Published online Sep 14, 2022. doi: 10.3748/wjg.v28.i34.4973
Peer-review started: August 2, 2021
First decision: October 2, 2021
Revised: October 29, 2021
Accepted: August 23, 2022
Article in press: August 23, 2022
Published online: September 14, 2022
Processing time: 401 Days and 6.4 Hours
Long noncoding RNA (lncRNA) ZNFX1-AS1 (ZFAS1) is a newly discovered lncRNA, but its diagnostic value in gastric cancer is unclear.
To investigate the potential role of ZFAS1 in gastric cancer and to evaluate the clinical significance of ZFAS1 as a biomarker for gastric cancer screening.
Quantitative real-time polymerase chain reaction (qRT-PCR) was used to screen for gastric cancer-associated lncRNAs in gastric cancer patients, gastric stromal tumor patients, gastritis or gastric ulcer patients, and healthy controls. Correlations between ZFAS1 expression and clinicopathological features were analyzed. The biological effects of ZFAS1 on the proliferation, migration, and invasion of gastric cancer cells were studied by MTT, colony formation, and transwell mi-gration assays. The potential mechanism of ZFAS1 was demonstrated using enzyme-linked immunosorbent assay and qRT-PCR. The relationship between ZFAS1 and tumorigenesis was demonstrated using in vivo tumor formation assays.
The plasma level of lncRNA ZFAS1 was significantly higher in preoperative patients with gastric cancer than in individuals in the other 4 groups. Increased expression of ZFAS1 was significantly associated with lymph node metastasis, advanced TNM stage, and poor prognosis. ZFAS1 regulated the proliferation, migration, and invasion of gastric cancer cells and regulated the growth of gastric cancer cells in vivo. LIN28 and CAPRIN1 were identified as key downstream mediators of ZFAS1 in gastric cancer cells.
LncRNA ZFAS1 promoted the invasion and proliferation of gastric cancer cells by modulating LIN28 and CAPRIN1 expression, suggesting that ZFAS1 can be used as a potential diagnostic and prognostic biomarker in gastric cancer.
Core Tip: Long noncoding RNA (lncRNA) ZNFX1-AS1 (ZFAS1) is a newly discovered lncRNA, but its diagnostic value in gastric cancer is unclear. This study aimed to investigate the potential role of ZFAS1 in gastric cancer and to evaluate the clinical significance of ZFAS1 as a biomarker for gastric cancer screening. LncRNA ZFAS1 promoted invasion and proliferation of gastric cancer cells by modulating LIN28 and CAPRIN1, suggesting that ZFAS1 can be used as a potential biomarker for the diagnosis and prognosis of gastric cancer.
- Citation: Zhuo ZL, Xian HP, Sun YJ, Long Y, Liu C, Liang B, Zhao XT. Long noncoding RNA ZNFX1-AS1 promotes the invasion and proliferation of gastric cancer cells by regulating LIN28 and CAPR1N1. World J Gastroenterol 2022; 28(34): 4973-4992
- URL: https://www.wjgnet.com/1007-9327/full/v28/i34/4973.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i34.4973
Gastric carcinoma is the 2nd primary cause of death due to carcinoma and the 6th most common cancer worldwide. Approximately 1000000 new cases of gastric cancer are diagnosed each year, and approximately 800000 patients die from the disease, accounting for approximately 8.2% of global cancer-related mortality[1]. The progression-free survival (PFS) rate and prognosis of gastric carcinoma highly depend on the TNM stage of the disease. The high mortality is associated with the faultiness of standard screening protocols and lack of overt early symptoms[1]. Therefore, early detection and optimal treatment are necessary to improve the prognosis of gastric carcinoma. However, current biologic markers, such as carbohydrate antigen 199 (CA-199) and carcinoembryonic antigen (CEA), have low sensitivity and specificity[2]. Therefore, there is an urgent need to find novel biomarkers for early diagnosis of gastric cancer.
Long noncoding RNAs (lncRNAs) are a class of non-protein-coding RNA molecules > 200 nucleotides in length[3]. Recently, some studies have reported that lncRNAs are involved in protein modification and gene expression and that their dysregulation leads to a variety of genetic diseases[4-6]. Published articles have also shown that lncRNAs exhibit significant regulatory effects on transcription patterns in malignant tumors, some of which involve tumor cell invasion and metastasis, with a poor prognosis[6,7]. For example, H19 and SUMO1 pseudogene 3 (SUMO1P3) are upregulated in gastric cancer, while gastric cancer-associated transcript 1 is downregulated[7-11]. Additionally, HOTAIR overexpression may be associated with tumor escape mechanisms[12]. These studies strongly suggest that lncRNAs underlie the molecular etiology of gastric carcinoma. It is worth noting that the detection of circulating lncRNAs provides a new gastric biomarker for gastric cancer that is expected to be useful for moni-toring and screening patients with gastric cancer[13].
Here, we selected 15 candidate cancer-associated lncRNAs. Among these lncRNAs, lncRNA ZNFX1-AS1 (ZFAS1) was found to be upregulated in the plasma of preoperative gastric cancer patients compared with healthy controls, and expression of ZFAS1 was significantly associated with lymphatic invasion, advanced TNM stage, and poor prognosis. We then investigated the biological effects of ZFAS1 on the survival, proliferation, and migration of gastric cancer cells. The underlying mechanism of ZFAS1 and the relationship between ZFAS1 and tumorigenesis were identified. These results showed that lncRNA ZFAS1 is a potential biomarker for gastric cancer.
All samples were obtained from Peking University People’s Hospital between July 2015 and June 2016. Seventy-five matched (pre- and postoperative) whole blood and serum samples were collected from patients who underwent gastrointestinal surgery for gastric resection. In addition, intraperitoneal free cancer cell (IFCC) samples were collected from 60 of these patients. Other whole blood and serum specimens were collected from 60 gastric stromal tumor patients (before surgery), 60 gastritis or ulcer patients (before treatment), and 75 healthy controls. All specimens were collected with the corresponding patient information, such as age, sex, and collection time. In addition, the tumor size, tumor location (evaluated according to the National Comprehensive Cancer Network Clinical Practice Guidelines in Oncology, Version II[14]), pathological type, degree of differentiation (based on the World Health Organization Classification of Tumors of the Digestive System, 4th edition[15]), TNM stage and lymphatic invasion status (based on the American Joint Committee on Cancer Staging Manual, 7th edition[16]) of the corresponding patients were collected.
Plasma and serum samples were collected in BD vacutainer ethylenediamine tetraacetic acid tubes and BD vacutainer somatostatin tubes (BD Biosciences, NJ, United States), respectively. Preoperative samples of patients with gastric cancer were obtained at least 2 mo before surgery or after radiotherapy or chemotherapy. Postoperative samples from these patients were collected 7-10 d after surgery. Samples from patients with gastric stromal tumor or gastritis/peptic ulcer were collected before any treatment was administered. The healthy control samples were randomly collected from normal subjects. For plasma samples, a special centrifugation protocol (2348 × g for 30 min at 4 °C; 4696 × g for 5 min at 4 °C; 10733 × g for 5 min at 4 °C) was performed to prevent contamination with cellular nucleic acids. Plasma and IFCC samples were stored at -80 °C in 3 volumes of TRIzol® reagent (Qiagen, CA, United States) for further analysis. Serum samples were analyzed directly[17]. This study was approved by the Research Ethics Committee of Peking University People’s Hospital. Patient data and samples were treated according to the ethical and legal criteria adopted in the 2013 Declaration of Helsinki. Written informed consent for ethical approval and patient consent was obtained from all participants.
This method is the same as that described in our previously published article[17]. LncRNA was extracted from plasma using a Direct-zol™ RNA MiniPrep R2050i Kit (Zymo Research, CA, United States) according to the manufacturer’s protocol. The concentration and purity of total RNA were measured using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, MA, United States). Total RNA was then solubilized using RNase-free water, and reverse transcription was immediately performed using PrimeScript RT Master Mix (Takara Bio, Kyushu, Japan) according to the manu-facturer’s instructions. After reverse transcription, quantitative polymerase chain reaction (qPCR) was performed in a LightCycler 480 Instrument II (Roche Diagnostics, Basel, Switzerland) using TransStart Green qPCR SuperMix (TransGen, Beijing, China) according to the supplier’s instructions. The primers used for qPCR were designed with Primer Premier V 5.0 (Premier Biosoft International, CA, United States) and were synthesized by Beijing Sunbiotech Co., Ltd. (Beijing, China). The sequences of all primers, including those specific for GAPDH, are listed in Table 1. PCR was carried out with initial denaturation at 95 °C for 3 min followed by 40 cycles of 1 min at 95 °C and 1 min at 60 °C with subsequent detection. Due to the stability of GAPDH in plasma, it was chosen as the internal control for data standardization. For the calculations, the equation ΔCq = Cq selected lncRNA - Cq GAPDH lncRNA was used, where ΔCq was defined as the difference in the quantification cycle (Cq) values. Every specimen was analyzed in triplicate, and the experiment was repeated 3 times.
| Name | Forward-primer | Reverse-primer |
| GAPDH | ACCCACTCCTCCACCTTTGAC | TGTTGCTGTAGCCAAATTCGTT |
| H19 | TACAACCACTGCACTACCTG | TGGAATGCTTGAAGGCTGCT |
| CCAT1 | CATTGGGAAAGGTGCCGAGA | ACGCTTAGCCATACAGAGCC |
| HOTAIR | GGTAGAAAAAGCAACCACGAAGC | ACATAAACCTCTGTCTGTGAGTGCC |
| LINC00152 | CTCCAGCACCTCTACCTGTTG | GGACAAGGGATTAAGACACACA |
| ZNFX1-AS1 | CCAGTTCCACAAGGTTAC | GCAGGTAGGCAGTTAGAA |
| PVT1 | CTTGAGAACTGTCCTTACG | CAGATGAACCAGGTGAAC |
| GAS5 | CACAGGCATTAGACAGAA | AGGAGCAGAACCATTAAG |
| SNHG12 | GACTTCCGGGGTAATGACAG | GCCTTCTGCTTCCCATAGAG |
| TUG1 | TAGCAGTTCCCCAATCCTTG | CACAAATTCCCATCATTCCC |
| CHET1 | CCCCACAAATGAAGACACT | TTCCCAACACCCTATAAGAT |
| SUMP1P3 | ACTGGGAATGGAGGAAGA | TGAGAAAGGATTGAGGGAAAAG |
| GACAT3 | GGGGGCTTGTTTCTTTGTGTAG | CATTCGGCTCTGACCTCTCAC |
| ABHD11-AS1 | GAACGGGATGAAGCCATTG | GCTGATTCTGGACCTGCTG |
| GACAT2 | TGGATGCTTACAAAGGACTGG | CTGCAATTACGGAAAGAGCTG |
| uc001lsz | GACGGCACCTACTACACCTT | GCTGACCACCTTGTTGTTGAA |
The human gastric cancer cell lines BGC-823 and SGC-7901 were a gift from Peking University People’s Hospital Gastrointestinal Surgery Laboratory and were maintained in RPMI 1640 medium (Invitrogen, CA, United States) supplemented with 10% fetal bovine serum (FBS) (Gibco, CA, United States). The flask was incubated in a humidified incubator at 37 °C under 5% CO2 in air.
