Published online May 21, 2023. doi: 10.3748/wjg.v29.i19.2932
Peer-review started: November 28, 2022
First decision: February 15, 2023
Revised: March 6, 2023
Accepted: April 11, 2023
Article in press: April 11, 2023
Published online: May 21, 2023
Processing time: 168 Days and 17.7 Hours
Gastrointestinal stromal tumor (GIST) is a common neoplasm with high rates of recurrence and metastasis, and its therapeutic efficacy is still not ideal. There is an unmet need to find new molecular therapeutic targets for GIST. TATA-box-binding protein-associated factor 15 (TAF15) contributes to the progress of various tumors, while the role and molecular mechanism of TAF15 in GIST progression are still unknown.
To explore new molecular therapeutic targets for GIST and understand the biological role and underlying mechanisms of TAF15 in GIST progression.
Proteomic analysis was performed to explore the differentially expressed proteins in GIST. Western blotting and immunohistochemical analysis were used to verify the expression level of TAF15 in GIST tissues and cell lines. Cell counting kit-8, colony formation, wound-healing and transwell assay were executed to detect the ability of TAF15 on cell proliferation, migration and invasion. A xenograft mouse model was applied to explore the role of TAF15 in the progression of GIST. Western blotting was used to detect the phosphorylation level and total level of RAF1, MEK and ERK1/2.
A total of 1669 proteins were identified as differentially expressed proteins with 762 upregulated and 907 downregulated in GIST. TAF15 was selected for the further study because of its important role in cell proliferation and migration. TAF15 was significantly over expressed in GIST tissues and cell lines. Overexpression of TAF15 was associated with larger tumor size and higher risk stage of GIST. TAF15 knockdown significantly inhibited the cell proliferation and migration of GIST in vitro and suppressed tumor growth in vivo. Moreover, the inhibition of TAF15 expression significantly decreased the phosphorylation level of RAF1, MEK and ERK1/2 in GIST cells and xenograft tissues, while the total RAF1, MEK and ERK1/2 had no significant change.
TAF15 is over expressed in GIST tissues and cell lines. Overexpression of TAF15 was associated with a poor prognosis of GIST patients. TAF15 promotes cell proliferation and migration in GIST via the activation of the RAF1/MEK/ERK signaling pathway. Thus, TAF15 is expected to be a novel latent molecular biomarker or therapeutic target of GIST.
Core Tip: TATA-box-binding protein-associated factor 15 (TAF15) was upregulated in gastrointestinal stromal tumor (GIST) cells and tissues and was associated with a poor prognosis in GIST patients. TAF15 promotes cell proliferation and migration of GIST in vitro and tumor growth in vivo via the activation of the RAF1/MEK/ERK signaling pathway. Therefore, TAF15 is expected to be a novel potential molecular biomarker or therapeutic target of GIST.
- Citation: Guo CM, Tang L, Li X, Huang LY. TATA-box-binding protein-associated factor 15 is a novel biomarker that promotes cell proliferation and migration in gastrointestinal stromal tumor. World J Gastroenterol 2023; 29(19): 2932-2949
- URL: https://www.wjgnet.com/1007-9327/full/v29/i19/2932.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i19.2932
Gastrointestinal stromal tumor (GIST) is a common neoplasm that arises from the mesenchymal tissue of the gastrointestinal tract and likely originates from the interstitial cells of Cajal[1,2]. Mutations in mast/stem cell growth factor receptor (KIT) and platelet-derived growth factor receptor A (PDGFRA) are considered the primary causes in the pathogenesis of GIST[3]. GIST most commonly occurs in the stomach (50%-60%) and small intestine (30%-35%) but rarely occurs in the colorectum (5%) and esophagus (< 1%)[4]. Previous studies have indicated that GIST accounts for roughly 0.1%-3.0% of gastrointestinal malignances, and 15%-50% of patients present with metastasis at the time of diagnosis[5].
In clinical practice, GIST is liable to recur and distantly metastasize even after the initial tumor excision[6]. In fact, about 40% of patients develop tumor metastasis within 15 years after surgery[7], with a strong tendency to metastasize to the liver and peritoneal surfaces primarily and the lymph nodes and lung less frequently[4,8]. Based on tumor size, mitotic count, tumor site, and tumor rupture, the modified National Institutes of Health (NIH) consensus criteria stratifies GIST into four subgroups: Very low (NIH-VL), low (NIH-L), intermediate (NIH-I), and high risk (NIH-H)[9]. GIST patients with large sized tumors and high mitotic counts are more likely to metastasize to the liver and abdominal cavity and frequently have poor disease-free survival ratios[10].
Before the clinical application of imatinib, a tyrosine kinase inhibitor, surgical resection was the only accepted therapy that could prolong the life of GIST patients who had developed metastasis. Even then, the 5-year overall survival rate of patients was only 27%–34%[11]. Improved treatment with imatinib has been highly effective in advanced or metastatic GIST patients, and approximately 80% of them can reach complete or partial recovery[12,13]. Unfortunately, high rates of drug resistance due to secondary mutations of KIT have rendered imatinib therapy ineffective in recent years[14]. Taken together, the current research shows that the therapeutic efficacy of GIST treatments are still not ideal. Therefore, finding new molecular therapeutic targets for GIST is critical.
In this study, we implemented a proteomic analysis in GIST patients using tandem mass tag (TMT) labeling 10-plex kits to explore potential molecular therapeutic targets for GIST. A total of 4111 proteins were quantified in the GIST samples. Of these, 1669 were identified as significant differentially expressed proteins (DEPs), with 762 upregulated and 907 downregulated proteins. After a systematic analysis of the top 10 upregulated or downregulated proteins, we selected TATA-box-binding protein-associated factor 15 (TAF15) for further study because of its important role in cell proliferation and migration[15]. TAF15 is a member of the FUS/Ewing sarcoma protein (EWS)/TAF15 (FET) family of RNA- and DNA-binding proteins, which plays an essential role in mRNA transcription, RNA splicing and polyadenylation[16-18].
Previous studies have demonstrated that TAF15 can influence a large number of genes that are closely related to the cell cycle and cell death and that overexpression of TAF15 can promote proliferation in sarcomas, human neuroblastoma and leukemia cells[17,19]. TAF15 mediates resistance to radiation therapy through inhibiting tumor suppressors p53 and p21, and upregulation of TAF15 is closely related to worsened survival in non-small cell lung cancer patients[20]. Additionally, a prior study has described a human anti-TAF15 antibody, PAT-BA4, that targeted a tumor-specific TAF15 antigen and then inhibited cell adhesion and spreading in gastric cancer[21]. Moreover, in a recent study, TAF15 activated the mitogen-activated protein kinase (MAPK) signaling pathway by upregulating the expression of MAPK6 in lung squamous cell carcinoma[22]. Taken together, TAF15 may play an important role in the progression of human cancers. However, the latent functions and molecular mechanism of TAF15 in GIST progression are still unclear and must be further studied.
One hundred and sixty one patients with GIST were recruited from the Department of Gastrointestinal Surgery, Yantai Yu Huang Ding Hospital of Qing Dao University from March 2020 to June 2022, with ethical permission from the Biomedical Ethics Committee (No.2019–410). All patients were diagnosed with GIST by two independent pathologists based on Chinese clinical guidelines[23]. No patients had received chemotherapy before surgery. Out of the 161 cases, 18 cases had matched tumor (T) and normal (N) tissue samples, and the clinicopathologic features of the paired GIST samples were documented in Table 1. In the remaining 143 cases, only tissue from the tumor site was available. In the beginning we only collected 12 matched samples, which were sent for proteomic analysis, followed by another 6 matched samples. All specimens were immediately frozen in liquid nitrogen within 30 min after excision. All patients provided written informed consent.
