Published online Feb 28, 2022. doi: 10.3748/wjg.v28.i8.825
Peer-review started: October 17, 2021
First decision: December 12, 2021
Revised: December 24, 2021
Accepted: January 22, 2022
Article in press: January 22, 2022
Published online: February 28, 2022
Processing time: 129 Days and 19.8 Hours
Patients with colorectal cancer (CRC) undergo surgery, as well as perioperative chemoradiation or adjuvant chemotherapy primarily based on the tumor–node– metastasis (TNM) cancer staging system. However, treatment responses and prognostic outcomes of patients within the same stage vary markedly. The potential use of novel biomarkers can improve prognostication and shared decision making before implementation into certain therapies.
To investigate whether SUMF2, ADAMTS5, and PXDN methylation status could be associated with CRC prognosis.
We conducted a Taiwan region cohort study involving 208 patients with CRC recruited from Tri-Service General Hospital and applied the candidate gene approach to identify three genes involved in oncogenesis pathways. A methylation-specific polymerase chain reaction (MS-PCR) and EpiTYPER DNA methylation analysis were employed to detect methylation status and to quantify the methylation level of candidate genes in tumor tissue and adjacent normal tissue from participants. We evaluated SUMF2, ADAMTS5, and PXDN methylation as predictors of prognosis, including recurrence-free survival (RFS), progression-free survival (PFS), and overall survival (OS), using a Cox regression model and Kaplan–Meier analysis.
We revealed various outcomes related to methylation and prognosis. Significantly shorter PFS and OS were associated with the CpG_3+CpG_7 hypermethylation of SUMF2 from tumor tissue compared with CpG_3+CpG_7 hypomethylation [hazard ratio (HR) = 2.24, 95% confidence interval (CI) = 1.03-4.85 for PFS, HR = 2.56 and 95%CI = 1.08-6.04 for OS]. By contrast, a significantly longer RFS was associated with CpG_2 and CpG_13 hypermethylation of ADAMTS5 from normal tissue compared with CpG_2 and CpG_13 hypomethylation [HR (95%CI) = 0.15 (0.03-0.71) for CpG_2 and 0.20 (0.04-0.97) for CpG_13]. The relationship between the methylation status of PXDN and the prognosis of CRC did not reach statistical significance.
Our study found that CpG_3+CpG_7 hypermethylation of SUMF2 from tumor tissue was associated with significantly shorter PFS and OS compared with CpG_3+CpG_7 hypomethylation. CpG_2 and CpG_13 hypermethylation of ADAMTS5 from normal tissue was associated with a significantly longer RFS compared with CpG_2 and CpG_13 hypomethylation. These methylation-related biomarkers which have implications for CRC prognosis prediction may aid physicians in clinical decision-making.
Core Tip: Our research revealed that differential DNA methylation of candidate genes in tumor tissue and adjacent normal tissue can be used to evaluate colorectal cancer prognosis. Certain CpG sites and the methylation status of SUMF2 and ADAMTS5 were significantly associated with colorectal cancer recurrence, progression, and survival. We recommend using our findings to investigate prognostic biomarkers applicable to patients with colorectal cancer.
- Citation: Su JQ, Lai PY, Hu PH, Hu JM, Chang PK, Chen CY, Wu JJ, Lin YJ, Sun CA, Yang T, Hsu CH, Lin HC, Chou YC. Differential DNA methylation analysis of SUMF2, ADAMTS5, and PXDN provides novel insights into colorectal cancer prognosis prediction in Taiwan. World J Gastroenterol 2022; 28(8): 825-839
- URL: https://www.wjgnet.com/1007-9327/full/v28/i8/825.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i8.825
Colorectal cancer (CRC) is the third most commonly diagnosed malignancy and the fourth leading cause of cancer-related deaths worldwide[1]. According to global estimations, in 2018, 1.8 million new cases of CRC were diagnosed, and 0.8 million people died from CRC that year[2]. By 2030, the worldwide CRC burden is predicted to increase by 60%; in 2030, it is expected that 2.2 million patients will be newly diagnosed as having CRC and 1.1 million CRC-related deaths will occur worldwide[1]. The treatment and survival of patients with CRC are closely related to cancer staging systems. The classification of CRC stages is based on the tumor– node–metastasis (TNM; T, size of the primary tumor; N, nearby affected lymph nodes; M, distant metastasis) staging system from the Eighth Edition of the American Joint Committee on Cancer Staging Manual[3]. However, the heterogeneity of CRC means that patients with the same CRC stage may have different treatment responses and survival times[4]. Although breakthroughs in surgery, chemotherapy, radiotherapy, targeted therapy, and immunotherapy have improved the survival of patients with CRC, the 5-year survival of patients with stage IV disease is as low as 14% owing to CRC’s heterogeneous nature and association with diverse molecular alterations, which are involved in cancer cell progression[5]. Intensive postoperative surveillance programs are proposed after tumor resection to detect asymptomatic recurrence in advance and prolong the survival patients who may be suited to further curative therapy[6]. Therefore, identifying molecular biomarkers for predicting patient prognoses or monitoring cancer relapse is crucial.
Several studies have focused on identifying new prognostic indicators for CRC[7]. Prognostic biomarkers can be employed as personalized indicators to predict disease progression, such as early recurrence, metastasis, and mortality[8]. These biomarkers are associated with molecular patterns of genomic mutations and epigenetic alterations that lead to CRC carcinogenesis[9]. DNA methylation alterations play a pivotal role in CRC progression and metastasis[10]. Promoter DNA hypermethylation silences genes such as MLH1, CDKN2A, MGMT, RUNX3, TPEF, VIM, and SFRP1/2/4/5, which play crucial roles in the cell cycle, DNA repair, and signal transduction[11]. Consequently, DNA methylation of genes may be a novel epigenetic indicator of patient prognosis.
SUMF2 was regarded as one of the frequently mutated genes in CRC, and SUMF2 mutation frequently altered pathways in the tumorigenesis of CRC[12]. ADAMTS5 was found upregulated in CRC, which was associated with tumor progression and even unfavorable clinical outcomes[13]. To determine the effect of the DNA methylation of selected genes on CRC prognosis over 5 years, we examined methylation status and extent in tumor tissue and tumor-free areas adjacent to such tissue. We propose that the differential DNA methylation of candidate genes in tumor samples and in matched adjacent normal tissue could assist in prognosis prediction and the optimization of CRC treatment.
In this retrospective cohort study, we analyzed the data of patients diagnosed as having CRC from 2006 to 2010 and who underwent surgical treatment at Tri-Service General Hospital (TSGH), Taiwan, to assess their 5-year prognosis. All participants signed informed consent forms before their involvement in this research. Then surgeons gathered specimens in patients including colorectal cancer tissue and adjacent normal regions during surgery. The tissue were deposited at -80 ℃ ultra-low temperature freezers for further analysis. The study was approved by the TSGH Institutional Review Board (TSGHIRB approval numbers 098-05-292 and 2-105-05-129). According to the clinical practice guidelines of the Division of Colon and Rectal Surgery of TSGH, patients with CRC should return to the outpatient department for a follow-up every 3 mo in the first year after surgery and once every 3 to 6 mo afterward. The clinical and demographic characteristics of enrolled patients, including sex, age at surgery, clinical staging, tumor size, histological grade, lymph node count, tumor location, and adjuvant chemotherapy as well as follow-up information on recurrence, metastasis, and survival were acquired from the cancer registration database of TSGH.
Recurrence-free survival (RFS), progression-free survival (PFS), and overall survival (OS) were calculated from the date of surgery to disease progression (inclusive of cancer recurrence or metastasis), death from any cause, or until the final follow-up date before December 31, 2010. In total, 208 patients who met the inclusion criteria were enrolled. A flow diagram of the study’s design is presented in Figure 1.
