Cheng C, Li B. MicroRNA-1 suppresses proliferation, migration and invasion by targeting Notch2 in esophageal squamous cell carcinoma. Sci Rep. 2018;8:5183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 148. | Qi B, Wang Y, Chen ZJ, Li XN, Qi Y, Yang Y, Cui GH, Guo HZ, Li WH, Zhao S. Down-regulation of miR-30a-3p/5p promotes esophageal squamous cell carcinoma cell proliferation by activating the Wnt signaling pathway. World J Gastroenterol. 2017;23:7965-7977. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 36] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 149. | Chen C, Yang C, Tian X, Liang Y, Wang S, Wang X, Shou Y, Li H, Xiao Q, Shu J, Sun M, Chen K. Downregulation of miR-100-5p in cancer-associated fibroblast-derived exosomes facilitates lymphangiogenesis in esophageal squamous cell carcinoma. Cancer Med. 2023;12:14468-14483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 28] [Reference Citation Analysis (0)] |
| 150. | Xie R, Wu SN, Gao CC, Yang XZ, Wang HG, Zhang JL, Yan W, Ma TH. MicroRNA-30d inhibits the migration and invasion of human esophageal squamous cell carcinoma cells via the posttranscriptional regulation of enhancer of zeste homolog 2. Oncol Rep. 2017;37:1682-1690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 151. | Li W, Zhao W, Lu Z, Zhang W, Yang X. Long Noncoding RNA GAS5 Promotes Proliferation, Migration, and Invasion by Regulation of miR-301a in Esophageal Cancer. Oncol Res. 2018;26:1285-1294. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 152. | Sharma P, Saini N, Sharma R. miR-107 functions as a tumor suppressor in human esophageal squamous cell carcinoma and targets Cdc42. Oncol Rep. 2017;37:3116-3127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 153. | Men Y, Zhai Y, Wu L, Liu L, Zhang W, Jiang W, Bi N, Song Y, Hui Z, Wang L. MiR-323a-3p acts as a tumor suppressor by suppressing FMR1 and predicts better esophageal squamous cell carcinoma outcome. Cancer Cell Int. 2022;22:140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 154. | Xu ML, Liu TC, Dong FX, Meng LX, Ling AX, Liu S. Exosomal lncRNA LINC01711 facilitates metastasis of esophageal squamous cell carcinoma via the miR-326/FSCN1 axis. Aging (Albany NY). 2021;13:19776-19788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 155. | Han N, Zhao W, Zhang Z, Zheng P. MiR-328 suppresses the survival of esophageal cancer cells by targeting PLCE1. Biochem Biophys Res Commun. 2016;470:175-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 156. | Tian Z, Li Z, Zhu Y, Meng L, Liu F, Sang M, Wang G. Hypermethylation-mediated inactivation of miR-124 predicts poor prognosis and promotes tumor growth at least partially through targeting EZH2/H3K27me3 in ESCC. Clin Exp Metastasis. 2019;36:381-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 157. | Zhao Y, Ma K, Yang S, Zhang X, Wang F, Liu H, Fan Q. MicroRNA-125a-5p enhances the sensitivity of esophageal squamous cell carcinoma cells to cisplatin by suppressing the activation of the STAT3 signaling pathway. Int J Oncol. 2018;53:644-658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 158. | Suyal G, Pandey P, Saraya A, Sharma R. Tumour suppressor role of microRNA-335-5p in esophageal squamous cell carcinoma by targeting TTK (Mps1). Exp Mol Pathol. 2022;124:104738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 159. | Takashima Y, Komatsu S, Ohashi T, Kiuchi J, Nishibeppu K, Kamiya H, Arakawa H, Ishida R, Shimizu H, Arita T, Konishi H, Shiozaki A, Kubota T, Fujiwara H, Otsuji E. Plasma miR-1254 as a predictive biomarker of chemosensitivity and a target of nucleic acid therapy in esophageal cancer. Cancer Sci. 2023;114:3027-3040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 160. | Cui D, Zhu Y, Yan D, Lee NPY, Han L, Law S, Tsao GSW, Cheung ALM. Dual inhibition of cMET and EGFR by microRNA-338-5p suppresses metastasis of esophageal squamous cell carcinoma. Carcinogenesis. 2021;42:995-1007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 161. | Zuo J, Zhu K, Wang Y, Yu Z. MicroRNA-34a suppresses invasion and metastatic in esophageal squamous cell carcinoma by regulating CD44. Mol Cell Biochem. 2018;443:139-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 162. | Zhang Z, Liang X, Ren L, Zhang S, Li S, Wan T, Xu D, Lv S. LINC00662 promotes cell viability and metastasis in esophageal squamous cell carcinoma by sponging miR-340-5p and upregulating HOXB2. Thorac Cancer. 2020;11:2306-2315. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 163. | Tang R, Zhou Q, Xu Q, Lu L, Zhou Y. Circular RNA circ_0006948 Promotes Esophageal Squamous Cell Carcinoma Progression by Regulating microRNA-3612/LASP1 Axis. Dig Dis Sci. 2022;67:2158-2172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 164. | Kang M, Li Y, Liu W, Wang R, Tang A, Hao H, Liu Z, Ou H. miR-129-2 suppresses proliferation and migration of esophageal carcinoma cells through downregulation of SOX4 expression. Int J Mol Med. 2013;32:51-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 61] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 165. | Sun C, Zhang X, Chen Y, Jia Q, Yang J, Shu Y. MicroRNA-365 suppresses cell growth and invasion in esophageal squamous cell carcinoma by modulating phosphoserine aminotransferase 1. Cancer Manag Res. 2018;10:4581-4590. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 166. | Peng X, Wu X, Wu G, Peng C, Huang B, Huang M, Ding J, Mao C, Zhang H. MiR-129-2-3p Inhibits Esophageal Carcinoma Cell Proliferation, Migration, and Invasion via Targeting DNMT3B. Curr Mol Pharmacol. 