Published online Sep 7, 2022. doi: 10.3748/wjg.v28.i33.4812
Peer-review started: April 20, 2022
First decision: June 2, 2022
Revised: June 15, 2022
Accepted: August 16, 2022
Article in press: August 16, 2022
Published online: September 7, 2022
Processing time: 132 Days and 13.5 Hours
The etiology of pancreatic cancer remains unclear. This limits the possibility of prevention and effective treatment. Hepatitis B virus (HBV) is responsible for the development of different types of cancer, but its role in pancreatic cancer is still being discussed.
To assess the prevalence of previous HBV infection and to identify viral biomarkers in patients with pancreatic ductal adenocarcinoma (PDAC) to support the role of the virus in etiology of this cancer.
The data of 130 hepatitis B surface antigen-negative subjects were available for the final analysis, including 60 patients with PDAC confirmed by cytology or histology and 70 sex- and age-matched controls. All the participants were tested for HBV biomarkers in blood [antibody to hepatitis B core antigen (anti-HBc), antibody to hepatitis B surface antigen (anti-HBs) and HBV DNA], and for those with PDAC, biomarkers in resected pancreatic tissues were tested (HBV DNA, HBV pregenomic RNA and covalently closed circular DNA). We performed immunohistochemistry staining of pancreatic tissues for hepatitis B virus X antigen and Ki-67 protein. Non-parametric statistics were used for the analysis.
Anti-HBc was detected in 18/60 (30%) patients with PDAC and in 9/70 (13%) participants in the control group (P = 0.029). Accordingly, the odds of PDAC in anti-HBc-positive subjects were higher compared to those with no previous HBV infection (odds ratio: 2.905, 95% confidence interval: 1.191-7.084, standard error 0.455). HBV DNA was detected in 8 cases of PDAC and in 6 of them in the pancreatic tumor tissue samples only (all patients were anti-HBc positive). Blood HBV DNA was negative in all subjects of the control group with positive results of the serum anti-HBc test. Among 9 patients with PDAC, 5 revealed signs of replicative competence of the virus (covalently closed circular DNA with or without pregenomic RNA) in the pancreatic tumor tissue samples. Hepatitis B virus X antigen expression and active cell proliferation was revealed by immunohistochemistry in 4 patients with PDAC in the pancreatic tumor tissue samples.
We found significantly higher risks of PDAC in anti-HBc-positive patients. Detection of viral replication and hepatitis B virus X protein expression in the tumor tissue prove involvement of HBV infection in pancreatic cancer development.
Core Tip: Hepatitis B viral (HBV) infection is responsible for different types of cancer. Its role in pancreatic ductal adenocarcinoma (PDAC) remains unclear. This study assessed the prevalence of previous HBV infection and to identify viral biomarkers in patients with PDAC to support the role of the virus in the etiology of this cancer. Anti-HBc-positive subjects had an almost 3-fold greater chance of PDAC compared to the controls. Detection of viral replication and hepatitis B virus X protein expression in the tumor tissue show a possible involvement of HBV infection in pancreatic cancer development. Previous HBV infection is currently an underestimated cause of PDAC.
- Citation: Batskikh S, Morozov S, Dorofeev A, Borunova Z, Kostyushev D, Brezgin S, Kostyusheva A, Chulanov V. Previous hepatitis B viral infection–an underestimated cause of pancreatic cancer. World J Gastroenterol 2022; 28(33): 4812-4822
- URL: https://www.wjgnet.com/1007-9327/full/v28/i33/4812.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i33.4812
Pancreatic cancer (PC) is an aggressive gastrointestinal malignancy with a low rate of early detection, poor survival and a limited number of therapeutic options. It causes more than 430000 deaths yearly worldwide[1]. This makes PC the third leading cause of cancer-related deaths in the United States and the fourth in the European Union[2,3]. The incidence rate of PC is growing, while the improvement in the survival rates is negligible[4]. Pancreatic ductal adenocarcinoma (PDAC) is the most prevalent type of PC and is found in 85% of cases[5]. The etiology of PC remains unclear, which limits the possibility for prevention and effective treatment. Early detection of PC remains a challenge. Therefore, it is still relevant to explore etiological factors of PDAC further and to identify subjects at risk of the disease. Numerous risk factors for the disease have been identified (smoking, excessive alcohol intake, history of chronic pancreatitis, obesity and diabetes, etc)[6]. Some viruses, including hepatitis B virus (HBV), are responsible for the development of different types of cancer, but their role in PC is still being discussed[7].
In endemic regions, like Southeast Asia, the blood markers of current HBV infection are commonly found in subjects with PC[8]. Some authors from endemic regions report that HBV infection is not associated with the risk of developing PC after adjusting for age, sex, diabetes and smoking[9].
Cohort studies from Northern Europe (Denmark, Sweden), where HBV infection is not widespread, showed conflicting results and made an association between PC and HBV infection questionable[10-12]. In most of these studies, association of PC with previous HBV infection (PBI) was not considered. However, it may be important as the risk of liver cancer development perseveres even after hepatitis B surface antigen (HBsAg) loss[13-15].
Cancerogenic mechanisms of HBV infection may be explained by the integration of viral DNA fragments into the genome of host cells or persistence of the viral genome as a covalently closed circular DNA (cccDNA), which plays the role of viral reservoir and template for life-long synthesis of new virions. Both are responsible for preserved expression of viral proteins (especially HBx), which can lead to potentially oncogenic mutations[16-18].