ZFAS1-siRNA against ZFAS1 was synthesized by GenePharma (Shanghai, China). BGC-823 and SGC7901 cells were grown in 6-well plates (2 × 105 cells/well) and transfected for 36 h using Lipo-fectamine RNAiMAX Transfection Reagent (Invitrogen, CA, United States). The ZFAS1 siRNA sequences were as follows: ZFAS1 siRNA1, 5’-AGACGCGAAAGAACGAAUGTT-3’; ZFAS1 siRNA2, 5’-UUACAAGGCAGACUGAAUCTT-3’; and ZFAS1 siRNA3, 5’-UAUGCAGGUAGGCAGUUAGTT-3’. The GAPDH siRNA and negative control siRNA sequences were as follows: GAPDH siRNA, 5'-ACGUGACACGUUCGGAGAATT-3' and negative control siRNA, 5'-ACGUGCAC AGUACU
The ZFAS1 overexpression short hairpin RNA (ZFAS1-shRNA1), sh-ZFAS1 directed against ZFAS1 short hairpin RNA (ZFAS1-shRNA2), and NC-shRNA short hairpin RNA constructs were synthesized by GenePharma (Shanghai, China). The sequences of ZFAS1-shRNA1, ZFAS1-shRNA2, and NC-shRNA are shown in Table 2. BGC-823 and SGC7901 cells were grown in 6-well plates (2 × 105 cells/well) and transduced for 36 h using Polybrene (GenePharma, REVG0001) and Enhanced Infection Solution (GenePharma, REVG0002).
| Sequence | |
| ZFAS1-shRNA1 | ACCGGTCGGGGGCCCAGGGTGGAGAGCACGAGGGCCTGGCCCAGGCACGGCCGGCGCCTCCGCCCTCGA |
| ZFAS1-shRNA2 | TTACAAGGCAGACTGAATCTT |
| NC-shRNA | TTCTCCGAACGTGTCACGT |
After transduction with NC-shRNA, ZFAS1-shRNA1, and ZFAS1-shRNA2, BGC823 and SGC7901 cells were trypsinized, and six replicates of 12 × 103 cells per well were seeded in 96-well plates, with wells without cells used as blank wells. Proliferation was measured by an MTT (Sigma Aldrich, St. Louis, MO, United States) transformation assay. Briefly, 20 mL of MTT solution (5 mg/mL) was added to each well, and cells were grown for 4 h at 37 °C. After adding 100 mL of the dissolved solution (10% SDS in 0.01 M HCl solution), cells were further cultured at 37 °C for 3 h. The specific optical density of all wells was then measured at 490 nm.
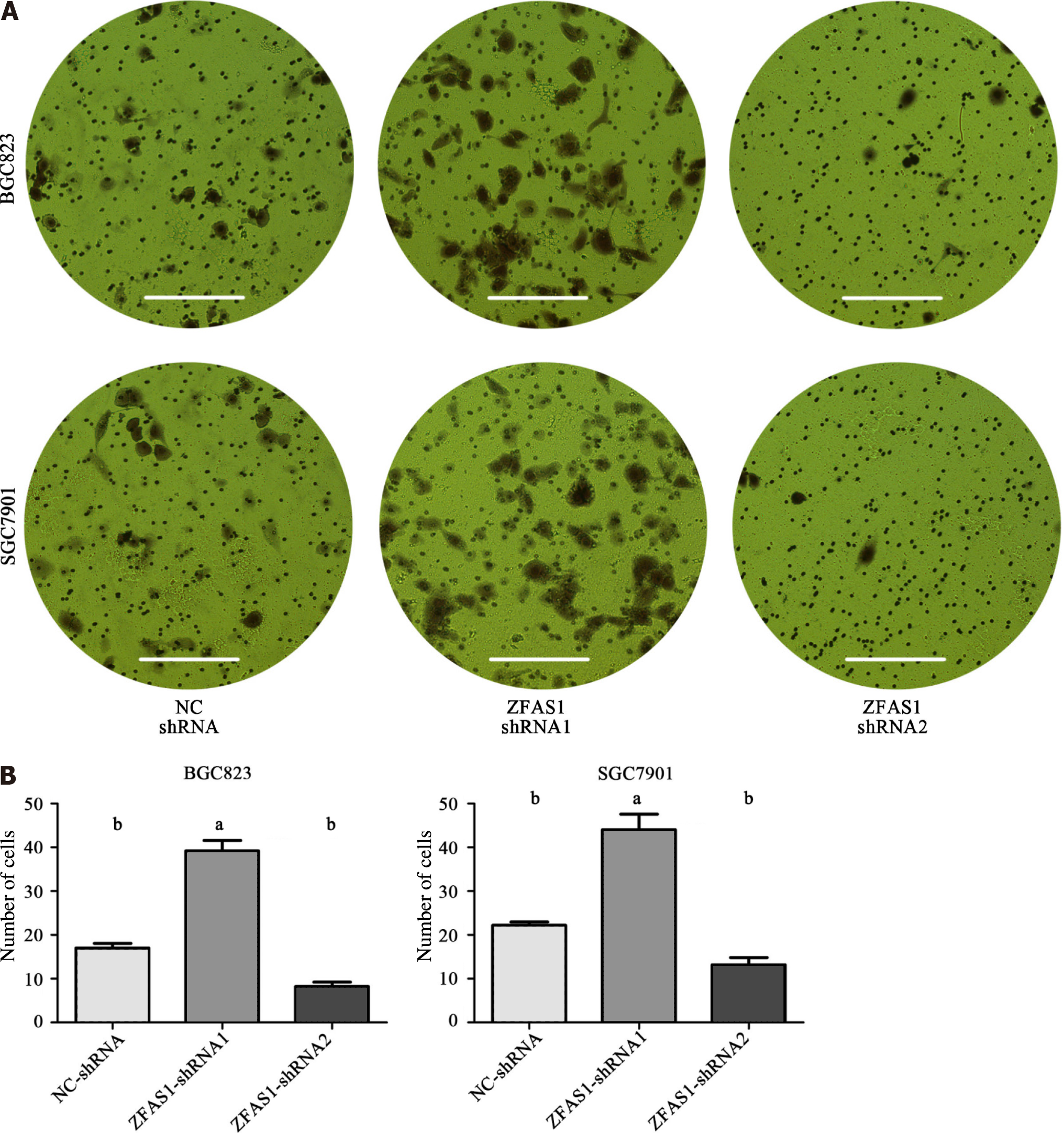
BGC823 and SGC7901 cells were suspended in serum-free medium after transduction with NC-shRNA, ZFAS1-shRNA1, and ZFAS1-shRNA2 and inoculated into transwell chambers with inserts of 8 μm pore size. Medium containing 10% FBS was placed in the bottom chamber. After 36 h of incubation, cells that migrated through the membrane to the lower surface were fixed with paraformaldehyde, stained with crystal violet, counted, and photographed. The results are shown as the average of three independent experiments.
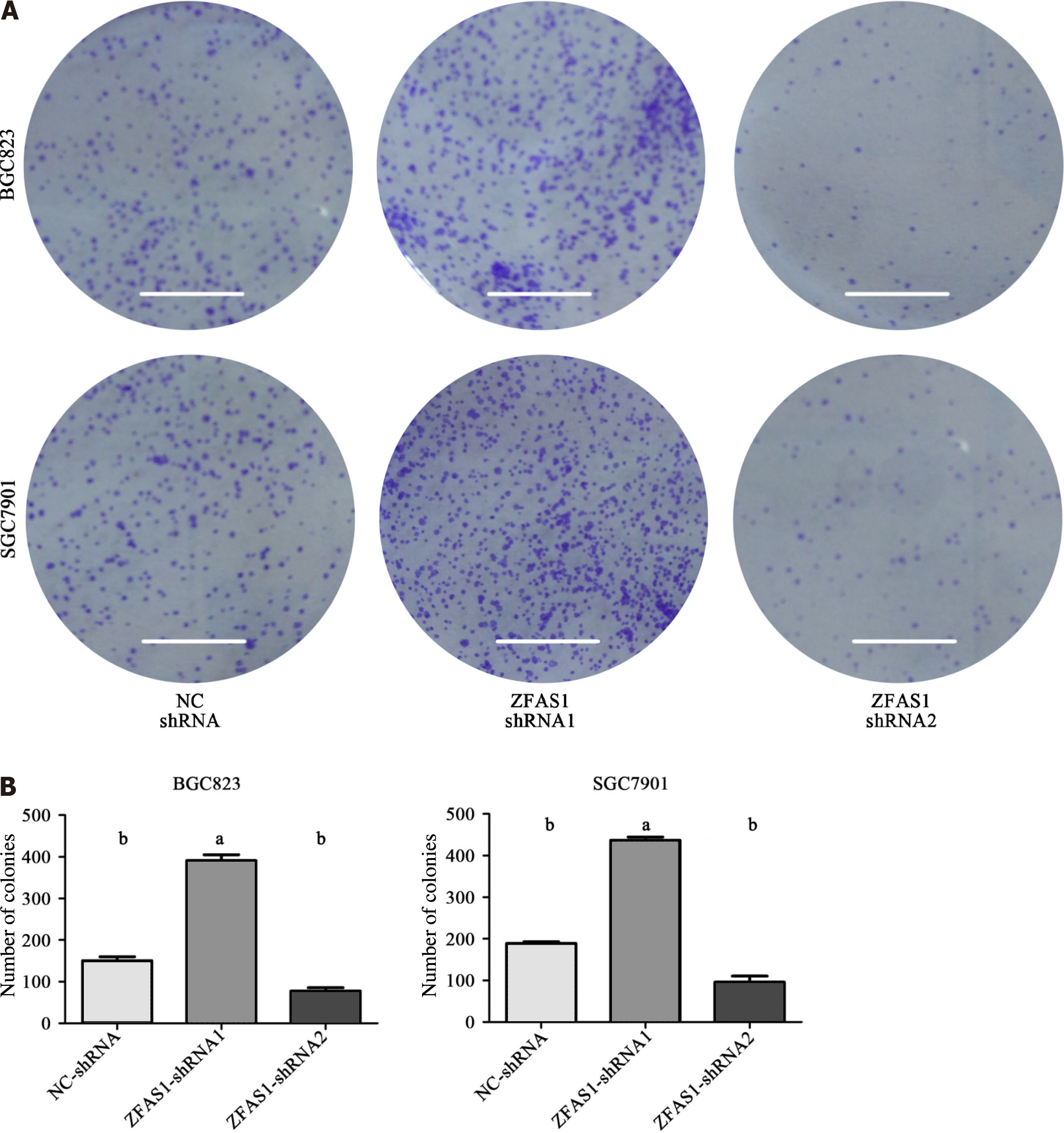
BGC823 and SGC7901 cells transduced with NC-shRNA, ZFAS1-shRNA1, and ZFAS1-shRNA2 were seeded into 6-well plates (1000 cells per well) and cultured in a humidified incubator at 37 °C in 5% CO2 for 10 d. The medium was changed every 3 d. At the end of the incubation period, the cultured cells were fixed with 4% paraformaldehyde and stained with crystal violet to assess the number of colonies. The results are shown as the average of three independent experiments.