The candidate gene approach was applied for genetic association studies, which has been widely used to investigate novel prognostic biomarkers related to CRC[14]. First, we searched for genes whose expression might influence CRC prognosis by browsing PubMed (https://pubmed.ncbi.nlm.nih.gov/), University of California, Santa Cruz Genome Browser (https://genome.ucsc.edu/) and Prediction of Clinical Outcomes from Genomic Profiles (https://precog.stanford.edu/). Subsequently, we confirmed gene methylation differences in CRC tissue and normal tissue using the Shiny Methylation Analysis Resource Tool website (http://bioinfo-zs.com/smartapp/). We then searched PubMed (https://pubmed.ncbi.nlm.nih.gov/) to review related literature with the keyword "gene name + colorectal cancer". If the number of results was less than 30, it was thought that the gene had been rarely studied for colorectal cancer. Those genes which met the above three conditions were included in this study. Therefore, we chose 16 candidate genes include CFLAR, RBM44, ABCG1, WDR74, ZNF292, EFHA2, PXDN, TEC, CDH2, ADAMTS5, COL4A2, PCGF2, EMID2, GRPEL2, DKK2 and SUMF2. Because of the limited resources and experimental results, we narrowed down to SUMF2, ADAMTS5, and PXDN. These three candidate genes involved in the pathways associated with cancer stages and prognosis, such as inflammation, epithelial–mesenchymal transition, tumor migration, and angiogenesis.
In accordance with the manufacturer’s instructions, cellulose-coated magnetic beads were used to extract genomic DNA from the samples by using the Genomic DNA Tissue Kit (Catalog No. 69504; Qiagen, Taipei, Taiwan). We used sterile blades to cut about 10 mg tissue minced into the 1.5 mL microcentrifuge tube, and 200 μL Lysis Buffer and 6 μL Proteinase K were prepared and added. The 1.5 mL microcentrifuge tube was placed at 56℃ water bath machine for 8-10 h after orbital shaker and centrifuge use. We confirmed that tissues were dissolved as clarified liquid, and then the tissue would have a vortex and centrifugation. Next, we used pipette to put tissue lysate into the 96 Deep Well Plate of the MagCore Compact Automated Nucleic Acid Extractor (Catalog No. MCA0801; RBC Bioscience, Taipei, Taiwan). Subsequently, we used the EZ DNA Methylation Kit (Zymo Research Corporation, Orange, CA, United States) to modify isolated DNA using sodium bisulfite.
The gene methylation statuses of SUMF2, ADAMTS5, and PXDN were evaluated through a methylation-specific polymerase chain reaction (MS-PCR). The total volume of the reaction solution was 20 μL, and it contained HotStart Taq Premix (RBC Bioscience, Taipei, Taiwan) (9 μL for SUMF2 and PXDN; 10 μL for ADAMTS5), 0.5 μL of forward and reverse primers, 1 μL of bisulfite-modified DNA, and pure water (9 μL for SUMF2 and PXDN; 8 μL for ADAMTS5).
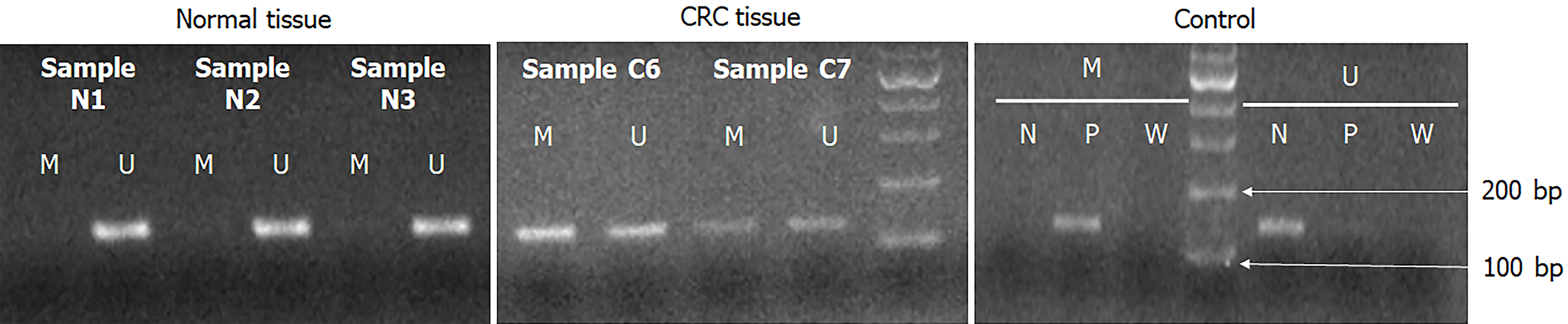
For MS-PCR, the oligonucleotide primers, annealing temperature of each primer used for amplification, and PCR product sizes were described in Table 1. PCR cycling was performed as follows: 10 min at 95 °C, 38 cycles of denaturation for 30 s at 95 °C, 30-s annealing at a gene-appropriate temperature, 30-s elongation at 72 °C, final extension for 7 min at 72 °C, and holding at 4 °C. After amplification, PCR products were mixed with a loading buffer, electrophoresed (100 V for 28–30 min) on 2.75%-4% agarose gels using 1–2 μL of gel-stained dye, and visualized using an ultraviolet transilluminator. To confirm our experiment results were without error, we used SssI-treated DNA as positive control, and sterile water as negative control. Figure 2 showed that the methylation-specific polymerase chain reaction (MS-PCR) results of PXDN gene in CRC patients. Negative control, positive control and sterile water represent unmethylation-specific reaction, methylation-specific reaction and no contaminant reaction for PCR, respectively.
| Genes | Forward primer (5’ → 3’) | Annealing temperature (°C) | Product size (bp) | |
| SUMF2 | M | F:TTTGATTATGGTCGGTTTTGC | 59.4 | 191 |
| R:GACTACTTACAACTCCCCTAACGAC | ||||
| U | F:TTTTTTGATTATGGTTGGTTTTGTG | 60.6 | 198 | |
| R:CCCAACTACTTACAACTCCCCTAACA | ||||
| Q | F:TTTGTTATAGAGGGATGGGAGATAG aggaagagag | 60 | 232 | |
| R:CAAAATAAACAACACTCCAAATTCA cagtaatacgactcactatagggagaaggct | ||||
| ADAMTS5 | M | F:GTTATTGTCGTGGAGCGTTAGC | 59.4 | 170 |
| R:CCTACCTCCCGTACTTCCCG | ||||
| U | F:TTATTGTTGTGGAGTGTTAGTGTTT | 59.4 | 169 | |
| R:CCTACCTCCCATACTTCCCACAT | ||||
| Q | F:aggaagagagTTGAAATTGTTATTGTAGGATGGTATG | 61.3 | 245 | |
| R:cagtaatacgactcactatagggagaaggctAATTAAAACAAAAATACAAAAAAACAACC | ||||
| PXDN | M | F:TATGCGGGACGAGAACGAGA | 61.6 | 137 |
| R:ACTTAAACAACTCCGTAACAATACGAT | ||||
| U | F:GTGTATGTGGGATGAGAATGAGAG | 60.4 | 142 | |
| R:CAACTTAAACAACTCCATAACAATACAA | ||||
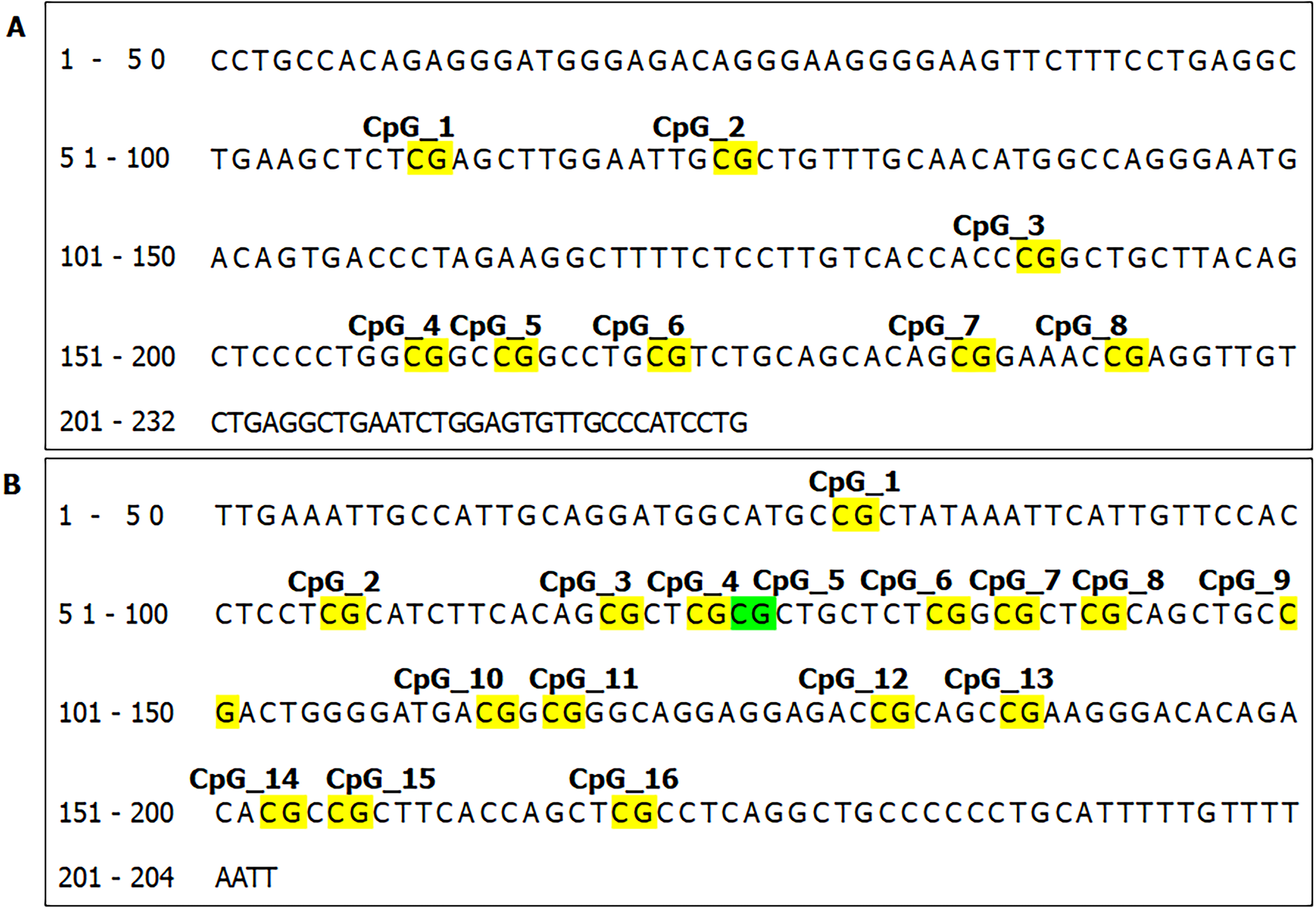
We further identified the CpG sites of SUMF2 and ADAMTS5 (Figure 3) and analyzed DNA methylation changes by using an Agena Bioscience MassARRAY system with EpiTYPER biochemistry (Agena Bioscience, San Diego, CA), an advanced method for quantitative DNA analysis.