Thus, not only active hepatitis B, but also previous HBV infection may contribute to PC development. Detection of HBV DNA and viral antigens in the pancreatic tumor tissues may provide direct evidence of the involvement of the virus in the etiology of this cancer. However, only a few studies demonstrated the presence of HBV DNA (cccDNA) and/or viral antigens in pancreatic tumor tissue. Therefore, the aim of our study was to assess the prevalence of PBI and to identify viral biomarkers in patients with PDAC to support the role of the virus in the etiology of this cancer.
The study was based on the data of complex examination of patients that applied for PC treatment to Moscow Clinical Scientific Center named after A.S. Loginov from January 2019 to November 2020. Subjects of the control group were also recruited. The study (registered АААА-А18-118021590196-1, АААА-А20-120051990006-1 at www.rosrid.ru) was approved by the Local Ethics Committee and was conducted in accordance with the Declaration of Helsinki (1968) and its consequent revisions. All subjects signed a written informed consent form before the enrollment.
Patients of both sexes, older than 18 years, willing to participate in the study were eligible.
In the group of PC, we enrolled patients with histologically or cytologically confirmed PDAC. In the control group we enrolled generally healthy subjects who applied for routine check-ups or treatment of other non-malignant gastrointestinal conditions and whose data of abdominal ultrasound and/or computed tomography revealed no signs of focal lesions in the pancreas.
Other/indeterminate types of PC or non-malignant lesions beside PDAC (for the main group); positive blood test for HBsAg, hepatitis C virus or HIV antibodies; past surgery for PC; current or previous treatment with interferons, nucleos(t)ide analogues for HBV infection or other reasons; clinically significant diseases or health disorders, making it impossible to perform procedures required by the study protocol.
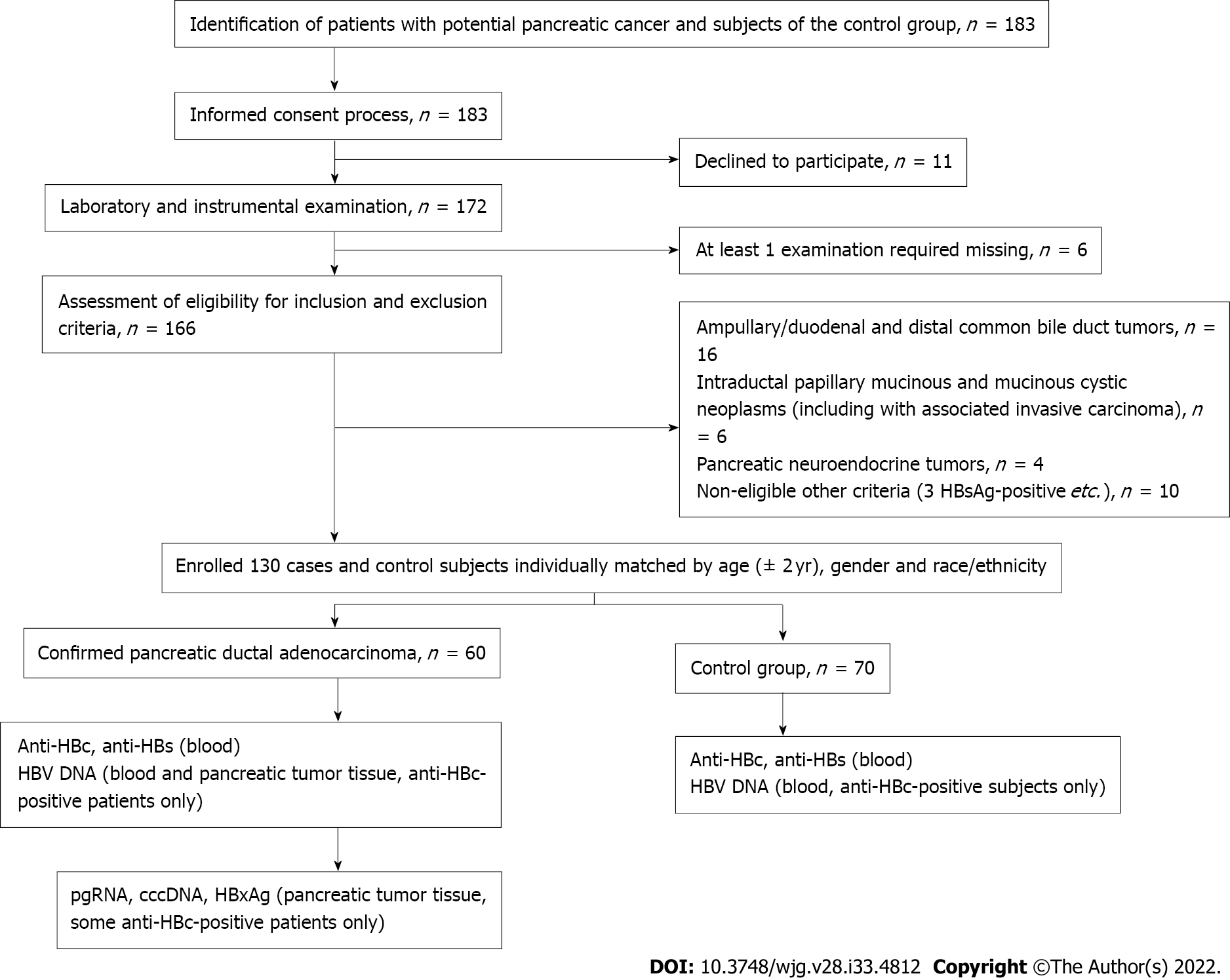
PBI was defined as the presence of antibody to hepatitis B core antigen (anti-HBc) with or without antibody to hepatitis B surface antigen (anti-HBs) or HBV DNA in serum[19]. Control subjects were matched for age (within 2 years), sex and race/ethnicity with the PDAC patients. Study design is shown in Figure 1.