| Case No. | Sex | Age in yr | Location | Tumor size length × width × height in cm | Mitosis count, per 50 HFPs | Risk classification based on NIH |
| 1 | Male | 74 | Stomach | 2.8 × 2.8 × 2.0 | > 5 | High |
| 2 | Female | 24 | Stomach | 6.0 × 4.0 × 5.0 | > 5 | High |
| 3 | Male | 69 | Stomach | 7.5 × 5.5 × 4.0 | > 10 | High |
| 4 | Male | 46 | Small intestine | 6.5 × 5.0 × 4.0 | > 5 | High |
| 5 | Female | 65 | Small intestine | 6.5 × 5.5 × 5.0 | > 5 | High |
| 6 | Female | 61 | Stomach | 5.5 × 3.5 × 4.0 | > 5 | High |
| 7 | Female | 49 | Stomach | 8.5 × 8.0 × 7.0 | < 5 | Intermediate |
| 8 | Male | 77 | Stomach | 6.0 × 6.0 × 5.0 | < 5 | Intermediate |
| 9 | Female | 54 | Stomach | 6.0 × 4.5 × 3.5 | < 5 | Intermediate |
| 10 | Female | 43 | Stomach | 5.5 × 4.5 × 3.0 | < 5 | Intermediate |
| 11 | Male | 70 | Stomach | 7.0 × 7.0 × 5.5 | < 5 | Intermediate |
| 12 | Female | 55 | Stomach | 8.0 × 8.0 × 7.0 | < 5 | Intermediate |
| 13 | Male | 63 | Small intestine | 5.0 × 3.5 × 3.0 | < 5 | Low |
| 14 | Male | 67 | Stomach | 2.3 × 1.5 × 1.5 | < 5 | Low |
| 15 | Male | 44 | Stomach | 3.0 × 2.7 × 1.3 | < 5 | Low |
| 16 | Female | 49 | Stomach | 1.0 × 0.8 × 0.8 | < 5 | Very low |
| 17 | Female | 66 | Stomach | 2.0 × 1.8 × 1.0 | < 5 | Very low |
| 18 | Female | 57 | Stomach | 2.0 × 1.8 × 1.5 | < 5 | Very low |
Tissues from GIST patients were homogenized in RIPA lysis buffer containing protease inhibitors using a mortar and pestle. After homogenizing lightly for 1 min on ice, the lysate was centrifuged twice at 13000 RPM for 10 min at 4 °C, and then the supernatants were transferred to a fresh tube. A BCA assay kit (catalog no.P0010, Beyotime Biotechnology, Shanghai, China) was used to measure the protein concentration of each sample. The protein content of each sample was reduced using 5 mmol/L DL-dithiothreitol at 55 °C for 30 min. Samples were then incubated with 10 mmol/L of iodoacetamide at room temperature for 15 min in the dark to alkylate the protein. Each protein sample was dissolved using triethylammonium bicarbonate buffer and then digested overnight at 37 °C using trypsin.
Tryptic peptides from each sample were labeled with TMT (Thermo Fisher Scientific, Waltham, MA, United States) kits following the manufacturer’s protocol. After quenching the reaction using 5% hydroxylamine (Sigma-Aldrich, Saint Louis, MO, United States), the TMT-labeled peptide solution was fractionated using basic pH reversed phase high performance liquid chromatography through an Agilent Zorbax Extend C18 column (5 μm, 150 mm × 2.1 mm). The mobile phase consisted of Buffer A (98% water with 2% acetonitrile) and Buffer B (90% acetonitrile with 10% water). The procedure gradient was run as follows: 0%-8% Buffer B for 0-10 min; 8%-35% Buffer B for 10-80 min; 35%-60% Buffer B for 80-95 min; 60%-70% Buffer B for 95-105 min; 70%-100% Buffer B for 105-120 min. The flow rate was set at 1 mL/min. TMT labeled peptides were separated into 10 fractions by reversed phase high performance liquid chromatography.
Liquid chromatography-tandem mass spectrometry (MS) analysis was conducted on a Q Exactive Mass Spectrometer (Thermo Scientific) coupled to an EASY-nanoLC System (Thermo Fisher Scientific). Peptides were loaded onto a reverse-phase C18 column (Thermo Scientific Easy Column, 10 cm long, 75 μm inner diameter, 3μm resin) in Buffer A (0.1% formic acid) and separated with a linear gradient of Buffer B (84% acetonitrile and 0.1% formic acid) at a flow rate of 300 nL/min. The MS was utilized in positive ion mode. MS data were acquired following a data-dependent top10 method, dynamically selecting the most abundant precursor ions from the survey scan (300–1800 m/z). The automatic gain control target was set to 3e6 with maximum inject time to 10 ms. Survey scans were acquired from a resolution of 70000 at m/z 200 with an isolation width of 2 m/z.
Data for each sample were processed using the Swiss-Prot Homo sapiens database (20413 entries, January 14, 2017). The parameters of the procedures were as follows: Cysteine carbamidomethylation was set to fixed modification; oxidation was set to variable modification; peptide mass tolerance was 20 ppm, trypsin digestion, max 2 missed cleavages; and the false discovery rate was less than 1%. Proteins with a fold change of > 1.2 or < 0.83 and a P value < 0.05 were identified as significantly upregulated or downregulated.
Western blot analysis was performed using 10% sodium dodecyl sulfonate-polyacrylamide gel electrophoresis to separate equal amounts (25 μg/load) of protein samples. Protein samples were transferred onto polyvinylidenedifluoride membranes. The membranes were blocked at room temperature for 1 h with 5% nonfat milk in TBS-T buffer and subsequently incubated with primary antibodies at 4 °C overnight. Secondary antibodies were incubated for 1 hat room temperature. Protein samples were visualized using ECL reagents (MA0186, Meilunbio, Dalian, China) using the ChemiScope 6200 Touch Imaging System (CLINX, Shanghai, China). Some membranes were recycled using stripping buffer (SL1340, Coolaber, Beijing, China) and incubated with other primary antibodies. The primary antibodies were used and obtained as follows: TAF15 (1:3000, ab134916, Abcam, United States); GAPDH (1:5000, D110016, Sangon Biotech, Shanghai, China); RAF1 (1:500, D155090, Sangon Biotech, Shanghai, China); p-RAF1 (1:1000, 66592-1-lg, Proteintech, Wuhan, China); MEK1/2 (1:1000, 11049-1-AP, Proteintech, Wuhan, China); p-MEK1 (1:1000, 28930-1-AP, Proteintech, Wuhan, China); ERK1/2 (1:1000, 16443-1-AP, Proteintech, Wuhan, China); and p-ERK1/2 (1:1000, 28733-1-AP, Proteintech, Wuhan, China). The secondary antibodies were used and obtained as follows: HRP goat anti-rabbit immunoglobulin G (IgG) (1:3000, D110058, Sangon Biotech, shanghai, China); and HRP goat anti-mouse IgG (1:3000, D110058, Sangon Biotech, Shanghai, China). Three independent tests were performed.
Tissue specimens were cut into 4 μm sections and processed by deparaffinization and rehydration. Slides were incubated with 3% hydrogen peroxide for 20 min at room temperature to block endogenous peroxidase activity. Tissue sections were incubated with primary antibody TAF15 (1:100, ab134916, Abcam, United States) at 4 °C in a wet chamber overnight and then incubated with secondary antibody HRP goat anti-rabbit IgG (1:100, D110058, Sangon Biotech, Shanghai, China) for 1 h in a wet chamber at room temperature. Color was developed using diaminobenzidine and hematoxylin. Two independent pathologists evaluated the samples by randomly selecting and evaluating five × 400 microscopic fields per slide. The percentage of stained tumor cells was scored as follows: 0, < 5% positive cells; 1, 5%-24% positive cells; 2, 25%-49% positive cells; 3, 50%-74% positive cells; and 4, ≥ 75% positive cells. The staining intensity was scored as follows: 0 for absence; 1 for weak; 2 for moderate; and 3 for strong. The total stained index was equivalent to intensity score × positive score. A total score ≥ 4 was considered positive expression, and < 4 was treated as negative expression.
GIST-882 and GIST-T1 cell lines were obtained from the Shanghai Cancer Institute, and GIST-48 cell lines were kindly provided by Professor Jonathan Fletcher (Harvard Medical School, United States). GIST-882 and GIST-T1 cells were cultured with RPMI 1640 medium (Gibco, United States), and GIST-48 cells were cultured with DMEM (BioInd, Israel). Both medium preparations contained 10% fetal bovine serum (FBS) (Gibco, United States) and 1% penicillin-streptomycin in a humidified incubator at 37 °C with 5% CO2.
The lentivirus for knocking down TAF15 (shRNA1, shRNA2, shRNA3) and control scrambled lentivirus (sh-scrambled) were purchased from Genomeditech (Shanghai, China). GIST-882 and GIST-T1 cells were seeded in a 12-well plate(Corning Life Sciences) at 1 × 105 cells per well, and the transfection was performed when the cell density reached about 50%. Cells were cultured in fresh medium after being infected with lentivirus for 72 h. The multiplicity of infection of lentivirus to GIST-882 and GIST-T1 was 30. Stable cell lines were generated through selection with puromycin (2 μg/mL). Transfection efficiency was detected using western blot analysis.
