Published online Apr 14, 2023. doi: 10.3748/wjg.v29.i14.2064
Peer-review started: September 28, 2022
First decision: October 17, 2022
Revised: October 23, 2022
Accepted: March 21, 2023
Article in press: March 21, 2023
Published online: April 14, 2023
Processing time: 196 Days and 20.2 Hours
As of June 2022, more than 530 million people worldwide have become ill with coronavirus disease 2019 (COVID-19). Although COVID-19 is most commonly associated with respiratory distress (severe acute respiratory syndrome), meta-analysis have indicated that liver dysfunction also occurs in patients with severe symptoms. Current studies revealed distinctive patterning in the receptors on the hepatic cells that helps in viral invasion through the expression of angiotensin-converting enzyme receptors. It has also been reported that in some patients with COVID-19, therapeutic strategies, including repurposed drugs (mitifovir, lopinavir/ritonavir, tocilizumab, etc.) triggered liver injury and cholestatic toxicity. Several proven indicators support cytokine storm-induced hepatic damage. Because there are 1.5 billion patients with chronic liver disease world
Core Tip: Several review articles have contributed to the pathophysiology, therapeutic strategies, vaccine development, and clinical trials of coronavirus disease 2019 (COVID-19). Since the liver is the primary site of immune protein synthesis, any liver defect may compromise the immune system. Patients with chronic liver disease are at a higher risk of severe COVID-19. This review article demonstrated the pathophysiology and molecular mechanisms responsible for more severe outcomes in patients with hepatic defects. Further, we critically evaluated the molecular mechanisms concerning hepatotropism in patients with COVID-19, which could lead to the development of new therapeutics.
- Citation: Khullar N, Bhatti JS, Singh S, Thukral B, Reddy PH, Bhatti GK. Insight into the liver dysfunction in COVID-19 patients: Molecular mechanisms and possible therapeutic strategies. World J Gastroenterol 2023; 29(14): 2064-2077
- URL: https://www.wjgnet.com/1007-9327/full/v29/i14/2064.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i14.2064
On December 31, 2019, reports surfaced of patients with an unusual severe acute respiratory syndrome (pneumonia) in Wuhan, China. On January 7, 2020, the causal agent responsible for the mysterious deaths was branded as coronavirus disease 2019 (COVID-19) by the International Classification Committee of Viruses[1]. A severe global pandemic was declared on March 11, 2020. Since then, there have been 623893894 confirmed cases of COVID-19, including 6553936 deaths as of October 21, 2022[2].
The clinical manifestation of COVID-19 is usually interpreted as severe lung infection (acute respiratory distress syndrome), causing turmoil in the patient’s respiratory system and even death[3]. Many individuals admitted to intensive care units were known to have hepatic and heart-related complications[4]. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in patients with chronic liver disease leads to an elevated and dysregulated immune response[5]. Even healthy individuals infected with COVID-19 displayed liver dysfunction, increasing morbidity and mortality of such patients[6].
Hepatologists have been working to deduce the relationship between COVID-19 and the liver. About 300 million patients with chronic liver diseases in China alone had severe COVID-19 symptoms[7]. It was advocated that SARS-CoV-2 invasion might trigger reactivation of existing liver disorders in the patient, causing hepatotoxicity[8]. In addition, hepatic patients are more prone to COVID-19 infection[9]. Some COVID-19 cases have reported multiorgan failure significantly increasing cytokine levels, including vascular endothelial growth factor, interleukin (IL)-6, macrophage inflammatory protein 1α, and macrophage inflammatory protein 1β[9-11].
Since the liver is a primary site of the synthesis of proteins associated with immunity, it inhibits infectious microbes from entering the bloodstream from the gut. Any liver defect would thus cause a compromised immune system[12]. Increased levels of hepatic biomarkers including alanine transaminase and aspartate transaminase indicate a close pathophysiological association between the liver and COVID-19. Obesity predisposes individuals to metabolic disorders, diabetes, and insulin resistance, which can lead to chronic liver disease and may culminate into cirrhosis, fibrosis, and even hepatocellular carcinoma[13]. This indicates a dire need to investigate the pathophysiology and molecular mechanisms responsible for hindering the immune system, which leads to more severe outcomes in patients with cardiovascular and hepatic defects[14]. In this article, we comprehensively evaluated the invasion and spread of SARS-CoV-2 as a tool to improve therapeutic strategies against liver damage in patients with COVID-19.
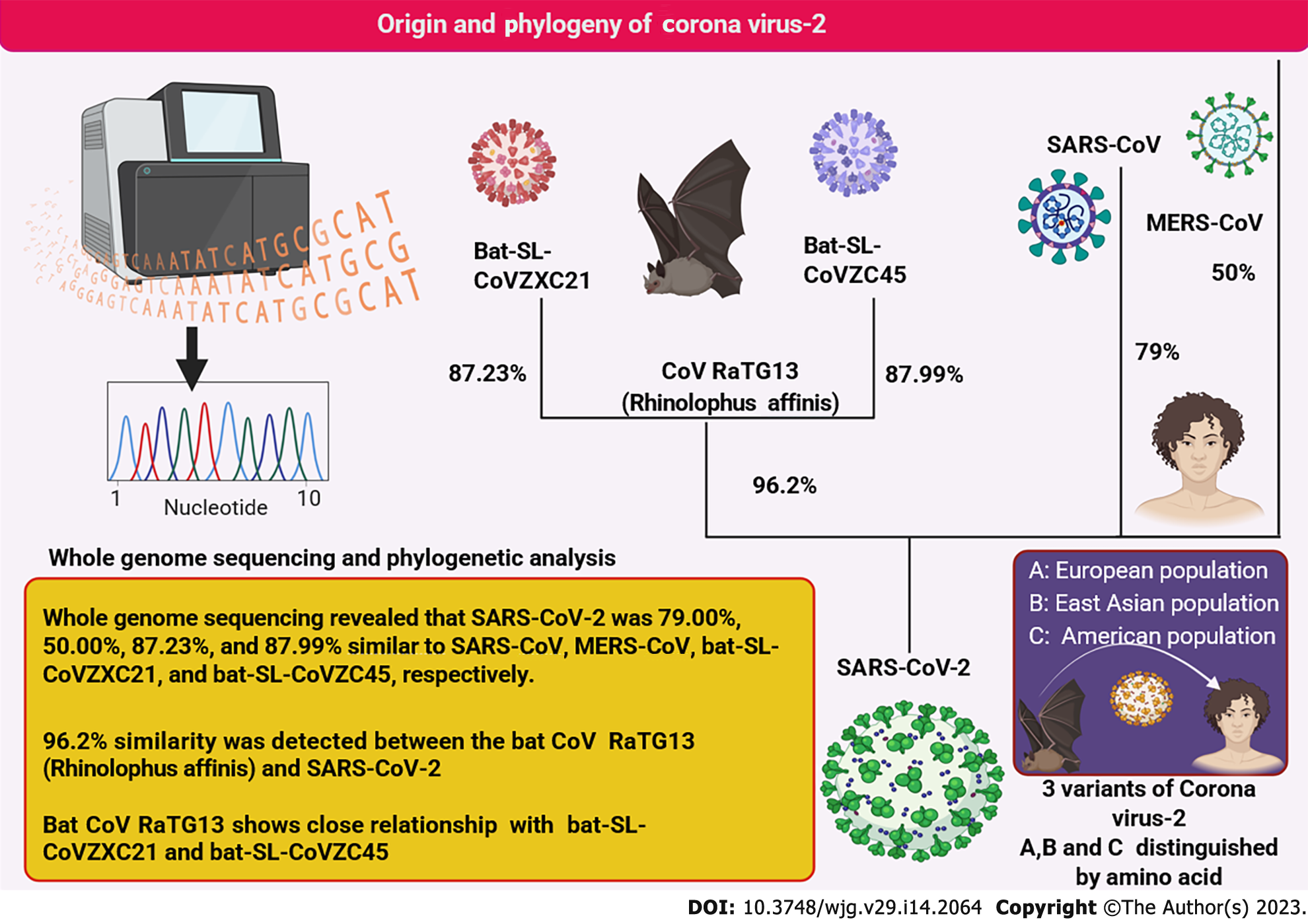
The beta coronavirus isolated from COVID-19 patients was identified using whole genome sequencing and matrix representation with parsimony phylogenetic analysis[15,16]. This virus is the closest relative of Rhinolophus affinis virus (bat CoV RaTG13) with more than 96% similarity[17,18]. Thus, the bat is hypothesized to be its most suitable natural host. The phylogenetic interrelationship is depicted in Figure 1. Whole genome studies revealed the presence of three SARS-CoV-2 strains (A, B, and C) among various human populations[19]. Originally strain A was found in the European population and was regarded as the ancestral strain closest to the bat coronavirus[20,21]. Strain B was primarily found in East Asian populations[22]. Strain C, however, was more common among Americans, thus defining and tracking their outbreak areas[23]. Although the phylogenetic relationship of SARS-CoV-2 is still under investigation, it is well established that the preferred reservoir host of this virus is the bat and was transferred to humans from consumption of wild bats[18,24,25]. These are undoubtedly among the most prevalent RNA viruses (positive sense) that can infect a wide range of hosts[6,26]. Whole genome sequencing demonstrated that SARS-CoV-2 is most closely related to the CoV RaTG13 virus[27,28].
After almost three years since the discovery of SARS-CoV-2, the mechanism that allows SARS-CoV-2 to jump to a human host and invade hepatocytes remains unclear. Upregulated transaminases in patients with COVID-19 indicate a strong link between SARS-CoV-2 and liver injury[29,30]. The genetic algorithm for detecting recombination demonstrated that SARS-CoV-2 possesses a mosaic genome comparable to five coronaviruses (probable donor strains), namely Rhinolophus affinis RaTG13 coronavirus, Rhinolophus pusillus RpYN06, Rhinolophus pusillus BANAL-103, Rhinolophus malayanus RmYN02, and Rhinolophus malayanus BANAL-52[31-33]. The spike protein of SARS-CoV-2 has more similarity with Rhinolophus affinis RaTG13, and human angiotensin-converting enzyme 2 (ACE2) interaction is closely related to Rhinolophus malayanus BANAL-52. Another noteworthy feature is the absence of furin cleavage sites in all of these viruses[34].
