Published online Mar 15, 1997. doi: 10.3748/wjg.v3.i1.19
Revised: September 30, 1996
Accepted: January 31, 1997
Published online: March 15, 1997
AIM: To study the relationship between the point mutation of ras oncogenes and the prognosis of patients with gastric cancer.
METHODS: The point mutations at codon 12 and 61 of c-Ha-ras, at codon 12 and 13 of K-ras, and at codon 12 of N-ras were studied with PCR-RFLP in 88 formalin fixed and paraffin embedded specimens of gastric cancer.
RESULTS: It was found that the overall rate of point mutation of ras oncogenes was 18.2% and the positivity of the point mutation of ras oncogenes was related to the cancerous invasion of the serosa, the status of lymph node metastasis, the stage of cancer and the survival time after surgery.
CONCLUSION: The findings suggest that the determination of point mutations of ras oncogenes can be used to determine the prognosis of patients with gastric cancer.
- Citation: Fang DC, Luo YH, Lu R, Liu WW. Studies on the relationship between the point mutation of ras oncogenes and the prognosis of patients with gastric cancer. World J Gastroenterol 1997; 3(1): 19-21
- URL: https://www.wjgnet.com/1007-9327/full/v3/i1/19.htm
- DOI: https://dx.doi.org/10.3748/wjg.v3.i1.19
In recent years, the study of oncogenes has become one of the hot subjects in cancer research. It has been reported that the point mutation of ras oncogenes plays an important role in the development of certain cancers[1-3]. This study deals with the point mutation of c-Ha-ras, K-ras and N-ras in 88 formalin fixed and paraffin embedded specimens of gastric cancer. In addition, the relationship between the point mutation of the ras oncogenes and the prognosis of patients with gastric cancer was studied.
Eighty-eight specimens of gastric cancer were obtained during surgery in our hospital during the period from 1986 to 1991. All the specimens were fixed with 10% formalin and embedded in paraffin. Normal control tissue was collected from uninvolved gastric mucosa in all the patients. A 5 μm thick section was made, HE stained and examined under an optical microscope. Five similar sections were made from every specimen for DNA extraction.
For the extraction of DNA, 5 sections from each lesion were put into Eppendorf tubes and the paraffin was removed with xylene. After the tissue of the specimen was hydrolyzed and digested with proteinase K, the DNA content was extracted with the phenol chloroform method[4]. The samples of the purified DNA were diluted with TE buffer solution and their absorption value was determined on an ultraviolet ray spectrometer at the wavelength of 260 and 280 nm. The concentration and purity of nucleic acid were calculated.
The primers for the polymerase chain reactions (PCR) were synthesized by the Center of Science of Human Life of the Beijing University. The primer sets for ras mutation analysis were as follows:
P1A 5’-CAGGGCCCTCCTTGGCAGG-3’
P1B 5’-GTCGTAGGCGTCCACAAAATGG-3’
P2A 5’-ACGTGCCTGTTGGACATCCT-3’
P2B 5’-CACACAGGAAGCCCTCCCCG-3’
P3A 5’ ACTGAATATAAACTTGTGGTAGTTGGACCT-3’
P3B 5’-TCAAAGAATGGTCCTGGACC-3’
P4A 5’-AACTGGTGGTGGTTGGACCA-3’
P4B 5’-CTCTATGGTGGGATCATATTC-3’
P1A and P1B is the primer to analyze codon 12 of c-Ha-ras. The amplified fragment had a length of 170 bp. After the cleavage with restriction endonuclease Hpa II or Msp I, 3 fragments with the length of 66, 56 and 48 bp were obtained in the wild type and 2 fragments with the length of 122 and 48 bp in the mutative type; P2A and P2B are the primers to analyze the point mutation at codon 61 of c-Ha-ras. The amplified fragment had a length of 95 bp. After the cleavage with restriction endonuclease Bst N1, 3 fragments with the length of 15, 20 and 60 bp were obtained in the wild type and 2 fragments with the length of 20 and 75 bp were obtained in the mutative type. The primers for the analysis of the nonrestriction endonuclease point of the codon 12 and 13 of K-ras, P3A and P3B, were designed with the base mispairing method[5]. The amplified fragment was 157 bp in length. Endonuclease Bst N1 was used to analyze codon 12. The wild type was cleaved into 3 fragments of 114, 29 and 14 bp in length while the mutative type was cleaved into 2 fragments of 143 and 14 bp. Endonuclease Hph I was used to analyze codon 13. The fragment of the wild type was 157 bp in length while the mutative type was cleaved into 2 fragments of 114 and 43 bp; codon 12 of N-ras is also a nonrestriction endonuclease point and the primer was designed with the base mispairing method. The amplified fragment was 98 bp in length. The wild type was cleaved with Bst N1 into 2 fragments of 19 and 79 bp and the mutative type remained as a fragment of 98 bp.