Binding proteins of lncRNA ZFAS1 were predicted based on the starBase V2.0 database (http://starbase.sysu.edu.cn). The primers specific for the mRNA sequences of the genes encoding the binding proteins were designed according to the nucleic acid sequence information for the corresponding proteins in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) with a primer design tool (http://www.ncbi.nlm.nih.gov/tools/primer-Blast). The sequences of all primers, including those specific for GAPDH, are listed in Table 3.
| Name | Forward-primer (5’ to 3’) | Reverse-primer (5’to 3’) |
| GAPDH | ACCCACTCCTCCACCTTTGAC | TGTTGCTGTAGCCAAATTCGTT |
| UPF1 | GGTCCCTGATAATTATGGCGATG | ACGGCATAAACCTGGGAGTG |
| eIF4AIII | GGCATCTACGCTTACGGTTT | CAGCCAACTCTCTTGTGGGA |
| IGF2BP1 | AGCTCCTTTATGCAGGCTCC | GAGCCTTGAATTGGGCCTCT |
| FMRP | CTCAAGGCTTGGCAGGGTATG | CCGTGCCCCCTATTTCTGTA |
| LIN28 | AGATCAAAAGGAGACAGGTGCT | AATAGCCCCCACCCATTGTG |
| IGF2BP2 | ACCCTCTCGGGTAAAGTGGA | GTTGACAACGGCGGTTTCTG |
| FUS | GCAAGATGGATTCCAGGGGTG | TCCAGGAAAGTGAAAAGGGGG |
| ZC3H7B | TGTGCAAAGGAGGAGATCGAC | ACAGACGGAGAGTCCTTGGT |
| IGF2BP3 | CCTGGTGAAGACTGGCTACG | CCAGCACCTCCCACTGTAAAT |
| CAPRIN1 | CTGCTGGCTGGCTAAGTCC | GGCCGAGGGCATCGTG |
NC-shRNA, ZFAS1-shRNA1, and ZFAS1-shRNA2 were transduced into BGC823 and SGC7901 cells, and mRNA was then reverse transcribed. The mRNA expression levels of the genes encoding the candidate lncRNA ZFAS 1 binding proteins were determined by quantitative real-time-PCR (qRT-PCR) using the primers listed in Table 3. The experimental method was the same as that described previously.
Stably transfected cells, as well as tumor/lymph node tissue from BALB/c nude mice, were lysed with NP40 cell lysis buffer (Invitrogen, CA, United States) supplemented with SigmaFASTTM protease inhibitor tablets (Sigma Aldrich, St. Louis, MO, United States). LIN28, CAPRIN1, CEA, and CK20 were detected by enzyme-linked immunosorbent assay (ELISA) using a Mouse Protein lin-28 homolog A (Lin28a) ELISA Kit (ELISAGenie, Dublin, Ireland), a Human Caprin 1 (CAPRIN1) ELISA Kit (Abbexa, Cambridge, United Kingdom), a CEA ELISA Kit (Bioss, United Kingdom) and CK20 ELISA kits (FKBIO, Chengdu, China), respectively.
Four-week-old female athymic BALB/c nude mice were maintained under specific pathogen-free conditions and operated on according to the protocol approved by the Beijing Medical Laboratory Animal Management Committee. NC-shRNA-, ZFAS1-shRNA1-, and ZFAS1-shRNA2-transduced BGC823 cells were harvested. For tumor formation assays, 107 cells were injected subcutaneously into one side of each mouse. Tumor growth was examined every week, and the tumor volume was calculated using the equation V = 0.5 × D × d2 (V, volume; D, longitudinal diameter; d, latitudinal diameter). The expression of lncRNA ZFAS1 in BALB/c mouse blood was detected by qRT-PCR every week, and the primer sequences are listed in Table 1. The expression of LIN28 and CAPRIN1 proteins in BALB/c nude mouse tumors and the expression of CEA and CK20 in BALB/c nude mouse lymph nodes were detected by ELISA after four weeks. This study was conducted in strict accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals. The program was approved by the Animal Experiment Ethics Committee of Peking University People’s Hospital.
The formula 2-ΔΔCt were used to calculate the levels of relative lncRNA expression in plasma[18]. Based on this formula, all relative lncRNA expression levels in plasma were evaluated. Continuous variables were described by the mean ± SD (normal distribution), or the median and interquartile range M (P25, P75) (non-normal distribution). The t-test or Mann-Whitney U test was used according to the data distribution to identify statistically significant differences. For categorical variables, the percentages of patients in each category were calculated using the χ2 test or Fisher’s exact test. In addition, receiver operating characteristic (ROC) curve analysis was performed to assess the diagnostic value of circulating lncRNA and traditional tumor markers in distinguishing between gastric carcinoma patients and healthy subjects. Statistical analysis was performed using SPSS software version 19.0 (IBM Corp., Armonk, NY, United States). P values less than 0.05 were considered significant.
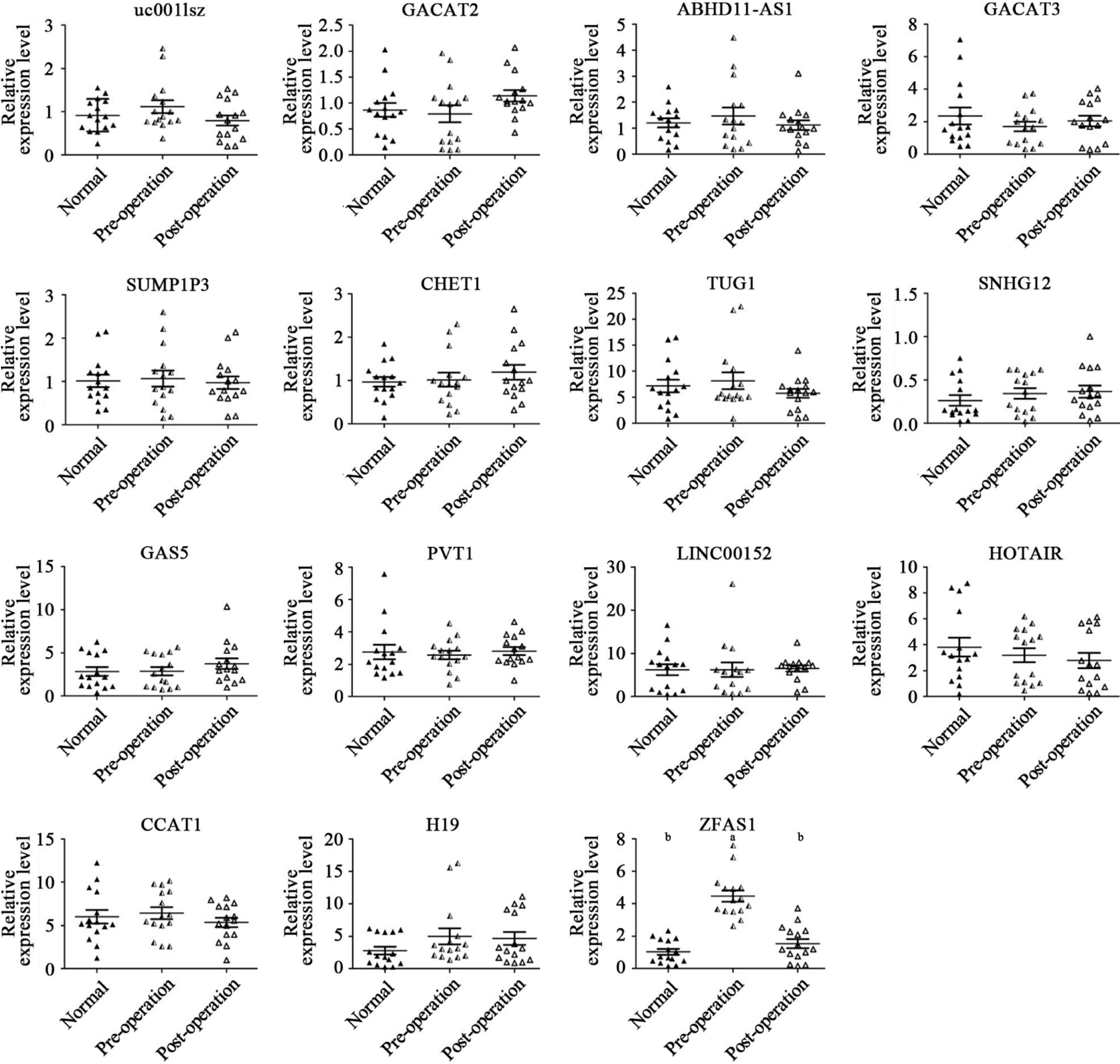
Fifteen matched samples from preoperative and postoperative gastric cancer patients and samples from 15 healthy control subjects were used as the sample cohort. The general characteristics and clinicopathological features of all subjects are provided in Table 4. The relative expression of the 15 lncRNAs, namely, uc001 Lsz/HOTAIR/CCAT1/H19/GACAT2/ABHD11-AS1/GACAT3 /SUMO1P3/ CHET1/TUG1/SNHG12/GAS5/PVT1/LINC00152, and ZFAS1, was evaluated in the plasma of all subjects. As mentioned in Figure 1, the levels of lncRNA ZFAS1 in preoperative patient plasma were significantly higher than those in postoperative patient and healthy control subject plasma (P < 0.01).