SARS-CoV-2 has a deadly combination of disease severity and transmissibility[35,36]. Coronaviruses are remarkable entities (125 nm in size) with one of the heftiest viral RNA genomes, accounting for 30000 nucleotides and equipped with an extraordinary ability to correctly repair drug-induced mutations[37]. The chances of zoonotic transmission or accidental spill-over through exponential proliferation are also possible. In addition, a new host like a human would lack previous immunity to the pathogen, which provides a compatible incapacitating host defense mechanism[38]. Above all, the increasing human-animal interactions due to deforestation, hunting, domestication, wet market, and wild animals as a food source in many countries further increases the chance of viruses to adapt to infect humans[39]. Five SARS-CoV-2 strains are currently prevalent[40-44] (Table 1).
| Sr. No. | Strain | Mutation | Host entry | Location | Time of first report | Ref. |
| 1 | Alpha (B.1.1.7) | N501Y mutation on the RBD | The affinity between RBD and ACE2 is significantly increased | United Kingdom | December 2020 | [40-42] |
| 2 | Beta (B.1.351) | N501Y mutation on the RBD N417/K848/Y501 | The affinity between RBD and ACE2 is significantly increased | South Africa | December 2020 | [41-43] |
| 3 | Gamma (P.1) | N501Y mutation on the RBD N417/K848/Y501 | The affinity between RBD and ACE2 is significantly increased | Brazil | January 2021 | [41,42,44] |
| 4 | Delta (B.1.617.2) | Absence of N501Y mutation | No effect | India | December 2020 | [41,42] |
| 5 | Omicron (B.1.1.529) | N501Y mutation on the RBD | The affinity between RBD and ACE2 is significantly increased | South Africa | November 2021 | [41,42] |
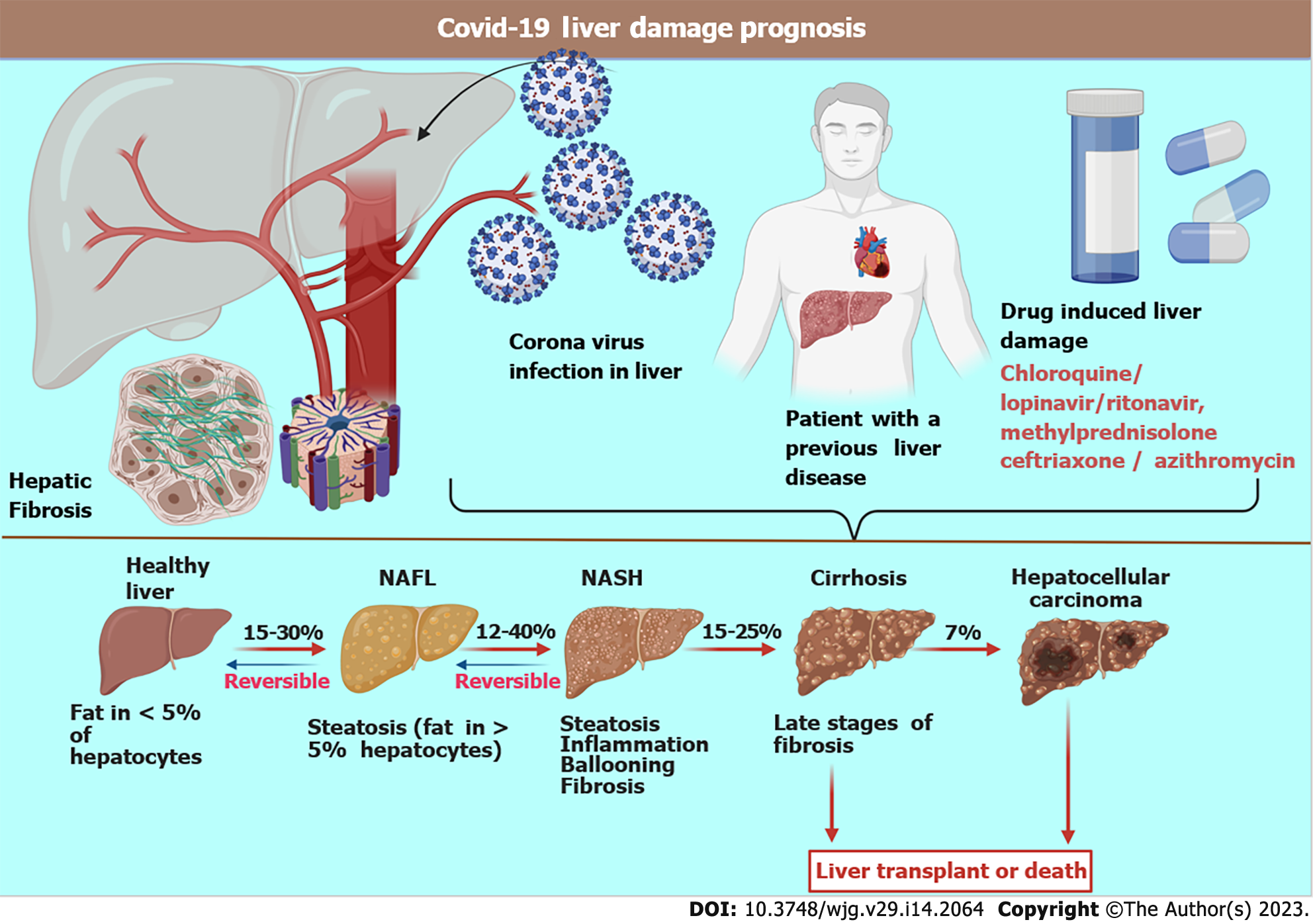
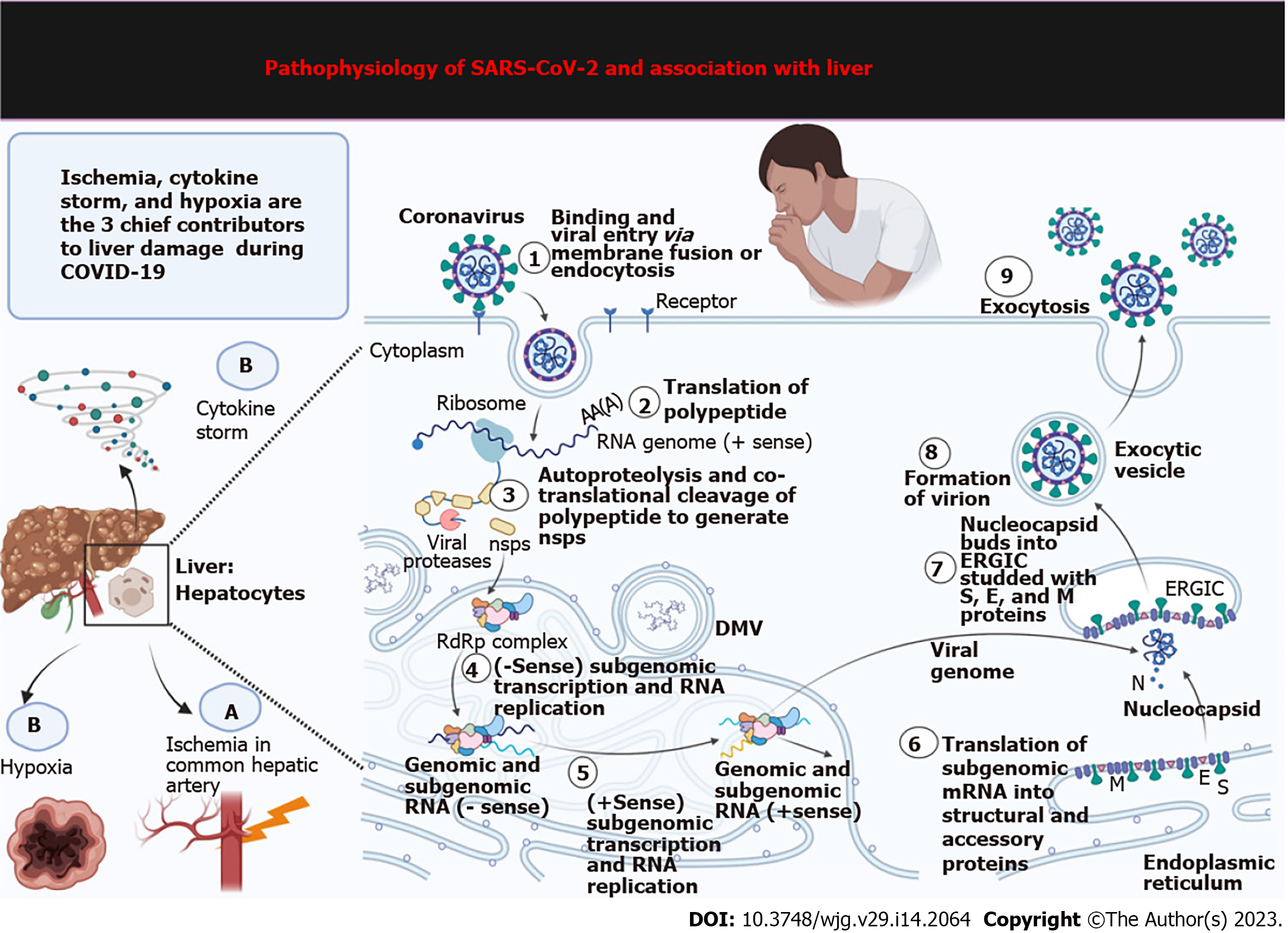
Various factors govern the prognosis of this disease based upon pre-existing health conditions, comorbidity, age, course of treatment adopted, and the response to treatment. The overall fatality rate is above 2%. Like most pathogens, the liver serves as one of the preferred sites of proliferation spots for coronaviruses because it is a common gateway for the blood[45,46]. It has been observed in many studies that one-third of patients with COVID-19 develop liver dysfunction, which was more frequent among elderly, male patients[47]. A survey conducted on 4000 seriously ill COVID-19 patients confirmed an inflated mortality rate after 3 mo, causing death in 31% of the patients[48]. Autopsies and post-mortem biopsies of the liver revealed infection of cholangiocytes, hepatocytes, and endothelial cells with SARS-CoV-2, which led to severe liver damage. Mechanisms of injury include major hepatocyte ballooning, eosinophilic action creating a cytokine storm, hypoxia, and ischemia leading to liver necrosis (Figure 2)[49-51].