The oligonucleotide primers, annealing temperature of each primer used for amplification, and PCR product sizes used in the EpiTyper assay were described in Table 1. The PCR cycling protocols were as follows: 15 min at 95 °C, 38 cycles of denaturation for 30 s at 95 °C, 30-s annealing at a gene-appropriate temperature, 30-s elongation at 72 °C, final extension for 10 min at 72 °C, and holding at 4 °C. In vitro transcription and base-specific cleavage were performed using the MassCLEAVE kit (Agena Bioscience). In total, 0.22 μL of T cleavage mix, 3.14 mmol/L DTT, 20 U of T7 RNA and DNA polymerase, and 0.09 mg/mL RNase A were prepared and added to a 7 μL reaction solution with shrimp alkaline phosphatase inactive PCR product. The final product was stored at 37 °C for 3 h. After the addition of a cation exchange resin to remove salt that remained after the reactions, the EpiTYPER reaction products were loaded onto the matrix pad of a SpectroCHIP Array (Agena Bioscience). The size and mass of the resulting cleavage products varied based on sequence changes. The fragment contained CpG sites had two kinds of molecular weight (GC or AC) because of the methylation status. There are 16 Daltons of signal divergence between the methylated and unmethylated sequence, which was observed by the mass spectrometry. These differences provided quantitative information on each target fragment, which was acquired using the MassARRAY Analyzer 4 (Agena Bioscience). Signal shifts of fragments indicated methylation events at single CpGs or small groups of CpGs (CpG units), and signal intensity was linked to DNA methylation extent, which was analyzed using EpiTYPER software (Agena Bioscience).
For each candidate gene, different DNA methylation statuses related to RFS, PFS, and OS were investigated using univariate Cox proportional hazards analyses. The resultant multivariate Cox proportional hazards regression model was then used to verify the independent prognostic effects of gene methylation status after adjustment for various clinical variables (sex, age at surgery, stage, and lymph node counts). The 5-year RFS, PFS, and OS curves for the methylation status of the selected genes were presented using a Kaplan–Meier survival analysis and compared using a log-rank test. We used SPSS version 23 (IBM SPSS Statistics 23) for statistical analyses. All P values were calculated from two - sided tests, and P value below 0.05 represented statistical significance.
In total, 208 patients diagnosed as having CRC at TSGH were recruited to participate. We analyzed tumor samples and adjacent nontumorous tissue of enrollees. The correlation of the candidate genes’ methylation status and extent with the demographic and clinicopathological characteristics of patients was assessed, and the results are presented in Table 2. Normal tissue with SUMF2 methylation significantly belonged to poor or undifferentiated histological grade. Tumor tissue with PXDN methylation significantly belonged to lower stages and had less lymphovascular invasion. No other associations were found between the methylation statuses of these three genes and the demographic or clinicopathological variables of the participants.
| Characteristics | Total | Methylation status | |||||
| SUMF2 | ADAMTS5 | PXDN | |||||
| Normal | Tumor | Normal | Tumor | Normal | Tumor | ||
| Sex | |||||||
| Male | 103 (49.5) | 20 (60.6) | 27 (55.1) | 28 (60.9) | 44 (77.2) | 35 (76.1) | 44 (77.2) |
| Female | 105 (50.5) | 17 (45.9) | 25 (45.5) | 38 (64.4) | 49 (73.1) | 41 (69.5) | 53 (79.1) |
| χ2 (P value) | 0.97 (0.324) | 0.62 (0.432) | 0.03 (0.866) | 0.1 (0.755) | 0.28 (0.596) | < 0.01 (0.969) | |
| Age at surgery | |||||||
| mean ± SD | 64.3 ± 14.6 | 64.9 ± 14.2 | 66.9 ± 15.8 | 66.4 ± 14.8 | 67.7 ± 15.5 | 65.0 ± 14.4 | 66.2 ± 15.0 |
| < 65 | 103 (49.5) | 17 (51.5) | 26 (53.1) | 28 (58.3) | 43 (72.9) | 34 (70.8) | 46 (78.0) |
| ≥ 65 | 105 (50.5) | 20 (54.1) | 26 (47.3) | 38 (66.7) | 50 (76.9) | 42 (73.7) | 51 (78.5) |
| χ2 (P value) | < 0.01 (1.00) | 0.15 (0.694) | 0.46 (0.498) | 0.1 (0.755) | 0.01 (0.915) | < 0.01(1.00) | |
| Stage | |||||||
| I | 29 (13.9) | 7 (58.3) | 7 (50.0) | 9 (50.0) | 12 (63.2) | 12 (66.7) | 17 (89.5) |
| II | 77 (37.0) | 13 (48.1) | 17 (45.9) | 21 (56.8) | 33 (75.0) | 29 (78.4) | 39 (88.6) |
| III | 68 (32.7) | 13 (65.0) | 21 (63.6) | 22 (68.8) | 31 (75.6) | 22 (68.8) | 27 (65.9) |
| IV | 34 (16.3) | 4 (36.4) | 7 (35.0) | 14 (77.8) | 17 (85.0) | 13 (72.2) | 14 (70.0) |
| χ2 (P value) | 2.77 (0.429) | 4.5 (0.212) | 4.06 (0.255) | 2.5 (0.476) | 1.17 (0.760) | 8.69a (0.034) | |
| 5-yr recurrence1 | |||||||
| No | 141 (82.0) | 28 (51.9) | 35 (49.3) | 43 (56.6) | 61 (70.9) | 54 (71.1) | 71 (82.6) |
| Yes | 31 (18.0) | 4 (40.0) | 9 (60.0) | 12 (80.0) | 14 (82.4) | 11 (73.3) | 12 (70.6) |
| χ2 (P value) | 0.12 (0.731) | 0.22 (0.639) | 1.98 (0.160) | 0.45 (0.504) | < 0.01 (1.00) | 0.65 (0.421) | |
| 5-yr all-cause death | |||||||
| No | 168 (80.8) | 29 (54.7) | 43 (51.8) | 50 (61.0) | 79 (78.2) | 59 (72.0) | 77 (76.2) |
| Yes | 40 (19.2) | 8 (47.1) | 9 (42.9) | 16 (69.6) | 14 (60.9) | 17 (73.9) | 20 (87.0) |
| χ2 (P value) | 0.07 (0.786) | 0.24 (0.625) | 0.26 (0.611) | 2.15 (0.142) | < 0.01 (1.00) | 0.71 (0.399) | |
| 5-yr progression | |||||||
| No | 155 (74.5) | 28 (56.0) | 39 (50.0) | 45 (58.4) | 73 (76.8) | 56 (72.7) | 74 (77.9) |