Transfected cells were cultured in 12-well plates. When the cell density reached 50%-65%, cells were fixed using 4% paraformaldehyde for 30 min to make slides. 0.5% Triton X-100 was added to slides to allow for cell penetration, and 10% goat serum was used to block cells for 2 h at room temperature. Slides were then incubated with the TAF15 primary antibody (1:500, ab134916, Abcam, United States) at 4 °C overnight and then incubated with secondary antibodies for 1 h at room temperature. DAPI was used to stain the cell nuclei, and images were photographed using a fluorescent microscope. The fluorescent intensity of cells were measured using Image J software (version 3.2.0.8).
The cell counting kit (CCK)-8 (Dojindo, Tokyo, Japan) assay was performed according to the manufacturer’s instructions. Briefly, different groups of cells were seeded onto 96-well plates at 1 × 103 cells per well. The CCK-8 reagent was added (10 μL/well) into the individual wells during the last hour. A microplate reader (Bio-Rad) was used to measure the optical density of the samples at 450 nm, as a measure of cell proliferation. With respect to the colony formation assay, different groups of cells were cultured in 6-well plates (500 cells/well) for 2 wk. Cell colonies were fixed with 4% paraformaldehyde for 30 min and stained with 1% crystal violet for 15 min at the room temperature. Colonies consisting of 50 or more cells were counted.
GIST-882 and GIST-T1 cells (3 × 104 cells/well) were inoculated into the upper polycarbonate membrane chambers of a 12-well plate without FBS, while the lower chamber contained RPMI 1640 medium with 10% FBS. To detect the cell invasion ability, matrix gel was in advance added to the polycarbonate membrane chambers. After being cultured for 48 h, the cells attached to the lower side were fixed with 4% paraformaldehyde for 30 min and stained with 1% crystal violet for 15 min at the room temperature. Five fields were selected randomly to count using an inverted microscope.
GIST-882 and GIST-T1 cells were cultured up to 90% confluency and then serum-starved for 12 h. The cell monolayer was scratched using a 1 mL plastic pipette tip. Culture medium was used to wash away the cell debris twice, and then cells were incubated in respective culture medium containing 10% FBS. Samples were photographed at 0 h, 12 h and 24 h post scratch using an Olympus microscope. The percentage of wound healing was evaluated using Image J software.
Nude mice aged 4 wk were purchased from the Beijing Vital River Laboratory Animal Technology Co., Ltd (Beijing, China). All animal care and processing were performed according to the Institutional Animal Care and Use Committee of Qingdao University guidelines. Twenty mice were divided randomly into two groups (10 mice per group) and then subcutaneously injected in the axillary region with stable cells (2 × 106 cells) transfected with sh-TAF15 or sh-scrambled. The volume (volume = length × width × width/2) of tumors were measured every 2 d. The mice were humanely sacrificed using the cervical dislocation method after anesthesia with 0.7% sodium pentobarbital. Tumor tissues were then excised, photographed and weighed.
Statistical analyses were performed using GraphPad Prism 9.3.1. The data analyses were conducted using Student's t-test, χ2 test, one-way analysis of variance or Mann-Whitney U test. Data were expressed as mean ± standard deviation. P < 0.05 was considered statistically significant, and significance was indicated in each graph.
In total, 4111 proteins were quantified with a false discovery rate < 0.01 in all 12 GIST matched samples. Of these, 1669 were identified as significant DEPs on the basis of the filtering criteria of fold change > 1.2 or < 0.83 and P value < 0.05, including 762 upregulated proteins and 907 downregulated proteins (Figure 1A and Supplementary Table 1). Notably, the key proteins used for diagnosing GIST, including KIT, DOG1 and hematopoietic progenitor cell antigen CD34 (CD34), were all contained in the upregulated protein category (Supplementary Table 1), suggesting that our proteomic data were reliable. Moreover, 667, 686, 840 and 974 significant DEPs were identified in the NIH-H, NIH-I, NIH-L and NIH-VL subgroups, respectively (Figure 1B and Supplemental Table 2). After our systematic analysis of the top 10 upregulated and downregulated proteins (Table 2), we decided to focus on the protein TAF15 due to its important role in cell proliferation and migration.
| Category | Accession | Protein name | FC,T vs N | P value |
| Up | P29400 | COL4A5 | 3.129881192 | 0.001086699 |
| O94772 | LY6H | 2.995133385 | 0.012292301 | |
| P05204 | HMGN2 | 2.92528705 | 2.10765E-07 | |
| O00479 | HMGN4 | 2.900071252 | 3.22924E-09 | |
| P20930 | FLG | 2.87152332 | 0.002555182 | |
| P52926 | HMGA2 | 2.687785701 | 0.008942412 | |
| Q6UX72 | B3GNT9 | 2.576105848 | 4.91404E-09 | |
| Q92804 | TAF15 | 2.456058544 | 1.54458E-05 | |
| O95081 | AGFG2 | 2.441304592 | 2.93054E-06 | |
| P09936 | UCHL1 | 2.157884674 | 7.498E-07 | |
| Down | Q96BQ1 | FAM3D | 0.225135868 | 0.022611851 |
| Q5DID0 | UMODL1 | 0.242287446 | 0.00020958 | |
| P25815 | S100P | 0.267045455 | 3.77116E-06 | |
| P13640 | MT1G | 0.274242 | 0.0028833 | |
| P42330 | AKR1C3 | 0.290893518 | 6.40059E-07 | |
| Q9UBU3 | GHRL | 0.293081189 | 0.000379102 | |
| Q9NS71 | GKN1 | 0.311488849 | 6.5931E-06 | |
| O95994 | AGR2 | 0.330784665 | 9.59846E-06 | |
| P19075 | TSPAN8 | 0.350779233 | 7.40703E-09 | |
| P55087 | AQP4 | 0.352590936 | 0.035544439 |
Western blotting was used to further identify the expression levels of TAF15 in GIST. We detected the extracted proteins from tumor tissues (T) and matched normal tissues (N) of 18 GIST patients, including 6 NIH-H, 6 NIH-I, 3 NIH-L, and 3 NIH-VL cases. The results showed that the expression levels of TAF15 were significantly increased in tumor tissues compared with matched normal tissues (Figure 1C and D), which were in agreement with the results of our quantitative proteomics analysis (Table 2 and Supplemental Table 2).
We evaluated the expression level of TAF15 in GIST patients using immunohistochemical (IHC) analysis of tumor tissues (T) from 161 patients and 18 matched normal tissues (N). We found significantly higher levels of expression of TAF15 in tumor tissues compared to matched normal tissues (red arrows, Figure 1E and F, Supplemental Table 3). Moreover, subgroup analyses showed that the expression level of TAF15 was significantly higher in the NIH-H subgroup than in the NIH-I, NIH-L and NIH-VL subgroups and that the expression level of TAF15 was significantly higher in the NIH-I subgroup than the NIH-VL subgroup (Figure 1G and Supplemental Table 3). We found no significant difference in the expression of TAF15 between the NIH-L and NIH-VL subgroups and between the NIH-I and NIH-L subgroups. Furthermore, based on our IHC data, we found that the overexpression level of TAF15 was positively correlated with tumor size and GIST risk stage but was not significantly correlated with gender, age or tumor metastasis (Table 3). These results provided evidence that the higher expression of TAF15 may contribute to malignant progression of GIST and that TAF15 may be a latent promoter or biomarker of GIST.
