Published online Jun 7, 2023. doi: 10.3748/wjg.v29.i21.3302
Peer-review started: January 20, 2023
First decision: February 15, 2023
Revised: April 6, 2023
Accepted: May 4, 2023
Article in press: May 4, 2023
Published online: June 7, 2023
Processing time: 132 Days and 6.9 Hours
Peutz-Jeghers syndrome (PJS) is an autosomal dominant genetic disease with skin mucosal pigment spots and gastrointestinal (GI) multiple hamartoma polyps as clinical characteristics. At present, it is considered that the germline mutation of STK11 gene is the genetic cause of PJS. However, not all PJS patients can be detected STK11 germline mutations. The specific clinical characteristics of these PJS patients without STK11 mutation is an interesting clinical question. Or, like wild type GI stromal tumor, whether these PJS without STK11 mutation are also called PJS is worth discussing. Therefore, we designed the study to understand the clinical characteristics of these PJS patients without STK11 mutation.
To investigates whether PJS patients with known STK11 mutations have a more severe spectrum of clinical phenotypes compared to those without.
A total of 92 patients with PJS admitted to the Air Force Medical Center from 2010 to 2022 were randomly selected for study. Genomic DNA samples were extracted from peripheral blood samples, and pathogenic germline mutations of STK11 were detected by high-throughput next-generation gene sequencing. Clinical-pathologic manifestations of patients with and without STK11/LKB1 mutations were compared.
STK11 germline mutations were observed in 73 patients with PJS. Among 19 patients with no detectable STK11 mutations, six had no pathogenic germline mutations of other genes, while 13 had other genetic mutations. Compared with PJS patients with STK11 mutations, those without tended to be older at the age of initial treatment, age of first intussusception and age of initial surgery. They also had a lower number of total hospitalizations relating to intussusception or intestinal obstruction, and a lower load of small intestine polyps.
PJS patients without STK11 mutations might have less severe clinical-pathologic manifestations than those with.
Core Tip: Peutz-Jeghers syndrome (PJS) is an autosomal dominant genetic disease with skin mucosal pigment spots and gastrointestinal (GI) multiple hamartoma polyps as clinical characteristics. At present, it is considered that the germline mutation of STK11 gene is the genetic cause of PJS. However, not all PJS patients can be detected STK11 germline mutations. The specific clinical characteristics of these PJS patients without STK11 mutation is an interesting clinical question. Or, like wild type GI stromal tumor, whether these PJS without STK11 mutation are also called PJS is worth discussing. Therefore, we designed the study to understand the clinical characteristics of these PJS patients without STK11 mutation. Final results found that PJS patients without STK11 mutations might have less severe clinical-pathologic manifestations than those with.
- Citation: Jiang LX, Chen YR, Xu ZX, Zhang YH, Zhang Z, Yu PF, Dong ZW, Yang HR, Gu GL. Peutz-Jeghers syndrome without STK11 mutation may correlate with less severe clinical manifestations in Chinese patients. World J Gastroenterol 2023; 29(21): 3302-3317
- URL: https://www.wjgnet.com/1007-9327/full/v29/i21/3302.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i21.3302
Peutz-Jeghers syndrome (PJS) is an autosomal dominant disorder which is mainly characterized by mucocutaneous pigmentation and hamartomatous polyps of the gastrointestinal (GI) tract[1,2]. While PJS is rare, with an estimated prevalence of 1:200000 births[3], the continuous growth of multiple GI polyps predisposes patients to serious complications including intussusception, intestinal obstruction, GI bleeding and malignancies. PJS patients also have a markedly increased risk of developing various neoplasms in extraintestinal sites such as the lungs, liver and breast[4].
Germline mutations in the STK11 gene (also named LKB1), which is located on Chromosome 19p13.3 and encodes a serine/threonine protein kinase[5], have been identified as the major cause of PJS[6,7]. STK11 is a tumor suppressor gene comprised of 433 amino acids with nine coding exons and one non-coding exon[8]. Depending on the screening method, STK11 variants can be detected in over 80%-90% of PJS cases[8,9]. Previous studies have largely focused on exploring the phenotypic landscape of STK11 variants based on their type or location. Due to the rare nature of PJS, very few studies have attempted to examine the correlations between STK11 mutations and overall severity of PJS phenotype in terms of the earlier onset of GI pathology arising from the polyps, such as intussusception or earlier onset malignancy. Understanding the phenotypic differences between PJS patients with and without STK11 variants could facilitate more personalized care for PJS patients and their families via appropriate counseling, risk stratification and targeted cancer screening[10].
A total of 92 PJS patients admitted to the Air Force Medical Center between February 2010 and February 2022 were randomly selected for inclusion in the study, and their peripheral venous blood was collected for high-throughput next-generation gene sequencing (NGS). 73 cases in which STK11 gene mutations were detected were named mutant-type, and 19 cases in which no STK11 gene mutations were detected were named wild-type. In this retrospective study, we aimed to investigate the differences in clinical phenotypes between wild-type and mutant-type STK11 gene, and provide a theoretical basis for a more precise medical monitoring and follow-up strategy for different types of PJS patients.
A total of 92 patients with PJS admitted to the Air Force Medical Center, PLA between February 2010 and February 2022 were randomly selected for inclusion in the study, and all patients met the diagnostic criteria for PJS recommended by the NCCN guidelines[11]. All enrolled patients and their guardians were aware of the purpose and process of the study, and had signed informed consent agreements. All data and information collection for this study followed the ethical principles of the Universal Declaration on the Human Genome and Human Rights, Declaration of Helsinki and Statement of the Human Genome Organisation Ethics Committee on DNA Sampling, Control and Access.
Inclusion criteria: All enrolled patients met the clinical diagnostic criteria of PJS[12-14] (in accordance with any of the following): (1) Two or more histologically confirmed PJS polyps; (2) Any number of PJS polyps in an individual with a family history of PJS in close relative(s); (3) Characteristic mucocutaneous pigmentation in an individual with a family history of PJS in close relative(s); and (4) Any number of PJS polyps in an individual with characteristic mucocutaneous pigmentation. Peripheral venous blood was retained and genomic DNA extracted, and the sequence of the coding region of the STK11 gene was detected using polymerase chain reaction (PCR) amplification and NGS sequencing.
Exclusion criteria: Patients who could not meet both of the above two inclusion criteria, could not provide experimental specimens or did not agree to participate in this study.
Observational index: The general information, diagnosis and treatment history, pathology, times of examination and other clinical data of the 92 enrolled PJS patients were collected for statistical analysis. The observed indices were as follows: (1) General patient information: Origin, gender, personal marital status, family history and ABO/RH blood groups; (2) History of diagnosis and treatment: Age of initial treatment, age of mucocutaneous pigmentation appearance, order of mucocutaneous pigmentation appearance, time interval from mucocutaneous pigmentation appearance to abdominal symptoms (abdominal pain, intestinal obstruction, GI bleeding, etc.), location of GI polyps, load and maximum diameter of GI polyps, pathology of polyps, carcinogenesis, total hospitalizations, number of operations and final age of follow-up; (3) Examinations: Endoscopic examinations and times of GI imaging examinations; and (4) Other: Comorbidities.