Hepatic fat accumulation indicating microvesicular steatosis is a clear consequence of SARS-CoV-2[52]. Additionally, lobular inflammation and fibrosis cause severe liver cirrhosis. Cirrhosis further reduces blood flow through the liver, thereby increasing blood pressure in the hepatic vein, which is supplied by the intestines and spleen. Once SARS-CoV-2 enters the human body, major histocompatibility complexes ensure the release of proinflammatory (acquired and innate)[53-55]. SARS-CoV-2 binds to the ACE2 receptor to enter the target cells[56]. These receptors are typically expressed on the bile duct epithelial cells, sinusoidal and capillary endothelial cells, and hepatocytes[57-59]. The viral antigen epitopes are recognized by antigen-presenting cells (macrophages and dendritic cells). They are presented to CD4+ T helper cells, lymphocytes, and natural killer cells, which activate B cells and CD8+ cytotoxic T cells[60-63].
Several patients with COVID-19 have abnormal biochemistry, displaying fluctuating levels of vital enzymes and biomolecules concerning hepatobiliary manifestations[64]. Patients with COVID-19 have decreased hepatic functions, typically displayed as increased liver enzymes and alanine transaminase and aspartate transaminase levels[5,65-67]. Several case studies observed these findings in more than 50% of critically ill COVID-19 patients[68]. Liver injury, chronic congestion, and nodular proliferation in patients with COVID-19 may occur, with more than 70% of patients developing steatosis[69,70]. The pathogenesis of hepatic damage in patients with COVID-19 is multifactorial, including pre-existing hepatic disease, hypoxia, ACE2 aided viral invasion and damage, ischemia, and drug-induced liver injury[71,72] (Figure 3).
The specificity of the virus to preferably invade hepatocytes is considered hepatotropism[65,73-75]. According to current research, SARS-CoV-2 does not specifically display hepatotropism but shows a preference for hepatocytes in patients with a pre-existing liver disease or a compromised immune system[76,77]. Patients with an existing liver disease display severe and prolonged symptoms of SARS-CoV-2 because the immune system dysfunction displays a more pronounced effect[45]. A meta-analysis of 90000 patients with COVID-19 pertaining to 40 case studies in the United States and China provided strong evidence for hepatic deterioration[78]. Hepatic frailty makes liver cells more susceptible and sensitive to COVID-19[79,80]. COVID-19 and comorbidities of hepatic diseases is a global phenomenon[80,81].
The invasion of SARS-CoV-2 virus in the human body, its genomic single-stranded RNAs, and the replicative double-stranded RNAs are sensed by cytosolic RNA sensors. These are then identified and bound to NOD-like receptors, endosomal toll-like receptors, melanoma differentiation-associated gene 5, and retinoic acid-inducible gene-I-like receptors[82]. These receptors then stimulate the next set of effectors molecules downstream interferon (IFN) regulatory factor 3/7 (IRF3/7); activator protein-1; and nuclear factor-κB (NF-κB). The next step encompasses the synthesis of pro-inflammatory cytokines, namely, interleukin (IL)-2, IL-10, IL-6, IL-8; and IFN-I, by activating their transcription. These IFN-I molecules are thus supposed to be the first line of defense to combat and clear the viral particles from the body; these thus induce signal transducer molecules the Janus kinase 1 (JAK1)/tyrosine kinase 2 and transcription 1/2 (STAT1/2), turning on the JAK1/TYK2-STAT1/2 pathway. This generates STAT1/2/IRF9 complex that additionally triggers the transcription of IFN-stimulated genes. Thus, a cascade of events leads to the massive synthesis of antiviral chemicals: Procalcitonin; IL-6, CCL-5, IL-1, IFN-alpha, CXCL10, and CXCL-8, C-reactive protein[83]. Many studies have hinted at the unconventional triggering of certain supplementary systemic inflammatory responses leading to uncontrolled immune responses signaled by a storm of cytokines produced due to the activation of NF-κB and mitogen-activated protein kinase (MAPK) pathways[82]. This is commonly known as systemic inflammatory response syndrome, where a horde of immune cells (B cells, T-cells, natural killer cells, dendritic cells, neutrophils, and macrophages) bring about a cumulative and exaggerated response[84]. Apoptosis and cell death remain the culminating stage regulated by the MAPK pathway. Pyroptosis is a specialized mechanism induced by coronaviruses to prevent viral spread leading to an inflammatory caspase-1-dependent cell death in patients in response to rapid viral replication within infected cells[85]. In this activation process, the virus secures its persistence through the PI3 kinase/Akt pathway[86].
Scientists worldwide could conceive fairly early the devastating effects of SARS-CoV-2, and social distancing was the only way out since impending outcomes were far from the view[87,88]. As an opportunist virus, it intimidated the whole world, shutting down everyday life and hampering the economy and health worldwide. Though we have successfully tamed this dangerous and feral pathogen, the efficacy of existing vaccines and drug therapies in preventing SARS-CoV-2 variants is still a matter of concern[87-89]. Vaccines were tracked on the plan to target spike proteins of the SARS-CoV-2, which the virus variants inventively cons[90-92]. Several novel vaccines, as well as drugs, have ardently helped in tackling these viruses. Since the virus shows high transmissibility and the future modulation in these viruses is erratic and unforeseen[90,92,93], prevention and management strategies should entail a multi-omic, closed-loop follow-up and holistic approach comprising scientists, government authorities, clinicians, pharmacists, and as the general public. Thus, prevention and management, including pharmacologic therapies against COVID-19, have been worked out under different approaches certified under Emergency Use Authorization[94-96]. Several therapeutic strategies are followed depending on the patient’s condition as diagnosed by the clinician[97,98].
More appropriate to provide in the later stage of COVID-19 infection. Clinical trials conducted on 113 COVID-19 patients critically suffering from this disease, with both Baricitinib (inhibitor of Janus kinase) and Anakinra (IL-1 antagonists on 52 COVID-19 patients), have shown promising results in the case of COVID-19 patients facing hyperinflammation (cytokine storm). These offer a dual inhibitory effect by preventing both entries of SARS-CoV-2 and preventing an exaggerated cytokine response[99-102]. Such trials have attested to the efficacy of critical-stage COVID-19 patients, especially those with hepatic complaints[103,104]. Table 2 shows the currently used effective vaccines developed and successfully reduced morbidity and mortality across the world[105-112].
| Sr. No. | Name of vaccine | FDA approval | Type of vaccine | Manufactured by | Efficacy | No. of doses | Safety profile | Ref.
|
| 1 | NVX-CoV2373 vaccine | December 20, 2021 | Recombinant SARS-CoV-2 nanoparticle | Novavax | 92.6% | 2 | Safe till date | [105] |
| 2 | BNT162b2 vaccine | FDA issued a EUA on December 11, 2020 | mRNA-based | BioNTech/Pfizer | 95% | 2 | Safe till date | [106] |
| 3 | mRNA-1273 vaccine | FDA issued a EUA on December 18, 2020 | mRNA-1273 based | Moderna | 94.1% | 2 | Safe till date | [107] |
| 4 | ChAdOx1 nCoV-19 vaccine | Not yet received a EUA or approval from the FDA | Recombinant spike protein vaccine | Serum institute of India, private limited | 70.4% | 2 | Vaccine-induced immune thrombotic thrombocytopenia | [108] |
| 5 | Ad26.COV2 | EUA by the FDA on February 27, 2021 | Recombinant vaccine | Janssen-Cilag International, NV Belgium | 73.1% | 1 | Vaccine-induced immune thrombotic thrombocytopenia | [109] |
| 6 | Covaxin | EUA for adults | Whole inactivated virus-based | Bharat Biotech in collaboration with ICMR and NIV, India | 64% | 2 | Safe till date | [110] |
| 7 | Sputnik V | EUA qualified | Human adenovirus vector | Russian direct investment fund | 97.2% | 2 | Safe till date | [111] |
| 8 | CoronaVac | FDA issued under EUA | Inactivated virus alum-adjuvanted candidate vaccine | Sinovac biotech, China | 51% against symptomatic SARS-CoV-2 infection, 100% against severe COVID-19 | 2 | Safe till date | [112] |
These are more suitable during the early phase of Corona infection. Molnupiravir, a very effective drug that reduces both morbidity and mortality; paxlovid: Reduced 89% mortality (trial conducted on 1219 patients); remdesivir, hydroxychloroquine, lopinavir/ritonavir, ivermectin, and chloroquine are all Food and Drug Administration approved, but show little or no effect over Coronavirus; it is even not effective against corona variants[113,114]. Therefore, they are not recommended in case of patients with hepatic trouble during COVID-19. Also, some of these drugs (lopinavir/ritonavir, mitifovir, and tocilizumab) are not recommended and prescribed to patients with pre-existing liver diseases as they are known to cause cholestatic toxicity and hepatic injury[115-117].