| Characteristics | TAF15 expression | P value | |
| Negative | Positive | ||
| Total number | 31 | 130 | |
| Sex | 0.2730a | ||
| Male | 16 (9.9) | 53 (32.9) | |
| Female | 15 (9.3) | 77 (58.7) | |
| Age in yr | 0.8228a | ||
| ≤ 60 | 15 (9.3) | 60 (37.2) | |
| > 60 | 16 (9.9) | 70 (43.5) | |
| Tumor size | |||
| Maximum diameter in cm | |||
| ≤ 5 | 29 (18.0) | 74 (46.0) | 0.0001a |
| > 5 | 2 (1.2) | 56 (34.8) | |
| GIST risk stage | 0.0021b | ||
| Very low risk | 15 (9.3) | 30 (18.6) | |
| Low risk | 11 (6.8) | 31 (19.3) | |
| Intermediate risk | 3 (1.9) | 29 (18.0) | |
| High risk | 2 (1.2) | 40 (24.8) | |
| Metastasis | 0.1316a | ||
| - | 31 (19.3) | 121 (75.2) | |
| + | 0 (0.0) | 9 (5.6) | |
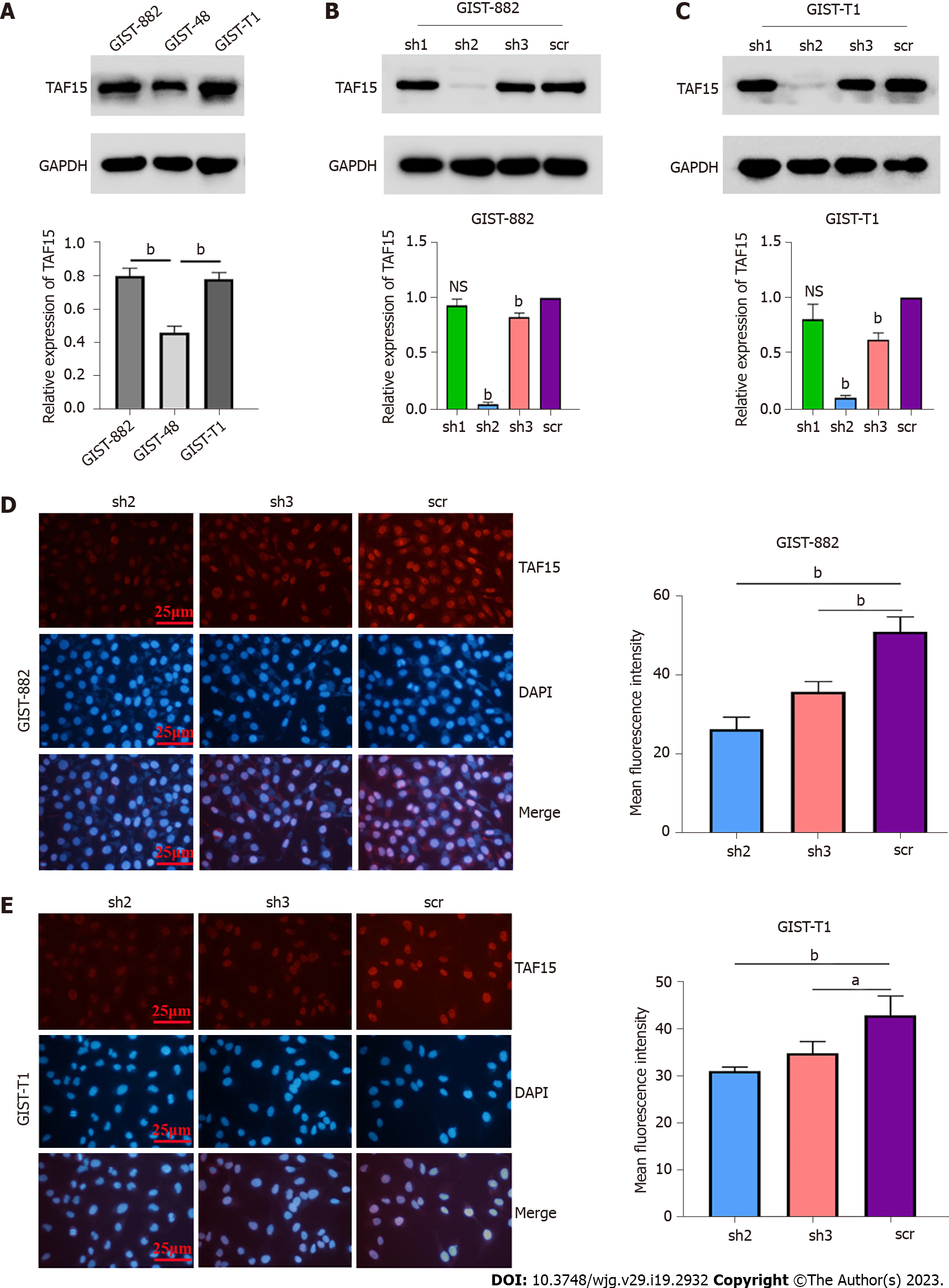
In order to understand the real role of TAF15 in GIST progression, short-hairpin RNAs (shRNAs) were used to knock down TAF15 in GIST-882 and GIST-T1 cells. GIST-882 and GIST-T1 cells were used instead of GIST-48 cells, as the expression level of TAF15 was higher in the former cell lines (Figure 2A). Three shRNAs targeting TAF15 (sh1, sh2 and sh3) were applied to knock down TAF15 protein. The results showed that sh2 and sh3 could significantly reduce the expression of TAF15 when compared to scrambled shRNA (scr), as tested by western blotting analysis (Figure 2B and C) and cell immunofluorescence assays (Figure 2D and E). Therefore, sh2 and sh3 were used to perform further experiments.
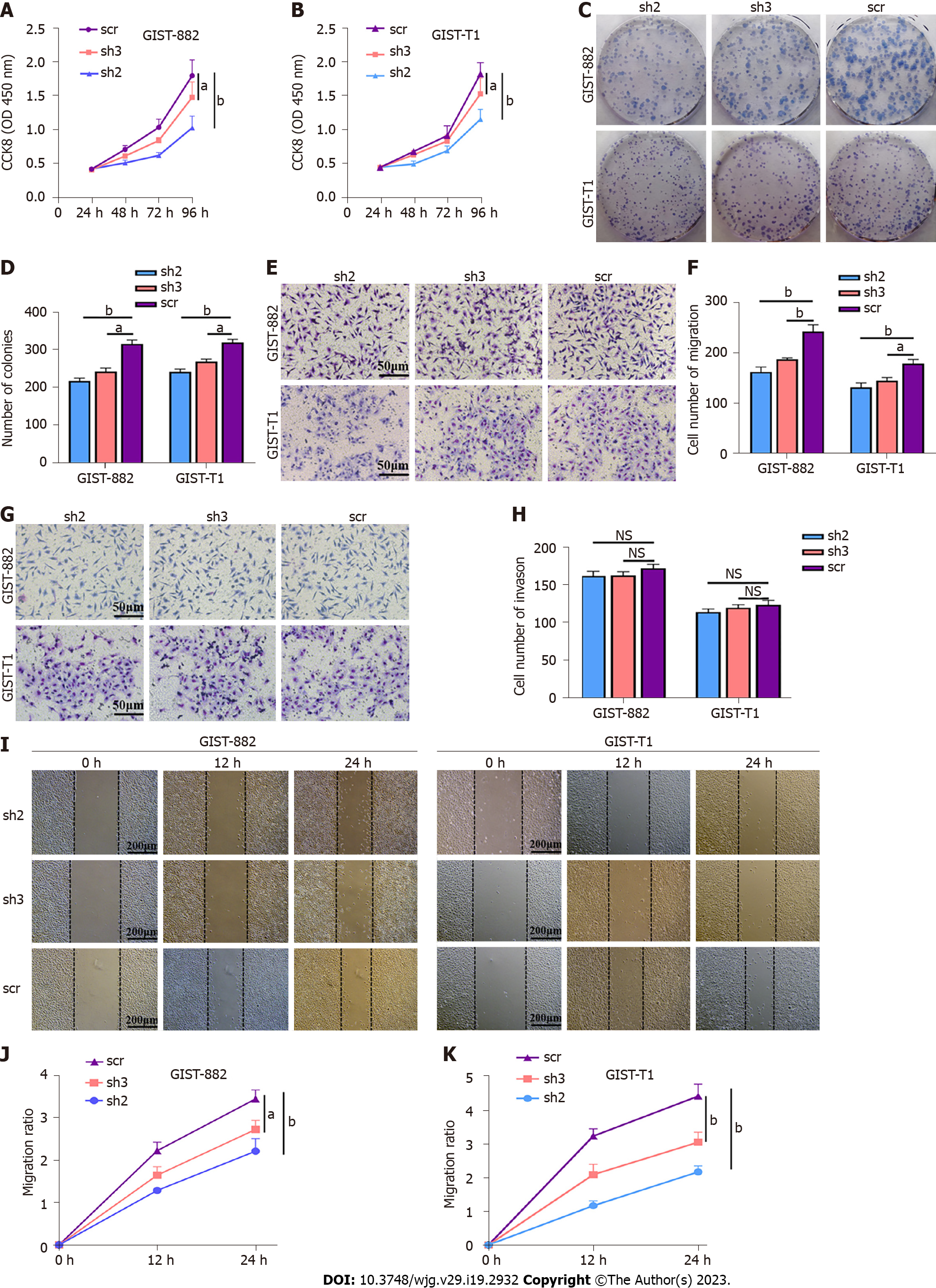
The results of the CCK-8 assay and the colony formation assay indicated that TAF15 knockdown (sh2 and sh3) exhibited a significant decrease in proliferative and colony forming ability in GIST-882 and GIST-T1 cells when compared with the scr (Figure 3A-D). Transwell assay showed that the sh-TAF15 group (sh2 and sh3) exhibited significantly weaker migratory abilities compared to the scr in GIST-882 and GIST-T1 cells (Figure 3E and F). However, there was no significant difference in the invasion ability of GIST-882 and GIST-T1 cells between the sh-TAF15 group (sh2 and sh3) and sh-scrambled group (Figure 3G and H). The results of the wound healing assay also showed that migratory abilities significantly decreased in the sh-TAF15 group (sh2 and sh3) compared to the scr in GIST-882 and GIST-T1 cells (Figure 3I-K). These findings indicated that TAF15 knockdown can significantly inhibit GIST cell proliferation and migration.