2023;16:116-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 167. | Chen M, Xia Y, Tan Y, Jiang G, Jin H, Chen Y. Downregulation of microRNA-370 in esophageal squamous-cell carcinoma is associated with cancer progression and promotes cancer cell proliferation via upregulating PIN1. Gene. 2018;661:68-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 168. | Zhang Y, Xiong W, Yang C, Li P, Tong H. Circ-FNDC3B Functions as an Oncogenic Factor in Esophageal Squamous Cell Carcinoma via Upregulating MYO5A by Absorbing miR-136-5p and miR-370-3p. Biochem Genet. 2023;61:1917-1936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 169. | Wang W, Wu D, He X, Hu X, Hu C, Shen Z, Lin J, Pan Z, He Z, Lin H, Wang M. CCL18-induced HOTAIR upregulation promotes malignant progression in esophageal squamous cell carcinoma through the miR-130a-5p-ZEB1 axis. Cancer Lett. 2019;460:18-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 170. | Li S, Qin X, Li Y, Zhang X, Niu R, Zhang H, Cui A, An W, Wang X. MiR-133a suppresses the migration and invasion of esophageal cancer cells by targeting the EMT regulator SOX4. Am J Transl Res. 2015;7:1390-1403. [PubMed] |
| 171. | Fang N, Shi Y, Fan Y, Long T, Shu Y, Zhou J. Circ_0072088 Promotes Proliferation, Migration, and Invasion of Esophageal Squamous Cell Cancer by Absorbing miR-377. J Oncol. 2020;2020:8967126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 172. | Jin W, Wang L, Cheng S, Lv H. Prognostic value of microRNA-378 in esophageal cancer and its regulatory effect on tumor progression. Exp Ther Med. 2021;22:704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 173. | Wu Y, Zhi L, Zhao Y, Yang L, Cai F. Knockdown of circular RNA UBAP2 inhibits the malignant behaviours of esophageal squamous cell carcinoma by microRNA-422a/Rab10 axis. Clin Exp Pharmacol Physiol. 2020;47:1283-1290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 174. | Xu S, Li X, Li L, Wang Y, Geng C, Guo F, Zhang T, Du A, Lu Z, Hui H, Wang Q. CTCF-silenced miR-137 contributes to EMT and radioresistance in esophageal squamous cell carcinoma. Cancer Cell Int. 2021;21:155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 175. | Wang Y, Li M, Zang W, Ma Y, Wang N, Li P, Wang T, Zhao G. MiR-429 up-regulation induces apoptosis and suppresses invasion by targeting Bcl-2 and SP-1 in esophageal carcinoma. Cell Oncol (Dordr). 2013;36:385-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 66] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 176. | Chen X, Jiang J, Zhao Y, Wang X, Zhang C, Zhuan L, Zhang D, Zheng Y. Circular RNA circNTRK2 facilitates the progression of esophageal squamous cell carcinoma through up-regulating NRIP1 expression via miR-140-3p. J Exp Clin Cancer Res. 2020;39:133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 177. | Li T, Li S. Circ_0023984 Facilitates Esophageal Squamous Cell Carcinoma Progression by Regulating miR-433-3p/REV3L Axis. Dig Dis Sci. 2022;67:892-903. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 178. | Li W, Jiang G, Zhou J, Wang H, Gong Z, Zhang Z, Min K, Zhu H, Tan Y. Down-regulation of miR-140 induces EMT and promotes invasion by targeting Slug in esophageal cancer. Cell Physiol Biochem. 2014;34:1466-1476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 179. | Jiang T, Zhao R, Du Y, Shi Z, Cheng H. MiR-449a-5p regulates the proliferation of esophageal carcinoma cell by targeting B-cell lymphoma 2. Transl Cancer Res. 2020;9:7236-7247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 180. | Liu H, Zheng M, Zhao Y, Zhang S. miR-143 inhibits migration and invasion through regulating LASP1 in human esophageal cancer. Int J Clin Exp Pathol. 2019;12:466-476. [PubMed] |
| 181. | Yang H, Wei YN, Zhou J, Hao TT, Liu XL. MiR-455-3p acts as a prognostic marker and inhibits the proliferation and invasion of esophageal squamous cell carcinoma by targeting FAM83F. Eur Rev Med Pharmacol Sci. 2017;21:3200-3206. [PubMed] |
| 182. | Jin W, Luo W, Fang W, Wang Y, Wang L, Shen Q, Liu W, Zhang H. miR-145 expression level in tissue predicts prognosis of patients with esophageal squamous cell carcinoma. Pathol Res Pract. 2019;215:152401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 183. | Li Y, Tang C, Huang S, He J, An J, Liu L, Lao S, Zhang J. MicroRNA -145 suppresses esophageal squamous cell carcinoma by targeting phospholipase C epsilon 1. Zhonghua Shiyan Waike Zazhi. 2017;34:1472-1475. [DOI] [Full Text] |
| 184. | Liu Y, Tang Y, Li P. Inhibitory effect of microRNA-455-5p on biological functions of esophageal squamous cell carcinoma Eca109 cells via Rab31. Exp Ther Med. 2018;16:4959-4966. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 185. | Chen G, Teng Z, Zhu Z, Li X. miR-145-3p Hampers the Malignant Progression of Esophageal Carcinoma via CXCL5 Downregulation. Anal Cell Pathol (Amst). 2022;2022:5418356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 186. | Zhou PL, Wu Z, Zhang W, Xu M, Ren J, Zhang Q, Sun Z, Han X. Circular RNA hsa_circ_0000277 sequesters miR-4766-5p to upregulate LAMA1 and promote esophageal carcinoma progression. Cell Death Dis. 2021;12:676. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 187. | Mei LL, Wang WJ, Qiu YT, Xie XF, Bai J, Shi ZZ. miR-145-5p Suppresses Tumor Cell Migration, Invasion and Epithelial to Mesenchymal Transition by Regulating the Sp1/NF-κB Signaling Pathway in Esophageal Squamous Cell Carcinoma. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 80] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 188. | Zhao R, Shan Y, Zhou X, Zhang C, Zhao R, Zhao L, Shan B. MicroRNA4855p suppresses the progression of esophageal squamous cell carcinoma by targeting flotillin1 and inhibits the epithelialmesenchymal transition. Oncol Rep. 2021;45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 189. | Chang HY, Lee CH, Li YS, Huang JT, Lan SH, Wang YF, Lai WW, Wang YC, Lin YJ, Liu HS, Cheng HC. MicroRNA-146a suppresses tumor malignancy via targeting vimentin in esophageal squamous cell carcinoma cells with lower fibronectin membrane assembly. J Biomed Sci. 2020;27:102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 190. | Yi Y, Lu X, Chen J, Jiao C, Zhong J, Song Z, Yu X, Lin B. Downregulated miR-486-5p acts as a tumor suppressor in esophageal squamous cell carcinoma. Exp Ther Med. 2016;12:3411-3416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 191. | Liu J, Wang C, Liu X, Wang Y, Liu H, Ren G, Zhu L, Sun Z, Chen Z. Low expression of miR-1469 predicts disease progression and unfavorable post-surgical clinical outcomes in patients with esophageal squamous cell cancer. Oncol Lett. 2017;13:4469-4474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 192. | Pan Z, Lin J, Wu D, He X, Wang W, Hu X, Zhang L, Wang M. Hsa_circ_0006948 enhances cancer progression and epithelial-mesenchymal transition through the miR-490-3p/HMGA2 axis in esophageal squamous cell carcinoma. Aging (Albany NY). 2019;11:11937-11954. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 72] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 193. | Xu F, Zhang J. Long non-coding RNA HOTAIR functions as miRNA sponge to promote the epithelial to mesenchymal transition in esophageal cancer. Biomed Pharmacother. 2017;90:888-896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 74] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 194. | Chen S, Liu Z, Feng Q, Zhou J, Huang J, Yu J. Circular RNA circ-AGFG1 contributes to esophageal squamous cell carcinoma progression and glutamine catabolism by targeting microRNA-497-5p/solute carrier family 1 member 5 axis. Anticancer Drugs. 2023;34:195-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 195. | Yokobori T, Suzuki S, Tanaka N, Inose T, Sohda M, Sano A, Sakai M, Nakajima M, Miyazaki T, Kato H, Kuwano H. MiR-150 is associated with poor prognosis in esophageal squamous cell carcinoma via targeting the EMT inducer ZEB1. Cancer Sci. 2013;104:48-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 115] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 196. | Jin G, Yang Y, Tuo G, Wang W, Zhu Z. LncRNA TUG1 promotes tumor growth and metastasis of esophageal squamous cell carcinoma by regulating XBP1 via competitively binding to miR-498. Neoplasma. 2020;67:751-761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 197. | Osako Y, Seki N, Koshizuka K, Okato A, Idichi T, Arai T, Omoto I, Sasaki K, Uchikado Y, Kita Y, Kurahara H, Maemura K, Natsugoe S. Regulation of SPOCK1 by dual strands of pre-miR-150 inhibit cancer cell migration and invasion in esophageal squamous cell carcinoma. J Hum Genet. 2017;62:935-944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 198. | Zang B, Zhao J, Chen C. LncRNA PCAT-1 Promoted ESCC Progression via Regulating ANXA10 Expression by Sponging miR-508-3p. Cancer Manag Res. 2019;11:10841-10849. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 199. | Wang M, Huang C, Gao W, Zhu Y, Zhang F, Li Z, Tian Z. MicroRNA-181a-5p prevents the progression of esophageal squamous cell carcinoma in vivo and in vitro via the MEK1-mediated ERK-MMP signaling pathway. Aging (Albany NY). 2022;14:3540-3553. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 200. | Zhao Y, Wang Y, Xing G. miR-516b functions as a tumor suppressor by directly modulating CCNG1 expression in esophageal squamous cell carcinoma. Biomed Pharmacother. 2018;106:1650-1660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 201. | Retraction statement: miR-518b is down-regulated, and involved in cell proliferation and invasion by targeting Rap1b in esophageal squamous cell carcinoma. FEBS Lett. 2023;597:2699. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 202. | Sun N, Ye L, Chang T, Li X. microRNA-195-Cdc42 axis acts as a prognostic factor of esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:6871-6879. [PubMed] |
| 203. | Zhou S, Guo Z, Zhou C, Zhang Y, Wang S. circ_NRIP1 is oncogenic in malignant development of esophageal squamous cell carcinoma (ESCC) via miR-595/SEMA4D axis and PI3K/AKT pathway. Cancer Cell Int. 2021;21:250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 204. | Shi Y, Fang N, Li Y, Guo Z, Jiang W, He Y, Ma Z, Chen Y. Circular RNA LPAR3 sponges microRNA-198 to facilitate esophageal cancer migration, invasion, and metastasis. Cancer Sci. 2020;111:2824-2836. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 100] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 205. | Wang C, Tang D, Wang H, Hu G, Hu S, Li L, Min B, Wang Y. Circular RNA hsa_circ_0030018 acts as a sponge of miR-599 to aggravate esophageal carcinoma progression by regulating ENAH expression. J Cell Biochem. 2020;121:3730-3738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 206. | Ma G, Zhang F, Dong X, Wang X, Ren Y. Low expression of microRNA-202 is associated with the metastasis of esophageal squamous cell carcinoma. Exp Ther Med. 2016;11:951-956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 207. | Yang B, Xie R, Wu SN, Gao CC, Yang XZ, Zhou JF. MicroRNA-615-5p targets insulin-like growth factor 2 and exerts tumor-suppressing functions in human esophageal squamous cell carcinoma. Oncol Rep. 2018;39:255-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 208. | Zhu Y, Li F, Wan Y, Liang H, Li S, Peng B, Shao L, Xu Y, Jiang D. Cancer-Secreted Exosomal MiR-620 Inhibits ESCC Aerobic Glycolysis via FOXM1/HER2 Pathway and Promotes Metastasis. Front Oncol. 