| Yes | 53 (25.5) | 9 (45.0) | 13 (50.0) | 21 (75.0) | 20 (69.0) | 20 (71.4) | 23 (79.3) |
| χ2 (P value) | 0.32 (0.570) | < 0.01(1.00) | 1.75 (0.185) | 0.38 (0.540) | < 0.01 (1.00) | < 0.01 (1.00) | |
| Lymphovascular invasion1 | |||||||
| No | 106 (52.5) | 20 (48.8) | 25 (48.1) | 32 (56.1) | 46 (73.0) | 41 (71.9) | 55 (87.3) |
| Yes | 96 (47.5) | 16 (57.1) | 27 (51.9) | 34 (72.3) | 47 (78.3) | 34 (72.3) | 41 (68.3) |
| χ2 (P value) | 0.19 (0.662) | 0.04 (0.845) | 2.26 (0.133) | 0.23 (0.634) | < 0.01 (1.00) | 5.44a (0.02) | |
| Histological grade1 | |||||||
| Well or moderately | 156 (89.7) | 27 (48.2) | 35 (47.3) | 52 (64.2) | 67 (74.4) | 58 (71.6) | 67 (74.4) |
| Poor or undifferentiated | 18 (10.3) | 6 (100.0) | 9 (64.3) | 7 (70.0) | 14 (82.4) | 9 (90.0) | 13 (76.5) |
| χ2 (P value) | 3.94a (0.047) | 0.76 (0.382) | < 0.01 (0.99) | 0.15 (0.697) | 0.75 (0.387) | < 0.01 (1.00) | |
| Lymph node counts1 | |||||||
| 0-11 | 34 (18.4) | 7 (63.6) | 7 (50.0) | 12 (80.0) | 15 (83.3) | 13 (86.7) | 16 (88.9) |
| ≥ 12 | 151 (81.6) | 29 (52.7) | 40 (50.0) | 50 (61.7) | 70 (73.7) | 58 (71.6) | 70 (73.7) |
| χ2 (P value) | 0.07 (0.786) | < 0.01 (1.00) | 1.14 (0.287) | 0.33 (0.568) | 0.81 (0.368) | 1.18 (0.278) | |
| Tumor location1 | |||||||
| Colon | 147 (79.9) | 28 (50.0) | 38 (50.7) | 53 (67.9) | 62 (70.5) | 60 (76.9) | 68 (77.3) |
| Rectum | 37 (20.1) | 8 (80.0) | 9 (47.4) | 9 (50.0) | 23 (92.0) | 11 (61.1) | 18 (72.0) |
| χ2 (P value) | 1.99 (0.158) | < 0.01(1.00) | 1.35 (0.245) | 3.76 (0.052) | 1.17 (0.280) | 0.08 (0.780) | |
| Adjuvant chemotherapy1 | |||||||
| No | 54 (29.3) | 10 (58.8) | 12 (42.9) | 15 (55.6) | 25 (73.5) | 20 (74.1) | 29 (85.3) |
| Yes | 130 (70.7) | 26 (53.1) | 35 (53.0) | 47 (68.1) | 60 (75.9) | 51 (73.9) | 57 (72.2) |
| χ2 (P value) | 0.02 (0.898) | 0.46 (0.499) | 0.85 (0.358) | 0.01 (0.971) | < 0.01(1.00) | 1.59 (0.207) | |
Although the selected genes were methylated in both tumor tissue and matched normal tissue (SUMF2, 50% vs 52.9%; ADAMTS5, 75.0% vs 62.9%; PXDN, 78.2% vs 72.4%), we observed that the differential DNA methylation of candidate genes, especially SUMF2 and ADAMTS5, tended to affect patient prognosis according to the Kaplan–Meier method. To further investigate the extent and patterns of gene methylation in patients with CRC, we analyzed SUMF2 and ADAMTS5 methylation levels in primary CRC tissue samples and in adjacent normal samples. A higher percentage of SUMF2 genes methylation at loci CpG_1, CpG_2, CpG_3 and CpG_7 was detected in primary CRC samples compared with adjacent normal tissue. We also found a higher level of ADAMTS5 genes methylation at loci CpG_1, CpG_2, CpG_9, CpG_10.11, CpG_12, CpG_13, CpG_14.15, and CpG_16 in CRC tissue than in adjacent normal samples. Both findings reached statistical significance (Table 3).
| Normal | Tumor | P value | |||||
| n1 | Median | mean ± SD2 | n1 | Median | mean ± SD2 | ||
| SUMF2 | |||||||
| CpG_1 | 69 | 0.40 | 0.43 ± 0.15 | 104 | 0.53 | 0.55 ± 0.17 | < 0.001 |
| CpG_2 | 70 | 0.56 | 0.56 ± 0.11 | 104 | 0.76 | 0.73 ± 0.13 | < 0.001 |
| CpG_3 | 70 | 0.38 | 0.39 ± 0.11 | 104 | 0.54 | 0.54 ± 0.17 | < 0.001 |
| CpG_7 | 48 | 0.64 | 0.64 ± 0.15 | 80 | 0.87 | 0.81 ± 0.20 | 0.001 |
| ADAMTS5 | |||||||
| CpG_1 | 66 | 0.06 | 0.08 ± 0.07 | 95 | 0.19 | 0.25 ± 0.20 | < 0.001 |
| CpG_2 | 70 | 0.06 | 0.08 ± 0.06 | 105 | 0.20 | 0.24 ± 0.18 | < 0.001 |
| CpG_9 | 69 | 0.06 | 0.07 ± 0.06 | 91 | 0.21 | 0.25 ± 0.18 | < 0.001 |
| CpG_10.11 | 69 | 0.08 | 0.10 ± 0.09 | 103 | 0.30 | 0.34 ± 0.22 | < 0.001 |
| CpG_12 | 65 | 0.09 | 0.10 ± 0.08 | 98 | 0.19 | 0.24 ± 0.21 | 0.001 |
| CpG_13 | 63 | 0.07 | 0.12 ± 0.14 | 94 | 0.15 | 0.22 ± 0.22 | 0.009 |
| CpG_14.15 | 71 | 0.32 | 0.33 ± 0.05 | 105 | 0.43 | 0.44 ± 0.11 | < 0.001 |
| CpG_16 | 71 | 0.10 | 0.11 ± 0.04 | 105 | 0.19 | 0.22 ± 0.12 | < 0.001 |
The correlation between the methylation status of each gene and the 5-year RFS, PFS, and OS of patients with CRC was analyzed. We did not observe a significant correlation of the SUMF2, ADAMTS5, and PXDN methylation statuses of normal and tumor tissue and with patients’ 5-year RFS, PFS, and OS in univariate and multivariate Cox proportional hazards regression analyses. The methylation of SUMF2 in normal tissue was associated with poorer 5-year PFS and OS in a Kaplan–Meier survival analyses (P = 0.500 and 0.260, respectively). In a univariate analysis, we observed that patients with a stage III+IV disease had a poor RFS, PFS, and OS [hazard ratio (HR) = 4.68 and 95% confidence interval (95%CI) = 2.09-10.46 for RFS, 2.21 (1.25-3.91) for PFS, and 1.90 (1.00-3.61) for OS], even after adjustment for confounding factors in the multivariable analysis of tumor tissue [HR (95%CI) = 9.85 (2.20-44.15) for RFS, 4.08 (1.49-11.14) for PFS, and 4.70 (1.50-14.74) for OS]. Methylation of ADAMTS5 in normal or tumor tissue was associated with a poor 5-year RFS (P = 0.144 for normal tissue and 0.332 for tumor tissue), and ADAMTS5 methylation in normal tissue was associated with a poor 5-year PFS (P = 0.176). Patients with a stage III+IV disease had a poor RFS, PFS, and OS even after adjustment for confounding factors in the multivariable analysis [HR (95%CI) = 4.82 (1.33-17.48) for RFS (normal tissue), 7.37 (2.11-25.83) for RFS (tumor tissue), 2.83 (1.19-6.74) for PFS (tumor tissue), and 2.63 (1.02-6.81) for OS (tumor tissue)]. Finally, no relationship was observed between PXDN methylation status and CRC prognosis.