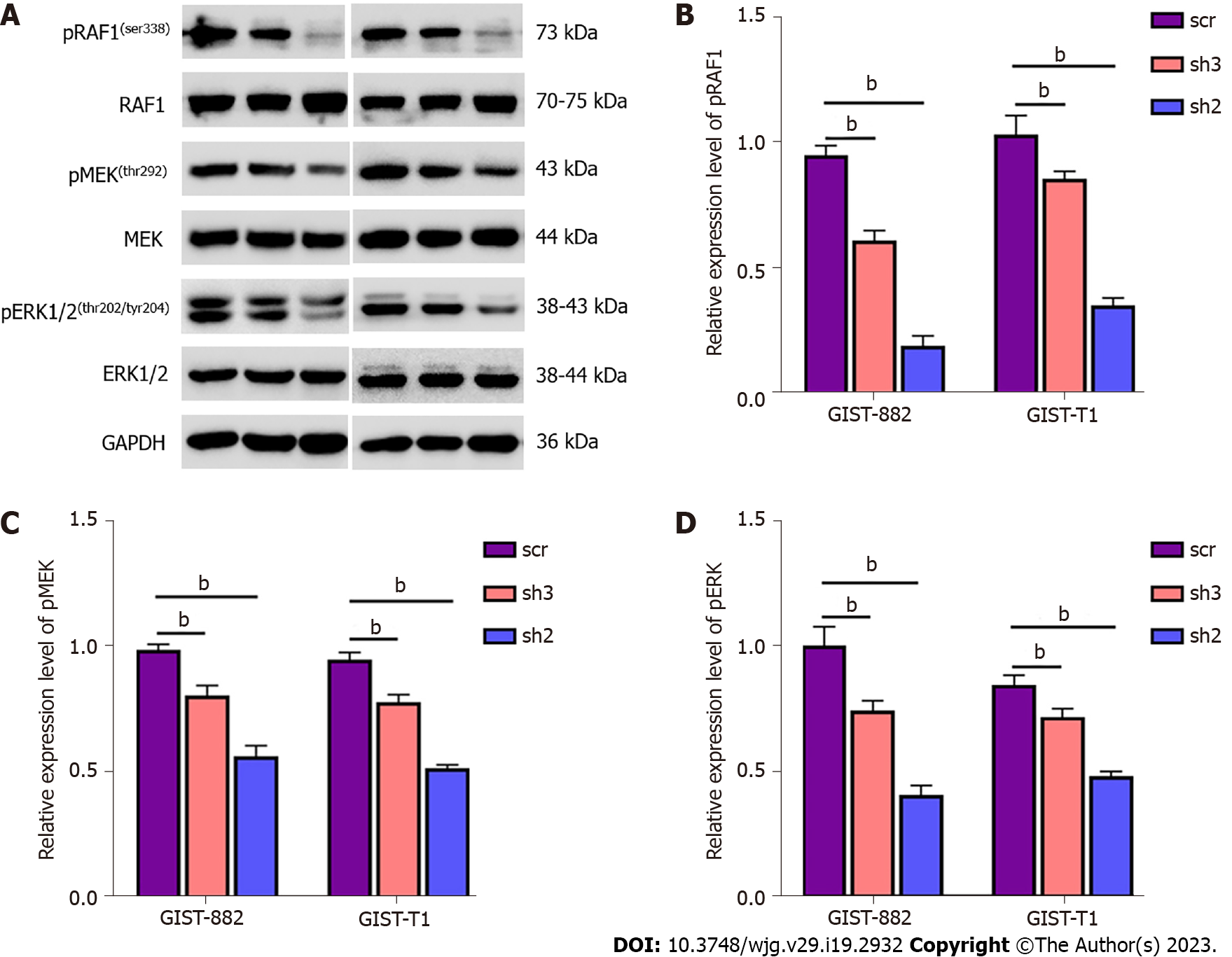
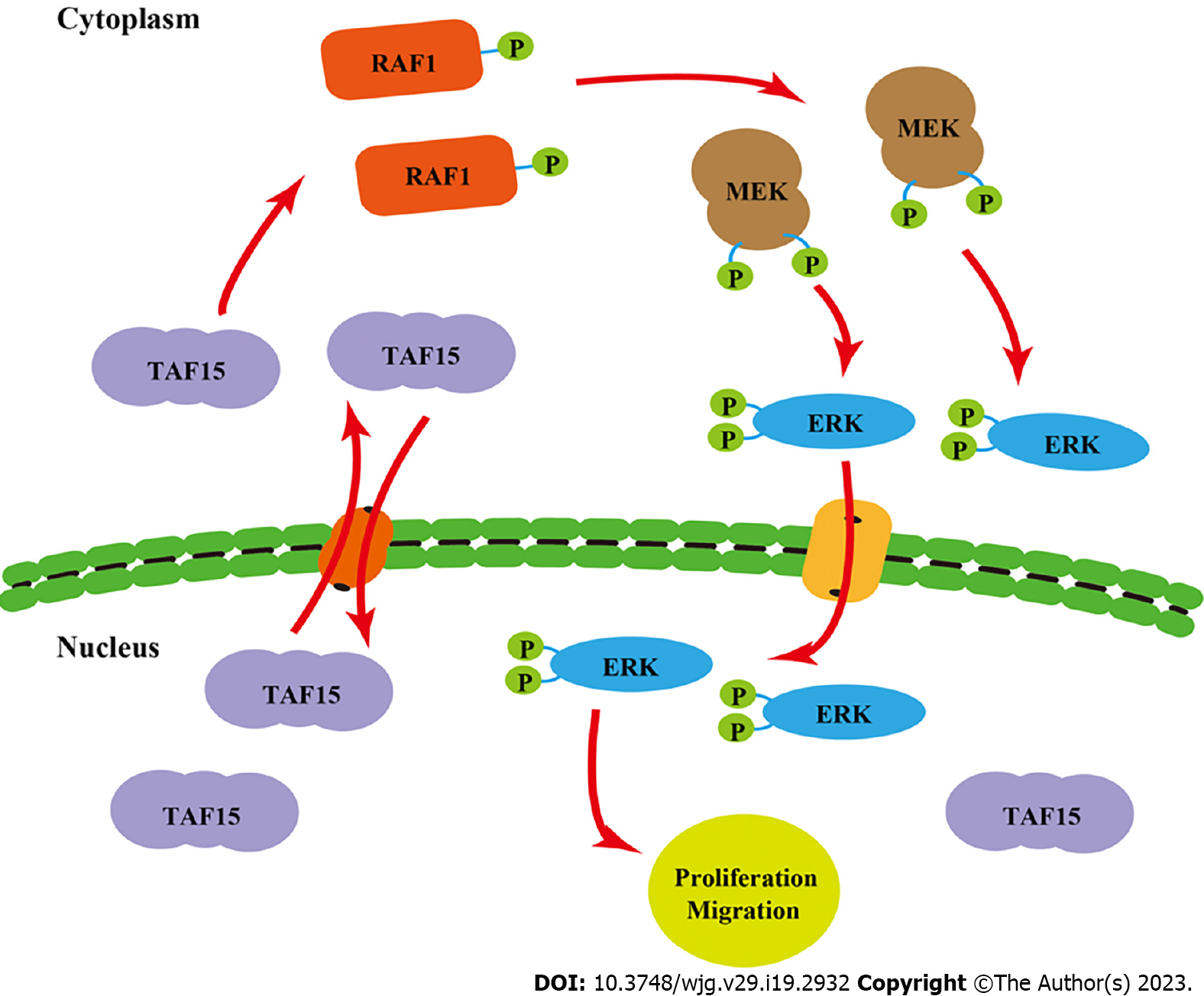
Previous studies have shown that EWS, one of the members of the FET family proteins, forms a fusion oncoprotein EWS-FLI1 whose expression leads to the activation of ERK1/2 signaling in zebrafish embryos and adult tumors[24]. In consideration of the important role of the ERK signaling pathway in many human tumors[25], we speculated that TAF15 could promote cell proliferation and migration directly through the activation of the ERK signaling pathway in GIST. Therefore, we assessed the phosphorylation level of RAF1, MEK and ERK1/2 in GIST-882 and GIST-T1 by western blotting. The results showed that the phosphorylation levels of RAF1, MEK and ERK1/2 were significantly decreased following TAF15 knockdown (sh2 and sh3). However, there was no significant change in levels of total RAF1, MEK and ERK1/2 (Figure 4). Collectively, these findings suggest that TAF15 may promote cell proliferation and migration through the activation of the RAF1/MEK/ERK signaling pathway in GIST.