2022;12:756109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 209. | Jiang M, Shi H, Xu Y, Bai W, Wang P, Ju Q. miR-203a-3p regulates the cellular processes of esophageal cancer cells via targeting CtBP2. Transl Cancer Res. 2019;8:2791-2802. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 210. | Zhang H, Wu X, Sui Z, Ma Z, Gong L, Meng B, Tang P, Yu Z. High-mobility group AT-hook 2 promotes growth and metastasis and is regulated by miR-204-5p in oesophageal squamous cell carcinoma. Eur J Clin Invest. 2021;51:e13563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 211. | Wang Z, Qiao Q, Chen M, Li X, Wang Z, Liu C, Xie Z. miR-625 down-regulation promotes proliferation and invasion in esophageal cancer by targeting Sox2. FEBS Lett. 2014;588:915-921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 212. | Li C, Li DC, Che SS, Ma K, Wang YJ, Xia LH, Dai XM, Zhang GT, Shen Y, Jiao WJ, Tian KH. The decreased expression of miR-625 predicts poor prognosis of esophageal squamous cell carcinoma. Int J Clin Exp Med. 2015;8:9560-9564. [PubMed] |
| 213. | Dong S, Yin H, Lyu Y, Meng W, Dong C, He F. Expression and clinical significance of miRNA-205 in esophageal squamous cell carcinoma. Disan Junyi Daxue Xuebao. 2016;38:85-88. |
| 214. | Jin L, Yi J, Gao Y, Han S, He Z, Chen L, Song H. MiR-630 inhibits invasion and metastasis in esophageal squamous cell carcinoma. Acta Biochim Biophys Sin (Shanghai). 2016;48:810-819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 215. | Wu Z, Yu B, Jiang L. MiR-212-3p mediates apoptosis and invasion of esophageal squamous cell carcinoma through inhibition of the Wnt/β-catenin signaling pathway by targeting SOX4. J Thorac Dis. 2020;12:4357-4367. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 216. | Huang SD, Yuan Y, Zhuang CW, Li BL, Gong DJ, Wang SG, Zeng ZY, Cheng HZ. MicroRNA-98 and microRNA-214 post-transcriptionally regulate enhancer of zeste homolog 2 and inhibit migration and invasion in human esophageal squamous cell carcinoma. Mol Cancer. 2012;11:51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 96] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 217. | Zhao H, Diao C, Wang X, Xie Y, Liu Y, Gao X, Han J, Li S. LncRNA BDNF-AS inhibits proliferation, migration, invasion and EMT in oesophageal cancer cells by targeting miR-214. J Cell Mol Med. 2018;22:3729-3739. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 218. | Li R, Chai L, Lei L, Guo R, Wen X. MiR-671-5p sponging activity of circMMP1 promotes esophageal squamous cancer progression. Thorac Cancer. 2023;14:2924-2933. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 219. | Phatak P, Burrows WM, Creed TM, Youssef M, Lee G, Donahue JM. MiR-214-3p targets Ras-related protein 14 (RAB14) to inhibit cellular migration and invasion in esophageal Cancer cells. BMC Cancer. 2022;22:1265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 220. | Dong S, Yin H, Dong C, Sun K, Lv P, Meng W, Ming L, He F. Predictive Value of Plasma MicroRNA-216a/b in the Diagnosis of Esophageal Squamous Cell Carcinoma. Dis Markers. 2016;2016:1857067. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 221. | Wang T, Chen T, Niu H, Li C, Xu C, Li Y, Huang R, Zhao J, Wu S. MicroRNA-218 inhibits the proliferation and metastasis of esophageal squamous cell carcinoma cells by targeting BMI1. Int J Mol Med. 2015;36:93-102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 222. | Zhang L, Yu S, Yin X, Tu M, Cai L, Zhang Y, Yu L, Zhang S, Pan X, Huang Y. MiR-942-5p inhibits tumor migration and invasion through targeting CST1 in esophageal squamous cell carcinoma. PLoS One. 2023;18:e0277006. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 223. | Yang C, Zheng S, Liu T, Liu Q, Dai F, Zhou J, Chen Y, Sheyhidin I, Lu X. Down-regulated miR-26a promotes proliferation, migration, and invasion via negative regulation of MTDH in esophageal squamous cell carcinoma. FASEB J. 2017;31:2114-2122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 224. | Mei LL, Qiu YT, Huang MB, Wang WJ, Bai J, Shi ZZ. MiR-99a suppresses proliferation, migration and invasion of esophageal squamous cell carcinoma cells through inhibiting the IGF1R signaling pathway. Cancer Biomark. 2017;20:527-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 225. | Han M, Li N, Li F, Wang H, Ma L. MiR-27b-3p exerts tumor suppressor effects in esophageal squamous cell carcinoma by targeting Nrf2. Hum Cell. 2020;33:641-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 226. | Liu Q, Lv GD, Qin X, Gen YH, Zheng ST, Liu T, Lu XM. Role of microRNA let-7 and effect to HMGA2 in esophageal squamous cell carcinoma. Mol Biol Rep. 2012;39:1239-1246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 227. | Ling R, Zhou Y, Zhou L, Dai D, Wu D, Mi L, Mao C, Chen D. Lin28/microRNA-let-7a promotes metastasis under circumstances of hyperactive Wnt signaling in esophageal squamous cell carcinoma. Mol Med Rep. 2018;17:5265-5271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 228. | Wang H, Fu L, Wei D, Wang B, Zhang C, Zhu T, Ma Z, Li Z, Wu Y, Yu G. MiR-29c-3p Suppresses the Migration, Invasion and Cell Cycle in Esophageal Carcinoma via CCNA2/p53 Axis. Front Bioeng Biotechnol. 2020;8:75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 229. | Tian Y, Luo A, Cai Y, Su Q, Ding F, Chen H, Liu Z. MicroRNA-10b promotes migration and invasion through KLF4 in human esophageal cancer cell lines. J Biol Chem. 2010;285:7986-7994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 244] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 230. | Zhu Y, Liu J, Fan L, Wang F, Bu L, Ma T, Du Y, Zhang C, Wang H, Sun G. Serum expression and significance of MicroRNA-30d-5p in esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2017;10:8677-8685. [PubMed] |