Antibodies naturally produced by the body of recovering patients or stimulated through vaccination can block the attachment and hence the entry of an enveloped viral pathogen inside the cell, conferring lifelong immunity[118-121]. Convalescent plasma transfusion therapy with a high anti-SARS-CoV-2 immunoglobulin G (IgG) titer effectively lowered the mortality of critical COVID-19 patients[122-124]. The bamlanivimab and etesevimab antibody combination has been found to be super effective in COVID-19 patients with 87% lower death rate[125]. Another antibody cocktail, REGN-COV2, constitutes a group of two IgG1 antibodies (casirivimab and imdevimab) that target the receptor binding domain of SARS-CoV-2 and thereby reduce both morbidity and mortality of COVID-19 patients by 70%[126].
Extraordinary efforts have reaped fruits. As per the World Health Organization report on COVID-19 Vaccine Implementation Analysis & Insights, 63.4% of the World’s population today stands vaccinated against COVID-19 in September 2022 (https://www.who.int/publications/m/item/covid-19-vaccine-implementation-analysis-insights-2-september-2022), and India alone proudly puts up 68% of its population in the list of a fully vaccinated cluster. This was made possible due to the untiring efforts of clinicians and researchers braced and heavily funded by the government and private agencies to curb this callous pandemic. It is anticipated that once 100% global vaccination is achieved, the virus will no longer be felonious. However, there are reasons to negate this notion. One explanation is that despite marshy governmental efforts, many people are vaccine-hesitant for inexplicable motives which may hamper virus block[127,128]. Even if this temper is somehow overcome, the dynamics of remodelling human immunity to ongoing viral mutations and evolution is worth consideration. The co-evolution may equip the virus with new immune strategies to escape the human immune defense mechanism and maintain its virulence. According to the United States Centres for Disease Control and Prevention, viruses with new mutations are specifically a matter of concern and shall not be considered lightly[128]. Lifestyle, assess to wet markets, climate change, and increased animal-human interactions offer preferred gateways and richer niches to these evolving viruses[129]. The armchair experts in virology, immunology, and genetics contribute substantially to future mitigation strategies. It can no longer be one bug, one drug approach. Herd immunity is expectantly looked upon but is short-term and modulates the virus with new attacking feats. What is good to know is that though the future is unseen, this COVID-19 pandemic has taught us valuable lessons and equipped health agencies, clinical experts, and the general public to face the subsequent pandemic terror.
| 1. | Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536-544. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5202] [Cited by in RCA: 4713] [Article Influence: 785.5] [Reference Citation Analysis (11)] |
| 2. | World Health Organization. Coronavirus disease (COVID-19). [cited 21 October 2022]. Available from: https://covid19.who.int/. |
| 3. | Aquino-Martinez R, Hernández-Vigueras S. Severe COVID-19 Lung Infection in Older People and Periodontitis. J Clin Med. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 4. | Kunutsor SK, Laukkanen JA. Hepatic manifestations and complications of COVID-19: A systematic review and meta-analysis. J Infect. 2020;81:e72-e74. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 5. | Martinez MA, Franco S. Impact of COVID-19 in Liver Disease Progression. Hepatol Commun. 2021;5:1138-1150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 6. | Li G, Fan Y, Lai Y, Han T, Li Z, Zhou P, Pan P, Wang W, Hu D, Liu X, Zhang Q, Wu J. Coronavirus infections and immune responses. J Med Virol. 2020;92:424-432. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1056] [Cited by in RCA: 1187] [Article Influence: 197.8] [Reference Citation Analysis (3)] |
| 7. | Kovalic AJ, Huang G, Thuluvath PJ, Satapathy SK. Elevated Liver Biochemistries in Hospitalized Chinese Patients With Severe COVID-19: Systematic Review and Meta-analysis. Hepatology. 2021;73:1521-1530. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 8. | Yu D, Du Q, Yan S, Guo XG, He Y, Zhu G, Zhao K, Ouyang S. Liver injury in COVID-19: clinical features and treatment management. Virol J. 2021;18:121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 76] [Article Influence: 15.2] [Reference Citation Analysis (1)] |
| 9. | Sun J, Aghemo A, Forner A, Valenti L. COVID-19 and liver disease. Liver Int. 2020;40:1278-1281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 222] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 10. | Moore JB, June CH. Cytokine release syndrome in severe COVID-19. Science. 2020;368:473-474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1185] [Cited by in RCA: 1401] [Article Influence: 233.5] [Reference Citation Analysis (0)] |
| 11. | Recovery Collaborative Group; Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, Linsell L, Staplin N, Brightling C, Ustianowski A, Elmahi E, Prudon B, Green C, Felton T, Chadwick D, Rege K, Fegan C, Chappell LC, Faust SN, Jaki T, Jeffery K, Montgomery A, Rowan K, Juszczak E, Baillie JK, Haynes R, Landray MJ. Dexamethasone in Hospitalized Patients with Covid-19. N Engl J Med. 2021;384:693-704. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6762] [Cited by in RCA: 7599] [Article Influence: 1519.8] [Reference Citation Analysis (7)] |
| 12. | Racanelli V, Rehermann B. The liver as an immunological organ. Hepatology. 2006;43:S54-S62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 870] [Cited by in RCA: 997] [Article Influence: 49.9] [Reference Citation Analysis (0)] |
| 13. | GBD 2017 Cirrhosis Collaborators. The global, regional, and national burden of cirrhosis by cause in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2020;5:245-266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1184] [Cited by in RCA: 1123] [Article Influence: 187.2] [Reference Citation Analysis (5)] |
| 14. | Wong YJ, Tan M, Zheng Q, Li JW, Kumar R, Fock KM, Teo EK, Ang TL. A systematic review and meta-analysis of the COVID-19 associated liver injury. Ann Hepatol. 2020;19:627-634. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 15. | Xu X, Chen P, Wang J, Feng J, Zhou H, Li X, Zhong W, Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63:457-460. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1252] [Cited by in RCA: 1325] [Article Influence: 220.8] [Reference Citation Analysis (0)] |
| 16. | Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, Tan KS, Wang DY, Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res. 2020;7:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1854] [Cited by in RCA: 2029] [Article Influence: 338.2] [Reference Citation Analysis (0)] |
| 17. | Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565-574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8473] [Cited by in RCA: 7698] [Article Influence: 1283.0] [Reference Citation Analysis (0)] |
| 18. | Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270-273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15248] [Cited by in RCA: 14361] [Article Influence: 2393.5] [Reference Citation Analysis (10)] |
| 19. | Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A; COVID-19 Genomics UK (COG-UK) Consortium, Peacock SJ, Robertson DL. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19:409-424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2349] [Cited by in RCA: 2499] [Article Influence: 499.8] [Reference Citation Analysis (0)] |
| 20. | Yamamoto N, Bauer G. Apparent difference in fatalities between Central Europe and East Asia due to SARS-COV-2 and COVID-19: Four hypotheses for possible explanation. Med Hypotheses. 2020;144:110160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 21. | Delanghe JR, Speeckaert MM, De Buyzere ML. COVID-19 infections are also affected by human ACE1 D/I polymorphism. Clin Chem Lab Med. 2020;58:1125-1126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 89] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 22. | Vijgen L, Keyaerts E, Moës E, Thoelen I, Wollants E, Lemey P, Vandamme AM, Van Ranst M. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol. 2005;79:1595-1604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 368] [Cited by in RCA: 432] [Article Influence: 20.6] [Reference Citation Analysis (8)] |
| 23. | Yamamoto N, Ariumi Y, Nishida N, Yamamoto R, Bauer G, Gojobori T, Shimotohno K, Mizokami M. SARS-CoV-2 infections and COVID-19 mortalities strongly correlate with ACE1 I/D genotype. Gene. 2020;758:144944. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 116] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 24. | Latinne A, Hu B, Olival KJ, Zhu G, Zhang L, Li H, Chmura AA, Field HE, Zambrana-Torrelio C, Epstein JH, Li B, Zhang W, Wang LF, Shi ZL, Daszak P. Origin and cross-species transmission of bat coronaviruses in China. Nat Commun. 2020;11:4235. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 259] [Cited by in RCA: 236] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 25. | Hu B, Ge X, Wang LF, Shi Z. Bat origin of human coronaviruses. Virol J. 2015;12:221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 247] [Cited by in RCA: 282] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 26. | Zhang RH, Ai X, Liu Y, Li CH, Zhang HL. Genomic characterization and phylogenetic evolution of the SARS-CoV-2. Acta Virol. 2020;64:496-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 27. | Abdelrahman Z, Li M, Wang X. Comparative Review of SARS-CoV-2, SARS-CoV, MERS-CoV, and Influenza A Respiratory Viruses. Front Immunol. 2020;11:552909. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 340] [Cited by in RCA: 304] [Article Influence: 50.7] [Reference Citation Analysis (0)] |