To exclude health conditions able to affect results of the study, all patients underwent routine diagnostic procedures (including but not limited to blood tests, electrocardiogram, abdominal ultrasound and chest X-ray) within standards of care.
All the participants were tested for HBV biomarkers in blood (HBsAg, anti-HBc, anti-HBs). Those HBsAg-negative with positive anti-HBc result were tested for HBV DNA in blood. Anti-HBc-positive patients with PDAC were examined for HBV DNA in the pancreatic tumor tissue.
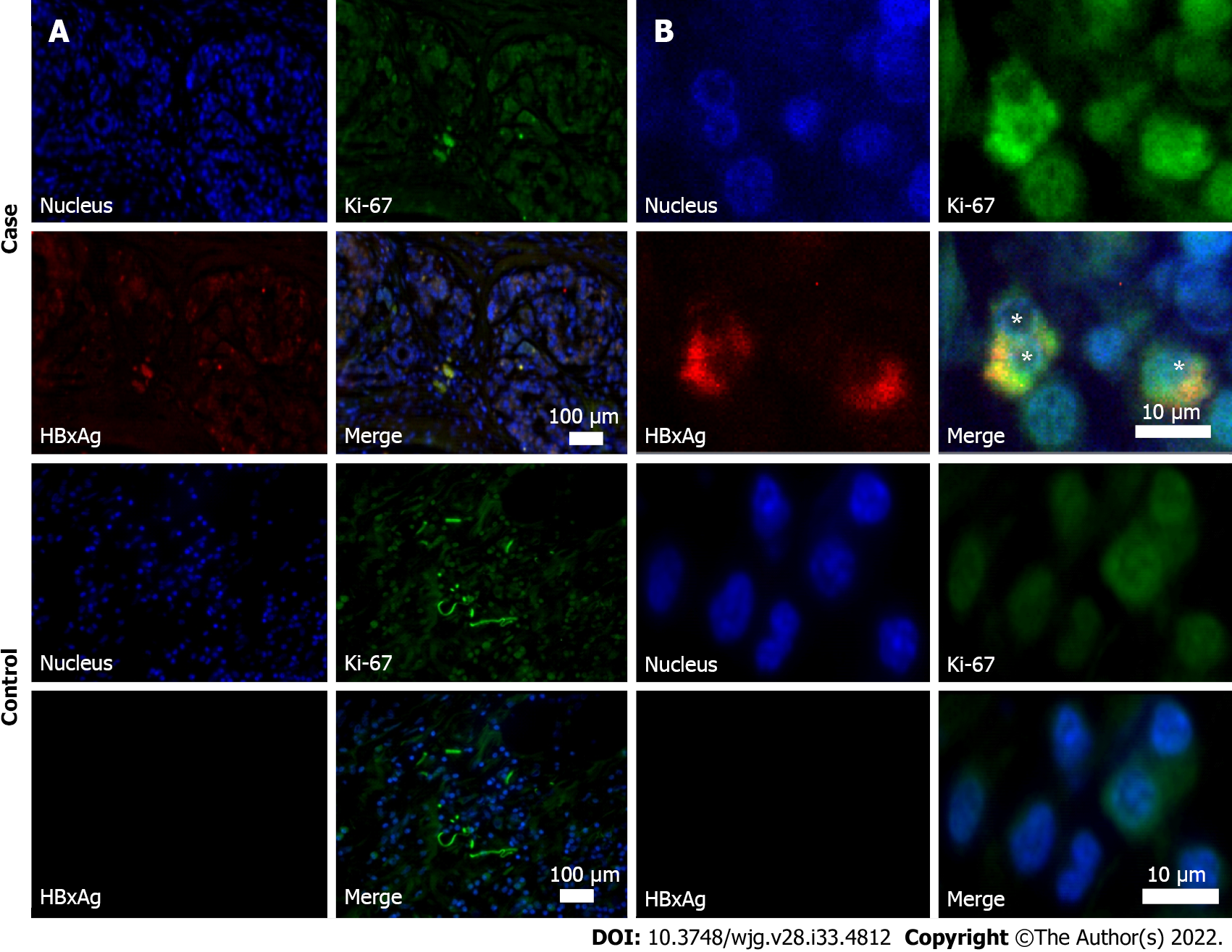
Tumor tissues of anti-HBc-positive patients with PDAC underwent examination for HBV biomarkers (HBV pregenomic RNA and cccDNA) and immunohistochemistry staining for hepatitis B virus X antigen (HBxAg) and Ki-67 protein in cases of the signed informed consent for these tests and sufficient quantity and good quality of the samples.
All anti-HBc-positive participants were tested for the presence of HBV DNA in the blood. In addition, all 18 anti-HBc-positive patients with PDAC were tested for the presence of HBV DNA in pancreatic tumor tissue. In 8 of them, the quality and quantity of samples were suitable for additional testing for HBV pregenomic RNA and cccDNA. Five patients had eligible samples according to these criteria and gave additional consent for immunohistochemical staining for HBxAg and Ki-67 protein.