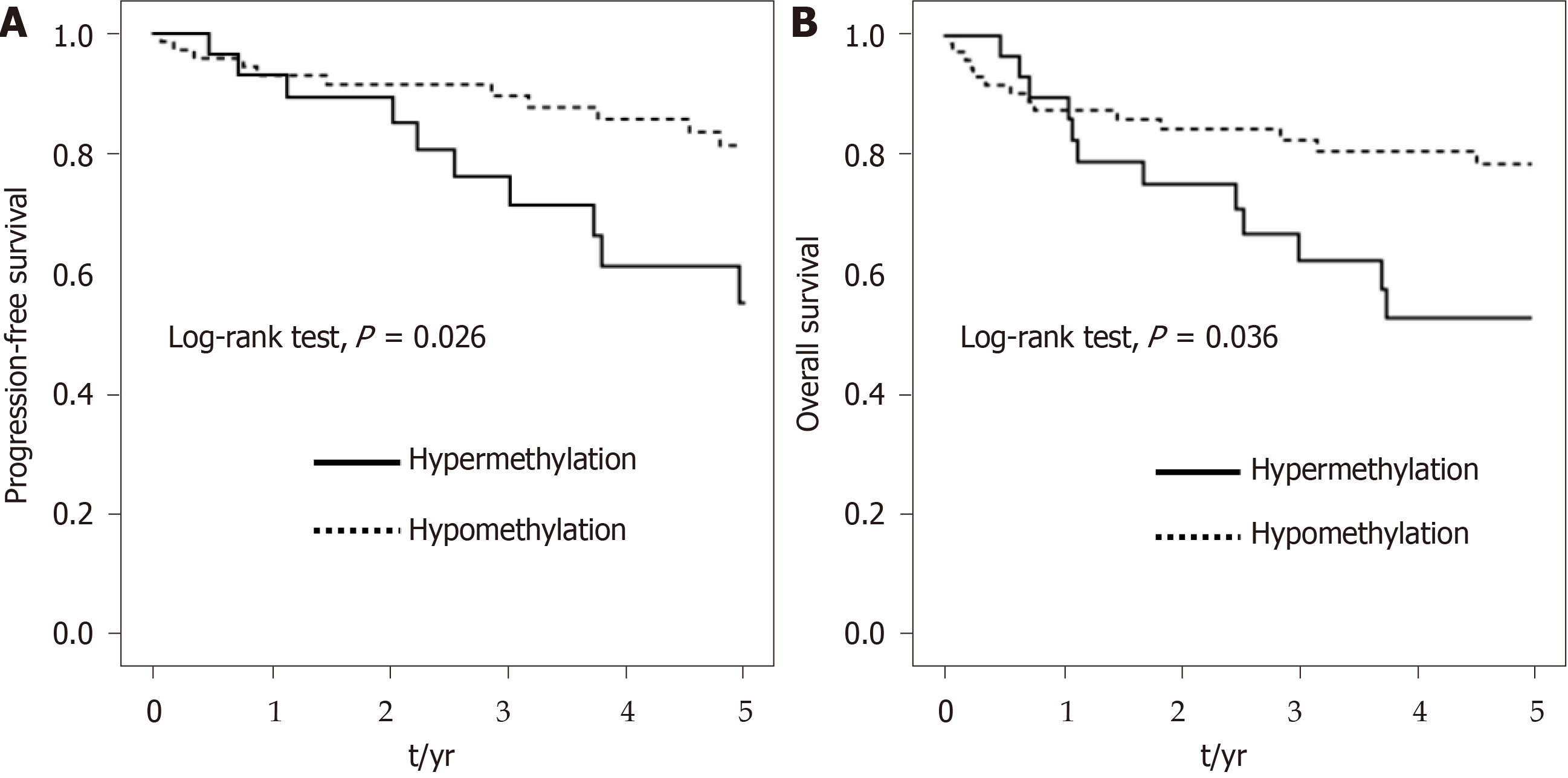
We assessed the associations of SUMF2 and ADAMTS5 with prognosis using a quantitative analysis (Table 4). To enhance statistical power, we classified the methylation level of candidate genes into hypermethylation and hypomethylation groups according to the median value shown in Table 3, which functioned as the threshold value. On the basis of consecutive 5-year RFS, PFS, and OS Kaplan–Meier plot validation, a clear trend of differences was observed between hypermethylation and hypomethylation of the SUMF2 gene at CpG_3 and CpG_7 Loci from tumor tissue. We further focused on the methylation statuses at CpG_3+CpG_7 to evaluate their relationship with patient prognosis. The 5-year PFS and OS curves revealed a significant difference between the hypermethylation and hypomethylation of the SUMF2 gene at CpG_3+CpG_7 Loci from tumor tissue (P = 0.026 for PFS and 0.036 for OS; Figure 4). Compared with the CpG_3+CpG_7 hypomethylation of tumor tissue, a significantly shorter PFS and OS were observed for CpG_3+CpG_7 hypermethylation [HR (95%CI) = 2.24 (1.03-4.85) for PFS and 2.56 (1.08-6.04) for OS]. After adjustment for confounders in the multivariable analysis, the shorter OS associated with CpG_3+CpG_7 hypermethylation remained significant [HR (95%CI) = 3.53 (1.35-9.26)], whereas the shorter PFS associated with CpG_3+CpG_7 hypermethylation did not [HR (95%CI) = 2.05 (0.91-4.62)] (Table 4).
| RFS | PFS | OS | ||||
| cHR (95%CI) | aHR (95%CI)1 | cHR (95%CI) | aHR (95%CI)1 | cHR (95%CI) | aHR (95%CI)2 | |
| SUMF2 in tumor tissue | ||||||
| CpG_3+CpG_7 | ||||||
| Hypomethylation | 1.00 (Reference) | 1.00 (Reference) | 1.00 (Reference) | 1.00 (Reference) | 1.00 (Reference) | 1.00 (Reference) |
| Hypermethylation | 2.37 (0.86-6.55) | 1.64 (0.55-4.89) | 2.24 (1.03-4.85)a | 2.05 (0.91-4.62) | 2.56 (1.08-6.04)a | 3.53 (1.35-9.26)a |
| ADAMTS5 in normal tissue | ||||||
| CpG_2 | ||||||
| Hypomethylation | 1.00 (Reference) | 1.00 (Reference) | 1.00 (Reference) | 1.00 (Reference) | 1.00 (Reference) | 1.00 (Reference) |
| Hypermethylation | 0.15 (0.03-0.71)a | 0.17 (0.03-0.95)a | 0.57 (0.24-1.37) | 0.54 (0.21-1.41) | 0.82 (0.31-2.16) | 0.94 (0.31-2.85) |
| CpG_13 | ||||||
| Hypomethylation | 1.00 (Reference) | 1.00 (Reference) | 1.00 (Reference) | 1.00 (Reference) | 1.00 (Reference) | 1.00 (Reference) |
| Hypermethylation | 0.20 (0.04-0.97)a | 0.16 (0.03-0.85)a | 0.48 (0.19-1.19) | 0.45 (0.17-1.18) | 0.50 (0.19-1.30) | 0.72 (0.24-2.15) |
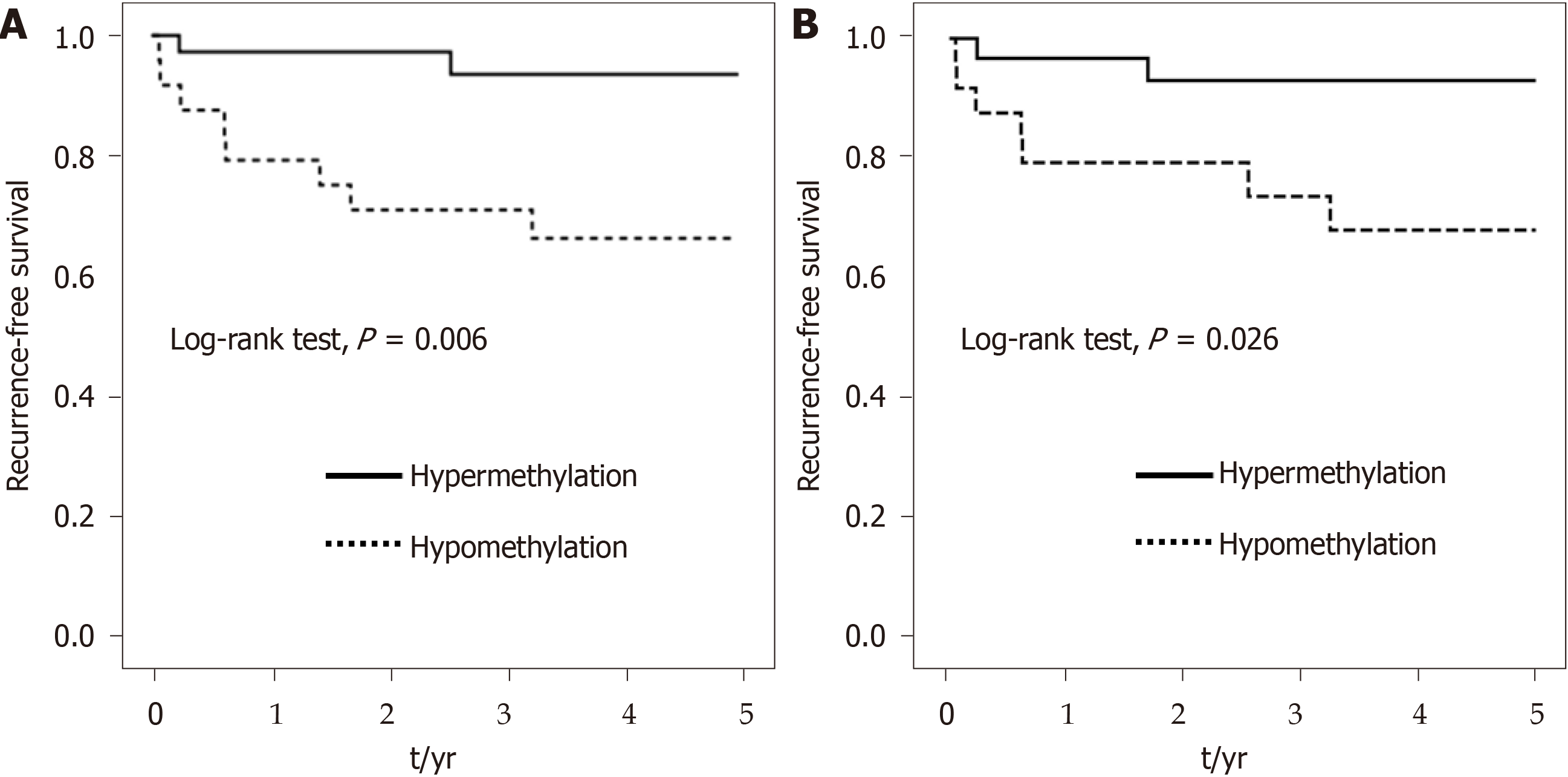
The 5-year RFS curves showed a significant difference between hypermethylation and hypomethylation of the ADAMTS5 gene at CpG_2 and CpG_13 from normal tissue (P = 0.006 for CpG_2 and 0.026 for CpG_13; Figure 5). Compared with CpG_2 and CpG_13 hypomethylation of normal tissue, a significantly longer RFS was observed for CpG_2 and CpG_13 hypermethylation [HR (95%CI) = 0.15 (0.03-0.71) for CpG_2 and 0.20 (0.04-0.97) for CpG_13]. After adjustment for confounders in the multivariable analysis, the longer RFS associated with CpG_2 and CpG_13 hypermethylation remained significant [HR (95%CI) = 0.17 (0.03-0.95) for CpG_2 and 0.16 (0.03-0.85) for CpG_13] (Table 4).
The TNM staging system based on tumor depth, nodal status, and metastasis guides treatment strategies and improves the accuracy of predicting prognosis. However, the etiologically heterogeneous characteristics of CRC contributes to differences in survival between patients with the same TNM stage of CRC. Because of this heterogeneity, TNM staging of CRC requires further classification for better disease management. Therefore, combining other classifications related to several biologic mechanisms with TNM staging is required[15].
Aberrant DNA methylation of certain loci is involved in the aberrant expression of oncogenes through the hypomethylation of CpG islands in promoters. On the other hand, transcriptional silencing by the hypermethylation of CpG islands in promoters is observed in tumor-suppressor genes[16]. Aberrant DNA methylation elicits apoptosis[17], metastasis[18], cell adherence[19], tumor progression[18,20], and resistance to current anticancer therapies[21]. Hypermethylation at CpG islands of genes has promise as a robust and valid diagnostic method for CRC[22]. Additionally, methylation-based molecular markers can be employed as prognostic or predictive biomarkers for CRC to improve clinical management; such markers can predict malignant tumor potential and survival outcomes[23].
Globally acquired DNA hypomethylation profiles represent a key step in the development and advancement of CRC because hypomethylation triggers genomic instability and the global loss of imprinting[24]. This epigenetic change contributes to the activation of oncogenes and mainly affects repetitive transposable elements, such as long interspersed nuclear element-1 (LINE-1 or L1), which represent 17% of the human genome[25]. LINE-1 is more hypomethylated in CRC tumors compared with adjacent normal tissue, and this is linked to metastasis. Moreover, LINE-1 is more hypermethylated in the neoplastic tissue of patients treated for CRC compared with untreated patients, and after neoadjuvant treatment, poor survival was observed in patients with tumor LINE-1 hypomethylation[26]. The results of a meta-analysis also support the idea that LINE-1 hypomethylation is considerably associated with the shortened OS and disease-free survival (DFS) of patients with CRC[27].