Genetic sequencing: EDTA anticoagulation tubes were used to extract 8 mL of peripheral venous blood from PJS patients. Leukocytes were isolated and DNA extracted, and the extracted DNA was interrupted by ultrasound using a Covaris M220 instrument to build a DNA library. The DNA library was then purified and hybridised by the probe library, which would bind specifically to the target DNA fragment through the principle of complementary binding of nucleic acid sequences. Magnetic beads conjugated with streptavidin were mixed with the hybridisation solution, and the streptavidin was tightly bound to the biotin. The captured exon target fragment was indirectly bound by the probe to the beads, which were adsorbed by a magnet, and the supernatant was discarded. The unbound DNA fragments were washed off and the desired DNA library eluted from the beads with the eluent. The eluted DNA library was then amplified using a PCR instrument. Lastly, NGS sequencing was used to detect full exons and associated single nucleotide polymorphism (SNP) and microsatellite instability sites for previously reported genes associated with hereditary GI tract tumors, including STK11 (Table 1).
| No. | Genes |
| 1 | AKT1 |
| 2 | BRAF |
| 3 | CYP2D6 |
| 4 | GALNT12 |
| 5 | MET |
| 6 | NRAS |
| 7 | POLD1 |
| 8 | SDHC |
| 9 | UGT1A1 |
| 10 | APC |
| 11 | BRCA1 |
| 12 | DPYD |
| 13 | GREM1 |
| 14 | MLH1 |
| 15 | PDGFRA |
| 16 | POLE |
| 17 | SDHD |
| 18 | ATM |
| 19 | BRCA2 |
| 20 | EGFR |
| 21 | HRAS |
| 22 | MSH2 |
| 23 | PIK3CA |
| 24 | PTCH1 |
| 25 | SMAD4 |
| 26 | BLM |
| 27 | CDH1 |
| 28 | EPCAM |
| 29 | KIT |
| 30 | MSH6 |
| 31 | PMS1 |
| 32 | PTEN |
| 33 | STK11 |
| 34 | BMPRA |
| 35 | CHEK2 |
| 36 | ERBB2 |
| 37 | KRAS |
| 38 | MUTYH |
| 39 | PMS2 |
| 40 | SDHB |
| 41 | TP53 |
Statistical analysis: Statistical analysis was carried out using the SPSS 26.0 software package: (1) Cases and proportion of qualitative data were presented as percentages (%), and comparisons between groups were conducted using the chi-square test or Fisher's exact test; and (2) The t-test was used for quantitative data that matched the normal distribution with equal variance, the t-test was used for those with unequal variance and the rank sum test was used for skewed data; P < 0.05 was considered statistically significant.
The 73 of 92 patients with PJS (79.3%) in this group had STK11 gene mutations, of which 47 had STK11 gene mutations in combination with other mutations. 19 of the 92 PJS patients (20.7%) had no STK11 gene mutations, of which 6 (6.6%) had no other gene mutations and 13 (14.1%) had other gene mutations in the 41 genes group. By comparing the Human Gene Mutation Database (HGMD) (http://www.hgmd.cf.ac.uk/ac/index.php), dbSNP database (https://www.ncbi.nlm.nih.gov/snp/) and COSMIC database (https://cancer.sanger.ac.uk/cosmic/), a total of 582 STK11 gene mutant sites were included in HGMD as of March 1, 2022. We identified 21 new mutant sites in other genes (Table 2) and 26 new STK11 gene mutant sites (Table 3).
| Sample | Gene | Description | HGVSc | Mutation type |
| 1 | AKT1 | p.E135G | c.404A>G | Missense variant |
| 2 | APC | p.A41T | c.121G>A | Missense variant |
| 3 | APC | p.C417G | c.1249T>G | Missense variant |
| 4 | ATM | p.L27501 | c.8249T>G | Stop gained |
| 5 | ATM | p.A84S | c.250G>T | Missense variant |
| 6 | ATM | p.I1332M | c.3996T>G | Missense variant |
| 7 | BLM | p.E1035G | c.3104A>G | Missense variant |
| 8 | BRCA2 | p.D635E | c.1905T>A | Missense variant |
| 9 | BRCA2 | p.T1346N | c.4037C>A | Missense variant |
| 10 | CHEK2 | c.908+16T>G | c.908+16T>G | Intron variants |
| 11 | CDH1 | c.47G>A1 | c.47G>A1 | 3 prime UTR variant |
| 12 | CDH1 | p.S145Y | c.434C>A | Missense variant |
| 13 | CDH1 | p.883Yext?1 | c.2649G>C | Stop lost |
| 14 | GALNT12 | c.-6G>T | c.-6G>T | Upstream genetic variant |
| 15 | KIT | p.M289I | c.867G>C | Missense variant |
| 16 | MLH1 | p.T451R | c.1352C>G | Missense variant |
| 17 | PMS1 | p.D405E | c.1215T>A | Missense variant |
| 18 | POLE | p.R1556G | c.4666C>G | Missense variant |
| 19 | POLD1 | p.K486del | c.1456_1458del | Conservative inframe deletion |
| 20 | SDHC | p.L106V | c.316C>G | Missense variant |
| 21 | SMAD4 | p.A309V | c.926C>T | Missense variant |
| Sample | Mutation_type | Description | HGVSc | dbSNP RS | COSM_ID | Classification |
| 1 | Stop gained | p.Y601 | c.180C>G | / | COSM20874 | P |
| 2 | Splice acceptor variant | c.921-1G>A | c.921-1G>A | / | COSM49008 | LP |
| 3 | Splice acceptor variant | c.921-1G>C | c.921-1G>C | rs398123406 | / | P |
| 4 | Stop gained | p.K841 | c.250A>T | rs137853076 | COSM3388586; COSM3388585 | P |
| 5 | Splice acceptor variant | c.921-1G>C | c.921-1G>C | rs398123406 | / | P |
| 6 | Stop gained | p.Q1231 | c.367C>T | / | COSM5224269; COSM380443 | P |
| 7 | Missense variant | p.W239C | c.717G>T | / | COSM333593; COSM4278104 | LP |
| 8 | Missense variant | p.R297S | c.891G>T | rs730881984 | / | P |
| 9 | Stop gained | p.Q1001 | c.298C>T | / | / | LP |
| 10 | Stop gained | p.K841 | c.250A>T | rs137853076 | COSM3388586; COSM3388585 | P |
| 11 | Missense variant | p.R409W | c.1225C>T | rs368466538 | COSM25854 | VUS |
| 12 | Stop gained | p.Q1121 | c.334C>T | / | COSM3528680; COSM3528681 | LP |
| 13 | Missense variant | p.D176N | c.526G>A | rs730881979 | COSM4827691; COSM4827690 | P |
| 14 | Stop gained | p.K841 | c.250A>T | rs137853076 | COSM3388586; COSM3388585 | P |
| 15 | Missense variant | p.R304W | c.910C>T | rs786201090 | COSM29468 | LP |
| 16 | Stop gained | p.E1201 | c.358G>T | rs775595174 | COSM20875 | P |
| 17 | Stop gained | p.K841 | c.250A>T | rs137853076 | COSM3388586; COSM3388585 | P |
| 18 | Conservative inframe deletion | p.Y60fs | c.179dup | rs876661012 | COSM5219400; COSM1480565 | P |
| 19 | Stop gained | p.R861 | c.256C>T | / | COSM4767773; COSM4767772 | P |
| 20 | Stop gained | p.Q1701 | c.508C>T | rs121913323 | COSM20943 | LP |
| 21 | Stop gained | p.Q1701 | c.508C>T | rs121913323 | COSM20943 | LP |