| 28. | Yadav PD, Potdar VA, Choudhary ML, Nyayanit DA, Agrawal M, Jadhav SM, Majumdar TD, Shete-Aich A, Basu A, Abraham P, Cherian SS. Full-genome sequences of the first two SARS-CoV-2 viruses from India. Indian J Med Res. 2020;151:200-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 87] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 29. | Duan ZP, Chen Y, Zhang J, Zhao J, Lang ZW, Meng FK, Bao XL. [Clinical characteristics and mechanism of liver injury in patients with severe acute respiratory syndrome]. Zhonghua Gan Zang Bing Za Zhi. 2003;11:493-496. [PubMed] |
| 30. | Han MW, Wang M, Xu MY, Qi WP, Wang P, Xi D. Clinical features and potential mechanism of coronavirus disease 2019-associated liver injury. World J Clin Cases. 2021;9:528-539. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 11] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 31. | Zhou B, Thao TTN, Hoffmann D, Taddeo A, Ebert N, Labroussaa F, Pohlmann A, King J, Steiner S, Kelly JN, Portmann J, Halwe NJ, Ulrich L, Trüeb BS, Fan X, Hoffmann B, Wang L, Thomann L, Lin X, Stalder H, Pozzi B, de Brot S, Jiang N, Cui D, Hossain J, Wilson MM, Keller MW, Stark TJ, Barnes JR, Dijkman R, Jores J, Benarafa C, Wentworth DE, Thiel V, Beer M. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature. 2021;592:122-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 379] [Article Influence: 75.8] [Reference Citation Analysis (0)] |
| 32. | Deng X, Garcia-Knight MA, Khalid MM, Servellita V, Wang C, Morris MK, Sotomayor-González A, Glasner DR, Reyes KR, Gliwa AS, Reddy NP, Martin CSS, Federman S, Cheng J, Balcerek J, Taylor J, Streithorst JA, Miller S, Kumar GR, Sreekumar B, Chen PY, Schulze-Gahmen U, Taha TY, Hayashi J, Simoneau CR, McMahon S, Lidsky PV, Xiao Y, Hemarajata P, Green NM, Espinosa A, Kath C, Haw M, Bell J, Hacker JK, Hanson C, Wadford DA, Anaya C, Ferguson D, Lareau LF, Frankino PA, Shivram H, Wyman SK, Ott M, Andino R, Chiu CY. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. medRxiv. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 112] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 33. | Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI; Sheffield COVID-19 Genomics Group, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812-827.e19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3504] [Cited by in RCA: 2972] [Article Influence: 495.3] [Reference Citation Analysis (0)] |
| 34. | Temmam S, Vongphayloth K, Baquero E, Munier S, Bonomi M, Regnault B, Douangboubpha B, Karami Y, Chrétien D, Sanamxay D, Xayaphet V, Paphaphanh P, Lacoste V, Somlor S, Lakeomany K, Phommavanh N, Pérot P, Dehan O, Amara F, Donati F, Bigot T, Nilges M, Rey FA, van der Werf S, Brey PT, Eloit M. Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature. 2022;604:330-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 295] [Article Influence: 73.8] [Reference Citation Analysis (0)] |
| 35. | Pal M, Berhanu G, Desalegn C, Kandi V. Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2): An Update. Cureus. 2020;12:e7423. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 280] [Cited by in RCA: 331] [Article Influence: 55.2] [Reference Citation Analysis (0)] |
| 36. | Zheng J. SARS-CoV-2: an Emerging Coronavirus that Causes a Global Threat. Int J Biol Sci. 2020;16:1678-1685. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 650] [Cited by in RCA: 576] [Article Influence: 96.0] [Reference Citation Analysis (8)] |
| 37. | Cyranoski D. Profile of a killer: the complex biology powering the coronavirus pandemic. Nature. 2020;581:22-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 105] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 38. | Sauter D, Kirchhoff F. Key Viral Adaptations Preceding the AIDS Pandemic. Cell Host Microbe. 2019;25:27-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 81] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 39. | Furuse Y, Oshitani H. Viruses That Can and Cannot Coexist With Humans and the Future of SARS-CoV-2. Front Microbiol. 2020;11:583252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 40. | Sathian B, Asim M, Banerjee I, Roy B, Pizarro AB, Mancha MA, van Teijlingen ER, Kord-Varkaneh H, Mekkodathil AA, Subramanya SH, Borges do Nascimento IJ, Antony N, Menezes RG, Simkhada P, Al Hamad H. Development and implementation of a potential coronavirus disease 2019 (COVID-19) vaccine: A systematic review and meta-analysis of vaccine clinical trials. Nepal J Epidemiol. 2021;11:959-982. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 41. | Duong D. Alpha, Beta, Delta, Gamma: What's important to know about SARS-CoV-2 variants of concern? CMAJ. 2021;193:E1059-E1060. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 111] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 42. | Telenti A, Hodcroft EB, Robertson DL. The Evolution and Biology of SARS-CoV-2 Variants. Cold Spring Harb Perspect Med. 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 136] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 43. | Yadav PD, Sapkal GN, Abraham P, Ella R, Deshpande G, Patil DY, Nyayanit DA, Gupta N, Sahay RR, Shete AM, Panda S, Bhargava B, Mohan VK. Neutralization of Variant Under Investigation B.1.617.1 With Sera of BBV152 Vaccinees. Clin Infect Dis. 2022;74:366-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 140] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 44. | Moghadas SM, Vilches TN, Zhang K, Wells CR, Shoukat A, Singer BH, Meyers LA, Neuzil KM, Langley JM, Fitzpatrick MC, Galvani AP. The Impact of Vaccination on Coronavirus Disease 2019 (COVID-19) Outbreaks in the United States. Clin Infect Dis. 2021;73:2257-2264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 270] [Cited by in RCA: 332] [Article Influence: 66.4] [Reference Citation Analysis (0)] |
| 45. | Wu J, Song S, Cao HC, Li LJ. Liver diseases in COVID-19: Etiology, treatment and prognosis. World J Gastroenterol. 2020;26:2286-2293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 105] [Cited by in RCA: 99] [Article Influence: 16.5] [Reference Citation Analysis (1)] |
| 46. | Harisha EJ, Gosavi S, Rao AA, Sahana GV, Manjunath S, Meghana TC. Liver: Function and dysfunction in COVID-19. J Family Med Prim Care. 2022;11:758-761. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 47. | Portincasa P, Krawczyk M, Machill A, Lammert F, Di Ciaula A. Hepatic consequences of COVID-19 infection. Lapping or biting? Eur J Intern Med. 2020;77:18-24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 77] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 48. | Ni W, Yang X, Yang D, Bao J, Li R, Xiao Y, Hou C, Wang H, Liu J, Xu Y, Cao Z, Gao Z. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit Care. 2020;24:422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 843] [Cited by in RCA: 723] [Article Influence: 120.5] [Reference Citation Analysis (1)] |
| 49. | Sharma A, Jaiswal P, Kerakhan Y, Saravanan L, Murtaza Z, Zergham A, Honganur NS, Akbar A, Deol A, Francis B, Patel S, Mehta D, Jaiswal R, Singh J, Patel U, Malik P. Liver disease and outcomes among COVID-19 hospitalized patients - A systematic review and meta-analysis. Ann Hepatol. 2021;21:100273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 81] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 50. | Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450-454. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4113] [Cited by in RCA: 4648] [Article Influence: 202.1] [Reference Citation Analysis (0)] |
| 51. | Chau TN, Lee KC, Yao H, Tsang TY, Chow TC, Yeung YC, Choi KW, Tso YK, Lau T, Lai ST, Lai CL. SARS-associated viral hepatitis caused by a novel coronavirus: report of three cases. Hepatology. 2004;39:302-310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 307] [Cited by in RCA: 305] [Article Influence: 13.9] [Reference Citation Analysis (1)] |
| 52. | Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, Tai Y, Bai C, Gao T, Song J, Xia P, Dong J, Zhao J, Wang FS. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420-422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5228] [Cited by in RCA: 5836] [Article Influence: 972.7] [Reference Citation Analysis (3)] |
| 53. | Castelli EC, de Castro MV, Naslavsky MS, Scliar MO, Silva NSB, Andrade HS, Souza AS, Pereira RN, Castro CFB, Mendes-Junior CT, Meyer D, Nunes K, Matos LRB, Silva MVR, Wang JYT, Esposito J, Coria VR, Bortolin RH, Hirata MH, Magawa JY, Cunha-Neto E, Coelho V, Santos KS, Marin MLC, Kalil J, Mitne-Neto M, Maciel RMB, Passos-Bueno MR, Zatz M. MHC Variants Associated With Symptomatic Versus Asymptomatic SARS-CoV-2 Infection in Highly Exposed Individuals. Front Immunol. 2021;12:742881. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 54. | Yoo JS, Sasaki M, Cho SX, Kasuga Y, Zhu B, Ouda R, Orba Y, de Figueiredo P, Sawa H, Kobayashi KS. SARS-CoV-2 inhibits induction of the MHC class I pathway by targeting the STAT1-IRF1-NLRC5 axis. Nat Commun. 2021;12:6602. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 123] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 55. | de Sousa E, Ligeiro D, Lérias JR, Zhang C, Agrati C, Osman M, El-Kafrawy SA, Azhar EI, Ippolito G, Wang FS, Zumla A, Maeurer M. Mortality in COVID-19 disease patients: Correlating the association of major histocompatibility complex (MHC) with severe acute respiratory syndrome 2 (SARS-CoV-2) variants. Int J Infect Dis. 2020;98:454-459. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 56. | Shah VK, Firmal P, Alam A, Ganguly D, Chattopadhyay S. Overview of Immune Response During SARS-CoV-2 Infection: Lessons From the Past. Front Immunol. 2020;11:1949. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 342] [Cited by in RCA: 308] [Article Influence: 51.3] [Reference Citation Analysis (0)] |