In this study, we analyzed 208 of each tumor tissue samples and normal-appearing tissue samples from patients with CRC. CpG_3+CpG_7 hypermethylation of SUMF2 in tumor tissue was strongly related to poorer 5-year PFS and OS according to a Cox proportional hazards regression model and Kaplan–Meier curves. By contrast, CpG_2 and CpG_13 hypermethylation of ADAMTS5 in adjacent normal tissue was significantly associated with better 5-year RFS than CpG_2 and CpG_13 hypomethylation of ADAMTS5 in adjacent normal tissue. Finally, no correlation was observed between the promoter methylation status of PXDN and patient prognosis regardless of the tissue type analyzed.
Therefore, our findings indicate that SUMF2 could be a tumor-suppressor gene, whereas ADAMTS5 may be classified as a CRC oncogene. These findings can be applied along with current staging to modify the treatment of patients with CRC, and the genes discussed identified herein can serve as appropriate biomarkers to identify patients at a higher risk of having a poor prognosis and to indicate requirements for intensive follow-up.
We revealed that ADAMTS5 methylation status in tumor-free areas adjacent to tumors was significantly associated with the prognosis of patients with CRC. The progressive accumulation of genetic mutations and DNA methylation changes in normal tissue around tumors lead to the development and progression of adenomas, which might become adenocarcinomas. Such alterations are a result of field cancerization[28]. Studies have demonstrated that the aberrant methylation of cancer-related genes could serve as epigenetic markers for CRC risk owing to the field of susceptibility[29], and such findings are consistent with our finding that compared with abnormal DNA methylation in tumor tissue, abnormal DNA methylation in adjacent normal tissue is highly correlated with an unfavorable prognosis after surgical resection.
SUMF2, located in the luminal space of the endoplasmic reticulum, is a member of the formylglycine-generating enzyme family and regulates the activity of sulfatase and the formation of formylglycine[30]. According to a whole-genome microarray expression study, Ala62Thr changes in ZNF365 isoform D are related to the altered expression of SUMF2 in patients with Crohn disease[31]. According to Liang et al[32], SUMF2 was one of the main genes with significant mutation in CRC. No report has examined the association between SUMF2 methylation status and the prognosis of patients with CRC. ADAMTS family members containing disintegrin domains, metalloproteinases, and thrombospondin motifs likely contribute to malignant transformation such as cancer cell adhesion, fusion, migration, proliferation, and metastasis in CRC through their modification of the structure and function of the extracellular matrix (ECM)[33] and desmoplastic reactions (the overgrowth of fibrous connective tissue around carcinoma cells)[34]. The expression level of ADAMTS5 was increased in late CRC stages, and ADAMTS5 may have served as the fundamental component of tumor invasion through degrading ECM so as to promote tumor progression to more advanced stages of CRC[35]. The ratios of lymphatic invasion and lymph node metastasis were significantly higher in patients with CRC and high ADAMTS5 expression, but such expression did not affect OS and DFS[36]. However, Li et al[37] suggested that ADAMTS5 overexpression inhibited the invasion and migration of CRC, and ADAMTS5 was more hypermethylated in tumor tissue compared with normal tissue, corresponding to poor OS and DFS. This result was not observed in our research. PXDN, which regulates cell plasticity and remodels the ECM by encoding ECM protein with peroxidase activity[38], engages in epithelial mesenchymal transition(EMT)[39]. EMT is a process by which epithelial cells lose cell polarity and cell–cell adhesion and acquire migratory and invasive capabilities to become mesenchymal cells; this activity has shown to occur during the initiation of metastasis[40]. A few studies have studied PXDN in the context of cancer; for example, the increased expression of PXDN has been shown in patients with melanoma[41] as well as in patients with brain[42], breast[43], ovarian[44] or prostate[45] cancer. No reports have examined the correlation between PXDN expression and the prognosis of patients with CRC. Thus, we investigated whether PXDN methylation status had an impact on CRC. We did not identify any correlation of PXDN methylation status in tumor tissue or adjacent normal tissue with CRC prognosis.