| 231. | Shou Y, Wang X, Chen C, Liang Y, Yang C, Xiao Q, Li H, Wang S, Shu J, Tian X, Chen K. Exosomal miR-301a-3p from esophageal squamous cell carcinoma cells promotes angiogenesis by inducing M2 polarization of macrophages via the PTEN/PI3K/AKT signaling pathway. Cancer Cell Int. 2022;22:153. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 57] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 232. | Jiang L, Wang Y, Rong Y, Xu L, Chu Y, Zhang Y, Yao Y. miR-1179 promotes cell invasion through SLIT2/ROBO1 axis in esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2015;8:319-327. [PubMed] |
| 233. | Gao Y, Yi J, Zhang K, Bai F, Feng B, Wang R, Chu X, Chen L, Song H. Downregulation of MiR-31 stimulates expression of LATS2 via the hippo pathway and promotes epithelial-mesenchymal transition in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2017;36:161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 234. | Meng H, Wang K, Chen X, Guan X, Hu L, Xiong G, Li J, Bai Y. MicroRNA-330-3p functions as an oncogene in human esophageal cancer by targeting programmed cell death 4. Am J Cancer Res. 2015;5:1062-1075. [PubMed] |
| 235. | Jin Y, Meng Q, Zhang B, Xie C, Chen X, Tian B, Wang J, Shih TC, Zhang Y, Cao J, Yang Y, Chen S, Guan X, Hong A. Cancer-associated fibroblasts-derived exosomal miR-3656 promotes the development and progression of esophageal squamous cell carcinoma via the ACAP2/PI3K-AKT signaling pathway. Int J Biol Sci. 2021;17:3689-3701. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 236. | Chen J. Downregulation of MicroRNA-135b-5p Inhibits Esophageal Cancer Cell Progression via Targeting GNG7. J Biomater Tiss Eng. 2019;9:1222-1231. [RCA] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 237. | Lin RJ, Xiao DW, Liao LD, Chen T, Xie ZF, Huang WZ, Wang WS, Jiang TF, Wu BL, Li EM, Xu LY. MiR-142-3p as a potential prognostic biomarker for esophageal squamous cell carcinoma. J Surg Oncol. 2012;105:175-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 94] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 238. | Zhang M, Tian H, Li R, Yan W, Xu R. MicroRNA-4286 promotes esophageal carcinoma development by targeting INPP4A to evoke the JAK2/STAT3 pathway activation. Pharmazie. 2018;73:342-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 239. | Mei LL, Qiu YT, Wang WJ, Bai J, Shi ZZ. Overexpression of microRNA-1470 promotes proliferation and migration, and inhibits senescence of esophageal squamous carcinoma cells. Oncol Lett. 2017;14:7753-7758. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 240. | Gong T, Zheng S, Huang S, Fu S, Zhang X, Pan S, Yang T, Sun Y, Wang Y, Hui B, Guo J. PTENP1 inhibits the growth of esophageal squamous cell carcinoma by regulating SOCS6 expression and correlates with disease prognosis. Mol Carcinog. 2017;56:2610-2619. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 241. | Xu R, Ding P, Zhao X, Li Z, Liu F, Gu L, Zheng Y, Sang M, Meng L. Circular RNA circ-TNRC6B inhibits the proliferation and invasion of esophageal squamous cell carcinoma cells by regulating the miR-452-5p/DAG1 axis. Mol Oncol. 2023;17:1437-1452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 242. | Xu XL, Jiang YH, Feng JG, Su D, Chen PC, Mao WM. MicroRNA-17, microRNA-18a, and microRNA-19a are prognostic indicators in esophageal squamous cell carcinoma. Ann Thorac Surg. 2014;97:1037-1045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 75] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 243. | Ma J, Hong L, Xu G, Hao J, Wang R, Guo H, Liu J, Zhang Y, Nie Y, Fan D. miR-483-3p plays an oncogenic role in esophageal squamous cell carcinoma by targeting tumor suppressor EI24. Cell Biol Int. 2016;40:448-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 244. | Qi J, Fan W. miR-18a-5p and ATM Expression in Esophageal Squamous Cell Carcinoma and Their Correlations with Clinicopathological Features. Comput Math Methods Med. 2022;2022:5260608. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 245. | Li SP, Su HX, Zhao D, Guan QL. Plasma miRNA-506 as a Prognostic Biomarker for Esophageal Squamous Cell Carcinoma. Med Sci Monit. 2016;22:2195-2201. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 246. | Chen S, Ainiwaer B, Qing S, Liu T, Ma Z, Shi Y, Pang X, Zhang W, Li X. [Expression levels of miR-181c-3p and miR-5692b in esophageal cancer and their clinical significance]. Zhonghua Bing Li Xue Za Zhi. 2015;44:905-909. [PubMed] |