Collection of samples: Blood samples were taken after overnight fasting, coded and processed immediately at the local laboratory according to the standard instructions.
Pancreatic tumor tissue samples were obtained during surgery or diagnostic biopsy, coded and processed locally. They were stained with hematoxylin-eosin and assessed by a qualified morphologist.
Immunology: Serum samples were tested for HBsAg, anti-HBc IgG, anti-HBs, hepatitis C virus and HIV antibodies. These tests were performed with the use of a Sunrise analyzer (Tecan GmbH, Austria) and specific immunoassays kits (Vector-Best Co., Russia).
Analysis of HBV nucleic acids: Plasma HBV DNA was isolated using commercial AmpliSens Riboprep kit (AmpliSens Biotechnologies, Russia) according to manufacturer’s instructions and quantified using the PCR assay AmpliSens HBV-FL (AmpliSens Biotechnologies) kit (lower limit of detection of 10 IU/mL).
To isolate nucleic acids from biopsies, samples were first homogenized in the MagNA Lyser (Roche Diagnostics, Switzerland). HBV DNA was isolated by AmpliSens Riboprep kit (AmpliSens Biotechnologies) and quantified by AmpliSens HBV-FL (AmpliSens Biotechnologies) kit.
To quantify cccDNA, nucleic acids were first treated with T5 exonuclease (New England Biolabs, United Kingdom) at 37 ºC for 60 min and inactivation at 70 ºC for 20 min[20]. HBV cccDNA was quantified with specific sets of primers and probes and normalized to genomic β-globin.
Specific sets of primers (Table 1) and TaqMan fluorescent probes were used for PCR analysis to detect HBV DNA in pancreatic tissue samples.
| Target | Sequence | |
| cссDNA | Fw | CCGTGTGCACTTCGCTTCA |
| Rev | GCACAGCTTGGAGGCTTGA | |
| Probe | FAM-CATGGAGACCACCGTGAACGCCC-BHQ1 | |
| β-globin | - | V31-FEP-CE (АmpliSens) |
| HPV HCR-Screen (AmpliSens) | ||
To analyze pregenomic HBV RNA (pgRNA HBV), nucleic acids were treated with RNase-free DNase I (NEB) for 30 min at 37 ºC, purified by using AmpliSens Riboprep kit (AmpliSens Biotechnologies), reverse transcribed by AmpliSens Reverta-FL (AmpliSens Biotechnologies) and quantified by AmpliSens HBV-FL (AmpliSens Biotechnologies) kit. CFX96 Real-Time System (Bio-Rad, United States) PCR machine was used for the analysis of plasma and pancreatic tissue samples.
Immunohistochemistry of pancreatic tissues was performed after deparaffinization. Slides were fixed in 4% paraformaldehyde, washed three times in Tris-HCl (50 mM, pH 8.0) followed by incubation with a blocking buffer (0.02% of Triton X-100, 10% horse serum, and 150 mmol/L NaCl in Tris-HCl, 50 mmol/L, pH 8.0) for 30 min and 1 h staining with primary rabbit anti-HBx (ab39716) (Abcam, United Kingdom). Then, slides were washed three times for 5 min in a washing buffer (0.02% of Triton X-100 and 200 mmol/L NaCl in Tris-HCl, 50 mmol/L, pH 8.0) and incubated for 1 h with secondary Alexa Fluor 594 goat anti-rabbit antibodies (ab150080) (Abcam). After that, the slides were treated with primary tagged Alexa Fluor® 488 rabbit anti-Ki-67 (ab197234) and Hoechst 33342 (ab228551) for 1 h, washed three times for 5 min in washing buffer and finally mounted with a Fluoroshield reagent (Abcam). Images were captured using Thunder imaging systems (Leica Microsystems, Germany) with 10 × objectives. Ki-67 and HBxAg staining was analyzed using LAS X (Leica Microsystems). Ki-67 index was counted as the percentage of Ki-67-positive cells[21].
Statistica 12.0 software (StatSoft Inc., United States) was used for analysis of the data. Statistical processing of the obtained data was carried out using nonparametric statistics. Quantitative indicators were preliminarily assessed for compliance with the normal distribution using the Kolmogorov-Smirnov and Lilliefors tests. When quantitative indicators’ distributions differed from normal, we used medians and the interquartile ranges (25%-75%) for the description and processed the data using the Mann-Whitney U test. Nominal data were compared using Pearson χ2 test with Yates’s correction. P values < 0.05 were considered significant. Odds ratio (OR) and 95% confidence interval (95%CI) calculations were performed to assess the association between PDAC and PBI marker detection.
Data of 60 patients with PDAC and 70 participants of the control group were available for the final analysis. Demographic and viral characteristics of the participants are shown in Table 2.
| PDAC, n = 60 | Control, n = 70 | |
| Age in yr, Me (25%-75%) | 64 (59-69) | 62 (58-65) |
| Race/Ethnicity | ||
| White/Caucasian, n (%) | 56 (93.3) | 65 (92.9) |
| Asian, n (%) | 4 (6.7) | 5 (7.1) |
| Females, n (%) | 33 (55.0) | 45 (64.3) |
| Anti-HBc (total), n (%) | 18 (30.0)a | 9 (12.9) |
| Anti-HBc + anti-HBs, n (%) | 8 (13.3) | 5 (7.1) |
| Anti-HBc (only), n (%) | 10 (16.7) | 4 (5.7) |
| HBV DNA (blood and/or pancreatic tissue), n (%1) | 8 (44.4)a | 0 |
| HBV DNA (pancreatic tissue only), n (%1) | 6 (33.3)a | 0 |
In patients with PDAC, anti-HBc antibodies were found more often than in the control group (P = 0.029). Accordingly, the odds of PDAC in anti-HBc-positive subjects were significantly higher compared to those who had no PBI (OR: 2.905, 95%CI: 1.191-7.084, standard error 0.455).