| 22 | Missense variant | p.S240W | c.719C>G | rs730881976 | / | VUS |
| 23 | Splice acceptor variant | c.921-2A>G | c.921-2A>G | / | / | LP |
| 24 | Splice acceptor variant | c.921-1G>C | c.921-1G>C | rs398123406 | / | P |
| 25 | Conservative inframe deletion | p.P281fs | c.842del | rs121913321 | COSM4336438; COSM20871 | P |
| 26 | Splice acceptor variant | p.L245F | c.733C>T | / | COSM1523960; COSM4278108 | VUS |
| 27 | Stop gained | p.Q1371 | c.409C>T | rs730881970 | COSM48901 | P |
| 28 | Stop gained | p.Q1371 | c.409C>T | rs730881970 | COSM48901 | P |
| 29 | Stop gained | p.Q1231 | c.367C>T | / | COSM5224269; COSM380443 | P |
| 30 | Missense variant | p.D194N | c.580G>A | rs121913315 | COSM25847 | VUS |
| 31 | 3_prime_UTR_variant | c.201G>A1 | c.201G>A1 | rs528679025 | / | P |
| 32 | Splice acceptor variant | c.598-2A>G | c.598-2A>G | / | / | LP |
| 33 | Stop gained | p.Q1591 | c.475C>T | / | COSM5002233; COSM27316 | LP |
| 34 | Gene fusion | STK11-MIDN | / | / | / | LP |
| 35 | Conservative inframe deletion | p.D53fs | c.157del | / | COSM27282; COSM6048514 | VUS |
| 36 | Missense variant | p.D194N | c.580G>A | rs121913315 | COSM25847 | LP |
| 37 | Missense variant | p.P179Q | c.536C>A | / | COSM4822602; COSM4822601 | LP |
| 38 | Stop gained | p.W3081 | c.924G>A | / | / | LP |
| 39 | Stop gained | p.E651 | c.193G>T | / | COSM20876 | P |
| 40 | Splice acceptor variant | c.920+1G>C | c.920+1G>C | / | COSM4412472; COSM4412473 | LP |
| 41 | Conservative inframe deletion | p.C134fs | c.402_403del | rs587782424 | COSM5508976; COSM5508975 | P |
| 42 | Nonsense variant | p.Q220X | c.658C>T | / | COSM13480; COSM4278102 | P |
| 43 | Nonsense variant | p.Y60X | c.180del | / | COSM20874; COSM27322; COSM48900; COSM5490514 | P |
| 44 | Missense variant | p.D194N | c.580G>A | rs121913315 | / | LP |
| 45 | Missense variant | p.R297K | c.890G>A | / | COSM401786; COSM6149636 | LP |
| 46 | Splice acceptor variant | / | c.863-2A>G | / | / | LP |
| 47 | Nonsense variant | p.Q137X | c.409C>T | rs730881970 | / | P |
| 48 | Splice acceptor variant | c.735-6_735-2del | c.735-6_735-2del | rs759090799 | / | VUS |
| Conservative inframe deletion2 | p.L183fs | c.548del | / | / | LP | |
| 49 | 3 prime UTR variant | c.201G>A1 | c.201G>A1 | rs528679025 | / | LP |
| Conservative inframe deletion2 | p.C158fs | c.472del | / | / | VUS | |
| 50 | Stop gained2 | p.K811 | c.241A>T | / | / | LP |
| 51 | Missense variant2 | p.R304P | c.911G>C | / | / | P |
| 52 | Missense variant2 | p.R297K | c.890G>A | / | / | LP |
| 53 | Conservative inframe deletion2 | p.E145fs | c.426_448del | / | / | LP |
| 54 | Stop gained2 | p.Y2721 | c.816C>A | / | / | LP |
| 55 | Stop gained2 | p.Q1001 | c.298C>T | / | / | LP |
| 56 | Conservative inframe deletion2 | p.K64fs | c.190_191del | / | / | LP |
| 57 | Splice acceptor variant2 | c.921-2A>G | c.921-2A>G | / | / | LP |
| 58 | Stop gained2 | p.Y2921 | c.876C>G | / | / | LP |
| 59 | Conservative inframe deletion2 | p.T212fs | c.634del | / | / | LP |
| 60 | Stop gained2 | p.K971 | c.289A>T | / | / | P |
| 61 | Missense variant2 | p.H154P | c.461A>C | / | / | VUS |
| 62 | Missense variant2 | p.A153P | c.457G>C | / | / | VUS |
| 63 | Missense variant2 | p.L140P | c.419T>C | / | / | VUS |
| 64 | Conservative inframe deletion2 | p.F264fs | c.792del | / | / | P |
| 65 | Stop gained2 | p.W3081 | c.924G>A | / | / | P |
| 66 | Splice acceptor variant2 | c.598-2A>G | c.598-2A>G | / | / | LP |
| 67 | Conservative inframe deletion2 | p.F157fs | c.471_472del | / | / | LP |
| 68 | Conservative inframe deletion2 | p.S193fs | c.577_578del | / | / | LP |
| 69 | Splice acceptor variant2 | c.734+1G>A | c.734+1G>A | / | / | LP |
| 70 | Missense variant2 | p.L290H | c.869T>A | / | COSM20944; COSM25847; COSM4278092 | VUS |
| 71 | Nonsense variant2 | p.Y60X | c.179dup | / | / | P |
| 72 | Conservative inframe deletion2 | p.L282Afs | c.842dup | / | / | P |
| 73 | Conservative inframe deletion2 | p.V77Rfs | c.228dup | / | COSM48901 | LP |
Gene detection results and pathogenicity analysis of PJS with mutant-type STK11:STK11 gene detection results and pathogenicity analysis. STK11 gene mutations were detected in 73 PJS patients in our group. By making comparisons with the dbSNP and COSMIC databases, 49 PJS-related gene mutations were included and 26 STK11 gene mutation sites were newly identified by sequencing in this group (Table 3). Types of STK11 gene mutations included premature termination codons (24), frame shift mutations (14), splice site variants (13), missense mutations (17), nonsense mutations (4), mutations in the 3' untranslated region (2) and gene fusion (1).
The pathogenicity of STK11 gene mutations was determined by comparing the HGMD and ClinVar gene mutations in the disease database, and following the corresponding grading criteria (Table 3). By making conservative predictions of amino acid sequences, we made determinations about the pathogenicity of STK11 gene mutations according to the corresponding grading criteria (Table 3): Premature termination codons, frame shift mutations, splice site variants, missense mutations, nonsense mutations and mutations in the 3' untranslated region were associated with pathogenicity. Premature termination codons accounted for 58.3% (14/24) that were clearly pathogenic and 41.7% (10/24) that were probably pathogenic. Frameshift mutations accounted for 35.7% (5/14) that were clearly pathogenic, 50.0% (7/14) that were probably pathogenic and 14.3% (2/14) that were of uncertain significance. Splice site mutations accounted for 23.1% (3/13) that were clearly pathogenic, 61.5% (8/13) that were probably pathogenic and 15.4% (2/13) that were of uncertain significance. Missense mutations accounted for 17.6% (3/17) that were clearly pathogenic, 41.2% (7/17) that were probably pathogenic and 41.2% (7/17) that were of uncertain significance. 3' untranslated region mutations accounted for 50.0% (1/2) that were clearly pathogenic and 50.0% (1/2) that were probably pathogenic. All four nonsense mutations were clearly pathogenic (100.0%).