Our study has several advantages. The candidate gene approach was used in a rigorous approach comprising three steps. We further conducted quantitative analysis determined by what we observed from an MS-PCR. EpiTYPER DNA methylation analysis is a quantitative method of assessing methylation status. It also can confirm the degree of methylation at different CpG sites, which might be conductive to accurately identifying sites that affect CRC prognosis. This study was not without limitations. The results should be interpreted with caution on account of the small sample size. In the future, a larger prospective cohort study should be conducted to confirm our results. Furthermore, the participants were recruited from a single medical center in Taiwan. The effectiveness of the candidate genes as prognostic biomarkers for CRC should be validated in other ethnic populations. Finally, we did not detect the expression levels of selected genes, meaning we could not prove that the status or extent of methylation directly influenced gene expression.
Current clinical staging systems do not allow physicians to precisely evaluate the prognosis and outcomes of patients with CRC. Novel methylation biomarkers in tumor tissue and adjacent normal samples were identified in our study, providing a novel insight into how these markers could be used for CRC prognosis estimations. We anticipate that large-scale and independent cohort studies will clarify the utility of these novel markers and address whether clinical management can be adjusted based on supplementary information on the RFS, PFS, and OS of patients with CRC. We propose investigating these new biomarkers in patients with CRC to assist with clinical decision-making.
The tumor–node–metastasis (TNM) cancer staging system provides clinical guidelines for the classification of tumors and prediction of outcomes. However, patients within the same stage can have markedly different outcomes. For example, some patients with an early disease stage of colorectal cancer (CRC) experience relapse after surgical treatment. Prognostic factors related to relapse or progression should be considered to improve treatment selection. The combination of several novel prognostic biomarkers involving epigenetic changes may aid CRC prognosis predictions.
To investigate the impact of the differential DNA methylation of novel candidate genes on CRC prognosis.
This study focused on the association between CRC prognosis and the status and level of differential DNA methylation of candidate genes.
In total, 208 patients with CRC were recruited to assess the relationship between the methylation status of selected genes and clinical outcomes after surgical resection. The methylation statuses of SUMF2, ADAMTS5, and PXDN in tumor tissue and tumor-free adjacent areas were evaluated through a methylation-specific polymerase chain reaction (MS-PCR), and the methylation degrees of SUMF2 and ADAMTS5 were assessed using EpiTYPER DNA methylation analysis. The relationships of gene methylation with recurrence-free survival (RFS), progression-free survival (PFS), and overall survival (OS) were evaluated using a Cox proportional hazards model and Kaplan–Meier survival curves.
CpG_3+CpG_7 hypermethylation of SUMF2 from tumor tissue was associated with significantly shorter PFS and OS compared with CpG_3+CpG_7 hypomethylation. CpG_2 and CpG_13 hypermethylation of ADAMTS5 from normal tissue was associated with a significantly longer RFS compared with CpG_2 and CpG_13 hypomethylation. No significant difference was noted in the association between the methylation status of PXDN in both tissue types and CRC prognosis.
These results can be applied to develop useful prognostic biomarkers of CRC, especially the methylation of certain CpG islands of candidate genes. The results can add value to current cancer staging systems.
Examining the differential DNA methylation of candidate genes could aid in clinical decision-making related to CRC. Further validation and investigations involving larger cohorts are required to confirm the utility of these new epigenetic biomarkers and determine whether they can be used to improve the RFS, PFS, and OS of patients with CRC.
| 1. | Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3058] [Cited by in RCA: 3497] [Article Influence: 388.6] [Reference Citation Analysis (4)] |
| 2. | Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941-1953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3585] [Cited by in RCA: 5041] [Article Influence: 630.1] [Reference Citation Analysis (1)] |
| 3. | Amin MB, Edge S, Greene F, Byrd DR, Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR, Sullivan DC, Jessup JM, Brierley JD, Gaspar LE, Schilsky RL, Balch CM, Winchester DP, Asare EA, Madera M, Gress DM, Meyer LR. AJCC Cancer Staging Manual. 8 ed: Springer International Publishing, 2017. [DOI] [Full Text] |
| 4. | Dienstmann R, Mason MJ, Sinicrope FA, Phipps AI, Tejpar S, Nesbakken A, Danielsen SA, Sveen A, Buchanan DD, Clendenning M, Rosty C, Bot B, Alberts SR, Milburn Jessup J, Lothe RA, Delorenzi M, Newcomb PA, Sargent D, Guinney J. Prediction of overall survival in stage II and III colon cancer beyond TNM system: a retrospective, pooled biomarker study. Ann Oncol. 2017;28:1023-1031. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 176] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 5. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11573] [Cited by in RCA: 13298] [Article Influence: 1662.3] [Reference Citation Analysis (4)] |
| 6. | Broadbridge VT, Karapetis CS, Beeke C, Woodman RJ, Padbury R, Maddern G, Kim SW, Roder D, Hakendorf P, Price TJ. Do metastatic colorectal cancer patients who present with late relapse after curative surgery have a better survival? Br J Cancer. 2013;109:1338-1343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 7. | Koncina E, Haan S, Rauh S, Letellier E. Prognostic and Predictive Molecular Biomarkers for Colorectal Cancer: Updates and Challenges. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 170] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 8. | Ogunwobi OO, Mahmood F, Akingboye A. Biomarkers in Colorectal Cancer: Current Research and Future Prospects. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 145] [Cited by in RCA: 179] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 9. | Lech G, Słotwiński R, Słodkowski M, Krasnodębski IW. Colorectal cancer tumour markers and biomarkers: Recent therapeutic advances. World J Gastroenterol. 2016;22:1745-1755. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 234] [Cited by in RCA: 294] [Article Influence: 29.4] [Reference Citation Analysis (8)] |
| 10. | Ili C, Buchegger K, Demond H, Castillo-Fernandez J, Kelsey G, Zanella L, Abanto M, Riquelme I, López J, Viscarra T, García P, Bellolio E, Saavedra D, Brebi P. Landscape of Genome-Wide DNA Methylation of Colorectal Cancer Metastasis. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 11. | Weisenberger DJ, Liang G, Lenz HJ. DNA methylation aberrancies delineate clinically distinct subsets of colorectal cancer and provide novel targets for epigenetic therapies. Oncogene. 2018;37:566-577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 64] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 12. | Newsom KJ, Starostik P. Mapping the Mutation Landscape of Colorectal Cancer. Am J Med Sci. 2019;358:313-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 13. | Redondo-García S, Peris-Torres C, Caracuel-Peramos R, Rodríguez-Manzaneque JC. ADAMTS proteases and the tumor immune microenvironment: Lessons from substrates and pathologies. Matrix Biol Plus. 2021;9:100054. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 14. | O'Cathail SM, Wu CH, Lewis A, Holmes C, Hawkins MA, Maughan T. NRF2 metagene signature is a novel prognostic biomarker in colorectal cancer. Cancer Genet. 2020;248-249:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 15. | Yamano T, Yamauchi S, Igeta M, Takenaka Y, Song J, Kimura K, Yasuhara M, Babaya A, Kataoka K, Beppu N, Ikeda M, Tomita N, Sugihara K. Combination of preoperative tumour markers and lymphovascular invasion with TNM staging as a cost and labour efficient subtyping of colorectal cancer. Sci Rep. 2020;10:10238. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 16. | Romero-Garcia S, Prado-Garcia H, Carlos-Reyes A. Role of DNA Methylation in the Resistance to Therapy in Solid Tumors. Front Oncol. 