Overall, HBV DNA was found in 8 anti-HBc-positive patients with PDAC. In 2 subjects it was detected in both the blood and pancreatic tumor samples, whereas in the other 6 participants the testing gave positive results only in the pancreatic tumor tissues. No positive results for HBV DNA were obtained in the control group.
The data from special examination of the blood and pancreatic tissue samples are shown in Table 3. The markers confirming replicative competence of HBV (cccDNA with or without pgRNA) were found in the pancreatic tumor tissue samples in 5 patients with PDAC. In 1 subject with a positive test on HBV DNA in the pancreatic tissue examination on cccDNA and pgRNA was not performed.
| No. | Sex | Age in yr | Blood samples | Pancreatic tissue | |||||||
| HBsAg | Anti-HBc | Anti-HBs | HBV DNA, IU/mL | HBV DNA, IU/mL | pgRNA HBV, IU/mL | cccDNA, copies/cell × | HBxAg | HBx-positive cells1, % | |||
| 1 | M | 50 | Neg | Pos | Neg | Tnd | 653 | Tnd | 257 | Pos | 3.9 |
| 2 | F | 58 | Neg | Pos | Pos | Tnd | 415 | 797 | 1440 | Np | Np |
| 3 | M | 60 | Neg | Pos | Pos | Tnd | 1183 | Tnd | Tnd | Pos | 3.7 |
| 4 | M | 61 | Neg | Pos | Pos | Tnd | 364 | 520 | 314 | Pos | 3.4 |
| 5 | F | 63 | Neg | Pos | Pos | Tnd | Tnd | Tnd | Tnd | Neg | 0 |
| 6 | F | 66 | Neg | Pos | Pos | 134 | 868 | Np | Np | Np | Np |
| 7 | M | 69 | Neg | Pos | Pos | Tnd | 610 | Tnd | Tnd | Np | Np |
| 8 | M | 69 | Neg | Pos | Pos | Tnd | 302 | 1428 | 1384 | Np | Np |
| 9 | F | 70 | Neg | Pos | Pos | 126 | 834 | Tnd | 1026 | Pos | 9.9 |
In those with detectable HBV DNA, viral load in the pancreatic tissue was 632 (390-851) IU/mL [median (25%-75%)].
HBxAg expression and active cells’ proliferation was revealed by immunohistochemistry in 4 participants with PDAC in the pancreatic tumor tissue samples (Table 3 and Figure 2). The number of HBx-expressing cells in them did not exceed 10%.
Ki-67 proliferative index in subjects with PDAC in the cohort of special examination was 79.1 (45.2–86.4) [median (25%-75%)]. All HBx-expressing cells were also Ki-67 positive.
The results of the present study demonstrate the association of PBI with PDAC and provide direct molecular evidence for the presence of HBV biomarkers in the pancreatic tumor tissue. In 8 of our patients with PDAC, HBV DNA was detected in the pancreatic tumor tissue. In 5 of them, replicative competence of HBV DNA in the pancreas was supported by detection of cccDNA (with or without pgRNA). Identification of cccDNA and pgRNA (transcribed only from cccDNA) additionally suggests that these patients saved a silent replication of the virus in the pancreatic tissue. Detection of the virus nucleic acids in pancreatic tissue only (with no HBV DNA present in blood) in most of subjects excludes the possibility of artificial contamination of the tumor tissue samples.
Viral infections, including those caused by HBV were recognized among the modifiable risk factors of PC development[22,23]. The data of a meta-analysis of case-control and observational studies (number of subjects with PC: 5883) showed that the odds of PC were significantly higher in chronic or inactive HBsAg carriers [OR: 1.60 (95%CI: 1.26-2.05)] and anti-HBc-positive but anti-HBs-negative individuals [OR: 1.76 (95%CI: 1.05-2.93)] compared to those who were never exposed to HBV infection[7,24]. In our study, the odds of PDAC were even higher, and anti-HBc-positive subjects had an almost 3-fold greater chance of PDAC compared to the controls.
Only a few studies have demonstrated molecular evidence of possible HBV involvement in pancreatic tumor development by identifying HBV DNA (cccDNA) and/or its antigens in pancreatic tumor tissue, and only a limited number of subjects (especially HBsAg-negative but anti-HBc-positive) were involved[25,26]. Although certain pathogenetic mechanisms of PC associated with hepatitis B infection need to be explained in specially planned studies, some assumptions could be made.
HBV is a known carcinogen and is one of the main causes of hepatocellular carcinoma in endemic regions[8]. However, in HBV endemic areas, such as Africa and East Asia, there is a relatively low rate of PC-related deaths. Probably due to high mortality from other causes (including HBV-associated hepatocellular carcinoma), there is not enough time for the development of PC in people with PBI.