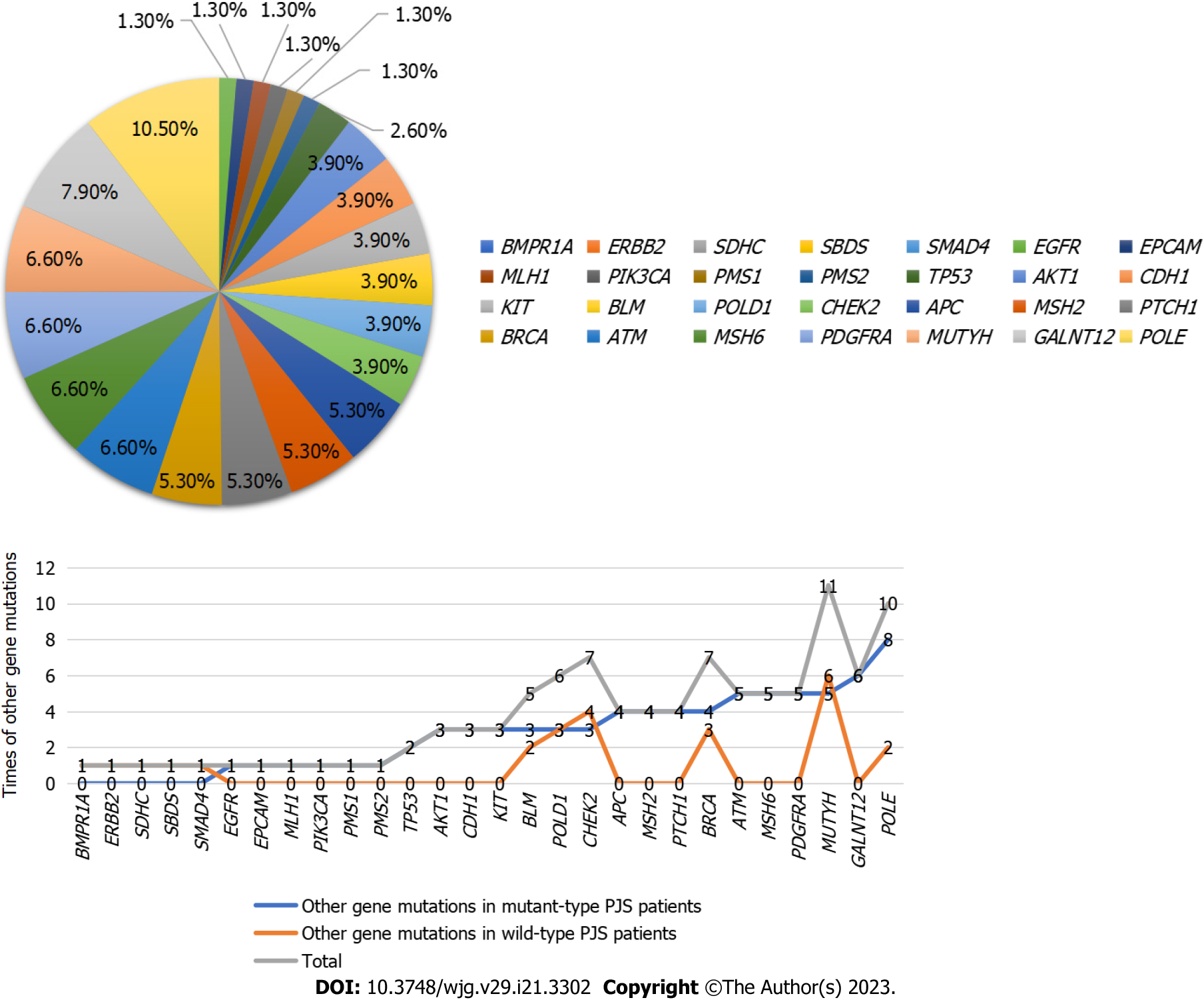
Other gene detection results and pathogenicity analysis in PJS patients with mutant-type STK11: Among the 73 PJS patients with mutant-type STK11, 47 were combined with other gene mutations: Combined with AKT1 mutation in 3 cases, APC mutation in 4 cases, ATM mutation in 5 cases, BLM mutation in 3 cases, BRCA2 mutation in 4 cases, CDH1 mutation in 3 cases, CHEK2 mutation in 3 cases, EGFR mutation in 1 case, EPCAM mutation in 1 case, GALNT12 mutation in 6 cases, KIT mutation in 3 cases, MUTYH mutation in 5 cases, MSH6 mutation in 5 cases, MSH2 mutation in 4 cases, MLH1 mutation in 1 case, PDGFRA mutation in 5 cases, PIK3CA mutation in 1 case, PMS1 mutation in 1 case, PMS2 mutation in 1 case, POLD1 mutation in 3 cases, PTCH1 mutation in 4 cases, POLE mutation in 8 cases and TP53 mutation in 2 cases.
Among the other gene mutations in PJS patients with mutant-type STK11, there were 52 missense mutations (68.4%), 5 intron mutations (6.6%), 11 mutations in the 3' untranslated region (14.5%), 3 splice site mutations (4.0%), 1 premature termination codon (1.3%), 1 frameshift deletion (1.3%), 1 mutation in the 5' untranslated region (1.3%), 1 upstream gene mutation (1.3%) and 1 case (1.3%) of stop gain. All mutations were of uncertain significance (100.0%) (Figure 1).
Gene detection results and pathogenicity analysis in PJS patients with wild-type STK11: Wild-type PJS gene mutations included BLM mutation in 2 cases, BMPR1A mutation in 1 case, POLD1 mutation in 3 cases, CHEK2 mutation in 4 cases, MUTYH mutation in 6 cases, SDHC mutation in 1 case, POLE mutation in 2 cases, BRCA mutation in 3 cases, APC mutation in 2 cases, CDH1 mutation in 1 case, ATM mutation in 3 cases, ERBB2 mutation in 1 case, SMAD4 mutation in 1 case and SBDS mutation in 1 case.
Among the other gene mutations in wild-type PJS patients, there were 23 missense mutations (74.2%), 5 intron mutations (16.1%), 1 splice site mutation (3.2%), 1 frameshift deletion (3.2%) and 1 mutation in the 3' untranslated region (3.2%). All mutations were of uncertain significance (Table 4).