PCR-RFLP was performed with the procedures as follows: DNA from 1 μg of tissue was dissolved in 50 μL PCR buffer solution containing 10 mmol/L Tris HCl, 50 mmol/ L KCl, 1.5 mmol/L MgCl2, 0.2 mmol/L dATP, dGTP, dCTP and dTTP, 25 pmol/L primer and 1.3 units Taq DNA polymerase. The cycle of denaturation at 95 °C for 70 s, renaturation at 57 °C for 70 s and extension at 72 °C for 70 s was repeated for 35 times, with the last elongation step lengthened to 10 min. 10 μL of the PCR product was cleaved with the corresponding endonuclease (purchased from the BRL Company of United States), underwent 8% polyacrylamide gel electrophoresis and was stained with ethidium bromide. Eventually the product was observed under ultraviolet light and photographed.
All 88 cases were followed up and the longest follow up lasted 13 years in order that the outcome of each case was obtained.
No point mutation (PM) of ras genes was found in specimens of normal gastric mucosa and 16 cases of the 88 with gastric cancer showed a PM rate of 18.2% (Figure 1). Among the 16 cases, PM at codon 12 of c-Ha-ras occurred in 12 cases (13.6%) and PM at codon 61 in 3 (3.4%); PM at codon 12 of K-ras in 5 (5.7%) and PM at codon 13 in none; and PM at codon 12 of N-ras in 1 (1.1%). PM at only 1 codon in the specimen was found in 11 cases and PM at 2 codons in the specimen was found in 5 cases. No case showed PM at 3 codons in the specimen simultaneously.
The 88 cases were divided into groups according to the pathological types of cancer, the size of cancer, the serosal invasion, the lymph node metastasis and the clinical staging of the case. It was found that the mutation rate was significantly higher in the group with serosal invasion than in that without (P < 0.01), in the group with lymph node metastasis than in that without it (P < 0.05) and in the group with clinical stage III and IV than in that with clinical stage I and II (P < 0.01) (Table 1).
| n | Point mutationnegative | Point mutationpositive (%) | |
| Histological types | |||
| Gland adenocarcinoma | 20 | 18 | 2 (10.0) |
| Low differentiated carcinoma | 29 | 22 | 7 (24.1) |
| Signet ring cell carcinoma | 23 | 18 | 5 (21.7) |
| Mucinous carcinoma | 16 | 14 | 2 (12.5) |
| Size | |||
| < 5 cm | 47 | 41 | 6 (12.8) |
| > 5 cm | 41 | 31 | 10 (24.4) |
| Serosal invasion | |||
| Without serosal invasion | 40 | 38 | 2 (5.0) |
| With serosal invasion | 48 | 34 | 14 (29.2)b |
| Lymph node metastases | |||
| Without lymph node metastases | 35 | 33 | 2 (5.7) |
| With lymph node metastases | 53 | 39 | 14 (29.2)a |
| Staging | |||
| Staging I and II | 43 | 42 | 1 (2.3) |
| Staging III and IV | 45 | 30 | 15 (33.3)c |
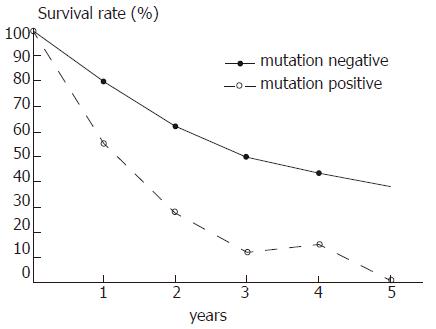
Survival analysis revealed a strong association between ras gene mutations of the tumor and patient survival time after surgery (Figure 1). The 5 year survival rate in the 72 cases without PM was 34.7% (25 cases) and compared with those with PM (0/16) the difference was significant (P < 0.01).
This study shows that the activation of ras oncogenes is the molecular basis of the development of certain cancers. PM is one of the important patterns of ras gene activation. Therefore, the detection of PM becomes one of the hot topics in cancer research.
PCR-RFLP, a technique developed in recent years, greatly simplifies the procedures to detect PM[6]. It can be used to detect the PM in the DNA extracted from fresh specimens of gastric cancer as well as in the DNA from paraffin embedded specimens. The simplified technique determining PM makes it possible to determine the prognosis for patients with gastric cancer.
PM at the codons 12, 13 and 61 of ras oncogenes can enable the cells to possess the capacity of transformation[7]. However, points for the restriction endonuclease to cleave at the codons 12 and 13 for K-ras and codon 12 of N-ras and PCR-RFLP can not be performed. The authors, according to the 3’ end region of the primers, introduced a mispaired base not affecting the amplification of PCR[5]. After the introduction of a mispaired base, a point to cleave is constituted for Bst N1. Thus, it is possible to study the PM at codon 12 and 13 of K-ras and codon 12 of N-ras.