| 57. | Salamanna F, Maglio M, Landini MP, Fini M. Body Localization of ACE-2: On the Trail of the Keyhole of SARS-CoV-2. Front Med (Lausanne). 2020;7:594495. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 166] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 58. | Kuba K, Yamaguchi T, Penninger JM. Angiotensin-Converting Enzyme 2 (ACE2) in the Pathogenesis of ARDS in COVID-19. Front Immunol. 2021;12:732690. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 47] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 59. | Beyerstedt S, Casaro EB, Rangel ÉB. COVID-19: angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur J Clin Microbiol Infect Dis. 2021;40:905-919. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 601] [Cited by in RCA: 533] [Article Influence: 106.6] [Reference Citation Analysis (0)] |
| 60. | Di Vito C, Calcaterra F, Coianiz N, Terzoli S, Voza A, Mikulak J, Della Bella S, Mavilio D. Natural Killer Cells in SARS-CoV-2 Infection: Pathophysiology and Therapeutic Implications. Front Immunol. 2022;13:888248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 61. | Maucourant C, Filipovic I, Ponzetta A, Aleman S, Cornillet M, Hertwig L, Strunz B, Lentini A, Reinius B, Brownlie D, Cuapio A, Ask EH, Hull RM, Haroun-Izquierdo A, Schaffer M, Klingström J, Folkesson E, Buggert M, Sandberg JK, Eriksson LI, Rooyackers O, Ljunggren HG, Malmberg KJ, Michaëlsson J, Marquardt N, Hammer Q, Strålin K, Björkström NK; Karolinska COVID-19 Study Group. Natural killer cell immunotypes related to COVID-19 disease severity. Sci Immunol. 2020;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 304] [Cited by in RCA: 365] [Article Influence: 60.8] [Reference Citation Analysis (0)] |
| 62. | Zuo W, Zhao X. Natural killer cells play an important role in virus infection control: Antiviral mechanism, subset expansion and clinical application. Clin Immunol. 2021;227:108727. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 56] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 63. | van Eeden C, Khan L, Osman MS, Cohen Tervaert JW. Natural Killer Cell Dysfunction and Its Role in COVID-19. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 124] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 64. | Behl T, Kaur I, Bungau S, Kumar A, Uddin MS, Kumar C, Pal G, Sahil, Shrivastava K, Zengin G, Arora S. The dual impact of ACE2 in COVID-19 and ironical actions in geriatrics and pediatrics with possible therapeutic solutions. Life Sci. 2020;257:118075. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 65. | Marjot T, Webb GJ, Barritt AS 4th, Moon AM, Stamataki Z, Wong VW, Barnes E. COVID-19 and liver disease: mechanistic and clinical perspectives. Nat Rev Gastroenterol Hepatol. 2021;18:348-364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 278] [Cited by in RCA: 290] [Article Influence: 58.0] [Reference Citation Analysis (2)] |
| 66. | Wang X, Lei J, Li Z, Yan L. Potential Effects of Coronaviruses on the Liver: An Update. Front Med (Lausanne). 2021;8:651658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 67. | Moon AM, Barritt AS 4th. Elevated Liver Enzymes in Patients with COVID-19: Look, but Not Too Hard. Dig Dis Sci. 2021;66:1767-1769. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 68. | Xu L, Liu J, Lu M, Yang D, Zheng X. Liver injury during highly pathogenic human coronavirus infections. Liver Int. 2020;40:998-1004. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 622] [Cited by in RCA: 577] [Article Influence: 96.2] [Reference Citation Analysis (1)] |
| 69. | Lax SF, Skok K, Zechner P, Kessler HH, Kaufmann N, Koelblinger C, Vander K, Bargfrieder U, Trauner M. Pulmonary Arterial Thrombosis in COVID-19 With Fatal Outcome: Results From a Prospective, Single-Center, Clinicopathologic Case Series. Ann Intern Med. 2020;173:350-361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 604] [Cited by in RCA: 623] [Article Influence: 103.8] [Reference Citation Analysis (0)] |
| 70. | Lindner D, Fitzek A, Bräuninger H, Aleshcheva G, Edler C, Meissner K, Scherschel K, Kirchhof P, Escher F, Schultheiss HP, Blankenberg S, Püschel K, Westermann D. Association of Cardiac Infection With SARS-CoV-2 in Confirmed COVID-19 Autopsy Cases. JAMA Cardiol. 2020;5:1281-1285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 428] [Cited by in RCA: 608] [Article Influence: 101.3] [Reference Citation Analysis (0)] |
| 71. | Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ; HLH Across Speciality Collaboration, UK. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033-1034. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6366] [Cited by in RCA: 6849] [Article Influence: 1141.5] [Reference Citation Analysis (1)] |
| 72. | Aleem A, Shah H. Gastrointestinal And Hepatic Manifestations Of Coronavirus (COVID-19). 2022 May 4. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2022 Jan-. [PubMed] |
| 73. | Chua RL, Lukassen S, Trump S, Hennig BP, Wendisch D, Pott F, Debnath O, Thürmann L, Kurth F, Völker MT, Kazmierski J, Timmermann B, Twardziok S, Schneider S, Machleidt F, Müller-Redetzky H, Maier M, Krannich A, Schmidt S, Balzer F, Liebig J, Loske J, Suttorp N, Eils J, Ishaque N, Liebert UG, von Kalle C, Hocke A, Witzenrath M, Goffinet C, Drosten C, Laudi S, Lehmann I, Conrad C, Sander LE, Eils R. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat Biotechnol. 2020;38:970-979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 596] [Cited by in RCA: 814] [Article Influence: 135.7] [Reference Citation Analysis (0)] |
| 74. | Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, Cao Y, Yousif AS, Bals J, Hauser BM, Feldman J, Muus C, Wadsworth MH 2nd, Kazer SW, Hughes TK, Doran B, Gatter GJ, Vukovic M, Taliaferro F, Mead BE, Guo Z, Wang JP, Gras D, Plaisant M, Ansari M, Angelidis I, Adler H, Sucre JMS, Taylor CJ, Lin B, Waghray A, Mitsialis V, Dwyer DF, Buchheit KM, Boyce JA, Barrett NA, Laidlaw TM, Carroll SL, Colonna L, Tkachev V, Peterson CW, Yu A, Zheng HB, Gideon HP, Winchell CG, Lin PL, Bingle CD, Snapper SB, Kropski JA, Theis FJ, Schiller HB, Zaragosi LE, Barbry P, Leslie A, Kiem HP, Flynn JL, Fortune SM, Berger B, Finberg RW, Kean LS, Garber M, Schmidt AG, Lingwood D, Shalek AK, Ordovas-Montanes J; HCA Lung Biological Network. Electronic address: lung-network@humancellatlas.org; HCA Lung Biological Network. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell. 2020;181:1016-1035.e19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1988] [Cited by in RCA: 1845] [Article Influence: 307.5] [Reference Citation Analysis (0)] |
| 75. | Paizis G, Tikellis C, Cooper ME, Schembri JM, Lew RA, Smith AI, Shaw T, Warner FJ, Zuilli A, Burrell LM, Angus PW. Chronic liver injury in rats and humans upregulates the novel enzyme angiotensin converting enzyme 2. Gut. 2005;54:1790-1796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 281] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 76. | Săbiescu DM, Kamal AM, Kamal CK, Alexandru DO, Mitruț P. Liver damage in the context of SARS-CoV-2. Covid-19 treatment and its effects on the liver. J Med Life. 2022;15:727-734. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 77. | Ali N. Relationship Between COVID-19 Infection and Liver Injury: A Review of Recent Data. Front Med (Lausanne). 2020;7:458. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (1)] |
| 78. | Nagarajan R, Krishnamoorthy Y, Rajaa S, Hariharan VS. COVID-19 Severity and Mortality Among Chronic Liver Disease Patients: A Systematic Review and Meta-Analysis. Prev Chronic Dis. 2022;19:E53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 79. | Portincasa P, Krawczyk M, Smyk W, Lammert F, Di Ciaula A. COVID-19 and non-alcoholic fatty liver disease: Two intersecting pandemics. Eur J Clin Invest. 2020;50:e13338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 101] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 80. | Ahmad A, Ishtiaq SM, Khan JA, Aslam R, Ali S, Arshad MI. COVID-19 and comorbidities of hepatic diseases in a global perspective. World J Gastroenterol. 2021;27:1296-1310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 12] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 81. | Ozkurt Z, Çınar Tanrıverdi E. COVID-19: Gastrointestinal manifestations, liver injury and recommendations. World J Clin Cases. 2022;10:1140-1163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 29] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (8)] |