| Variables | Gastric cancer (n = 15) | Healthy control (n = 15) |
| Gender | ||
| Male | 12 | 12 |
| Female | 3 | 3 |
| Age (yr) | ||
| Mean | 61 | 62 |
| Range | 48-70 | 47-80 |
| Tumor site1 | ||
| Upper third | 3 | - |
| Middle third | 4 | - |
| Lower third | 8 | - |
| Tumor size | ||
| ≥ 5 cm | 6 | - |
| < 5 cm | 9 | - |
| Pathological differentiation2 | ||
| Undifferentiated or poorly differentiated | 12 | - |
| Others | 3 | - |
| TNM classification3 | ||
| I + II | 4 | - |
| III + IV | 11 | - |
| Lymphatic invasion3 | ||
| Positive | 11 | - |
| Negative | 4 | - |
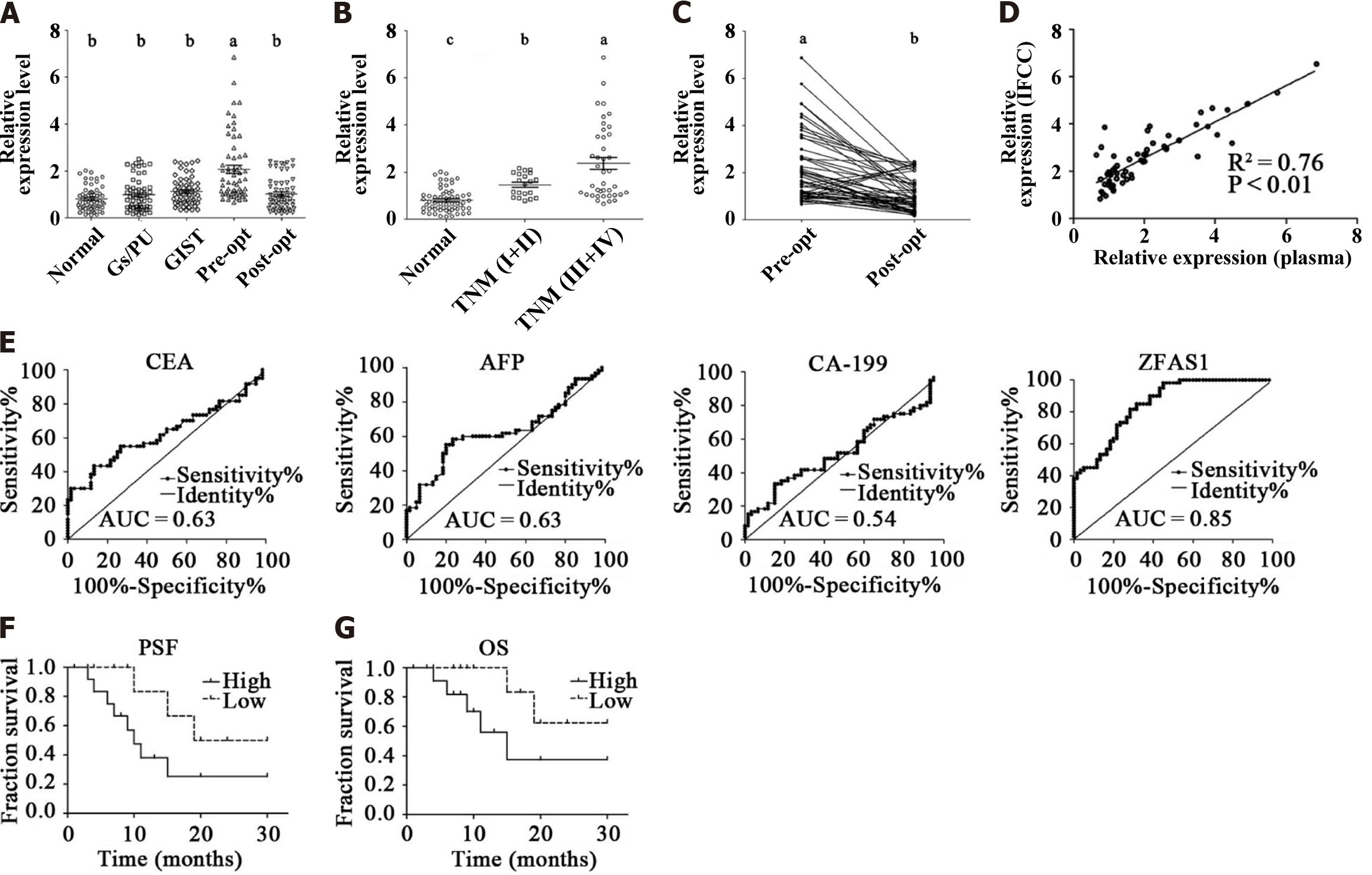
The general characteristics and clinicopathological features of 60 healthy control subjects, 60 gastritis/peptic ulcer patients, 60 GIST patients, and 60 patients with matched preoperative and postoperative data are summarized in Table 5. The relative levels of ZFAS1 in the plasma of all subjects were assessed by qRT-PCR. As shown in Figure 2A, the levels of lncRNA ZFAS1 in preoperative patient plasma were significantly higher than those in plasma from the individuals in the other four groups (P < 0.01).
| Variables | Healthy control (n = 60) | Gastritis/peptic ulcer (n = 60) | GIST (n = 60) | Gastric cancer (n = 60) |
| Gender | ||||
| Male | 46 | 37 | 35 | 46 |
| Female | 14 | 23 | 25 | 14 |
| Age (yr) | ||||
| Mean | 61 | 60 | 60 | 61 |
| Range | 37-91 | 37-89 | 30-85 | 37-91 |
| Tumor site1 | ||||
| Upper third | - | - | - | 13 |
| Middle third | - | - | - | 14 |
| Lower third | - | - | - | 33 |
| Tumor size | ||||
| ≥ 5 cm | - | - | - | 29 |
| < 5 cm | - | - | - | 31 |
| Pathological differentiation2 | ||||
| Undifferentiated or poorly differentiated | - | - | - | 45 |
| Others | - | - | - | 15 |
| TNM classification3 | ||||
| I + II | - | - | - | 20 |
| III + IV | - | - | - | 40 |
| Lymphatic invasion3 | ||||
| Positive | - | - | - | 43 |
| Negative | - | - | - | 17 |
We used the ∆∆Ct formula to calculate the relative levels of lncRNA ZFAS1 in the samples (plasma) of patients with different TNM stages, including 20 patients with stage I and II disease and 40 patients with stage III and IV disease. As mentioned in Figure 2B, the median relative levels of lncRNA ZFAS1 in healthy controls, patients with stage I and II disease, and patients with stage III and IV disease were 0.81, 1.61, and 2.52, respectively. The relative levels in early- and advanced-stage patients were significantly higher than that in healthy controls (P < 0.01).
Sixty matched pre-operative and post-operative samples (plasma) were included in our research. We found that the median relative levels of ZFAS1 before and after surgery were 2.22 and 1.01, respectively. The level decreased in 55 of the 60 gastric cancer patients (92%) approximately 10 d after surgery (P < 0.01; Figure 2C).
Sixty matched preoperative patient plasma and IFCC samples were collected. We used the ∆∆Ct method to evaluate the relative levels of ZFAS1 in plasma and IFCC. As shown in Figure 2D, the relative levels of ZFAS1 in plasma and IFCC were strongly positively correlated (R2= 0.76, P < 0.01).
ROC analysis revealed that the area under the curve (AUC) value of plasma ZFAS1 for discriminating gastric cancer patients from healthy control subjects was 0.85 (Figure 2E). The highest AUC value of any traditional serum biomarker was 0.63, while the AUC value of the other traditional biomarkers was lower than that of CEA. The highest accuracy of plasma ZFAS1 was obtained at a cutoff level of 1.00, where its sensitivity and specificity for identifying gastric carcinoma patients were 0.82 and 0.72, respectively.