| Sample | Gene | Description | HGVSc | Mutation_type | dbSNP RS | COSM_ID | Classification |
| 1 | BLM | p.I947V | c.2839A>G | Missense mutation | rs189925962 | NA | VUS |
| BMPR1A | p.A13T | c.37G>A | Missense mutation | rs200115604 | NA | VUS | |
| POLD1 | p.K486del | c.1456_1458del | Frameshift deletion | NA | NA | VUS | |
| 2 | CHEK2 | p.S252N | c.755G>A | Missense mutation | rs587781379 | COSM6004987; COSM6004988 | VUS |
| MUTYH | c.36+11C>T | c.36+11C>T | Intron mutations | rs2275602 | COSN17145138 | VUS | |
| SDHC | p.L106V | c.316C>G | Missense mutation | NA | NA | VUS | |
| 3 | CHEK2 | p.R181H | c.542G>A | Missense mutation | rs121908701 | NA | VUS |
| MUTYH | c.37_39del1 | c.37_39del1 | Mutation in the 3' untranslated region | rs373507005 | NA | VUS | |
| MUTYH | c.36+11C>T | c.36+11C>T | Intron mutations | rs2275602 | COSN17145138 | VUS | |
| MUTYH | p.G25D | c.74G>A | Missense mutation | rs75321043 | NA | VUS | |
| MUTYH | p.P18L | c.53C>T | Missense mutation | rs79777494 | NA | VUS | |
| POLE | c.3378+10A>G | c.3378+10A>G | Intron mutations | rs193075152 | NA | VUS | |
| 4 | BLM | p.M348I | c.1044G>A | Missense mutation | rs184657475 | COSM1580597 | VUS |
| 5 | BRCA1 | p.P1192L | c.3575C>T | Missense mutation | NA | COSM4991001; COSM4991000 | VUS |
| BRCA2 | p.F3328C | c.9983T>G | Missense mutation | rs770826575 | NA | VUS | |
| CHEK2 | p.H371Y | c.1111C>T | Missense mutation | rs531398630 | COSM4002125 | VUS | |
| 6 | APC | p.I1524R | c.4571T>G | Missense mutation | rs200803739 | NA | VUS |
| 7 | CDH1 | p.S145Y | c.434C>A | Missense mutation | NA | NA | VUS |
| POLE | c.3378+10A>G | c.3378+10A>G | Intron mutations | rs193075152 | NA | VUS | |
| 8 | ATM | c.3154-5C>T | c.3154-5C>T | Intron mutations | rs55719759 | NA | VUS |
| CHEK2 | p.S252N | c.755G>A | Missense mutation | rs587781379 | COSM6004987; COSM6004988 | VUS | |
| ERBB2 | p.V1253M | c.3757G>A | Missense mutation | rs36085723 | NA | VUS | |
| 9 | ATM | p.I1332M | c.3996T>G | Missense mutation | NA | NA | VUS |
| POLD1 | p.A532T | c.1594G>A | Missense mutation | rs765276497 | NA | VUS | |
| 10 | MUTYH | c.934-2A>G | c.934-2A>G | Splice receptor mutation | rs77542170 | NA | VUS |
| SMAD4 | p.A309V | c.926C>T | Missense mutation | NA | NA | VUS | |
| 11 | APC | p.A41T | c.121G>A | Missense mutation | NA | NA | VUS |
| POLD1 | p.R218H | c.653G>A | Missense mutation | rs150010804 | NA | VUS | |
| 12 | SBDS | p.K33R | c.98A>G | Missense mutation | rs373730800 | COSM4826086 | VUS |
| 13 | ATM | p.V519I | c.1555G>A | Missense mutation | NA | NA | VUS |
| BRCA2 | p.H523R | c.1568A>G | Missense mutation | rs80358443 | NA | VUS |
Through the comparison of related items between the two groups (Table 5), it can be found that there were significant differences between the two groups in the following items (P < 0.05). Such as Age of initial treatment, Total hospitalizations, Age of first intussusception, Frequency of intussusception, Age of initial surgery, Time interval from mucocutaneous pigmentation appearance to abdominal symptoms, Maximum diameter of gastric polyps, Load of duodenal intestine polyps, Distribution of colorectal polyps, Times of endoscopic examinations.
| Project | Wild-type (n = 19) | Mutant-type (n = 73) | P value |
| Gender | |||
| Male | 11 | 44 | 0.851 |
| Female | 8 | 29 | |
| Family history | |||
| No | 13 | 51 | 0.903 |
| Yes | 6 | 22 | |
| ABO blood group1 | |||
| A | 7 | 23 | 0.964 |
| B | 6 | 21 | |
| AB | 4 | 18 | |
| O | 2 | 9 | |
| Rh blood group1 | |||
| Negative | 0 | 0 | |
| Positive | 19 | 71 | |
| Age of initial treatment (years) | 18.474 ± 8.8089 | 12.973 ± 8.3881 | 0.021 |
| Final age of follow-up (years) | 30.842 ± 11.3101 | 27.425 ± 9.7680 | 0.239 |
| Total hospitalizations | 3 (1, 4) | 4 (3, 6) | 0.003 |
| Age of first intussusception (years) | 22 (14, 27) | 15 (9.25, 24) | 0.025 |
| Frequency of intussusception | 1 (1, 2) | 2 (1, 3) | 0.006 |
| Age of initial surgery (years) | 19 (14, 25) | 14 (8, 23.75) | 0.007 |
| Number of operations | 1 (1, 2) | 1 (1, 2) | 0.924 |
| Age of mucocutaneous pigmentation appearance (years) | 3 (0, 5) | 3 (0.5, 5) | 0.811 |
| Order of mucocutaneous pigmentation appearance | |||
| Lips | 2 | 17 | 0.213 |
| Lips and limbs | 1 | 46 | |
| Lips to limbs | 16 | 10 | |
| Time interval from mucocutaneous pigmentation appearance to abdominal symptoms (years) | 14.5 (8, 25.5) | 10 (5, 15) | 0.038 |
| Distribution of gastric polyps | |||
| Yes | 16 (84.2%) | 60 (82.2%) | 1 |
| No | 3 (15.8%) | 13 (17.8%) | |
| Load of gastric polyps | 5 (5, 10) | 5 (3.25, 10) | 0.111 |
| Maximum diameter of gastric polyps (mm) | 7 (4.25, 10) | 10 (6, 15) | 0.012 |
| Distribution of duodenal intestine polyps | |||
| Yes | 18 (94.7%) | 71 (97.3%) | 1 |
| No | 1 (5.3%) | 2 (2.7%) | |
| Load of duodenal intestine polyps | 3 (1, 6.5) | 7 (4, 15.5) | 0.013 |
| Maximum diameter of duodenal intestine polyps (mm) | 30 (15, 50) | 48 (30, 60) | 0.110 |
| Distribution of colorectal polyps | |||
| Yes | 6 (31.6%) | 52 (71.2%) | 0.001 |
| No | 13 (68.4%) | 21 (28.8%) | |
| Load of colorectal polyps | 4 (1.5, 12) | 3 (1, 10) | 0.864 |
| Maximum diameter of colorectal polyps (mm) | 30 (15, 50) | 25 (13.5, 40) | 0.664 |
| Carcinogenesis | |||
| Yes | 0 (0%) | 9 (9.52%) | 0.239 |
| No | 19 (100%) | 64 (90.48%) | |
| Pathology of polyps | |||
| Hamartoma | 12 | 35 | 0.344 |
| Adenoma | 2 | 8 | |
| Hamartoma + adenoma | 2 | 4 | |
| Carcinogenesis | 0 | 9 | |
| Deletion | 3 | 17 | |
| Times of endoscopic examinations | 2 (1, 2) | 2 (2, 4.75) | 0.012 |
Clinical phenotypic variations between PJS with mutant-type and wild-type STK11: (1) There were no statistical differences in gender, family history, ABO blood group or Rh blood group between PJS with mutant-type and wild-type STK11; (2) There were differences (P < 0.05) between wild-type and mutant-type in age of initial treatment, total hospitalizations, age of first intussusception, frequency of intussusception and age of initial surgery, with wild-type PJS having a much higher age of initial treatment, much smaller number of total hospitalizations, higher age of first intussusception and lower frequency of intussusception than mutant-type PJS. There was no statistical difference between wild-type and mutant-type PJS in number of operations; (3) There was no statistical difference between wild-type and mutant-type PJS in age or order of mucocutaneous pigmentation appearance, but there was a difference in time interval from mucocutaneous pigmentation appearance to abdominal symptoms (P < 0.05), which was longer in wild-type PJS than in the mutant-type PJS; (4) There was no statistical difference in distribution and load of gastric polyps between wild-type and mutant-type PJS, but the maximum diameter of gastric polyps was significantly lower in wild-type PJS than mutant-type PJS (P < 0.05). There was no statistical difference in distribution and maximum diameter of duodenal intestine polyps, but there was a difference in load of duodenal intestine polyps (P < 0.05), which was much lower in wild-type PJS than in mutant-type PJS. There was no statistical difference in load and maximum diameter of colorectal polyps, but distribution of colorectal polyps was significantly lower in wild-type PJS than in mutant-type PJS; and (5) There was no statistical difference in pathology or carcinogenesis of polyps between wild-type and mutant-type PJS. There was a difference in endoscopic examination between wild-type and mutant-type PJS (P < 0.05), with fewer times of endoscopic examination in wild-type than mutant-type PJS.