HBV integrates into the genome of infected cells, causes genomic aberrations, enhances expression of oncogenes or inhibits tumor suppressors and leads to the cancer development[14]. Similar mechanisms are possible in non-liver carcinogenesis, including the pancreas[25,27]. Pancreatic beta cells and hepatocytes develop from the ventral foregut endoderm during ontogenesis and thus may share characteristics that are favorable for HBV replication and virus-induced tumor development[28]. It seems that malignant transformation in the pancreas is not provoked by direct cellular damage and is caused by the integration of HBV DNA into the genome of pancreatic cells and subsequent disruption of the functions of anti-oncogenes or by stimulation of pro-oncogenes’ activity[29]. The replication of HBV in pancreatic tissue may decrease with time. However, DNA fragments of the virus integrated into the genome of host cells continue to express viral proteins (especially regulatory protein X) of HBV responsible for carcinogenesis. The expression capability of HBx from integrated fragments of the viral genome in tumor tissues when replication is absent was confirmed in hepatocellular carcinoma[30-32].
In our study, immunohistochemistry revealed expression of HBx in the pancreatic tumor tissue in 4 out of 5 HBsAg-negative and anti-HBc-positive patients with PDAC. Replicative competence of HBV (detected cccDNA) was found in 3 of them. This may mean that in 1 patient, expression of HBx was caused only by the integration of the virus into the genome of pancreatic cells. These fragments of viral DNA, which preserve the open reading frame and express HBx, may serve as a basis for carcinogenesis in subjects with PDAC. Although this mechanism may play a role in primary cancer development, its role in PC recurrence is not clear, and further studies are necessary. The low-grade replication may also play a role in HBV reactivation, especially in cases when immunodepressants are used. However, this question is insufficiently studied.
It is not clear whether the number of HBxAg expressing cells is important for cancer development. In hepatocellular carcinoma of HBsAg-negative HBV DNA-positive subjects, the relative number of HBxAg-expressing cells is about 30% within the tumor tissue and 20% in the rest of the liver tissue[33]. Similar data for PC are lacking. According to our results, the number of cells producing HBxAg in PDAC is about 4%. It seems that the number of cells producing HBx protein is less important than their presence, at least for PC development. This may be indirectly confirmed by the fact that in all HBxAg-positive subjects in our study proliferative index Ki-67 was significantly higher than 50%, whereas similarly high values of this marker were only found in about 12% of subjects with PDAC[21,34].
Detection of cccDNA in pancreatic tissue in HBsAg-negative subject supports the need for revision of the statements of the Taormina Workshop (2018), which defines occult HBV infection as the presence of replication-competent HBV DNA (i.e. cccDNA in the liver and/or HBV DNA in the blood of people who test negative for HBsAg by currently available assays)[16]. As extrahepatic HBV replication may occur in HBsAg-negative subjects (which was confirmed in the course of our study), it is reasonable not to indicate in the statement the specific organ for HBV DNA (cccDNA) detection.
Involvement of PBI in PC development requires revision of the ultimate targets of antiviral treatment. “Sterilizing cure” (undetectable HBsAg in blood in combination with the absence of DNA HBV in any tissues, including cccDNA and integrated viral DNA) was recognized unachievable in the near future[35]. However, “functional cure” (defined as sustained clearance of HBs with or without HBs-seroconversion and non-detectable HBV DNA in blood after the course of treatment) evidently cannot affect the expression of oncogenic proteins of HBV (especially HBx) and thus diminish the chances of cancer development. Although it is impossible to achieve eradication of cccDNA and integrated fragments of the viral genome with currently existing means, this should be stated as the ultimate goal for future therapy options.
The limitation of the study is a relatively small number of patients. Moreover, examination of the tumor tissues on cccDNA, pgRNA and HBxAg was possible for only a few of the 18 anti-HBc-positive subjects with PDAC due to the quality of the obtained specimens. Further randomized multicenter studies are necessary to confirm the obtained results, prove the role of HBV infection in the etiology of PC and clarify carcinogenic mechanisms in them.
An almost three-fold risk of PDAC was found in HBsAg-negative but anti-HBc-positive subjects. Detection of silent viral replication and pro-oncogenic HBx protein expression in the tumor tissue suggest involvement of HBV infection in PC development. PBI seems to be an underestimated cause of PDAC at the moment.
The etiology of pancreatic cancer is unclear. This limits possibilities for its prevention and effective treatment. Hepatitis B virus (HBV) is responsible for the development of hepatocellular carcinoma and different types of extrahepatic cancer, but its role in the etiology of pancreatic cancer is still being discussed.
The epidemiological relationship of previous HBV infection (PBI) with pancreatic cancer and identification of viral biomarkers within the tumor tissue may provide support for this. However, there is still a lack of such reports, especially from non-endemic regions for HBV infection.
In our study, we aimed to assess the prevalence of PBI and to identify viral biomarkers in patients with pancreatic ductal adenocarcinoma (PDAC) to support the role of the virus in the etiology of this cancer.
The data of 130 hepatitis B surface antigen-negative subjects were included in the final analysis (60 patients with PDAC confirmed by cytology or histology and 70 sex- and age-matched controls). All the participants were tested for HBV biomarkers in blood (antibody to hepatitis B core antigen, antibody to hepatitis B surface antigen and HBV DNA). Those with PDAC were tested for biomarkers in resected pancreatic tissues [HBV DNA, HBV pregenomic RNA and covalently closed circular DNA (cccDNA)]. Additionally, we performed immunohistochemistry staining of pancreatic tissues for hepatitis B virus X antigen and Ki-67 protein. Non-parametric statistics were used for the analysis.