To explore the correlations between the relative ZFAS1 level and gastric cancer clinicopathological characteristics, we divided the patients into groups by sex, age, tumor site/size/lymphatic invasion/pathological differentiation status and TNM classification (Table 6). Data analysis indicated that the relative ZFAS1 expression level was strongly associated with tumor size (P = 0.01), TNM classification (P = 0.02), and lymphatic invasion (P = 0.03). We then divided patients into two groups by the median relative ZFAS1 level. Kaplan-Meier survival analysis was used to assess the potential correlation between the relative ZFAS1 expression level and patient prognosis. As shown in Figures 2F and G, patients with a high ZFAS1 level had shorter overall survival (OS) and PFS times than those with a low ZFAS1 level. This result suggests that ZFAS1 is a potential prognostic biomarker in gastric cancer patients.
| Variables | Number of cases | P value |
| Gender | ||
| Male | 46 | 0.99 |
| Female | 14 | |
| Age (yr) | ||
| ≤ 61 | 28 | 0.26 |
| > 61 | 32 | |
| Tumor site1 | ||
| Upper third | 13 | 0.12 |
| Middle third | 14 | |
| Lower third | 33 | |
| Tumor size | ||
| ≥ 5 cm | 29 | 0.01 |
| < 5 cm | 31 | |
| Pathological differentiation2 | ||
| Undifferentiated or poorly differentiated | 45 | 0.55 |
| Others | 15 | |
| TNM classification3 | ||
| I + II | 20 | 0.02 |
| III + IV | 40 | |
| Distant metastasis3 | ||
| Positive | 5 | 0.97 |
| Negative | 55 | |
| Lymphatic invasion3 | ||
| Positive | 43 | 0.03 |
| Negative | 17 |
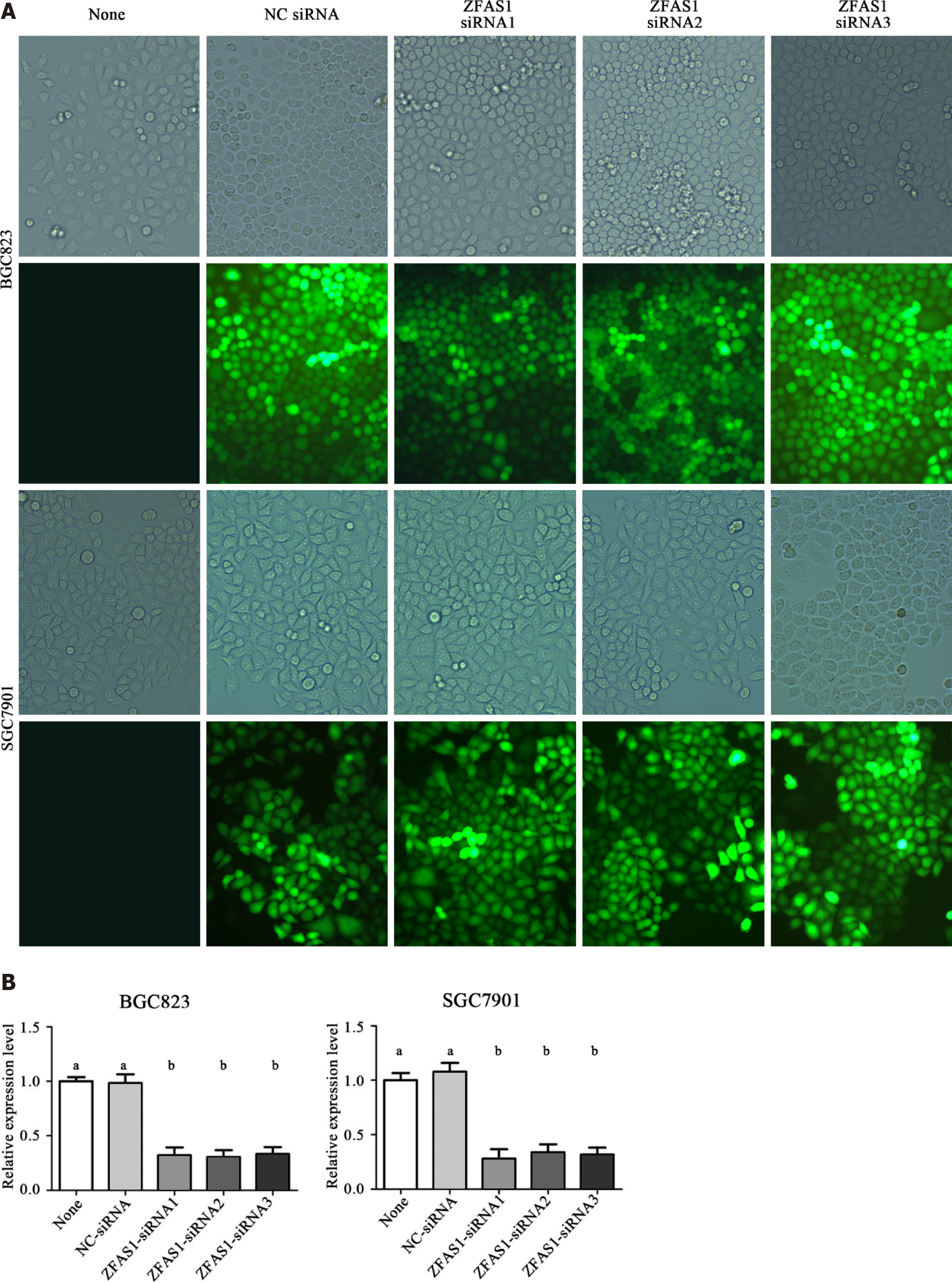
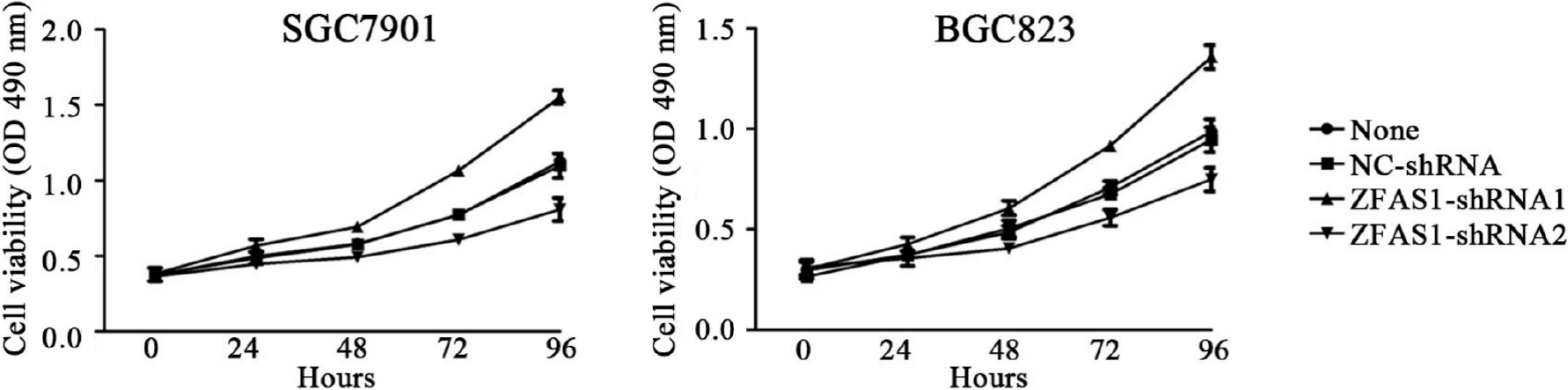
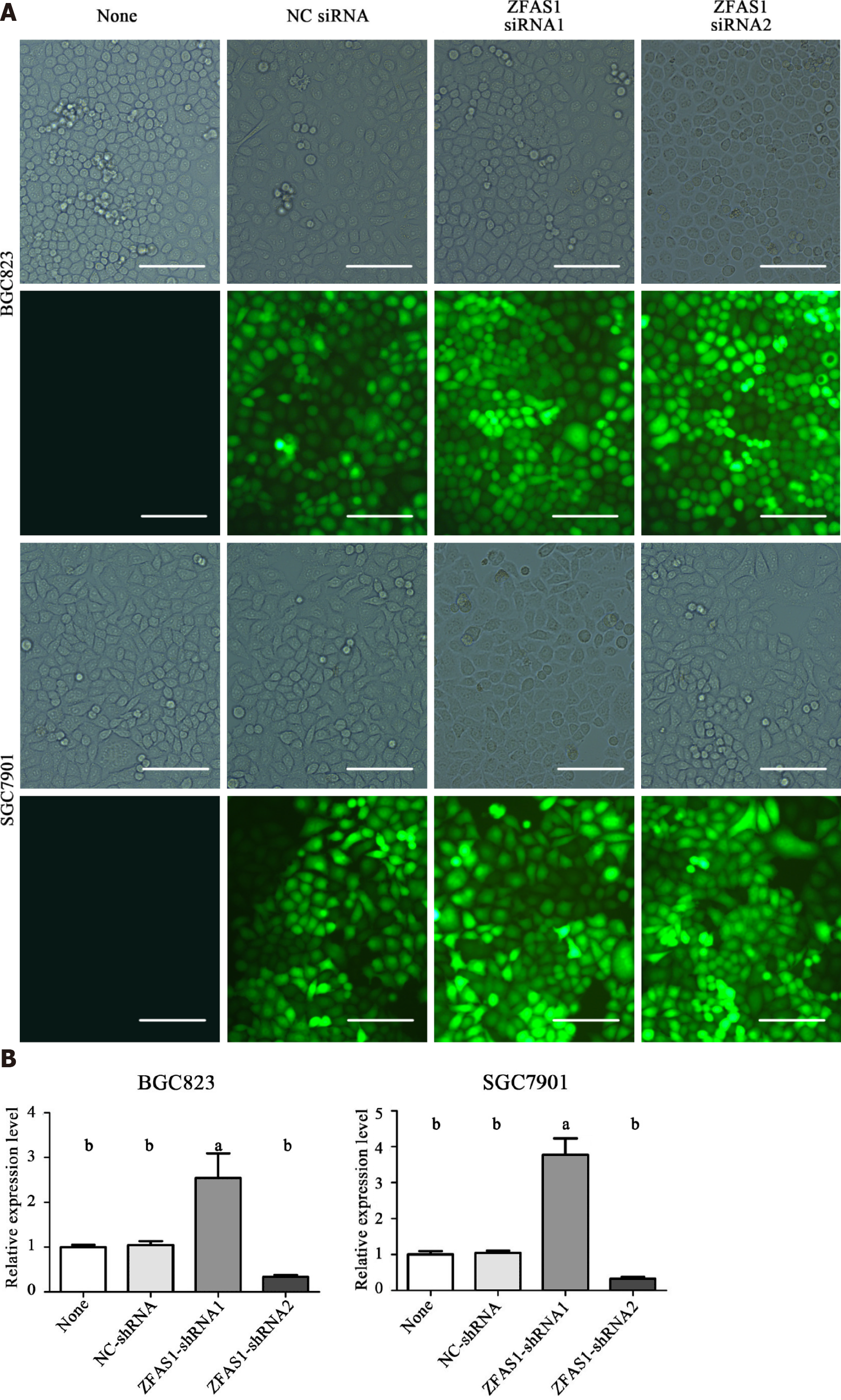
The expression level of ZFAS1 was positively correlated with lymph node metastasis, suggesting that ZFAS1 may be involved in tumor metastasis. Therefore, we studied the effect of ZFAS1 knockdown on gastric cancer cells. To verify the efficiency of ZFAS1 knockdown and prevent off-target effects, we first transfected three targeted siRNAs, ZFAS1-siRNA1, ZFAS1-siRNA2, and ZFAS1-siRNA3, into BGC823 and SGC7901 cells. Fluorescence microscopy was used to test the transfection efficiency. As shown in Figure 3, all three siRNAs significantly reduced the expression level of ZFAS1 in transfected BGC823 and SGC7901 cells. Next, we constructed ZFAS1-shRNA2 (with a sequence based on that of ZFAS1-siRNA2) and transduced it into BGC823 and SGC7901 cells. The MTT and colony formation assay results showed that compared to control cells transduced with NC-shRNA, BGC823 and SGC7901 cells with ZFAS1 knockdown showed significant decreases in viability and proliferation (Figures 4 and 5). The transwell migration assay showed that ZFAS1 knockdown in BGC823 and SGC7901 cells significantly inhibited cell migration (Figure 6). In summary, our data suggest that knockdown of ZFAS1 reduces the viability, proliferation, and migration of gastric cancer cells.