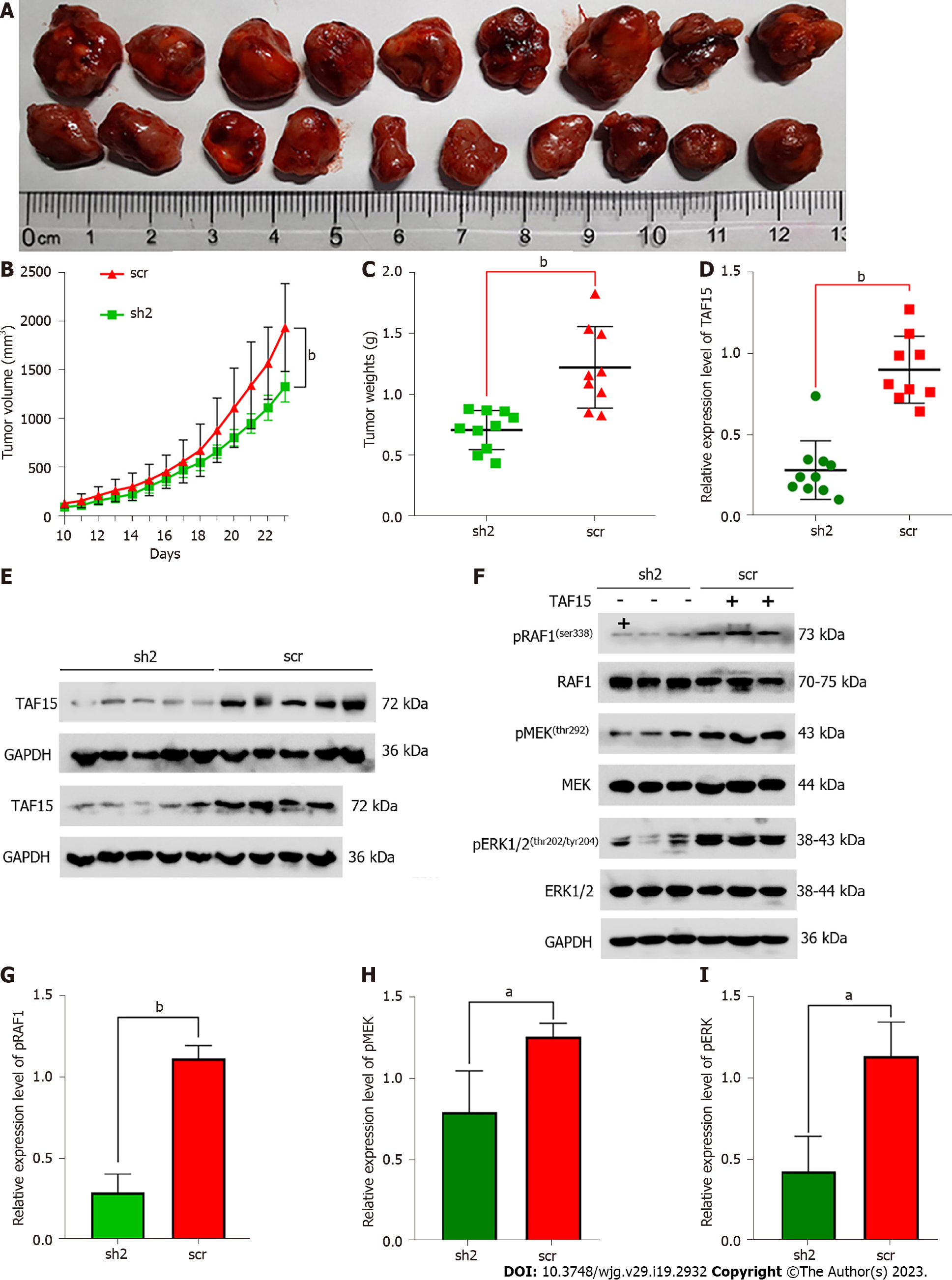
To further confirm the role of TAF15 in the progression of GIST, we established a xenograft mouse model utilizing GIST-882 cells due to their enhanced proliferative and migratory ability. sh2 was used in this experiment because of its higher knockdown efficiency, with scr as a control. Twenty nude female mice were randomly divided into two groups, and each mouse was then subcutaneously injected with either sh2 or scr cells. Unfortunately, one mouse in the scr group died on day 7 post-injection for an unknown reason. Ten days after injection, xenograft tumors in the remaining mice were detectable. The results showed that TAF15 knockdown significantly suppressed tumor growth and tumor weight (Figure 5A-C). Furthermore, western blotting analysis was used to confirm the level of TAF15 in the xenograft tumors. The results showed that the levels of TAF15 were significantly decreased in the sh2 group compared to the scr group (Figure 5D and E). Additionally, three xenograft tumors of the sh2 group and three xenograft tumors of the scr group were randomly selected to test the levels of RAF1, MEK and ERK1/2 by western blotting. The results indicated that the phosphorylation levels of RAF1, MEK and ERK also decreased significantly in the sh2 group, while the levels of total RAF1, MEK and ERK showed no significant change (Figure 5F-I). These data indicated that TAF15 may promote the growth of GIST in vivo via the activation of the RAF1/MEK/ERK signaling pathway.
KIT and PDGFRA mutations are considered to be two major factors in GIST pathogenesis[3,26], and drugs targeting KIT and PDGFRA pathways have achieved great success in the treatment of this disease[27,28]. However, GIST patients gradually become resistant to current targeted drug treatment in recent years[14,28]. Therefore, there is an unmet need to explore other latent targets.
An increasing number of biomarkers or targets have been reported to be important to GIST progression. For example, p53 is a biomarker and potential target in GIST; targeting the p53 pathway is a novel additional treatment strategy for GIST[29]. High expression of carbohydrate antigen 125 is an independent risk factor for GIST progression, and it is a significant biomarker in the overall management of GIST patients[30]. Brain-derived neurotrophic factor is over expressed in high-risk GIST patients, and it may serve as a significant prognostic factor[31].In the present study, proteomic analysis was performed in patients with GIST to explore the potential molecular biomarkers for prediction, early diagnosis or treatment.
A total of 4111 quantified proteins with 1669 DEPs (762 upregulated and 907 downregulated proteins) were revealed in our study. Among the top 10 DEPs identified, TAF15 was interesting due to its key role in promoting growth in other human tumors such as extraskeletalmyxoid sarcomas, leukemia, Ewing’s sarcomas, human neuroblastoma and lung squamous cell carcinoma[15,17,19,20,32]. TAF15 was first reported as an RNA or DNA binding protein, while more recent studies have shown that TAF15 also plays important roles in cellular stress responses, cell spreading and cell adhesion[33,34]. GAS5 binding to TAF15 could upregulate expression of HIF1A,thus promoting wound healing in diabetic foot ulcers[35].
We believe our study is the first to confirm that TAF15 is over expressed in GIST tissues and cell lines and that high expression levels of TAF15 are positively correlated with tumor size and GIST risk stage. Previous studies have shown that the inhibition of TAF15 expression could significantly reduce cellular proliferation in melanoma and lung cancer[20,21]. Similarly, our study demonstrated that TAF15 knockdown could significantly inhibit proliferation and migration of GIST cells in vitro and suppress tumor growth in vivo. These data provided evidence that overexpression of TAF15 may contribute to the malignant progression of GIST.
The latent molecular mechanism of TAF15 in GIST was also first investigated in this study. Previous studies have shown that FET family proteins are mainly present in the nucleus[34], but it is now known that they also shuttle between the nucleus and cytoplasm[36-38]. They have an extensive functionality including regulation and interaction with various proteins[39,40]. Moreover, recent work demonstrated that EWS-FLI1 fusion protein expression leads to activation of ERK1/2 signaling in zebrafish embryos and adult tumors[24]. Additionally, it is well known that the RAF/MEK/ERK signaling pathway plays an essential role in promoting cellular proliferation and survival[41].