| 82. | Jensen S, Thomsen AR. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J Virol. 2012;86:2900-2910. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 421] [Cited by in RCA: 480] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 83. | Premkumar M, Kedarisetty CK. Cytokine Storm of COVID-19 and Its Impact on Patients with and without Chronic Liver Disease. J Clin Transl Hepatol. 2021;9:256-264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 84. | Huang W, Berube J, McNamara M, Saksena S, Hartman M, Arshad T, Bornheimer SJ, O'Gorman M. Lymphocyte Subset Counts in COVID-19 Patients: A Meta-Analysis. Cytometry A. 2020;97:772-776. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 174] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 85. | Rios CI, Cassatt DR, Hollingsworth BA, Satyamitra MM, Tadesse YS, Taliaferro LP, Winters TA, DiCarlo AL. Commonalities Between COVID-19 and Radiation Injury. Radiat Res. 2021;195:1-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 86. | Battagello DS, Dragunas G, Klein MO, Ayub ALP, Velloso FJ, Correa RG. Unpuzzling COVID-19: tissue-related signaling pathways associated with SARS-CoV-2 infection and transmission. Clin Sci (Lond). 2020;134:2137-2160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 87. | Wang X, Pasco RF, Du Z, Petty M, Fox SJ, Galvani AP, Pignone M, Johnston SC, Meyers LA. Impact of Social Distancing Measures on Coronavirus Disease Healthcare Demand, Central Texas, USA. Emerg Infect Dis. 2020;26:2361-2369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 67] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 88. | Xu K, Chen Y, Yuan J, Yi P, Ding C, Wu W, Li Y, Ni Q, Zou R, Li X, Xu M, Zhang Y, Zhao H, Zhang X, Yu L, Su J, Lang G, Liu J, Wu X, Guo Y, Tao J, Shi D, Cao Q, Ruan B, Liu L, Wang Z, Xu Y, Liu Y, Sheng J, Li L. Factors Associated With Prolonged Viral RNA Shedding in Patients with Coronavirus Disease 2019 (COVID-19). Clin Infect Dis. 2020;71:799-806. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 348] [Cited by in RCA: 360] [Article Influence: 60.0] [Reference Citation Analysis (0)] |
| 89. | Harvey RA, Rassen JA, Kabelac CA, Turenne W, Leonard S, Klesh R, Meyer WA 3rd, Kaufman HW, Anderson S, Cohen O, Petkov VI, Cronin KA, Van Dyke AL, Lowy DR, Sharpless NE, Penberthy LT. Association of SARS-CoV-2 Seropositive Antibody Test With Risk of Future Infection. JAMA Intern Med. 2021;181:672-679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 222] [Cited by in RCA: 192] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 90. | Maher MC, Bartha I, Weaver S, di Iulio J, Ferri E, Soriaga L, Lempp FA, Hie BL, Bryson B, Berger B, Robertson DL, Snell G, Corti D, Virgin HW, Kosakovsky Pond SL, Telenti A. Predicting the mutational drivers of future SARS-CoV-2 variants of concern. Sci Transl Med. 2022;14:eabk3445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 111] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 91. | Saad-Roy CM, Metcalf CJE, Grenfell BT. Immuno-epidemiology and the predictability of viral evolution. Science. 2022;376:1161-1162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 92. | Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J. 2019;16:69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1277] [Cited by in RCA: 1202] [Article Influence: 171.7] [Reference Citation Analysis (10)] |
| 93. | Karunakaran KB, Balakrishnan N, Ganapathiraju MK. Interactome of SARS-CoV-2 Modulated Host Proteins With Computationally Predicted PPIs: Insights From Translational Systems Biology Studies. Front Syst Biol. 2022;2. [RCA] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 94. | Tran A, Witek TJ Jr. The Emergency Use Authorization of Pharmaceuticals: History and Utility During the COVID-19 Pandemic. Pharmaceut Med. 2021;35:203-213. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 95. | Ison MG, Wolfe C, Boucher HW. Emergency Use Authorization of Remdesivir: The Need for a Transparent Distribution Process. JAMA. 2020;323:2365-2366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 96. | Bhatti JS, Bhatti GK, Khullar N, Reddy AP, Reddy PH. Therapeutic Strategies in the Development of Anti-viral Drugs and Vaccines Against SARS-CoV-2 Infection. Mol Neurobiol. 2020;57:4856-4877. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 97. | Gandhi RT, Lynch JB, Del Rio C. Mild or Moderate Covid-19. N Engl J Med. 2020;383:1757-1766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 851] [Cited by in RCA: 954] [Article Influence: 159.0] [Reference Citation Analysis (0)] |
| 98. | Coopersmith CM, Antonelli M, Bauer SR, Deutschman CS, Evans LE, Ferrer R, Hellman J, Jog S, Kesecioglu J, Kissoon N, Martin-Loeches I, Nunnally ME, Prescott HC, Rhodes A, Talmor D, Tissieres P, De Backer D. The Surviving Sepsis Campaign: Research Priorities for Coronavirus Disease 2019 in Critical Illness. Crit Care Med. 2021;49:598-622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (1)] |
| 99. | Peter AE, Sandeep BV, Rao BG, Kalpana VL. Calming the Storm: Natural Immunosuppressants as Adjuvants to Target the Cytokine Storm in COVID-19. Front Pharmacol. 2020;11:583777. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 75] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 100. | Tang L, Yin Z, Hu Y, Mei H. Controlling Cytokine Storm Is Vital in COVID-19. Front Immunol. 2020;11:570993. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 122] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 101. | Zhao M. Cytokine storm and immunomodulatory therapy in COVID-19: Role of chloroquine and anti-IL-6 monoclonal antibodies. Int J Antimicrob Agents. 2020;55:105982. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 183] [Cited by in RCA: 185] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 102. | Zhang W, Zhao Y, Zhang F, Wang Q, Li T, Liu Z, Wang J, Qin Y, Zhang X, Yan X, Zeng X, Zhang S. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): The Perspectives of clinical immunologists from China. Clin Immunol. 2020;214:108393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 812] [Cited by in RCA: 864] [Article Influence: 144.0] [Reference Citation Analysis (0)] |
| 103. | Kalil AC, Patterson TF, Mehta AK, Tomashek KM, Wolfe CR, Ghazaryan V, Marconi VC, Ruiz-Palacios GM, Hsieh L, Kline S, Tapson V, Iovine NM, Jain MK, Sweeney DA, El Sahly HM, Branche AR, Regalado Pineda J, Lye DC, Sandkovsky U, Luetkemeyer AF, Cohen SH, Finberg RW, Jackson PEH, Taiwo B, Paules CI, Arguinchona H, Erdmann N, Ahuja N, Frank M, Oh MD, Kim ES, Tan SY, Mularski RA, Nielsen H, Ponce PO, Taylor BS, Larson L, Rouphael NG, Saklawi Y, Cantos VD, Ko ER, Engemann JJ, Amin AN, Watanabe M, Billings J, Elie MC, Davey RT, Burgess TH, Ferreira J, Green M, Makowski M, Cardoso A, de Bono S, Bonnett T, Proschan M, Deye GA, Dempsey W, Nayak SU, Dodd LE, Beigel JH; ACTT-2 Study Group Members. Baricitinib plus Remdesivir for Hospitalized Adults with Covid-19. N Engl J Med. 2021;384:795-807. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1159] [Cited by in RCA: 1381] [Article Influence: 276.2] [Reference Citation Analysis (1)] |
| 104. | Huet T, Beaussier H, Voisin O, Jouveshomme S, Dauriat G, Lazareth I, Sacco E, Naccache JM, Bézie Y, Laplanche S, Le Berre A, Le Pavec J, Salmeron S, Emmerich J, Mourad JJ, Chatellier G, Hayem G. Anakinra for severe forms of COVID-19: a cohort study. Lancet Rheumatol. 2020;2:e393-e400. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 525] [Cited by in RCA: 481] [Article Influence: 80.2] [Reference Citation Analysis (0)] |
| 105. | Heath PT, Galiza EP, Baxter DN, Boffito M, Browne D, Burns F, Chadwick DR, Clark R, Cosgrove C, Galloway J, Goodman AL, Heer A, Higham A, Iyengar S, Jamal A, Jeanes C, Kalra PA, Kyriakidou C, McAuley DF, Meyrick A, Minassian AM, Minton J, Moore P, Munsoor I, Nicholls H, Osanlou O, Packham J, Pretswell CH, San Francisco Ramos A, Saralaya D, Sheridan RP, Smith R, Soiza RL, Swift PA, Thomson EC, Turner J, Viljoen ME, Albert G, Cho I, Dubovsky F, Glenn G, Rivers J, Robertson A, Smith K, Toback S; 2019nCoV-302 Study Group. Safety and Efficacy of NVX-CoV2373 Covid-19 Vaccine. N Engl J Med. 2021;385:1172-1183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 659] [Cited by in RCA: 811] [Article Influence: 162.2] [Reference Citation Analysis (0)] |
| 106. | Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW Jr, Hammitt LL, Türeci Ö, Nell H, Schaefer A, Ünal S, Tresnan DB, Mather S, Dormitzer PR, Şahin U, Jansen KU, Gruber WC; C4591001 Clinical Trial Group. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med. 2020;383:2603-2615. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10556] [Cited by in RCA: 11179] [Article Influence: 1863.2] [Reference Citation Analysis (1)] |
| 107. | Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, McGettigan J, Khetan S, Segall N, Solis J, Brosz A, Fierro C, Schwartz H, Neuzil K, Corey L, Gilbert P, Janes H, Follmann D, Marovich M, Mascola J, Polakowski L, Ledgerwood J, Graham BS, Bennett H, Pajon R, Knightly C, Leav B, Deng W, Zhou H, Han S, Ivarsson M, Miller J, Zaks T; COVE Study Group. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med. 2021;384:403-416. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9565] [Cited by in RCA: 7963] [Article Influence: 1592.6] [Reference Citation Analysis (1)] |
| 108. | Voysey M, Clemens SAC, Madhi SA, Weckx LY, Folegatti PM, Aley PK, Angus B, Baillie VL, Barnabas SL, Bhorat QE, Bibi S, Briner C, Cicconi P, Collins AM, Colin-Jones R, Cutland CL, Darton TC, Dheda K, Duncan CJA, Emary KRW, Ewer KJ, Fairlie L, Faust SN, Feng S, Ferreira DM, Finn A, Goodman AL, Green CM, Green CA, Heath PT, Hill C, Hill H, Hirsch I, Hodgson SHC, Izu A, Jackson S, Jenkin D, Joe CCD, Kerridge S, Koen A, Kwatra G, Lazarus R, Lawrie AM, Lelliott A, Libri V, Lillie PJ, Mallory R, Mendes AVA, Milan EP, Minassian AM, McGregor A, Morrison H, Mujadidi YF, Nana A, O'Reilly PJ, Padayachee SD, Pittella A, Plested E, Pollock KM, Ramasamy MN, Rhead S, Schwarzbold AV, Singh N, Smith A, Song R, Snape MD, Sprinz E, Sutherland RK, Tarrant R, Thomson EC, Török ME, Toshner M, Turner DPJ, Vekemans J, Villafana TL, Watson MEE, Williams CJ, Douglas AD, Hill AVS, Lambe T, Gilbert SC, Pollard AJ; Oxford COVID Vaccine Trial Group. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99-111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3551] [Cited by in RCA: 3505] [Article Influence: 701.0] [Reference Citation Analysis (0)] |