To further investigate the function of ZFAS1 in gastric cancer, we transduced ZFAS1-shRNA1 into BGC823 and SGC7901 cells to overexpress ZFAS1 and evaluated the transduction efficiency by fluorescence microscopy. As shown in Figure 7, the expression level of ZFAS1 was significantly increased in transduced BGC823 and SGC7901 cells. The MTT and colony formation assay results showed that compared to control cells transfected with NC-shRNA, BGC823 and SGC7901 cells overexpressing ZFAS1 showed significant increases in viability and proliferation (Figures 4 and 5). The transwell migration assay showed that ZFAS1 overexpression in BGC823 and SGC7901 cells significantly promoted cell migration (Figure 6). In summary, our data indicate that overexpression of ZFAS1 enhances the viability, proliferation, and migration of gastric cancer cells.
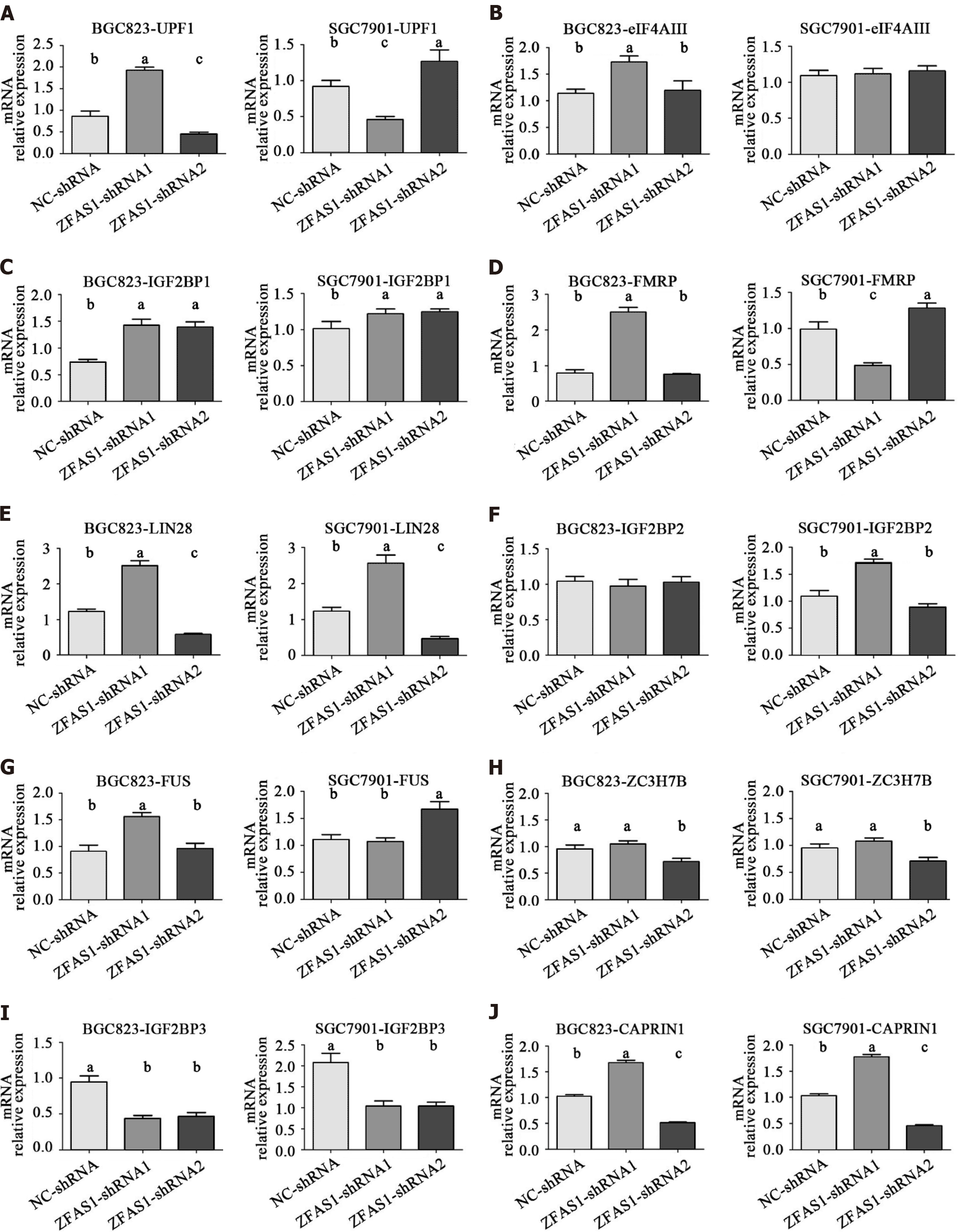
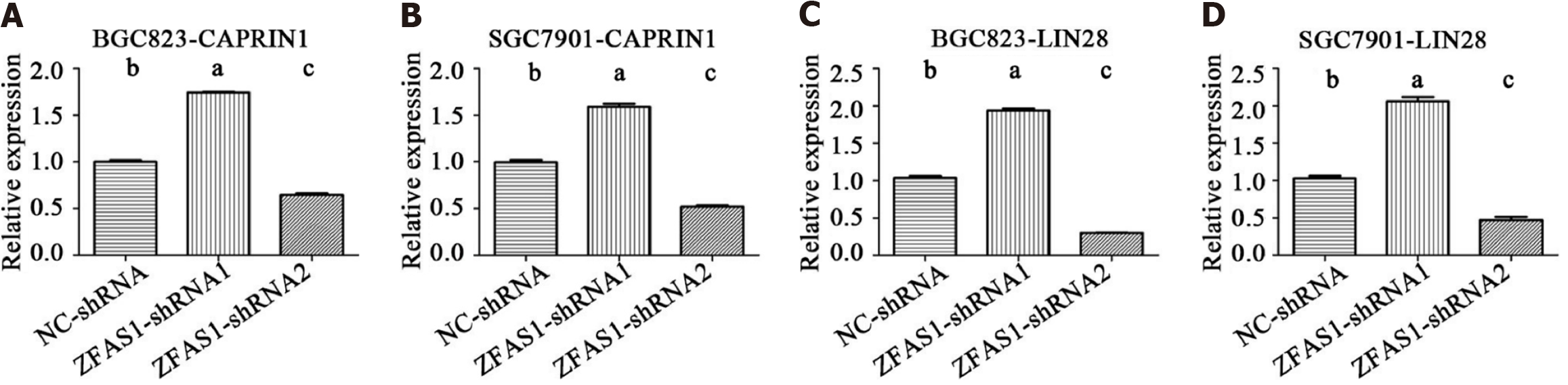
To further investigate the mechanism by which lncRNA ZFAS1 affects the aggressiveness of gastric cancer, we used the starBase V2.0 database to analyze binding proteins of lncRNA ZFAS1[19]. The results showed that a total of 10 proteins had more than three binding sites for lncRNA ZFAS1. These proteins were UPF1, eIF4AIII, IGF2BP1, FMRP, LIN28, IGF2BP2, FUS, ZC3H7B, IGF2BP3, and CAPRIN1. We then analyzed the mRNA expression levels of these proteins in BGC823 and SGC7901 cells transduced with ZFAS1-shRNA1 and ZFAS1-shRNA2. Our goal was to determine which proteins exhibited changes in expression consistent with the change in ZFAS1 expression. The qRT-PCR results showed that ZFAS1 overexpression significantly increased the expression of LIN28 and CAPRIN1 (P < 0.05) and that ZFAS1 knockdown significantly decreased the expression of LIN28 and CAPRIN1 (P < 0.05) (Figure 8). ELISA showed the same results (Figure 9).