In our study, we found TAF15 knockdown could significantly decrease the phosphorylation levels of RAF1, MEK and ERK, without significantly changing the total amounts of RAF1, MEK and ERK in GIST cell lines and xenograft tumors. Similarly, TAF15 is involved in the promotion of cell proliferation, migration and invasion of lung squamous cell carcinoma by activating the MAPK signaling pathway[22]. These results suggested that the activation of the ERK pathway may represent a new function of the TAF15 protein beyond DNA binding, mRNA splicing and mRNA transport. Taken together, these findings suggested that TAF15 shuttles between the nucleus and cytoplasm and may promote cell proliferation and migration directly through the activation of the RAF1/MEK/ERK signaling pathway in GIST (Figure 6). These observations greatly extend the functional repertoire of TAF15.
However, due to difficulty of obtaining matched tumor tissues after surgery[23], the number of paired samples used in this study for proteomic analysis, western blotting and IHC were small. Thus, we acknowledge that the accuracy of the proteomic analysis, western blotting and IHC in this research may be affected by the sample size and that the statistical analyses may be less robust due to the small sample size. Further studies will help validate some of the important and interesting findings of this study in larger GIST cohorts when available. Additional roles of TAF15 in GIST remain to be explored, as hundreds of genes lie downstream of TAF15[42].
In summary, this study utilized a proteomic analysis to find new molecular therapeutic targets for GIST and analyzed the expression level and underlying biological functions of TAF15 in GIST. Moreover, our study may be the first to demonstrate that TAF15 expression may contribute to malignant progression of GIST. TAF15promoted cell proliferation and migration in GIST via the RAF1/MEK/ERK signaling pathway. Therefore, TAF15 is expected to be a novel latent molecular biomarker or therapeutic target of GIST.
Gastrointestinal stromal tumors (GIST) are a common neoplasm with high rates of recurrence and metastasis, and its therapeutic efficacy is still not ideal. There is an unmet need to find new molecular therapeutic targets for GIST. TATA-box-binding protein-associated factor 15 (TAF15) contributes to the progress of various tumors, while the role and molecular mechanism of TAF15 in GIST progression are still unknown.
To explore the novel early diagnostic markers or therapeutic targets for the diagnosis and treatment of GIST.
To investigate new molecular therapeutic targets for GIST and explore the role and potential molecular mechanism of TAF15 in GIST progression.
Proteomic analysis was performed to explore the differentially expressed proteins in GIST. Western blotting and immunohistochemical analysis were used to verify expression levels of TAF15 in GIST tissues and cell lines. Cell counting kit-8, colony formation, transwell, wound healing and western blotting assays were applied to explore the effect and the molecular mechanism of TAF15 in GIST. The role of TAF15 in vivo was confirmed using a xenograft mouse model assay.
A total of 1669 proteins were identified as differentially expressed proteins with 762 upregulated and 907 downregulated in GIST. TAF15 was significantly upregulated in GIST tissues and cell lines. Overexpression of TAF15 was associated with larger tumor size and higher risk stage of GIST. TAF15 knockdown suppressed proliferation and migration of GIST cells in vitro and inhibited the growth of GIST in vivo. Moreover, the inhibition of TAF15 expression significantly decreased the phosphorylation level of RAF1, MEK and ERK1/2 in GIST cells and xenograft tissues, while the total levels of RAF1, MEK and ERK1/2 had no significant changes.
TAF15 may contribute to malignant progression of GIST and promote cell proliferation and migration in GIST via the activation of the RAF1/MEK/ERK signaling pathway.
TAF15 is expected to be a novel latent molecular biomarker or therapeutic target of GIST.
| 1. | Wu CE, Tzen CY, Wang SY, Yeh CN. Clinical Diagnosis of Gastrointestinal Stromal Tumor (GIST): From the Molecular Genetic Point of View. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (1)] |
| 2. | Casali PG, Abecassis N, Aro HT, Bauer S, Biagini R, Bielack S, Bonvalot S, Boukovinas I, BoveeJVMG, Brodowicz T, Broto JM, Buonadonna A, De Álava E, Dei Tos AP, Del MuroXG, Dileo P, Eriksson M, Fedenko A, Ferraresi V, Ferrari A, Ferrari S, Frezza AM, Gasperoni S, Gelderblom H, Gil T, Grignani G, Gronchi A, Haas RL, Hassan B, Hohenberger P, Issels R, Joensuu H, Jones RL, Judson I, Jutte P, Kaal S, Kasper B, Kopeckova K, Krákorová DA, Le Cesne A, Lugowska I, Merimsky O, Montemurro M, Pantaleo MA, Piana R, Picci P, Piperno-Neumann S, Pousa AL, Reichardt P, Robinson MH, Rutkowski P, Safwat AA, Schöffski P, Sleijfer S, Stacchiotti S, Sundby Hall K, Unk M, Van Coevorden F, van der Graaf WTA, Whelan J, Wardelmann E, Zaikova O, Blay JY; ESMO Guidelines Committee and EURACAN. Gastrointestinal stromal tumours: ESMO-EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29:iv267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 117] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 3. | Heinrich MC, Corless CL, Duensing A, McGreevey L, Chen CJ, Joseph N, Singer S, Griffith DJ, Haley A, Town A, Demetri GD, Fletcher CD, Fletcher JA. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708-710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1712] [Cited by in RCA: 1743] [Article Influence: 75.8] [Reference Citation Analysis (0)] |
| 4. | Miettinen M, Lasota J. Gastrointestinal stromal tumors. Gastroenterol Clin North Am. 2013;42:399-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 171] [Article Influence: 13.2] [Reference Citation Analysis (2)] |
| 5. | Xue A, Gao X, He Y, Shu P, Huang X, Sun J, Lu J, Hou Y, Fang Y, Shen K. Role of Surgery in the Management of Liver Metastases From Gastrointestinal Stromal Tumors. Front Oncol. 2022;12:903487. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 6. | Zhang D, He C, Guo Y, Li J, Li B, Zhao Y, Yu L, Chang Z, Pei H, Yang M, Li N, Zhang Q, He Y, Pan Y, Zhao ZJ, Zhang C, Chen Y. Efficacy of SCF drug conjugate targeting c-KIT in gastrointestinal stromal tumor. BMC Med. 2022;20:257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 7. | Joensuu H, Vehtari A, Riihimäki J, Nishida T, Steigen SE, Brabec P, Plank L, Nilsson B, Cirilli C, Braconi C, Bordoni A, Magnusson MK, Linke Z, Sufliarsky J, Federico M, Jonasson JG, Dei Tos AP, Rutkowski P. Risk of recurrence of gastrointestinal stromal tumour after surgery: an analysis of pooled population-based cohorts. Lancet Oncol. 2012;13:265-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 576] [Cited by in RCA: 690] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 8. | Fletcher CD, Berman JJ, Corless C, Gorstein F, Lasota J, Longley BJ, Miettinen M, O'Leary TJ, Remotti H, Rubin BP, Shmookler B, Sobin LH, Weiss SW. Diagnosis of gastrointestinal stromal tumors: A consensus approach. Hum Pathol. 2002;33:459-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2231] [Cited by in RCA: 2170] [Article Influence: 90.4] [Reference Citation Analysis (1)] |
| 9. | Li S, Chen D, Li S, Zhao Z, Yang H, Wang D, Zhang Z, Fu W. Novel Prognostic Nomogram for Recurrence-Free Survival of Patients With Primary Gastrointestinal Stromal Tumors After Surgical Resection: Combination of Prognostic Nutritional Index and Basic Variables. Front Oncol. 2020;10:581855. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 10. | Bang YH, Ryu MH, Kim HD, Lee HE, Kang YK. Clinical outcomes and prognostic factors for patients with high-risk gastrointestinal stromal tumors treated with 3-year adjuvant imatinib. Int J Cancer. 2022;151:1770-1777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 11. | Nunobe S, Sano T, Shimada K, Sakamoto Y, Kosuge T. Surgery including liver resection for metastatic gastrointestinal stromal tumors or gastrointestinal leiomyosarcomas. Jpn J Clin Oncol. 2005;35:338-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 41] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 12. | Keung EZ, Raut CP, Rutkowski P. The Landmark Series: Systemic Therapy for Resectable Gastrointestinal Stromal Tumors. Ann Surg Oncol. 2020;27:3659-3671. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 13. | Hemming ML, Heinrich MC, Bauer S, George S. Translational insights into gastrointestinal stromal tumor and current clinical advances. Ann Oncol. 2018;29:2037-2045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 14. | Joensuu H, Rutkowski P, Nishida T, Steigen SE, Brabec P, Plank L, Nilsson B, Braconi C, Bordoni A, Magnusson MK, Sufliarsky J, Federico M, Jonasson JG, Hostein I, Bringuier PP, Emile JF. KIT and PDGFRA mutations and the risk of GI stromal tumor recurrence. J Clin Oncol. 2015;33:634-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 130] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 15. | Lindén M, Vannas C, Österlund T, Andersson L, Osman A, Escobar M, Fagman H, Ståhlberg A, Åman P. FET fusion oncoproteins interact with BRD4 and SWI/SNF chromatin remodelling complex subtypes in sarcoma. Mol Oncol. 2022;16:2470-2495. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 16. | Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977-1986. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1145] [Cited by in RCA: 1078] [Article Influence: 59.9] [Reference Citation Analysis (0)] |