| 247. | Zhou P, Dong H, He S, Fang L, Jiang N, Sun Q. miR612 is associated with esophageal squamous cell carcinoma development and metastasis, mediated through TP53. Mol Med Rep. 2017;16:1855-1863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 248. | Xiao Q, Chen T, Wu Y, Wu W, Xu Y, Gong Z, Chen S. MicroRNA6753p promotes esophageal squamous cell cancer cell migration and invasion. Mol Med Rep. 2018;18:3631-3640. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 249. | Yu J, Chen S, Niu Y, Liu M, Zhang J, Yang Z, Gao P, Wang W, Han X, Sun G. Functional Significance and Therapeutic Potential of miRNA-20b-5p in Esophageal Squamous Cell Carcinoma. Mol Ther Nucleic Acids. 2020;21:315-331. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 250. | Kang N, Ou Y, Wang G, Chen J, Li D, Zhan Q. miR-875-5p exerts tumor-promoting function via down-regulation of CAPZA1 in esophageal squamous cell carcinoma. PeerJ. 2021;9:e10020. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 251. | Hao Z, Tang L, Deng Y, Cai Y, Zhang N, Fu X, Li W. Up-regulation of miR-221 in early-stage esophageal squamous cell cancer and its promotion effect on proliferation, migration and invasion of EC109 cells. Zhongliu. 2016;36:326-333. [DOI] [Full Text] |
| 252. | He X, Zhang Z, Li M, Li S, Ren L, Zhu H, Xiao B, Shi R. Expression and role of oncogenic miRNA-224 in esophageal squamous cell carcinoma. BMC Cancer. 2015;15:575. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 253. | Wang W, Fu S, Lin X, Zheng J, Pu J, Gu Y, Deng W, Liu Y, He Z, Liang W, Wang C. miR-92b-3p Functions As A Key Gene In Esophageal Squamous Cell Cancer As Determined By Co-Expression Analysis. Onco Targets Ther. 2019;12:8339-8353. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 254. | Shang M, Weng L, Wu S, Liu B, Yin X, Wang Z, Mao A. HP1BP3 promotes tumor growth and metastasis by upregulating miR-23a to target TRAF5 in esophageal squamous cell carcinoma. Am J Cancer Res. 2021;11:2928-2943. [PubMed] |
| 255. | Jin C, Zuo S, Wang X, Diao C, Xie Y, Li S, Li X. Expression of microRNA-935 in esophageal squamous cell carcinoma tissues and cells and its effects on cell proliferation and invasion. Zhonghua Shiyan Waike Zazhi. 2017;34:1466-1468. [DOI] [Full Text] |
| 256. | Li C, Ding C, Chen T, Chen J, Xu Z, Lei Z, Xu C, Zhao J. Micro ribonucleic acid-93 promotes proliferation and migration of esophageal squamous cell carcinoma by targeting disabled 2. Thorac Cancer. 2015;6:524-533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 257. | Meng F, Zhang X, Wang Y, Lin J, Tang Y, Zhang G, Qiu B, Zeng X, Liu W, He X. Hsa_circ_0021727 (circ-CD44) promotes ESCC progression by targeting miR-23b-5p to activate the TAB1/NFκB pathway. Cell Death Dis. 2023;14:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 258. | Ma J, Fan Y, Feng T, Chen F, Xu Z, Li S, Lin Q, He X, Shi W, Liu Y, Liu Z, Zhu B, Cao X. HOTAIR regulates HK2 expression by binding endogenous miR-125 and miR-143 in oesophageal squamous cell carcinoma progression. Oncotarget. 2017;8:86410-86422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 259. | Shi Z, Jiang T, Cao B, Sun X, Liu J. CAF-derived exosomes deliver LINC01410 to promote epithelial-mesenchymal transition of esophageal squamous cell carcinoma. Exp Cell Res. 2022;412:113033. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 260. | Zhang Z, Lin W, Gao L, Chen K, Yang C, Zhuang L, Peng S, Kang M, Lin J. Hsa_circ_0004370 promotes esophageal cancer progression through miR-1294/LASP1 pathway. Biosci Rep. 2019;39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 261. | Cheng J, Ma H, Yan M, Xing W. THAP9-AS1/miR-133b/SOX4 positive feedback loop facilitates the progression of esophageal squamous cell carcinoma. Cell Death Dis. 2021;12:401. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 262. | Shang M, Wang X, Zhang Y, Gao Z, Wang T, Liu R. LincRNA-ROR promotes metastasis and invasion of esophageal squamous cell carcinoma by regulating miR-145/FSCN1. Onco Targets Ther. 2018;11:639-649. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 263. | Zhu C, Bi W, Li H, Wang W. CircLONP2 Accelerates Esophageal Squamous Cell Carcinoma Progression via Direct MiR-27b-3p-ZEB1 Axis. Front Oncol. 2022;12:822839. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 264. | Du F, Guo T, Cao C. Silencing of Long Noncoding RNA SNHG6 Inhibits Esophageal Squamous Cell Carcinoma Progression via miR-186-5p/HIF1α Axis. Dig Dis Sci. 2020;65:2844-2852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 265. | Chen Y, Sheng HG, Deng FM, Cai LL. Downregulation of the long noncoding RNA SNHG1 inhibits tumor cell migration and invasion by sponging miR-195 through targeting Cdc42 in oesophageal cancer. Kaohsiung J Med Sci. 2021;37:181-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 266. | Shen S, Li K, Liu Y, Liu X, Liu B, Ba Y, Xing W. Silencing lncRNA AGAP2-AS1 Upregulates miR-195-5p to Repress Migration and Invasion of EC Cells via the Decrease of FOSL1 Expression. Mol Ther Nucleic Acids. 2020;20:331-344. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 267. | Zhang LW, Wang B, Yang JX, Yang H. Circ-PRMT5 stimulates migration in esophageal cancer by binding miR-203. Eur Rev Med Pharmacol Sci. 2020;24:9965-9972. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 268. | Liu HF, Zhen Q, Fan YK. LINC00963 predicts poor prognosis and promotes esophageal cancer cells invasion via targeting miR-214-5p/RAB14 axis. Eur Rev Med Pharmacol Sci. 2020;24:164-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 269. | Tang J, Xu H, Liu Q, Zheng J, Pan C, Li Z, Wen W, Wang J, Zhu Q, Wang Z, Chen L. LncRNA LOC146880 promotes esophageal squamous cell carcinoma progression via miR-328-5p/FSCN1/MAPK axis. Aging (Albany NY). 2021;13:14198-14218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 270. | Zhang C, Wang L, Yang J, Fu Y, Li H, Xie L, Cui Y. MicroRNA-33a-5p suppresses esophageal squamous cell carcinoma progression via regulation of lncRNA DANCR and ZEB1. Eur J Pharmacol. 