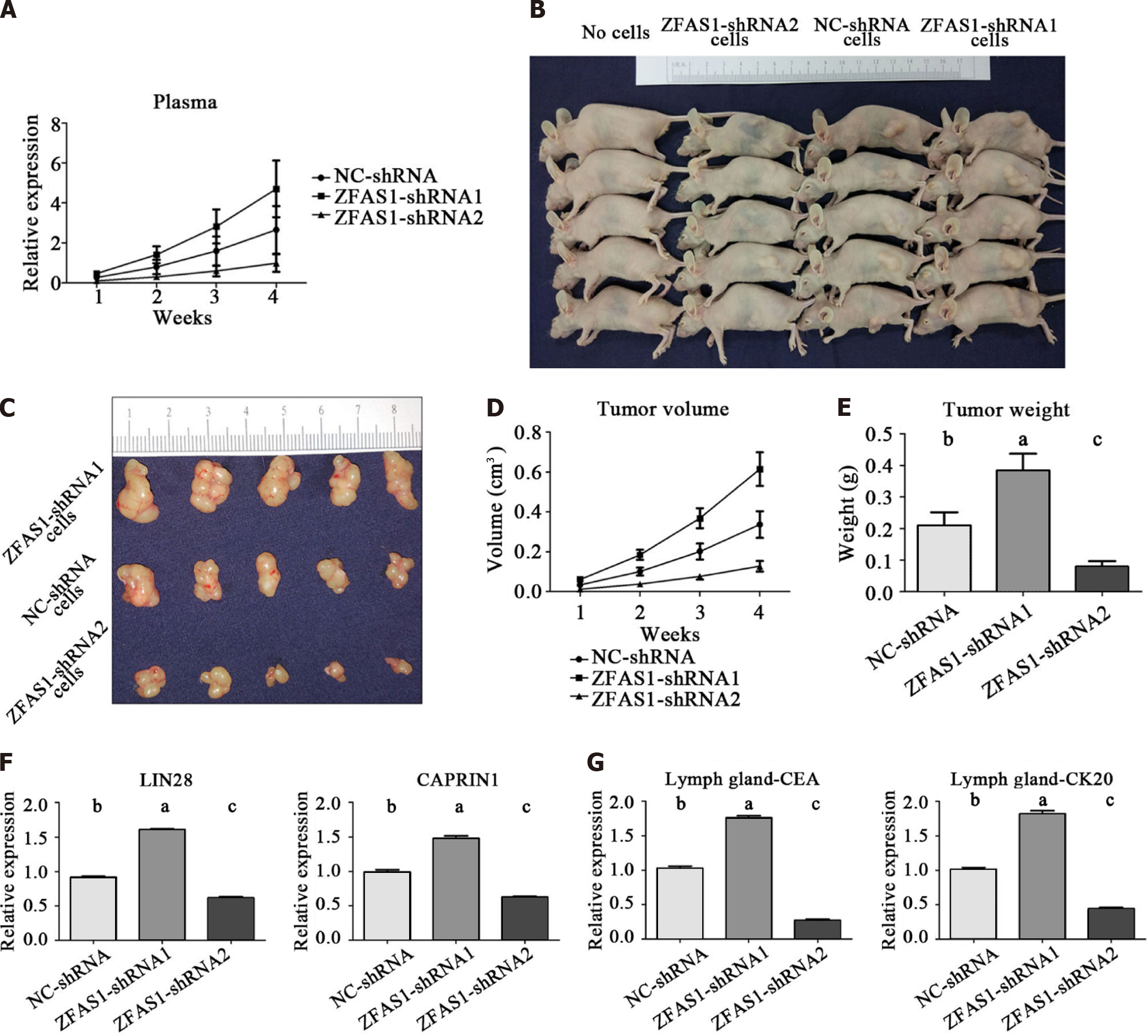
To further investigate whether ZFAS1 overexpression or knockdown affects tumor growth in vivo, BGC823 cells stably transduced with NC-shRNA, ZFAS1-shRNA1 and ZFAS1-shRNA2 were inoculated into 4-wk female athymic BALB/c nude mice. The expression of lncRNA ZFAS1 in nude mice was detected weekly by qRT-PCR. The results showed that the expression of lncRNA ZFAS1 in the three groups of nude mice increased over time. The expression level in the ZFAS1-shRNA1 group was higher than that in the control group, and the expression level in the ZFAS1-shRNA2 group was lower than that in the control group (Figure 10A). Images of tumors from nude mice are shown in Figures 10B and C. Tumor size and weight were measured weekly; compared with tumors in the control group, tumors in the ZFAS1-shRNA1 group were significantly larger and heavier, and tumors in the ZFAS1-shRNA2 group were significantly smaller and lighter (Figures 10D and E). Next, in tumor tissue, ELISA confirmed that the expression of LIN28 and CAPRIN1 was significantly increased in tumors in the ZFAS1-shRNA1 group compared with those in the control group (P < 0.05), while the expression of LIN28 and CAPRIN1 was significantly decreased in tumors in the ZFAS1-shRNA2 group (P < 0.05) (Figure 10F). In addition, ELISA showed significant increases in CEA and CK20 expression in lymph nodes in the ZFAS1-shRNA1 group compared with the control group (P < 0.05) and significant decreases in CEA and CK20 expression in lymph nodes in the ZFAS1-shRNA2 group (P < 0.05) (Figure 10G), confirming the relationship between lncRNA ZFAS1 expression and lymph node metastasis in gastric cancer cells. In conclusion, our results indicate that ZFAS1 overexpression promotes the growth of gastric cancer cells in vivo and that ZFAS1 knockdown inhibits the growth of gastric cancer cells in vivo.
Gastric cancer is a common malignancy that is particularly difficult to diagnose early[20]. However, traditional biomarkers have low specificity and low sensitivity. As lncRNA research has intensified, many lncRNAs have been determined to be involved in the development, progression, and prognosis of gastric cancer[21], but their diagnostic value as gastric cancer markers needs further study. By reviewing the literature, we selected 15 candidate lncRNAs that have high expression in gastric tumor tissues. Next, the relative levels of these 15 candidate lncRNAs were evaluated in the plasma of 15 normal subjects and 15 matched samples from preoperative and postoperative patients with gastric cancer. The results showed that the plasma level of only ZFAS1 was significantly higher in preoperative gastric cancer patients than in normal subjects and postoperative gastric cancer patients (Figure 1). Therefore, we selected ZFAS1 as a follow-up research object. Moreover, we confirmed that lncRNAs can exist stably in peripheral blood, consistent with the findings of Li et al[22]. Therefore, lncRNA ZFAS1 may be a potential plasma biomarker for gastric cancer.
We recollected plasma from 60 healthy control subjects, 60 gastritis/peptic ulcer patients, 60 GIST patients, and 60 matched preoperative and postoperative patients to further validate the aforementioned results. Our data indicated that the expression of ZFAS1 in the peripheral blood of patients with gastric cancer was significantly higher than that of healthy controls and patients with benign gastrointestinal disease (Figure 2A), consistent with previous reports. Our data also showed that ZFAS1 is significantly associated with advanced TNM stage and lymphatic invasion (Figure 2B). There was a sharp decrease in the relative level of ZFAS1 in peripheral blood after surgery (Figure 2C). We also evaluated traditional serological markers such as CEA, CA-199, and alpha fetoprotein in the 60 preoperative patients. Compared with traditional biomarkers, plasma lncRNA ZFAS1 had higher sensitivity and specificity (Figure 2E). In addition, the relative level of ZFAS1 in plasma was significantly correlated with that in IFCCs (Figure 2D). This indicates that lncRNA ZFAS1 is released into the peripheral blood by gastric tumors. This suggests that ZFAS1 may promote the progression of gastric cancer. Both the PFS and OS analysis results showed (Figures 2F and G) that a high level of lncRNAs in peripheral blood was not favorable for patients. Thus, lncRNAs may be used not only for early diagnosis but also for postoperative evaluation of patients.
In addition, ZFAS1 is reported to be involved in metabolism, prognosis, and cell proliferation in liver cancer[23,24]. It has been reported that the expression of ZFAS1 in gastric cancer tissues is significantly higher than that in adjacent tissues and that ZFAS1 expression is related to the prognosis of gastric cancer patients[25,26]. siRNA transfection techniques were used to determine the appropriate lncRNA ZFAS1 interference sequence. We used three types of siRNAs (ZFAS1 siRNA1, ZFAS1 siRNA2, and ZFAS1 siRNA3) to interfere with the expression of ZFAS1 in cell lines to prevent off-target effects. All these siRNAs significantly reduced the expression of lncRNA ZFAS1 in BGC823 cells and SGC7901 cells (Figure 3). We finally selected the ZFAS1 siRNA2 sequence to construct ZFAS1-shRNA2 and successfully generated BGC823 cells and SGC7901 cells with low expression of lncRNA ZFAS1. BGC823 cells and SGC7901 cells with stable knockdown of lncRNA ZFAS1 were also successfully cultured. We further employed loss-of-function and gain-of-function studies to assess the role of ZFAS1 in gastric cancer cell proliferation and migration by MTT, transwell, and colony formation assays (Figures 4, 5 and 6). We observed that ZFAS1 knockdown inhibited gastric cancer cell proliferation and migration, whereas ZFAS1 overexpression had the opposite effects.
We then studied the mechanism by which ZFAS1 affects the proliferation and invasion of gastric cancer cells. The starBase V2.0 database was used to analyze the lncRNA ZFAS1 binding proteins[19]. Only 10 proteins had more than three binding sites for lncRNA ZFAS1. We found that the changes in the mRNA expression of LIN28 and CAPRIN1 were consistent with the change in ZFAS1 expression in gastric cancer cells (Figure 8). The LIN28 protein is thought to play an important role in gastrointestinal tumors[27,28]. In colorectal cancer, LIN28 has been shown to enhance tumor cell invasion via the Wnt pathway, whereas overexpression of LIN28 also recruits the microRNA let-7 to enhance tumor cell metastasis[28]. In gastric cancer, LIN28 affects the human epidermal growth factor receptor 2 level through posttranscriptional regulation, which in turn affects the invasive ability of gastric cancer cells. In addition, LIN28 is an independent risk factor for the prognosis of gastric cancer. This result suggests that LIN28 may be a key protein downstream of lncRNA ZFAS1 and that lncRNA ZFAS1 may enhance the proliferation and invasion of gastric cancer cells by regulating LIN28. CAPRIN1 is thought to be involved in cell invasion in a variety of tumors[29-31]. In osteosarcoma, CAPRIN1 has been shown to cause cisplatin resistance and abnormal apoptosis of tumor cells via the Akt pathway and the ERK1/2 pathway[31], and it can also be used as an independent risk predictor for breast cancer. The changes in the mRNA expression of LIN28 and CAPRIN1 were consistent with knockdown and overexpression of ZFAS1, whereas LIN28 and CAPRIN1 were significantly associated with tumor invasion, which explains the mechanism connecting ZFAS1 with lymphatic invasion in tumor patients. ZFAS1 has been shown to silence the expression of Kruppel-like factor 2 and naked cuticle homolog 2 by binding to polycomb repressive complex 2 and lysine-specific demethylase 1, leading to the development of gastric cancer[25]. In addition, it promotes the development of liver cancer tumor cells[23]. CAPRIN1 may play a key role in the regulation of tumor cell proliferation and invasion via lncRNA ZFAS1.