| 109. | Sadoff J, Gray G, Vandebosch A, Cárdenas V, Shukarev G, Grinsztejn B, Goepfert PA, Truyers C, Fennema H, Spiessens B, Offergeld K, Scheper G, Taylor KL, Robb ML, Treanor J, Barouch DH, Stoddard J, Ryser MF, Marovich MA, Neuzil KM, Corey L, Cauwenberghs N, Tanner T, Hardt K, Ruiz-Guiñazú J, Le Gars M, Schuitemaker H, Van Hoof J, Struyf F, Douoguih M; ENSEMBLE Study Group. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against Covid-19. N Engl J Med. 2021;384:2187-2201. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2024] [Cited by in RCA: 1880] [Article Influence: 376.0] [Reference Citation Analysis (0)] |
| 110. | Behera P, Singh AK, Subba SH, Mc A, Sahu DP, Chandanshive PD, Pradhan SK, Parida SP, Mishra A, Patro BK, Batmanabane G. Effectiveness of COVID-19 vaccine (Covaxin) against breakthrough SARS-CoV-2 infection in India. Hum Vaccin Immunother. 2022;18:2034456. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 111. | Nogrady B. Mounting evidence suggests Sputnik COVID vaccine is safe and effective. Nature. 2021;595:339-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 112. | Florentino PTV, Alves FJO, Cerqueira-Silva T, Oliveira VA, Júnior JBS, Jantsch AG, Penna GO, Boaventura V, Werneck GL, Rodrigues LC, Pearce N, Barral-Netto M, Barreto ML, Paixão ES. Vaccine effectiveness of CoronaVac against COVID-19 among children in Brazil during the Omicron period. Nat Commun. 2022;13:4756. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 113. | Caly L, Druce JD, Catton MG, Jans DA, Wagstaff KM. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020;178:104787. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1171] [Cited by in RCA: 1318] [Article Influence: 219.7] [Reference Citation Analysis (0)] |
| 114. | Spinner CD, Gottlieb RL, Criner GJ, Arribas López JR, Cattelan AM, Soriano Viladomiu A, Ogbuagu O, Malhotra P, Mullane KM, Castagna A, Chai LYA, Roestenberg M, Tsang OTY, Bernasconi E, Le Turnier P, Chang SC, SenGupta D, Hyland RH, Osinusi AO, Cao H, Blair C, Wang H, Gaggar A, Brainard DM, McPhail MJ, Bhagani S, Ahn MY, Sanyal AJ, Huhn G, Marty FM; GS-US-540-5774 Investigators. Effect of Remdesivir vs Standard Care on Clinical Status at 11 Days in Patients With Moderate COVID-19: A Randomized Clinical Trial. JAMA. 2020;324:1048-1057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 986] [Cited by in RCA: 942] [Article Influence: 157.0] [Reference Citation Analysis (0)] |
| 115. | Goldman JD, Lye DCB, Hui DS, Marks KM, Bruno R, Montejano R, Spinner CD, Galli M, Ahn MY, Nahass RG, Chen YS, SenGupta D, Hyland RH, Osinusi AO, Cao H, Blair C, Wei X, Gaggar A, Brainard DM, Towner WJ, Muñoz J, Mullane KM, Marty FM, Tashima KT, Diaz G, Subramanian A; GS-US-540-5773 Investigators. Remdesivir for 5 or 10 Days in Patients with Severe Covid-19. N Engl J Med. 2020;383:1827-1837. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 987] [Cited by in RCA: 998] [Article Influence: 166.3] [Reference Citation Analysis (0)] |
| 116. | Boulware DR, Pullen MF, Bangdiwala AS, Pastick KA, Lofgren SM, Okafor EC, Skipper CP, Nascene AA, Nicol MR, Abassi M, Engen NW, Cheng MP, LaBar D, Lother SA, MacKenzie LJ, Drobot G, Marten N, Zarychanski R, Kelly LE, Schwartz IS, McDonald EG, Rajasingham R, Lee TC, Hullsiek KH. A Randomized Trial of Hydroxychloroquine as Postexposure Prophylaxis for Covid-19. N Engl J Med. 2020;383:517-525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1021] [Cited by in RCA: 928] [Article Influence: 154.7] [Reference Citation Analysis (0)] |
| 117. | Sodeifian F, Seyedalhosseini ZS, Kian N, Eftekhari M, Najari S, Mirsaeidi M, Farsi Y, Nasiri MJ. Drug-Induced Liver Injury in COVID-19 Patients: A Systematic Review. Front Med (Lausanne). 2021;8:731436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 118. | Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, Wang M, Yu J, Zhang B, Kwong PD, Graham BS, Mascola JR, Chang JY, Yin MT, Sobieszczyk M, Kyratsous CA, Shapiro L, Sheng Z, Huang Y, Ho DD. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. bioRxiv. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 177] [Cited by in RCA: 142] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 119. | Wu K, Werner AP, Moliva JI, Koch M, Choi A, Stewart-Jones GBE, Bennett H, Boyoglu-Barnum S, Shi W, Graham BS, Carfi A, Corbett KS, Seder RA, Edwards DK. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 223] [Article Influence: 44.6] [Reference Citation Analysis (0)] |
| 120. | Wang P, Casner RG, Nair MS, Wang M, Yu J, Cerutti G, Liu L, Kwong PD, Huang Y, Shapiro L, Ho DD. Increased Resistance of SARS-CoV-2 Variant P.1 to Antibody Neutralization. bioRxiv. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 121. | Yadav PD, Sapkal GN, Abraham P, Ella R, Deshpande G, Patil DY, Nyayanit DA, Gupta N, Sahay RR, Shete AM, Panda S, Bhargava B, Mohan VK. Neutralization of variant under investigation B.1.617 with sera of BBV152 vaccinees. 2021 Preprint. Available from: bioRxiv 2021: 2021.04.23.441101. [DOI] [Full Text] |
| 122. | Joyner MJ, Senefeld JW, Klassen SA, Mills JR, Johnson PW, Theel ES, Wiggins CC, Bruno KA, Klompas AM, Lesser ER, Kunze KL, Sexton MA, Diaz Soto JC, Baker SE, Shepherd JRA, van Helmond N, van Buskirk CM, Winters JL, Stubbs JR, Rea RF, Hodge DO, Herasevich V, Whelan ER, Clayburn AJ, Larson KF, Ripoll JG, Andersen KJ, Buras MR, Vogt MNP, Dennis JJ, Regimbal RJ, Bauer PR, Blair JE, Paneth NS, Fairweather D, Wright RS, Carter RE, Casadevall A. Effect of Convalescent Plasma on Mortality among Hospitalized Patients with COVID-19: Initial Three-Month Experience. medRxiv. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 149] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 123. | Joyner MJ, Bruno KA, Klassen SA, Kunze KL, Johnson PW, Lesser ER, Wiggins CC, Senefeld JW, Klompas AM, Hodge DO, Shepherd JRA, Rea RF, Whelan ER, Clayburn AJ, Spiegel MR, Baker SE, Larson KF, Ripoll JG, Andersen KJ, Buras MR, Vogt MNP, Herasevich V, Dennis JJ, Regimbal RJ, Bauer PR, Blair JE, van Buskirk CM, Winters JL, Stubbs JR, van Helmond N, Butterfield BP, Sexton MA, Diaz Soto JC, Paneth NS, Verdun NC, Marks P, Casadevall A, Fairweather D, Carter RE, Wright RS. Safety Update: COVID-19 Convalescent Plasma in 20,000 Hospitalized Patients. Mayo Clin Proc. 2020;95:1888-1897. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 330] [Cited by in RCA: 323] [Article Influence: 53.8] [Reference Citation Analysis (0)] |
| 124. | Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, Wang M, Yu J, Zhang B, Kwong PD, Graham BS, Mascola JR, Chang JY, Yin MT, Sobieszczyk M, Kyratsous CA, Shapiro L, Sheng Z, Huang Y, Ho DD. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1627] [Cited by in RCA: 1578] [Article Influence: 315.6] [Reference Citation Analysis (0)] |
| 125. | Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, Musser BJ, Soo Y, Rofail D, Im J, Perry C, Pan C, Hosain R, Mahmood A, Davis JD, Turner KC, Hooper AT, Hamilton JD, Baum A, Kyratsous CA, Kim Y, Cook A, Kampman W, Kohli A, Sachdeva Y, Graber X, Kowal B, DiCioccio T, Stahl N, Lipsich L, Braunstein N, Herman G, Yancopoulos GD; Trial Investigators. REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N Engl J Med. 2021;384:238-251. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1348] [Cited by in RCA: 1321] [Article Influence: 264.2] [Reference Citation Analysis (12)] |
| 126. | Baum A, Ajithdoss D, Copin R, Zhou A, Lanza K, Negron N, Ni M, Wei Y, Mohammadi K, Musser B, Atwal GS, Oyejide A, Goez-Gazi Y, Dutton J, Clemmons E, Staples HM, Bartley C, Klaffke B, Alfson K, Gazi M, Gonzalez O, Dick E Jr, Carrion R Jr, Pessaint L, Porto M, Cook A, Brown R, Ali V, Greenhouse J, Taylor T, Andersen H, Lewis MG, Stahl N, Murphy AJ, Yancopoulos GD, Kyratsous CA. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110-1115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 431] [Cited by in RCA: 427] [Article Influence: 71.2] [Reference Citation Analysis (9)] |
| 127. | de Figueiredo A, Simas C, Karafillakis E, Paterson P, Larson HJ. Mapping global trends in vaccine confidence and investigating barriers to vaccine uptake: a large-scale retrospective temporal modelling study. Lancet. 2020;396:898-908. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 648] [Cited by in RCA: 694] [Article Influence: 115.7] [Reference Citation Analysis (0)] |
| 128. | Fontanet A, Autran B, Lina B, Kieny MP, Karim SSA, Sridhar D. SARS-CoV-2 variants and ending the COVID-19 pandemic. Lancet. 2021;397:952-954. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 440] [Cited by in RCA: 372] [Article Influence: 74.4] [Reference Citation Analysis (0)] |
| 129. | Baker RE, Yang W, Vecchi GA, Metcalf CJE, Grenfell BT. Susceptible supply limits the role of climate in the early SARS-CoV-2 pandemic. Science. 2020;369:315-319. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 197] [Cited by in RCA: 207] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: India
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): E, E
P-Reviewer: Liakina V, Lithuania; Zhang XL, China; Zhang JW, China S-Editor: Wang JJ L-Editor: Filipodia P-Editor: Wang JJ