We have established that 18/60 (30%) patients with PDAC and 9/70 (13%) participants in the control group (P = 0.029) were anti-hepatitis B core antigen-positive. HBV DNA was detected in 8 anti-hepatitis B core antigen-positive patients of PDAC (in 6 of them—in the pancreatic tumor tissue samples only) but in neither subjects of the control group. In 5 patients with PDAC we revealed signs of replicative competence of the virus (cccDNA with or without pregenomic RNA) in the pancreatic tumor tissue samples. Hepatitis B virus X antigen expression and active cells’ proliferation was revealed by immunohistochemistry in 4 participants with PDAC in the pancreatic tumor tissue samples.
PBI seems to be an underestimated cause of PDAC.
Larger studies are necessary to assess risks of PDAC in subjects with PBI and define HBV-associated mechanisms of carcinogenesis in them.
The authors acknowledge all study participants.
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 56683] [Article Influence: 7085.4] [Reference Citation Analysis (135)] |
| 2. | NIH. National Cancer Institute. Cancer Stats Facts: Pancreatic Cancer. [cited 4 January 2022]. Available from: https://seer.cancer.gov/statfacts/html/pancreas.html. |
| 3. | European Commission. European Cancer Information System. [cited 6 January 2022]. Available from: https://ecis.jrc.ec.europa.eu/explorer.php. |
| 4. | Maisonneuve P. Epidemiology and burden of pancreatic cancer. Presse Med. 2019;48:e113-e123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 94] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 5. | Mostafa ME, Erbarut-Seven I, Pehlivanoglu B, Adsay V. Pathologic classification of "pancreatic cancers": current concepts and challenges. Chin Clin Oncol. 2017;6:59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 6. | Tempero MA, Malafa MP, Al-Hawary M, Behrman SW, Benson AB, Cardin DB, Chiorean EG, Chung V, Czito B, Del Chiaro M, Dillhoff M, Donahue TR, Dotan E, Ferrone CR, Fountzilas C, Hardacre J, Hawkins WG, Klute K, Ko AH, Kunstman JW, LoConte N, Lowy AM, Moravek C, Nakakura EK, Narang AK, Obando J, Polanco PM, Reddy S, Reyngold M, Scaife C, Shen J, Vollmer C, Wolff RA, Wolpin BM, Lynn B, George GV. Pancreatic Adenocarcinoma, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2021;19:439-457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 825] [Article Influence: 165.0] [Reference Citation Analysis (0)] |
| 7. | Liu X, Zhang ZH, Jiang F. Hepatitis B virus infection increases the risk of pancreatic cancer: a meta-analysis. Scand J Gastroenterol. 2021;56:252-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 8. | Arbuthnot P, Kew M. Hepatitis B virus and hepatocellular carcinoma. Int J Exp Pathol. 2001;82:77-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 89] [Reference Citation Analysis (0)] |
| 9. | Chang MC, Chen CH, Liang JD, Tien YW, Hsu C, Wong JM, Chang YT. Hepatitis B and C viruses are not risks for pancreatic adenocarcinoma. World J Gastroenterol. 2014;20:5060-5065. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 20] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 10. | Andersen ES, Omland LH, Jepsen P, Krarup H, Christensen PB, Obel N, Weis N; DANVIR Cohort Study. Risk of all-type cancer, hepatocellular carcinoma, non-Hodgkin lymphoma and pancreatic cancer in patients infected with hepatitis B virus. J Viral Hepat. 2015;22:828-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 11. | Huang J, Magnusson M, Törner A, Ye W, Duberg AS. Risk of pancreatic cancer among individuals with hepatitis C or hepatitis B virus infection: a nationwide study in Sweden. Br J Cancer. 2013;109:2917-2923. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 48] [Article Influence: 3.7] [Reference Citation Analysis (1)] |
| 12. | Sundquist K, Sundquist J, Ji J. Risk of hepatocellular carcinoma and cancers at other sites among patients diagnosed with chronic hepatitis B virus infection in Sweden. J Med Virol. 2014;86:18-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 13. | European Association for the Study of the Liver. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67:370-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3745] [Cited by in RCA: 4011] [Article Influence: 445.7] [Reference Citation Analysis (1)] |
| 14. | Brechot C, Pourcel C, Louise A, Rain B, Tiollais P. Presence of integrated hepatitis B virus DNA sequences in cellular DNA of human hepatocellular carcinoma. Nature. 1980;286:533-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 449] [Cited by in RCA: 424] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 15. | Zhao LH, Liu X, Yan HX, Li WY, Zeng X, Yang Y, Zhao J, Liu SP, Zhuang XH, Lin C, Qin CJ, Zhao Y, Pan ZY, Huang G, Liu H, Zhang J, Wang RY, Wen W, Lv GS, Zhang HL, Wu H, Huang S, Wang MD, Tang L, Cao HZ, Wang L, Lee TL, Jiang H, Tan YX, Yuan SX, Hou GJ, Tao QF, Xu QG, Zhang XQ, Wu MC, Xu X, Wang J, Yang HM, Zhou WP, Wang HY. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat Commun. 2016;7:12992. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 228] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 16. | Raimondo G, Locarnini S, Pollicino T, Levrero M, Zoulim F, Lok AS; Taormina Workshop on Occult HBV Infection Faculty Members. Update of the statements on biology and clinical impact of occult hepatitis B virus infection. J Hepatol. 2019;71:397-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 375] [Cited by in RCA: 400] [Article Influence: 57.1] [Reference Citation Analysis (0)] |
| 17. | Fanning GC, Zoulim F, Hou J, Bertoletti A. Therapeutic strategies for hepatitis B virus infection: towards a cure. Nat Rev Drug Discov. 2019;18:827-844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 426] [Article Influence: 60.9] [Reference Citation Analysis (1)] |