In this study, we validated the above hypothesis by establishing tumor-bearing mice. The expression level of lncRNA ZFAS1 in the plasma of mice injected with tumor cells with high expression of lncRNA ZFAS1 was significantly higher than that in mice in the other two groups (Figure 10A). The in vivo experiments also indicated that ZFAS1 overexpression promoted gastric cancer cell proliferation and migration, whereas ZFAS1 knockdown had the opposite effect (Figures 10B-E). The data also demonstrated that LIN28 and CAPRIN1 are still regulated by lncRNA ZFAS1 in mice (Figure 10F). CK20 and CEA are classical biomarkers of gastric cancer cells in tissues. The CEA and CK20 levels in the lymph nodes of nude mice injected with tumor cells with high expression of lncRNA ZFAS1 were significantly higher than those in mice in the other two groups (Figure 10G). This further demonstrated that high expression of lncRNA ZFAS1 may enhance the invasion of gastric cancer cells.
In conclusion, we demonstrated in this study that the lncRNA ZFAS1 level is increased in the plasma of gastric cancer patients. LncRNA ZFAS1 promotes the invasion and proliferation of gastric cancer cells by modulating LIN28 and CAPRIN1 expression. These findings indicate that ZFAS1 plays an oncogenic role in gastric cancer and can be used as a potential diagnostic biomarker and a new therapeutic target for gastric cancer.
Long noncoding RNA (lncRNA) ZNFX1-AS1 (ZFAS1) is a newly discovered lncRNA, but its diagnostic value in gastric cancer is unclear.
We investigated the biological effects of ZFAS1 on the survival, proliferation, and migration of gastric cancer cells.
This study aimed to investigate the potential role of ZFAS1 in gastric cancer and to evaluate the clinical significance of ZFAS1 as a biomarker for gastric cancer screening.
RNA extraction, quantitative realtime polymerase chain reaction, lncRNA silencing and overexpression, MTT assay, transwell migration assay, Colony formation assay, protein extraction, immunosorbent assay, and in vivo tumor formation assay were performed in this study.
ZFAS1 amplification was correlated with poor prognosis in gastric cancer. ZFAS1 knockdown inhibited the viability, migration, and proliferation of gastric cancer cells. ZFAS1 overexpression enhanced the viability, migration, and proliferation of gastric cancer cells. LIN28 and CAPRIN1 were the key downstream mediators of ZFAS1 in gastric cancer cells. ZFAS1 was associated with the tumorigenesis of gastric cancer cells in vivo.
LncRNA ZFAS1 is a potential biomarker for gastric cancer.
ZFAS1 plays an oncogenic role in gastric cancer and can be used as a potential diagnostic biomarker and a new therapeutic target for gastric cancer.
The authors would like to thank Peking University People’s Hospital Gastrointestinal Surgery Laboratory for providing the cell lines.
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 56668] [Article Influence: 7083.5] [Reference Citation Analysis (135)] |
| 2. | Riquelme I, Ili C, Roa JC, Brebi P. Long non-coding RNAs in gastric cancer: mechanisms and potential applications. Oncotarget. 2016;. [RCA] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 3. | Sun J, Song Y, Chen X, Zhao J, Gao P, Huang X, Xu H, Wang Z. Novel long non-coding RNA RP11-119F7.4 as a potential biomarker for the development and progression of gastric cancer. Oncol Lett. 2015;10:115-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 4. | Vance KW, Sansom SN, Lee S, Chalei V, Kong L, Cooper SE, Oliver PL, Ponting CP. The long non-coding RNA Paupar regulates the expression of both local and distal genes. EMBO J. 2014;33:296-311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 163] [Cited by in RCA: 184] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 5. | Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354-361. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1424] [Cited by in RCA: 1574] [Article Influence: 104.9] [Reference Citation Analysis (0)] |
| 6. | Qi P, Xu MD, Ni SJ, Huang D, Wei P, Tan C, Zhou XY, Du X. Low expression of LOC285194 is associated with poor prognosis in colorectal cancer. J Transl Med. 2013;11:122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 121] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 7. | Mei D, Song H, Wang K, Lou Y, Sun W, Liu Z, Ding X, Guo J. Up-regulation of SUMO1 pseudogene 3 (SUMO1P3) in gastric cancer and its clinical association. Med Oncol. 2013;30:709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 103] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 8. | Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159-3165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 379] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 9. | Sun W, Wu Y, Yu X, Liu Y, Song H, Xia T, Xiao B, Guo J. Decreased expression of long noncoding RNA AC096655.1-002 in gastric cancer and its clinical significance. Tumour Biol. 2013;34:2697-2701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 76] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 10. | Xiao B, Guo J. Long noncoding RNA AC096655.1-002 has been officially named as gastric cancer-associated transcript 1, GACAT1. Tumour Biol. 2013;34:3271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 11. | Song H, Sun W, Ye G, Ding X, Liu Z, Zhang S, Xia T, Xiao B, Xi Y, Guo J. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med. 2013;11:225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 198] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 12. | Song B, Guan Z, Liu F, Sun D, Wang K, Qu H. Long non-coding RNA HOTAIR promotes HLA-G expression via inhibiting miR-152 in gastric cancer cells. Biochem Biophys Res Commun. 2015;464:807-813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 67] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 13. | Arita T, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Shoda K, Kawaguchi T, Hirajima S, Nagata H, Kubota T, Fujiwara H, Okamoto K, Otsuji E. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013;33:3185-3193. [PubMed] |
| 14. | Levy MH, Back A, Benedetti C, Billings JA, Block S, Boston B, Bruera E, Dy S, Eberle C, Foley KM, Karver SB, Knight SJ, Misra S, Ritchie CS, Spiegel D, Sutton L, Urba S, Von Roenn JH, Weinstein SM. NCCN clinical practice guidelines in oncology: palliative care. J Natl Compr Canc Netw. 2009;7:436-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 153] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 15. | Nagtegaal ID, Odze RD, Klimstra D, Paradis V, Rugge M, Schirmacher P, Washington KM, Carneiro F, Cree IA; WHO Classification of Tumours Editorial Board. The 2019 WHO classification of tumours of the digestive system. Histopathology. 2020;76:182-188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2554] [Cited by in RCA: 2748] [Article Influence: 458.0] [Reference Citation Analysis (3)] |
| 16. | Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471-1474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5537] [Cited by in RCA: 6609] [Article Influence: 413.1] [Reference Citation Analysis (0)] |
| 17. | Xian HP, Zhuo ZL, Sun YJ, Liang B, Zhao XT. Circulating long non-coding RNAs HULC and ZNFX1-AS1 are potential biomarkers in patients with gastric cancer. Oncol Lett. 2018;16:4689-4698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 18. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149116] [Cited by in RCA: 139042] [Article Influence: 5561.7] [Reference Citation Analysis (3)] |
| 19. | Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92-D97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2607] [Cited by in RCA: 4304] [Article Influence: 331.1] [Reference Citation Analysis (0)] |
| 20. | Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20108] [Cited by in RCA: 20720] [Article Influence: 1883.6] [Reference Citation Analysis (23)] |
| 21. | Shao Y, Ye M, Jiang X, Sun W, Ding X, Liu Z, Ye G, Zhang X, Xiao B, Guo J. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer. 2014;120:3320-3328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 153] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 22. | Li Q, Shao Y, Zhang X, Zheng T, Miao M, Qin L, Wang B, Ye G, Xiao B, Guo J. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol. 2015;36:2007-2012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 323] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 23. | Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B, Peng C, Li H, Zhan Q, Zhu Z. Amplification of Long Noncoding RNA ZFAS1 Promotes Metastasis in Hepatocellular Carcinoma. Cancer Res. 2015;75:3181-3191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 252] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 24. | Askarian-Amiri ME, Crawford J, French JD, Smart CE, Smith MA, Clark MB, Ru K, Mercer TR, Thompson ER, Lakhani SR, Vargas AC, Campbell IG, Brown MA, Dinger ME, Mattick JS. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011;17:878-891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 295] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 25. | Nie F, Yu X, Huang M, Wang Y, Xie M, Ma H, Wang Z, De W, Sun M. Long noncoding RNA ZFAS1 promotes gastric cancer cells proliferation by epigenetically repressing KLF2 and NKD2 expression. Oncotarget. 2017;8:38227-38238. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 134] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 26. | Pan L, Liang W, Fu M, Huang ZH, Li X, Zhang W, Zhang P, Qian H, Jiang PC, Xu WR, Zhang X. Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J Cancer Res Clin Oncol. 2017;143:991-1004. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 265] [Cited by in RCA: 269] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 27. | Wang Q, Zhou J, Guo J, Teng R, Shen J, Huang Y, Xie S, Wei Q, Zhao W, Chen W, Yuan X, Chen Y, Wang L. Lin28 promotes Her2 expression and Lin28/Her2 predicts poorer survival in gastric cancer. Tumour Biol. 2014;35:11513-11521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 28. | Tu HC, Schwitalla S, Qian Z, LaPier GS, Yermalovich A, Ku YC, Chen SC, Viswanathan SR, Zhu H, Nishihara R, Inamura K, Kim SA, Morikawa T, Mima K, Sukawa Y, Yang J, Meredith G, Fuchs CS, Ogino S, Daley GQ. LIN28 cooperates with WNT signaling to drive invasive intestinal and colorectal adenocarcinoma in mice and humans. Genes Dev. 2015;29:1074-1086. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 89] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 29. | Bidet K, Dadlani D, Garcia-Blanco MA. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog. 2014;10:e1004242. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 229] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 30. | Gong B, Hu H, Chen J, Cao S, Yu J, Xue J, Chen F, Cai Y, He H, Zhang L. Caprin-1 is a novel microRNA-223 target for regulating the proliferation and invasion of human breast cancer cells. Biomed Pharmacother. 2013;67:629-636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 31. | Sabile AA, Arlt MJ, Muff R, Husmann K, Hess D, Bertz J, Langsam B, Aemisegger C, Ziegler U, Born W, Fuchs B. Caprin-1, a novel Cyr61-interacting protein, promotes osteosarcoma tumor growth and lung metastasis in mice. Biochim Biophys Acta. 2013;1832:1173-1182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kotelevets SM, Russia S-Editor: Wang JJ L-Editor: Webster JR P-Editor: Chen YX