Clinical phenotypic differences between mutant-type PJS combined with other mutations and not combined with other mutations: (1) MUYTH: There was a difference in ABO blood group between mutant-type PJS with and without MUYTH mutations (P < 0.05); (2) CHEK2: There were differences in ABO blood group, total hospitalizations and number of operations between mutant-type PJS with and without CHEK2 mutations (P < 0.05); (3) APC: There were differences in distribution, load and maximum diameter of gastric polyps between mutant-type PJS with and without APC mutations (P < 0.05); (4) CDH1: There were differences in load and maximum diameter of duodenal intestine polyps between mutant-type PJS with and without CDH1 mutations (P < 0.05); (5) GALNT12: There were differences in time interval from mucocutaneous pigmentation appearance to abdominal symptoms and maximum diameter of duodenal intestine polyps between mutant-type PJS with and without GALNT12 mutations (P < 0.05); (6) BRCA: There were differences in maximum diameter of duodenal intestine polyps between mutant-type PJS with and without BRCA mutations (P < 0.05); (7) KIT: There were differences in ABO blood group and age of first intussusception between mutant-type PJS with and without KIT mutations (P < 0.05); (8) MSH: There was a difference in distribution of colorectal polyps between mutant-type PJS with and without MSH mutations (P < 0.05); (9) PTCH1: There were differences in load of gastric polyps and maximum diameter of colorectal polyps between mutant-type PJS with and without PTCH1 mutations (P < 0.05); (10) ATM: There was a difference in age of mucocutaneous pigmentation appearance between mutant-type PJS with and without ATM mutations (P < 0.05); (11) PDGFRA: There was a difference in final age of follow-up between mutant-type PJS with and without PDGFRA mutations (P < 0.05); and (12) POLD1: There was a difference in age of first intussusception between mutant-type PJS with and without POLD1 mutations (P < 0.05).
PJS is an autosomal dominant disorder with a prevalence of approximately 1 in 200000[15,16]. Although PJS is a rare disease, the large population of China and the prolonged course of PJS lead to the accumulation of a great number of PJS patients in Chinese society. Germline mutations in the STK11 gene are recognized as the molecular genetic cause of PJS. The tumor suppressor gene STK11 is involved in multiple processes such as embryonic development, cell polarity, cell cycle arrest, apoptosis and metabolism, and its mutations have been detected in a variety of disseminated cancers. A related study[17] demonstrated that the lack of the STK11 gene resulted in a significant increase of intracellular reactive oxygen species levels and enhanced expression of phosphorylated histone γ-H2AX, which resulted in DNA damage, oxidative damage to the genome and an increased mutation rate. The rate of STK11 germline mutations detected in this group of PJS patients was 79.35%, which is generally consistent with the literature[18].
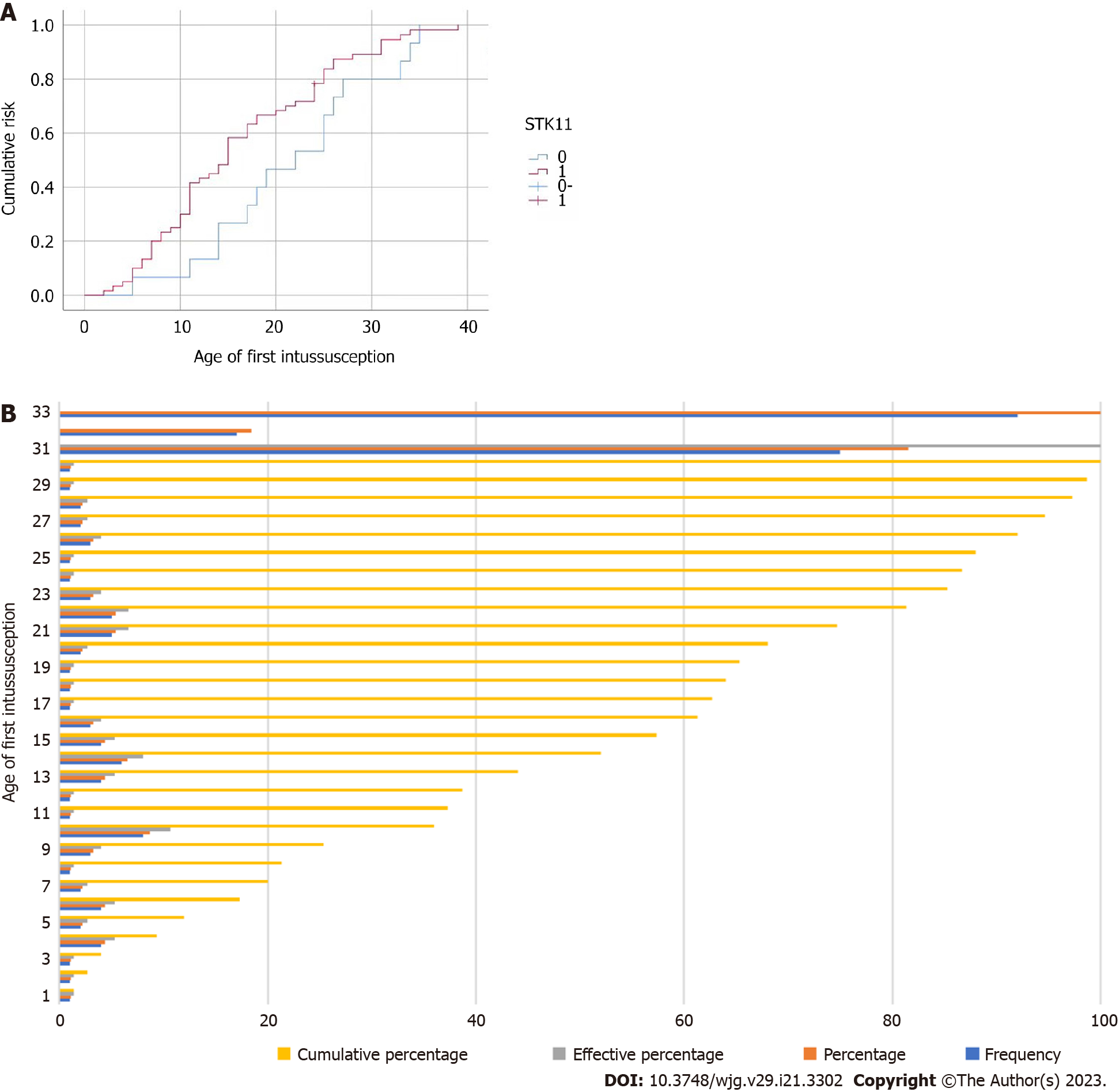
The target genome covered 41 pathogenic genes associated with digestive tract tumors, including STK11, and PJS patients were classified into wild-type or mutant-type based on the presence or absence of STK11 mutations. The results of the clinical characteristic analysis showed that there were significant differences between wild-type and mutant-types PJS in age of initial treatment, age of first intussusception, frequency of intussusception, age of initial surgery, time interval from mucocutaneous pigmentation appearance to abdominal symptoms, maximum diameter of gastric polyps, load of duodenal intestine polyps, distribution of colorectal polyps, times of hospitalization and times of endoscopic examinations. Mutant-type PJS typically has an earlier age of initial treatment and a shorter time interval from mucocutaneous pigmentation appearance to abdominal symptoms than wild-type PJS. They are often first admitted to hospital for serious complications such as intussusception and intestinal obstruction. Our study found that the age of first intussusception was significantly younger in mutant-type PJS than in wild-type PJS, and its cumulative risk of intussusception at the age of 20 years was 68.3%, which was significantly higher than that of wild-type PJS (Figure 2). This is consistent with the findings of domestic and international studies[19,20]. As a result, complications such as intestinal obstruction and intussusception are more likely to occur in mutant-type PJS than in wild-type, and occur earlier. The earlier intervention of treatment for GI polyps in mutant-type PJS patients would be beneficial in reducing the occurrence of these complications. In addition, mutant-type PJS has a higher distribution of colorectal polyps and a larger maximum diameter of gastric polyps than wild-type PJS, requiring more frequent endoscopic examinations and treatment in hospital.