2019;861:172590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 271. | Zhou WZ, Wang XW, Zhu J, Chen MZ, Jin H. LncRNA-CASC15 knockdown inhibits the progression of esophageal squamous cell carcinoma through targeting miR-33a-5p/PTGS2 axis. Histol Histopathol. 2023;38:223-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 272. | Li N, Wu J, Hu B, Lu H, Gao J, Zhu L, Zheng D. Upregulation of hsa_circ_0000977 participates in esophageal squamous cancer progression by sponging miR-874-3p. J Clin Lab Anal. 2022;36:e24458. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 273. | Chen W, Wang L, Li X, Zhao C, Shi L, Zhao H, Huang C. LncRNA SNHG17 regulates cell proliferation and invasion by targeting miR-338-3p/SOX4 axis in esophageal squamous cell carcinoma. Cell Death Dis. 2021;12:806. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 274. | Chu J, Jia J, Yang L, Qu Y, Yin H, Wan J, He F. LncRNA MIR31HG functions as a ceRNA to regulate c-Met function by sponging miR-34a in esophageal squamous cell carcinoma. Biomed Pharmacother. 2020;128:110313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 275. | Zhou Z, Huang F. Long Non-Coding RNA LINC00152 Regulates Cell Proliferation, Migration And Invasion In Esophageal Squamous Cell Carcinoma Via miR-107/Rab10 Axis. Onco Targets Ther. 2019;12:8553-8567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 276. | Chang Z, Fu Y, Jia Y, Gao M, Song L, Zhang W, Zhao R, Qin Y. Circ-SFMBT2 drives the malignant phenotypes of esophageal cancer by the miR-107-dependent regulation of SLC1A5. Cancer Cell Int. 2021;21:495. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 277. | Su M, Tang J, Zhang B, Yang D, Wu Z, Wu J, Zhou Y, Liao Q, Wang H, Wang W, Xiao Y. LncRNA GACAT3 promotes esophageal squamous cell carcinoma progression through regulation of miR-149/FOXM1. Cancer Cell Int. 2021;21:478. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 278. | Shen W, Yu L, Cong A, Yang S, Wang P, Han G, Gu B, Zhang W. Silencing lncRNA AFAP1-AS1 Inhibits the Progression of Esophageal Squamous Cell Carcinoma Cells via Regulating the miR-498/VEGFA Axis. Cancer Manag Res. 2020;12:6397-6409. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 279. | Zhang B, Chu W, Li Z, Zhang Y, Zhen Q, Lv B, Liu J, Lu C, Zhao X. Circ-ATIC Serves as a Sponge of miR-326 to Accelerate Esophageal Squamous Cell Carcinoma Progression by Targeting ID1. Biochem Genet. 2022;60:1585-1600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 280. | Du W, Xu P, Yin H. LncRNA NEAT1 regulates the growth, migration, and invasion of the human esophageal cancer cells via the miR-377/E2F3 axis. Acta Biochim Pol. 2022;69:731-736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 281. | He Y, Hua R, Yang Y, Li B, Guo X, Li Z. LncRNA JPX Promotes Esophageal Squamous Cell Carcinoma Progression by Targeting miR-516b-5p/VEGFA Axis. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 282. | Wang B, Hua P, Zhang L, Li J, Zhang Y. LncRNA-IUR up-regulates PTEN by sponging miR-21 to regulate cancer cell proliferation and apoptosis in esophageal squamous cell carcinoma. Esophagus. 2020;17:298-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 283. | Jiang Q, Wang H, Yuan D, Qian X, Ma X, Yan M, Xing W. Circular_0086414 induces SPARC like 1 (SPARCL1) production to inhibit esophageal cancer cell proliferation, invasion and glycolysis and induce cell apoptosis by sponging miR-1290. Bioengineered. 2022;13:12099-12114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 284. | Kano M, Seki N, Kikkawa N, Fujimura L, Hoshino I, Akutsu Y, Chiyomaru T, Enokida H, Nakagawa M, Matsubara H. miR-145, miR-133a and miR-133b: Tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer. 2010;127:2804-2814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 351] [Cited by in RCA: 393] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 285. | Li H, Jia J, Yang L, Chu J, Sheng J, Wang C, Meng W, Jia Z, Yin H, Wan J, He F. LncRNA MIR205HG Drives Esophageal Squamous Cell Carcinoma Progression by Regulating miR-214/SOX4 Axis. Onco Targets Ther. 2020;13:13097-13109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 286. | Jin YY, Chen QJ, Wei Y, Wang YL, Wang ZW, Xu K, He Y, Ma HB. Upregulation of microRNA-98 increases radiosensitivity in esophageal squamous cell carcinoma. J Radiat Res. 2016;57:468-476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 287. | Wang X, Fei X, Li C, Lu C, Chen H. MicroRNA-340-5p inhibits proliferation, migration and invasion of human esophageal squamous cell carcinoma cells by targeting inhibitors of DNA binding/differentiation 3. Disan Junyi Daxue Xuebao. 2021;42:1091-1097. [DOI] [Full Text] |
| 288. | Wang WW, Zhao ZH, Wang L, Li P, Chen KS, Zhang JY, Li WC, Jiang GZ, Li XN. MicroRNA-134 prevents the progression of esophageal squamous cell carcinoma via the PLXNA1-mediated MAPK signalling pathway. EBioMedicine. 2019;46:66-78. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 289. | Zong M, Liu Y, Zhang K, J Y, Chen L. The effects of miR-429 on cell migration and invasion by targeting Slug in esophageal squamous cell carcinoma. Pathol Res Pract. 2019;215:152526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 290. | Zhen C, Huang J, Lu J. MicroRNA-652 inhibits the biological characteristics of esophageal squamous cell carcinoma by directly targeting fibroblast growth factor receptor 1. Exp Ther Med. 2019;18:4473-4480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 291. | Shi Q, Wang Y, Mu Y, Wang X, Fan Q. MiR-433-3p Inhibits Proliferation and Invasion of Esophageal Squamous Cell Carcinoma by Targeting GRB2. Cell Physiol Biochem. 