It was reported that the mutation rate of ras genes was 90% in pancreatic cancer, 40% in colorectal cancer and below 10% in gastric cancer[8,9]. However, Deng et al[6] found the mutation rate at codon 12 of Ha-ras was as high as 41% in gastric cancer. In our study, 88 paraffin embedded specimens of gastric cancer were studied with PCR-RFLP and the mutation rate of ras genes was found to be 18.2%. Japanese authors reported that PM of K-ras predominated in gastric cancer[10,11]. The difference of the PM rate between China and Japan might result from different geographical and environmental conditions, race and detection methods.
It was also found that the PM of ras genes was related to some biological factors of gastric cancer. Different types of cancers and their size play an insignificant role in the PM rate of ras oncogenes while those cases with serosal invasion and lymph node metastasis showed a far higher PM rate than those without them.
The cases of gastric cancer in clinical stage III and IV showed a significantly higher PM rate than those in the clinical stage I and II.This finding is consistent with the result reported by the authors who obtained it through the study on the molecular mechanism of cancer infiltration and metastasis[7,11], which indicates the poor prognosis of the patients. Our results showed that the 5 year survival rate of patients without PM of ras genes was far higher than those with PM. Thus the determination of the PM of ras genes may be helpful for the prognosis of patients with gastric cancer and for the prescription of an efficient therapeutic plan for them.
The concept that cancer is a disease of genes has been widely accepted. Our study enables us to further understand the role played by the PM of genes in the development of certain cancers. On the basis of the data of our study that most cases of gastric cancer did not show any evidence of PM of ras oncogenes, it seems obvious that PM of ras oncogenes is not the only factor to induce gastric cancer and there must be some other factors which need to be further investigated and explored.
| 1. | Koh EH, Chung HC, Lee KB, Han EK, Oh SH, Min JS, Choi EM, Youn JK, Kim BS. Point mutation at codon 12 of the c-Ha-ras gene in human gastric cancers. J Korean Med Sci. 1992;7:110-115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 2. | Stemmermann G, Heffelfinger SC, Noffsinger A, Hui YZ, Miller MA, Fenoglio-Preiser CM. The molecular biology of esophageal and gastric cancer and their precursors: oncogenes, tumor suppressor genes, and growth factors. Hum Pathol. 1994;25:968-981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 80] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 3. | Kato M, Ito Y, Kobayashi S, Isono K. Detection of DCC and Ki-ras gene alterations in colorectal carcinoma tissue as prognostic markers for liver metastatic recurrence. Cancer. 1996;77:1729-1735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 4. | Rogers BB, Alpert LC, Hine EA, Buffone GJ. Analysis of DNA in fresh and fixed tissue by the polymerase chain reaction. Am J Pathol. 1990;136:541-548. [PubMed] |
| 5. | Jiang W, Kahn SM, Guillem JG, Lu SH, Weinstein IB. Rapid detection of ras oncogenes in human tumors: applications to colon, esophageal, and gastric cancer. Oncogene. 1989;4:923-928. [PubMed] |
| 6. | Deng GR, Liu XH, Wang JR. Correlation of mutations of oncogene C-Ha-ras at codon 12 with metastasis and survival of gastric cancer patients. Oncogene Res. 1991;6:33-38. [PubMed] |
| 7. | Ranzani GN, Pellegata NS, Previderè C, Saragoni A, Vio A, Maltoni M, Amadori D. Heterogeneous protooncogene amplification correlates with tumor progression and presence of metastases in gastric cancer patients. Cancer Res. 1990;50:7811-7814. [PubMed] |
| 8. | Nanus DM, Kelsen DP, Mentle IR, Altorki N, Albino AP. Infrequent point mutations of ras oncogenes in gastric cancers. Gastroenterology. 1990;98:955-960. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 51] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 9. | Wright PA, Williams GT. Molecular biology and gastric carcinoma. Gut. 1993;34:145-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 46] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 10. | Kihana T, Tsuda H, Hirota T, Shimosato Y, Sakamoto H, Terada M, Hirohashi S. Point mutation of c-Ki-ras oncogene in gastric adenoma and adenocarcinoma with tubular differentiation. Jpn J Cancer Res. 1991;82:308-314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 58] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 11. | Miki H, Ohmori M, Perantoni AO, Enomoto T. K-ras activation in gastric epithelial tumors in Japanese. Cancer Lett. 1991;58:107-113. [PubMed] |
| 12. | Neuman WL, Wasylyshyn ML, Jacoby R, Erroi F, Angriman I, Montag A, Brasitus T, Michelassi F, Westbrook CA. Evidence for a common molecular pathogenesis in colorectal, gastric, and pancreatic cancer. Genes Chromosomes Cancer. 1991;3:468-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
Original title:
S- Editor: Yang ZD L- Editor: Ma JY E- Editor: Liu WX