Overseas studies have shown that PJS patients have an increased risk of cancer at several sites, including the GI tract, breast, ovaries, testes and lungs[21]. PJS malignancy is a serious threat to the life and health of patients; studies have shown[21,22] that the death rate of PJS patients is as high as 32%, and malignancy is the main cause of death. The incidence of malignant tumors in PJS patients is 19%-32%, with an average age of 42-45 years, and a predominance of GI tract tumors (51%-69%), followed by gynecological tumors (22%-26%). The risk of GI malignancy is 50 times higher than that of the general population. The 19 cases of wild-type PJS in this study were not combined with malignant tumors. Among the 73 mutant-type patients, 8 had GI polyps and 4 had cancers of other sites: 1 adnexal cystadenocarcinoma, 1 ovarian mucinous tumor, 1 cervical adenocarcinoma and 1 nasopharyngeal carcinoma. It was found that patients with a detectable STK11 truncating mutation tend to develop more polyps and cancers, and require more surgical intervention[23]. There was no statistically significant difference in the cancer rate between mutant-type and wild-type in this study, which may be related to the shorter follow-up period of the enrolled patients. Germline mutations of STK11 are essential factors in GI tumourigenesis, and the cell types and signal pathways that lead to the malignant transformation of polyps are still unclear. A foreign study showed[24] that after the knockout of STK11 in mice, IL-11 inflammatory mediators mediated the activation of the JAK/STAT3 pathway due to STK11 deficiency in mouse stromal cells, which eventually resulted in the formation of polyp malignancy in mice, and the treatment of STK11-deficient mice with JAK1/2 inhibitors significantly reduced the occurrence of polyps. We can block the development and malignancy of polyps at the root by blocking the associated inflammatory mediators or transduction pathways, which provides a new idea for us in studying the pharmacological treatment of PJS patients. Previous studies have shown that PJS patients have a progression pathway of hamartoma-(adenoma)-carcinoma[25], which is corroborated by the presence in our study of PJS patients with malformation combined with adenomatous polyps and with adenoma combined with polyp carcinoma. Accordingly, the detection of STK11 mutations can be useful for guiding the assessment of polyp carcinogenesis risk in PJS patients and their relatives.
PJS patients have a prolonged disease course, with GI polyps growing larger with age and recurring more easily, and can result in intestinal obstruction, intussusception or even cancer, but the characteristics and severity of the disease vary significantly among PJS patients. The clinical presentation is distinctly heterogeneous. Clinical data and epidemiological data collected from more than 500 PJS patients at our center suggests that polyps grow fastest in adolescence, leading to serious complications such as intussusception, and the age of first surgery is significantly younger in mutant-type PJS than in wild-type PJS. Meanwhile, the incidence of tumors in the digestive tract and other organs of the body is significantly higher in middle-aged PJS patients.
Therefore, in the individualized treatment of PJS patients, we recommend that they should have the STK11 gene tested, and the genotypes classified into mutant-type and wild-type. Patients with mutant-type PJS should be strictly controlled in terms of treatment and follow-up strategies, while patients with wild-type PJS can be treated with relaxed treatment conditions and follow-up years. For the monitoring and treatment of PJS, we refer to the different follow-up strategies by age group proposed by a domestic study[26], and further refine the treatment and follow-up strategies for PJS based on this treatment strategy as follows (Table 6).
| Age (yr) | Mutant-type | Wild-type | ||
| Surveillance | Intervention | Surveillance | Intervention | |
| < 7 | Routine abdominal ultrasound surveillance is recommended every year | Removal of polyps | Abdominal ultrasound is recommended every 3-5 yr | Follow-up observation |
| 8-11 | Routine abdominal ultrasound surveillance is recommended every year. For symptomatic individuals with PJS, an abdominal ultrasound should be performed earlier | Removal of polyps | Abdominal ultrasound is recommended every 3-5 yr. For symptomatic individuals with PJS, an abdominal ultrasound should be performed earlier | Removal of polyps |
| 8-18 | Total GI surveillance every year (CT scan of small-bowel or enteroscopy/capsule endoscopy should be offered as options | Polyps > 10 mm should be removed | Total GI surveillance 2-3 yr | Removal of polyps |
| 19-30 | Routine total GI polyps surveillance every 2-3 yr and screening for systemic tumors | Removal of polyps and treatment of tumors | Routine total GI polyps surveillance every 2-3 yr | Removal of polyps |
| > 30 | Focus on detection of tumors in digestive tract and other organs | Treatment of tumors | Focus on detection of tumors in digestive tract and other organs | Treatment of tumors |
Peutz-Jeghers syndrome (PJS) is an autosomal dominant genetic disease with skin mucosal pigment spots and gastrointestinal multiple hamartoma polyps as clinical characteristics. At present, it is considered that the germline mutation of STK11 gene is the genetic cause of PJS. However, not all PJS patients can be detected STK11 germline mutations.
The specific clinical characteristics of these PJS patients without STK11 mutation is an interesting clinical question. Or, like wild type gastrointestinal stromal tumor (GIST), whether these PJS without STK11 mutation are also called PJS is worth discussing. Therefore, we designed the study to understand the clinical characteristics of these PJS patients without STK11 mutation.
To investigates whether PJS patients with known STK11 mutations have a more severe spectrum of clinical phenotypes compared to those without.
The general information, diagnosis and treatment history, pathology, times of examination and other clinical data of the 92 enrolled PJS patients were collected for statistical analysis. Genomic DNA samples were extracted from peripheral blood samples, and pathogenic germline mutations of STK11 were detected by high-throughput next-generation gene sequencing. Clinical-pathologic manifestations of patients with and without STK11/LKB1 mutations were compared.
Compared with PJS patients with STK11 mutations, those without tended to be older at the age of initial treatment, age of first intussusception and age of initial surgery. They also had a lower number of total hospitalizations relating to intussusception or intestinal obstruction, and a lower load of small intestine polyps. Final results found that PJS patients without STK11 mutations might have less severe clinical-pathologic manifestations than those with.
PJS patients without STK11 mutations might have less severe clinical-pathologic manifestations than those with.