2018;46:2187-2196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 292. | Wang M, Wang L, Zhang M, Li X, Zhu Z, Wang H. MiR-214 inhibits the proliferation and invasion of esophageal squamous cell carcinoma cells by targeting CDC25B. Biomed Pharmacother. 2017;95:1678-1683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 293. | Wang X, Gu M, Ju Y, Zhou J. PIK3C3 Acts as a Tumor Suppressor in Esophageal Squamous Cell Carcinoma and Was Regulated by MiR-340-5p. Med Sci Monit. 2020;26:e920642. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 294. | Zhang J, Fa X, Zhang Q. MicroRNA206 exerts antioncogenic functions in esophageal squamous cell carcinoma by suppressing the cMet/AKT/mTOR pathway. Mol Med Rep. 2019;19:1491-1500. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 295. | Fan Y, Bian X, Qian P, Wen J, Yan P, Luo Y, Wu J, Zhang Q. miRNA30a3p inhibits metastasis and enhances radiosensitivity in esophageal carcinoma by targeting insulinlike growth factor 1 receptor. Mol Med Rep. 2019;20:81-94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 296. | Kong KL, Kwong DL, Chan TH, Law SY, Chen L, Li Y, Qin YR, Guan XY. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut. 2012;61:33-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 198] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 297. | Li P, Mao WM, Zheng ZG, Dong ZM, Ling ZQ. Down-regulation of PTEN expression modulated by dysregulated miR-21 contributes to the progression of esophageal cancer. Dig Dis Sci. 2013;58:3483-3493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 56] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 298. | Zhang J, Chen D, Liang S, Wang J, Liu C, Nie C, Shan Z, Wang L, Fan Q, Wang F. miR-106b promotes cell invasion and metastasis via PTEN mediated EMT in ESCC. Oncol Lett. 2018;15:4619-4626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 299. | Zhou J, Zhu J, Jiang G, Feng J, Wang Q. Downregulation of microRNA-4324 promotes the EMT of esophageal squamous-cell carcinoma cells via upregulating FAK. Onco Targets Ther. 2019;12:4595-4604. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 300. | Meng X, Chen X, Lu P, Ma W, Yue D, Song L, Fan Q. MicroRNA-202 inhibits tumor progression by targeting LAMA1 in esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 2016;473:821-827. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 301. | Li Y, Wu D, Wang P, Li X, Shi G. miR-195 Regulates Proliferation and Apoptosis through Inhibiting the mTOR/p70s6k Signaling Pathway by Targeting HMGA2 in Esophageal Carcinoma Cells. Dis Markers. 2017;2017:8317913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 302. | Takeshita N, Mori M, Kano M, Hoshino I, Akutsu Y, Hanari N, Yoneyama Y, Ikeda N, Isozaki Y, Maruyama T, Akanuma N, Miyazawa Y, Matsubara H. miR-203 inhibits the migration and invasion of esophageal squamous cell carcinoma by regulating LASP1. Int J Oncol. 2012;41:1653-1661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 56] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 303. | Wang K, Li J, Xiong G, He G, Guan X, Yang K, Bai Y. Negative regulation of lncRNA GAS5 by miR-196a inhibits esophageal squamous cell carcinoma growth. Biochem Biophys Res Commun. 2018;495:1151-1157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 304. | Tang J, Li Z, Zhu Q, Wen W, Wang J, Xu J, Wu W, Zhu Y, Xu H, Chen L. miR-204-5p regulates cell proliferation, invasion, and apoptosis by targeting IL-11 in esophageal squamous cell carcinoma. J Cell Physiol. 2020;235:3043-3055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 305. | Ren S, Tan X, Fu MZ, Ren S, Wu X, Chen T, Latham PS, Lin P, Man YG, Fu SW. Downregulation of miR-375 contributes to ERBB2-mediated VEGFA overexpression in esophageal cancer. J Cancer. 2021;12:7138-7146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 306. | Bian W, Li Y, Zhu H, Gao S, Niu R, Wang C, Zhang H, Qin X, Li S. miR-493 by regulating of c-Jun targets Wnt5a/PD-L1-inducing esophageal cancer cell development. Thorac Cancer. 2021;12:1579-1588. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 307. | Zhou Y, Xu R, Luo J, Li X, Zhong Y, Sun Z. Dysregulation of miR-204-5p/APLN axis affects malignant progression and cell stemness of esophageal cancer. Mutat Res. 2022;825:111791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 308. | Zhang BX, Yu T, Yu Z, Yang XG. MicroRNA-148a regulates the MAPK/ERK signaling pathway and suppresses the development of esophagus squamous cell carcinoma via targeting MAP3K9. Eur Rev Med Pharmacol Sci. 2019;23:6497-6504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 309. | Zong M, Feng W, Wan L, Yu X, Yu W. miR-203 affects esophageal cancer cell proliferation, apoptosis and invasion by targeting MAP3K1. Oncol Lett. 2020;20:751-757. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 310. | Nie Z, Weng W, Li J, Xu Y, Li Z. Down-regulation of miR-126 in esophageal carcinoma tissues and its inhibition effect on proliferation and migration of esophageal carcinoma cell line EC109. Zhongliu. 2015;35:55-64. [DOI] [Full Text] |
| 311. | Yao L, Zhang Y, Zhu Q, Li X, Zhu S, Gong L, Han X, Lan M, Li S, Zhang W, Li Y. Downregulation of microRNA-1 in esophageal squamous cell carcinoma correlates with an advanced clinical stage and its overexpression inhibits cell migration and invasion. Int J Mol Med. 2015;35:1033-1041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Motlagh B, Iran S-Editor: Yan JP L-Editor: A P-Editor: Cai YX