2020;10:1152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 115] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 17. | Kang KA, Piao MJ, Hyun YJ, Zhen AX, Cho SJ, Ahn MJ, Yi JM, Hyun JW. Luteolin promotes apoptotic cell death via upregulation of Nrf2 expression by DNA demethylase and the interaction of Nrf2 with p53 in human colon cancer cells. Exp Mol Med. 2019;51:1-14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 90] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 18. | Casalino L, Verde P. Multifaceted Roles of DNA Methylation in Neoplastic Transformation, from Tumor Suppressors to EMT and Metastasis. Genes (Basel). 2020;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 19. | Liu J, Li H, Sun L, Wang Z, Xing C, Yuan Y. Aberrantly methylated-differentially expressed genes and pathways in colorectal cancer. Cancer Cell Int. 2017;17:75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 59] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 20. | Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN, Chen ZH, Zeng ZL, Wang F, Zheng J, Chen D, Li B, Kang TB, Xie D, Lin D, Ju HQ, Xu RH. METTL3 facilitates tumor progression via an m6A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18:112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 302] [Cited by in RCA: 623] [Article Influence: 89.0] [Reference Citation Analysis (0)] |
| 21. | Cheng Y, He C, Wang M, Ma X, Mo F, Yang S, Han J, Wei X. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct Target Ther. 2019;4:62. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 478] [Cited by in RCA: 747] [Article Influence: 106.7] [Reference Citation Analysis (2)] |
| 22. | Li D, Zhang L, Fu J, Huang H, Sun S, Zhang D, Zhao L, Ucheojor Onwuka J, Zhao Y, Cui B. SCTR hypermethylation is a diagnostic biomarker in colorectal cancer. Cancer Sci. 2020;111:4558-4566. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 23. | Kumar A, Gosipatala SB, Pandey A, Singh P. Prognostic Relevance of SFRP1 Gene Promoter Methylation in Colorectal Carcinoma. Asian Pac J Cancer Prev. 2019;20:1571-1577. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 24. | Porcellini E, Laprovitera N, Riefolo M, Ravaioli M, Garajova I, Ferracin M. Epigenetic and epitranscriptomic changes in colorectal cancer: Diagnostic, prognostic, and treatment implications. Cancer Lett. 2018;419:84-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 25. | Furlan D, Trapani D, Berrino E, Debernardi C, Panero M, Libera L, Sahnane N, Riva C, Tibiletti MG, Sessa F, Sapino A, Venesio T. Oxidative DNA damage induces hypomethylation in a compromised base excision repair colorectal tumourigenesis. Br J Cancer. 2017;116:793-801. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 26. | Boughanem H, Martin-Nuñez GM, Torres E, Arranz-Salas I, Alcaide J, Morcillo S, Tinahones FJ, Crujeiras AB, Macias-Gonzalez M. Impact of Tumor LINE-1 Methylation Level and Neoadjuvant Treatment and Its Association with Colorectal Cancer Survival. J Pers Med. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 27. | Ye D, Jiang D, Li Y, Jin M, Chen K. The role of LINE-1 methylation in predicting survival among colorectal cancer patients: a meta-analysis. Int J Clin Oncol. 2017;22:749-757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 28. | Rubio CA, Schmidt PT. Nondysplastic Crypts in Fission in Nonpolypoid Adenomas and in the Adjacent Mucosa Support Field Cancerization in the Colon. Anticancer Res. 2021;41:1515-1521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 29. | Wang T, Maden SK, Luebeck GE, Li CI, Newcomb PA, Ulrich CM, Joo JE, Buchanan DD, Milne RL, Southey MC, Carter KT, Willbanks AR, Luo Y, Yu M, Grady WM. Dysfunctional epigenetic aging of the normal colon and colorectal cancer risk. Clin Epigenetics. 2020;12:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 60] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 30. | Fang C, Li X, Liang H, Xue L, Liu L, Yang C, Gao G, Jiang X. Downregulation of SUMF2 gene in ovalbumin-induced rat model of allergic inflammation. Int J Clin Exp Pathol. 2015;8:12053-12063. [PubMed] |
| 31. | Haritunians T, Jones MR, McGovern DP, Shih DQ, Barrett RJ, Derkowski C, Dubinsky MC, Dutridge D, Fleshner PR, Ippoliti A, King L, Leshinsky-Silver E, Levine A, Melmed GY, Mengesha E, Vasilauskas EA, Ziaee S, Rotter JI, Targan SR, Taylor KD. Variants in ZNF365 isoform D are associated with Crohn's disease. Gut. 2011;60:1060-1067. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 32. | Liang Y, Jiang L, Zhong X, Hochwald SN, Wang Y, Huang L, Nie Q, Huang H, Xu JF. Discovery of Aberrant Alteration of Genome in Colorectal Cancer by Exome Sequencing. Am J Med Sci. 2019;358:340-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (1)] |
| 33. | Kelwick R, Desanlis I, Wheeler GN, Edwards DR. The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family. Genome Biol. 2015;16:113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 348] [Cited by in RCA: 484] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 34. | Mochizuki S, Ao T, Sugiura T, Yonemura K, Shiraishi T, Kajiwara Y, Okamoto K, Shinto E, Okada Y, Ueno H. Expression and Function of a Disintegrin and Metalloproteinases in Cancer-Associated Fibroblasts of Colorectal Cancer. Digestion. 2020;101:18-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 35. | Filou S, Korpetinou A, Kyriakopoulou D, Bounias D, Stavropoulos M, Ravazoula P, Papachristou DJ, Theocharis AD, Vynios DH. ADAMTS expression in colorectal cancer. PLoS One. 2015;10:e0121209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 36. | Haraguchi N, Ohara N, Koseki J, Takahashi H, Nishimura J, Hata T, Mizushima T, Yamamoto H, Ishii H, Doki Y, Mori M. High expression of ADAMTS5 is a potent marker for lymphatic invasion and lymph node metastasis in colorectal cancer. Mol Clin Oncol. 2017;6:130-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 37. | Li J, Liao Y, Huang J, Sun Y, Chen H, Chen C, Li S, Yang Z. Epigenetic silencing of ADAMTS5 is associated with increased invasiveness and poor survival in patients with colorectal cancer. J Cancer Res Clin Oncol. 2018;144:215-227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 38. | Tauber S, Jais A, Jeitler M, Haider S, Husa J, Lindroos J, Knöfler M, Mayerhofer M, Pehamberger H, Wagner O, Bilban M. Transcriptome analysis of human cancer reveals a functional role of heme oxygenase-1 in tumor cell adhesion. Mol Cancer. 2010;9:200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 39. | Sitole BN, Mavri-Damelin D. Peroxidasin is regulated by the epithelial-mesenchymal transition master transcription factor Snai1. Gene. 2018;646:195-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 40. | Ribatti D, Tamma R, Annese T. Epithelial-Mesenchymal Transition in Cancer: A Historical Overview. Transl Oncol. 2020;13:100773. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 185] [Cited by in RCA: 699] [Article Influence: 116.5] [Reference Citation Analysis (0)] |
| 41. | Jayachandran A, Prithviraj P, Lo PH, Walkiewicz M, Anaka M, Woods BL, Tan B, Behren A, Cebon J, McKeown SJ. Identifying and targeting determinants of melanoma cellular invasion. Oncotarget. 2016;7:41186-41202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 42. | Liu Y, Carson-Walter EB, Cooper A, Winans BN, Johnson MD, Walter KA. Vascular gene expression patterns are conserved in primary and metastatic brain tumors. J Neurooncol. 2010;99:13-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 76] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 43. | Briem E, Budkova Z, Sigurdardottir AK, Hilmarsdottir B, Kricker J, Timp W, Magnusson MK, Traustadottir GA, Gudjonsson T. MiR-203a is differentially expressed during branching morphogenesis and EMT in breast progenitor cells and is a repressor of peroxidasin. Mech Dev. 2019;155:34-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 44. | Zheng YZ, Liang L. High expression of PXDN is associated with poor prognosis and promotes proliferation, invasion as well as migration in ovarian cancer. Ann Diagn Pathol. 2018;34:161-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 45. | Dougan J, Hawsawi O, Burton LJ, Edwards G, Jones K, Zou J, Nagappan P, Wang G, Zhang Q, Danaher A, Bowen N, Hinton C, Odero-Marah VA. Proteomics-Metabolomics Combined Approach Identifies Peroxidasin as a Protector against Metabolic and Oxidative Stress in Prostate Cancer. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: Taiwan
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Batool SN, Jeong KY, Liu J, Tulassay Z S-Editor: Ma YJ L-Editor: A P-Editor: Ma YJ