At present, it is considered that the germline mutation of STK11 gene is the genetic cause of PJS. However, not all PJS patients can be detected STK11 germline mutations. The specific clinical characteristics of these PJS patients without STK11 mutation is an interesting clinical question. Or, like wild type GIST, whether these PJS without STK11 mutation are also called PJS is worth discussing.
| 1. | Sato E, Goto T, Honda H. Peutz-Jeghers Syndrome. JAMA Dermatol. 2022;158:1316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 2. | Alharbi ES, Alrumayh JS, Alzaghran RH, Algaith NK, Shaheen AN. A Case of Multiple Polyps Causing Intussusception in an Adult Patient With Peutz-Jeghers Syndrome. Cureus. 2022;14:e30532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 3. | Borun P, Bartkowiak A, Banasiewicz T, Nedoszytko B, Nowakowska D, Teisseyre M, Limon J, Lubinski J, Kubaszewski L, Walkowiak J, Czkwianianc E, Siolek M, Kedzia A, Krokowicz P, Cichy W, Plawski A. High Resolution Melting analysis as a rapid and efficient method of screening for small mutations in the STK11 gene in patients with Peutz-Jeghers syndrome. BMC Med Genet. 2013;14:58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 4. | Jelsig AM. Hamartomatous polyps - a clinical and molecular genetic study. Dan Med J. 2016;63. [PubMed] |
| 5. | Klimkowski S, Ibrahim M, Ibarra Rovira JJ, Elshikh M, Javadi S, Klekers AR, Abusaif AA, Moawad AW, Ali K, Elsayes KM. Peutz-Jeghers Syndrome and the Role of Imaging: Pathophysiology, Diagnosis, and Associated Cancers. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (1)] |
| 6. | Altamish M, Dahiya R, Singh AK, Mishra A, Aljabali AAA, Satija S, Mehta M, Dureja H, Prasher P, Negi P, Kapoor DN, Goyal R, Tambuwala MM, Chellappan DK, Dua K, Gupta G. Role of the Serine/Threonine Kinase 11 (STK11) or Liver Kinase B1 (LKB1) Gene in Peutz-Jeghers Syndrome. Crit Rev Eukaryot Gene Expr. 2020;30:245-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 7. | Kuroda Y, Saito T, Nagai J, Ida K, Naruto T, Masuno M, Kurosawa K. Microdeletion of 19p13.3 in a girl with Peutz-Jeghers syndrome, intellectual disability, hypotonia, and distinctive features. Am J Med Genet A. 2015;167A:389-393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 8. | Duan FX, Gu GL, Yang HR, Yu PF, Zhang Z. Must Peutz-Jeghers syndrome patients have the LKB1/STK11 gene mutation? A case report and review of the literature. World J Clin Cases. 2018;6:224-232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 8] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 9. | Byrjalsen A, Roos L, Diemer T, Karstensen JG, Løssl K, Jelsig AM. Preimplantation genetic testing in two Danish couples affected by Peutz-Jeghers syndrome. Scand J Gastroenterol. 2023;58:314-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 10. | Daniell J, Plazzer JP, Perera A, Macrae F. An exploration of genotype-phenotype link between Peutz-Jeghers syndrome and STK11: a review. Fam Cancer. 2018;17:421-427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 48] [Article Influence: 6.9] [Reference Citation Analysis (1)] |
| 11. | Ueki A, Hirasawa A. Molecular Features and Clinical Management of Hereditary Gynecological Cancers. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 12. | Boland CR, Idos GE, Durno C, Giardiello FM, Anderson JC, Burke CA, Dominitz JA, Gross S, Gupta S, Jacobson BC, Patel SG, Shaukat A, Syngal S, Robertson DJ. Diagnosis and management of cancer risk in the gastrointestinal hamartomatous polyposis syndromes: recommendations from the U.S. Multi-Society Task Force on Colorectal Cancer. Gastrointest Endosc. 2022;95:1025-1047. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 13. | Boland CR, Idos GE, Durno C, Giardiello FM, Anderson JC, Burke CA, Dominitz JA, Gross S, Gupta S, Jacobson BC, Patel SG, Shaukat A, Syngal S, Robertson DJ. Diagnosis and Management of Cancer Risk in the Gastrointestinal Hamartomatous Polyposis Syndromes: Recommendations From the US Multi-Society Task Force on Colorectal Cancer. Am J Gastroenterol. 2022;117:846-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 14. | Shaunak M, Side L, Afzal N, Davies JH. An atypical presentation of a pathogenic STK11 gene variant in siblings not fulfilling the clinical diagnostic criteria for Peutz-Jeghers Syndrome. J Pediatr Endocrinol Metab. 2022;35:131-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 15. | Wang R, Qi X, Liu X, Guo X. Peutz-Jeghers syndrome: Four cases in one family. Intractable Rare Dis Res. 2016;5:42-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 16. | Chae HD, Jeon CH. Peutz-Jeghers syndrome with germline mutation of STK11. Ann Surg Treat Res. 2014;86:325-330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 17. | Wang YS, Chen J, Cui F, Wang H, Wang S, Hang W, Zeng Q, Quan CS, Zhai YX, Wang JW, Shen XF, Jian YP, Zhao RX, Werle KD, Cui R, Liang J, Li YL, Xu ZX. LKB1 is a DNA damage response protein that regulates cellular sensitivity to PARP inhibitors. Oncotarget. 2016;7:73389-73401. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 18. | Jiang YL, Zhao ZY, Li BR, Wang H, Yu ED, Ning SB. STK11 gene analysis reveals a significant number of splice mutations in Chinese PJS patients. Cancer Genet. 2019;230:47-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 19. | Rycyk A, Kasztelan-Szczerbinska B, Cichoz-Lach H. A rare case of a juvenile polyp of patient with Peutz-Jeghers syndrome, complicated with intussusception of the small intestine. Folia Med (Plovdiv). 2022;64:693-696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 20. | Kariv R, Dahary D, Yaron Y, Petel-Galil Y, Malcov M, Rosner G. Whole Genome Sequencing Applied in Familial Hamartomatous Polyposis Identifies Novel Structural Variations. Genes (Basel). 2022;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 21. | Wu M, Krishnamurthy K. Peutz-Jeghers Syndrome. 2022 Aug 21. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2023 Jan-. [PubMed] |
| 22. | Latchford AR, Clark SK. Gastrointestinal aspects of Peutz-Jeghers syndrome. Best Pract Res Clin Gastroenterol. 2022;58-59:101789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (1)] |
| 23. | Gorji L, Huish G, Morgan J, Levy P. A case of sporadic Peutz-Jeghers syndrome presenting as multiple intussusceptions. J Surg Case Rep. 2022;2022:rjac070. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 24. | Ollila S, Domènech-Moreno E, Laajanen K, Wong IP, Tripathi S, Pentinmikko N, Gao Y, Yan Y, Niemelä EH, Wang TC, Viollet B, Leone G, Katajisto P, Vaahtomeri K, Mäkelä TP. Stromal Lkb1 deficiency leads to gastrointestinal tumorigenesis involving the IL-11-JAK/STAT3 pathway. J Clin Invest. 2018;128:402-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 25. | Ma H, Brosens LAA, Offerhaus GJA, Giardiello FM, de Leng WWJ, Montgomery EA. Pathology and genetics of hereditary colorectal cancer. Pathology. 2018;50:49-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 115] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 26. | Li BR, Sun T, Li J, Zhang YS, Ning SB, Jin XW, Zhu M, Mao GP. Primary experience of small bowel polypectomy with balloon-assisted enteroscopy in young pediatric Peutz-Jeghers syndrome patients. Eur J Pediatr. 2020;179:611-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Katagiri R, Japan; Lee EW, South Korea; Yashiro M, Japan S-Editor: Fan JR L-Editor: A P-Editor: Chen YX