| 18. | Fanning GC, Zoulim F, Hou J, Bertoletti A. Author Correction: Therapeutic strategies for hepatitis B virus infection: towards a cure. Nat Rev Drug Discov. 2020;19:291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 19. | Lok AS, Everhart JE, Di Bisceglie AM, Kim HY, Hussain M, Morgan TR; HALT-C Trial Group. Occult and previous hepatitis B virus infection are not associated with hepatocellular carcinoma in United States patients with chronic hepatitis C. Hepatology. 2011;54:434-442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 65] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 20. | Qu B, Ni Y, Lempp FA, Vondran FWR, Urban S. T5 Exonuclease Hydrolysis of Hepatitis B Virus Replicative Intermediates Allows Reliable Quantification and Fast Drug Efficacy Testing of Covalently Closed Circular DNA by PCR. J Virol. 2018;92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 21. | Pergolini I, Crippa S, Pagnanelli M, Belfiori G, Pucci A, Partelli S, Rubini C, Castelli P, Zamboni G, Falconi M. Prognostic impact of Ki-67 proliferative index in resectable pancreatic ductal adenocarcinoma. BJS Open. 2019;3:646-655. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 22. | Hassan MM, Li D, El-Deeb AS, Wolff RA, Bondy ML, Davila M, Abbruzzese JL. Association between hepatitis B virus and pancreatic cancer. J Clin Oncol. 2008;26:4557-4562. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 136] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 23. | Midha S, Chawla S, Garg PK. Modifiable and non-modifiable risk factors for pancreatic cancer: A review. Cancer Lett. 2016;381:269-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 197] [Article Influence: 19.7] [Reference Citation Analysis (3)] |
| 24. | Wang Y, Yang S, Song F, Cao S, Yin X, Xie J, Tu X, Xu J, Xu X, Dong X, Lu Z. Hepatitis B virus status and the risk of pancreatic cancer: a meta-analysis. Eur J Cancer Prev. 2013;22:328-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 25. | Song C, Lv J, Liu Y, Chen JG, Ge Z, Zhu J, Dai J, Du LB, Yu C, Guo Y, Bian Z, Yang L, Chen Y, Chen Z, Liu J, Jiang J, Zhu L, Zhai X, Jiang Y, Ma H, Jin G, Shen H, Li L, Hu Z; China Kadoorie Biobank Collaborative Group. Associations Between Hepatitis B Virus Infection and Risk of All Cancer Types. JAMA Netw Open. 2019;2:e195718. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 141] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 26. | Chen Y, Bai X, Zhang Q, Wen L, Su W, Fu Q, Sun X, Lou Y, Yang J, Zhang J, Chen Q, Wang J, Liang T. The hepatitis B virus X protein promotes pancreatic cancer through modulation of the PI3K/AKT signaling pathway. Cancer Lett. 2016;380:98-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 27. | Dejean A, Lugassy C, Zafrani S, Tiollais P, Brechot C. Detection of hepatitis B virus DNA in pancreas, kidney and skin of two human carriers of the virus. J Gen Virol. 1984;65 (Pt 3):651-655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 152] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 28. | Wandzioch E, Zaret KS. Dynamic signaling network for the specification of embryonic pancreas and liver progenitors. Science. 2009;324:1707-1710. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 202] [Cited by in RCA: 178] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 29. | Sherman M. Pancreatic cancer in chronic hepatitis B. Liver Int. 2010;30:339-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 30. | Tu T, Budzinska MA, Vondran FWR, Shackel NA, Urban S. Hepatitis B Virus DNA Integration Occurs Early in the Viral Life Cycle in an In Vitro Infection Model via Sodium Taurocholate Cotransporting Polypeptide-Dependent Uptake of Enveloped Virus Particles. J Virol. 2018;92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 150] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 31. | Kew MC. Hepatitis B virus x protein in the pathogenesis of hepatitis B virus-induced hepatocellular carcinoma. J Gastroenterol Hepatol. 2011;26 Suppl 1:144-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 217] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 32. | Geng M, Xin X, Bi LQ, Zhou LT, Liu XH. Molecular mechanism of hepatitis B virus X protein function in hepatocarcinogenesis. World J Gastroenterol. 2015;21:10732-10738. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 52] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 33. | Poussin K, Dienes H, Sirma H, Urban S, Beaugrand M, Franco D, Schirmacher P, Bréchot C, Paterlini Bréchot P. Expression of mutated hepatitis B virus X genes in human hepatocellular carcinomas. Int J Cancer. 1999;80:497-505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 34. | Kim H, Park CY, Lee JH, Kim JC, Cho CK, Kim HJ. Ki-67 and p53 expression as a predictive marker for early postoperative recurrence in pancreatic head cancer. Ann Surg Treat Res. 2015;88:200-207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 35. | Cornberg M, Lok AS, Terrault NA, Zoulim F; 2019 EASL-AASLD HBV Treatment Endpoints Conference Faculty. Guidance for design and endpoints of clinical trials in chronic hepatitis B – Report from the 2019 EASL-AASLD HBV Treatment Endpoints Conference‡. J Hepatol. 2020;72:539-557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 259] [Article Influence: 43.2] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Russia
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Pham TTT, Viet Nam; Sultana C, Romania; Tai DI, Taiwan S-Editor: Fan JR L-Editor: Filipodia P-Editor: Fan JR