| 17. | Zuo L, Zhang G, Massett M, Cheng J, Guo Z, Wang L, Gao Y, Li R, Huang X, Li P, Qi Z. Loci-specific phase separation of FET fusion oncoproteins promotes gene transcription. Nat Commun. 2021;12:1491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 107] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 18. | Wang J, Cai Y, Lu H, Zhang F, Zheng J. LncRNA APOA1-AS facilitates proliferation and migration and represses apoptosis of VSMCs through TAF15-mediated SMAD3 mRNA stabilization. Cell Cycle. 2021;20:1642-1652. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 19. | Ballarino M, Jobert L, Dembélé D, de la Grange P, Auboeuf D, Tora L. TAF15 is important for cellular proliferation and regulates the expression of a subset of cell cycle genes through miRNAs. Oncogene. 2013;32:4646-4655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 50] [Article Influence: 3.6] [Reference Citation Analysis (1)] |
| 20. | Singh AK, Kapoor V, Thotala D, Hallahan DE. TAF15 contributes to the radiation-inducible stress response in cancer. Oncotarget. 2020;11:2647-2659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 21. | Schatz N, Brändlein S, Rückl K, Hensel F, Vollmers HP. Diagnostic and therapeutic potential of a human antibody cloned from a cancer patient that binds to a tumor-specific variant of transcription factor TAF15. Cancer Res. 2010;70:398-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 22. | Zhu H, Liu Q, Yang X, Ding C, Wang Q, Xiong Y. LncRNA LINC00649 recruits TAF15 and enhances MAPK6 expression to promote the development of lung squamous cell carcinoma via activating MAPK signaling pathway. Cancer Gene Ther. 2022;29:1285-1295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (1)] |
| 23. | Li J, Ye Y, Wang J, Zhang B, Qin S, Shi Y, He Y, Liang X, Liu X, Zhou Y, Wu X, Zhang X, Wang M, Gao Z, Lin T, Cao H, Shen L; Chinese Society Of Clinical Oncology Csco Expert Committee On Gastrointestinal Stromal Tumor. Chinese consensus guidelines for diagnosis and management of gastrointestinal stromal tumor. Chin J Cancer Res. 2017;29:281-293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 142] [Cited by in RCA: 148] [Article Influence: 16.4] [Reference Citation Analysis (1)] |
| 24. | Vasileva E, Warren M, Triche TJ, Amatruda JF. Dysregulated heparan sulfate proteoglycan metabolism promotes Ewing sarcoma tumor growth. Elife. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 25. | Ullah R, Yin Q, Snell AH, Wan L. RAF-MEK-ERK pathway in cancer evolution and treatment. Semin Cancer Biol. 2022;85:123-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 342] [Article Influence: 68.4] [Reference Citation Analysis (0)] |
| 26. | Miettinen M, Sarlomo-Rikala M, Lasota J. Gastrointestinal stromal tumors: recent advances in understanding of their biology. Hum Pathol. 1999;30:1213-1220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 559] [Cited by in RCA: 522] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 27. | Serrano C, Mariño-Enríquez A, Tao DL, Ketzer J, Eilers G, Zhu M, Yu C, Mannan AM, Rubin BP, Demetri GD, Raut CP, Presnell A, McKinley A, Heinrich MC, Czaplinski JT, Sicinska E, Bauer S, George S, Fletcher JA. Complementary activity of tyrosine kinase inhibitors against secondary kit mutations in imatinib-resistant gastrointestinal stromal tumours. Br J Cancer. 2019;120:612-620. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 125] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 28. | Demetri GD, Reichardt P, Kang YK, Blay JY, Rutkowski P, Gelderblom H, Hohenberger P, Leahy M, von Mehren M, Joensuu H, Badalamenti G, Blackstein M, Le Cesne A, Schöffski P, Maki RG, Bauer S, Nguyen BB, Xu J, Nishida T, Chung J, Kappeler C, Kuss I, Laurent D, Casali PG; GRID study investigators. Efficacy and safety of regorafenib for advanced gastrointestinal stromal tumours after failure of imatinib and sunitinib (GRID): an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet. 2013;381:295-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 942] [Cited by in RCA: 1065] [Article Influence: 81.9] [Reference Citation Analysis (0)] |
| 29. | Wu CE, Chen CP, Huang WK, Pan YR, Aptullahoglu E, Yeh CN, Lunec J. p53 as a biomarker and potential target in gastrointestinal stromal tumors. Front Oncol. 2022;12:872202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 30. | Sui C, Lin C, Tao T, Guan W, Zhang H, Tao L, Wang M, Wang F. Prognostic significance of serum CA125 in the overall management for patients with gastrointestinal stromal tumors. BMC Gastroenterol. 2023;23:25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 31. | Tian GA, Xu WT, Sun Y, Wang J, Ke Q, Yuan MJ, Wang JJ, Zhuang C, Gong Q. BDNF expression in GISTs predicts poor prognosis when associated with PD-L1 positive tumor-infiltrating lymphocytes. Oncoimmunology. 2021;10:2003956. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 32. | Xiong Y, Yang C, Yang X, Ding C, Wang Q, Zhu H. LncRNA MIR9-3HG enhances LIMK1 mRNA and protein levels to contribute to the carcinogenesis of lung squamous cell carcinoma via sponging miR-138-5p and recruiting TAF15. Pathol Res Pract. 2022;237:153941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 33. | de Hoog CL, Foster LJ, Mann M. RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell. 2004;117:649-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 212] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 34. | Andersson MK, Ståhlberg A, Arvidsson Y, Olofsson A, Semb H, Stenman G, Nilsson O, Aman P. The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol. 2008;9:37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 233] [Cited by in RCA: 266] [Article Influence: 14.8] [Reference Citation Analysis (1)] |
| 35. | Peng WX, He PX, Liu LJ, Zhu T, Zhong YQ, Xiang L, Peng K, Yang JJ, Xiang GD. LncRNA GAS5 activates the HIF1A/VEGF pathway by binding to TAF15 to promote wound healing in diabetic foot ulcers. Lab Invest. 2021;101:1071-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 36. | Marko M, Vlassis A, Guialis A, Leichter M. Domains involved in TAF15 subcellular localisation: dependence on cell type and ongoing transcription. Gene. 2012;506:331-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 37. | Zinszner H, Sok J, Immanuel D, Yin Y, Ron D. TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J Cell Sci. 1997;110 ( Pt 15):1741-1750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 254] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 38. | Zakaryan RP, Gehring H. Identification and characterization of the nuclear localization/retention signal in the EWS proto-oncoprotein. J Mol Biol. 2006;363:27-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 80] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 39. | Law WJ, Cann KL, Hicks GG. TLS, EWS and TAF15: a model for transcriptional integration of gene expression. Brief Funct Genomic Proteomic. 2006;5:8-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 157] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 40. | Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173-1178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2192] [Cited by in RCA: 2091] [Article Influence: 99.6] [Reference Citation Analysis (0)] |
| 41. | Yaeger R, Corcoran RB. Targeting Alterations in the RAF-MEK Pathway. Cancer Discov. 2019;9:329-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 329] [Article Influence: 47.0] [Reference Citation Analysis (0)] |
| 42. | Kume K, Ikeda M, Miura S, Ito K, Sato KA, Ohmori Y, Endo F, Katagiri H, Ishida K, Ito C, Iwaya T, Nishizuka SS. α-Amanitin Restrains Cancer Relapse from Drug-Tolerant Cell Subpopulations via TAF15. Sci Rep. 2016;6:25895. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Li S, China; Mankotia DS, India S-Editor: Fan JR L-Editor: Filipodia P-Editor: Yu HG