Published online Aug 28, 2022. doi: 10.3748/wjg.v28.i32.4649
Peer-review started: February 28, 2022
First decision: May 9, 2022
Revised: June 10, 2022
Accepted: July 27, 2022
Article in press: July 27, 2022
Published online: August 28, 2022
Processing time: 178 Days and 9.5 Hours
Anoctamin 5 (ANO5)/transmembrane protein 16E belongs to the ANO/ tran
To examine the role of ANO5 in the regulation of tumor progression and clinico
Knockdown experiments using ANO5 small interfering RNA were conducted in human GC cell lines, and changes in cell proliferation, cell cycle progression, apoptosis, and cellular movement were assessed. The gene expression profiles of GC cells were investigated following ANO5 silencing by microarray analysis. Immunohistochemical staining of ANO5 was performed on 195 primary tumor samples obtained from patients with GC who underwent curative gastrectomy between 2011 and 2013 at our department.
Reverse transcription-quantitative polymerase chain reaction (PCR) and western blotting demonstrated high ANO5 mRNA and protein expression, respectively, in NUGC4 and MKN45 cells. In these cells, ANO5 silencing inhibited cell proliferation and induced apoptosis. In addition, the knockdown of ANO5 inhibited G1-S phase progression, invasion, and migration. The results of the microarray analysis revealed changes in the expression levels of several cyclin-associated genes, such as CDKN1A, CDK2/4/6, CCNE2, and E2F1, in ANO5-depleted NUGC4 cells. The expression of these genes was verified using reverse transcription-quantitative PCR. Immunohistochemical staining revealed that high ANO5 expression levels were associated with a poor prognosis. Multivariate analysis identified high ANO5 expression as an independent prognostic factor for 5-year survival in patients with GC (P = 0.0457).
ANO5 regulates the cell cycle progression by regulating the expression of cyclin-associated genes and affects the prognosis of patients with GC. These results may provide insights into the role of ANO5 as a key mediator in tumor progression and/or promising prognostic biomarker for GC.
Core Tip: The present study aimed to investigate the role of anoctamin 5 (ANO5) in the regulation of tumor progression and the clinicopathological significance of its expression in gastric cancer. Immunohistochemical staining revealed that high ANO5 expression levels were associated with a poor prognosis in patients with gastric cancer. Microarray analysis results suggest that ANO5 regulates cell cycle progression by regulating the expression of cyclin-associated genes. Our results provide insights into the role of ANO5 as a mediator of and/or biomarker for gastric cancer.
- Citation: Fukami T, Shiozaki A, Kosuga T, Kudou M, Shimizu H, Ohashi T, Arita T, Konishi H, Komatsu S, Kubota T, Fujiwara H, Okamoto K, Kishimoto M, Morinaga Y, Konishi E, Otsuji E. Anoctamin 5 regulates the cell cycle and affects prognosis in gastric cancer. World J Gastroenterol 2022; 28(32): 4649-4667
- URL: https://www.wjgnet.com/1007-9327/full/v28/i32/4649.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i32.4649
The anoctamin (ANO)/transmembrane protein 16 (TMEM16) family is present in numerous eukaryotes, and ten ANO paralogs, ANO1-ANO10 (TMEM16A-H, TMEM16J and K), have been identified in vertebrates[1]. Of these, several function as calcium-activated chloride channels. ANOs comprise a family of plasma membrane proteins that mediate ion transport, phospholipid scrambling, and other membrane protein regulation in numerous cell types[2-6]. Their expression has been detected in both epithelial and non-epithelial tissues types[4]. Although the regulation of ANOs has been extensively examined, the mechanisms by which increased intracellular calcium concentration activates chloride or cation conductance have not been elucidated.
Recent molecular and biochemical studies reported a role for ANOs in human carcinogenesis. For instance, the expression of ANO proteins is upregulated in cancer and associated with a poor patient prognosis[7]. A relationship has been demonstrated between ANO1 and patient prognosis in various cancer types, including gastric, esophageal, breast, lung, and head and neck cancer[8-12]. The upregulation of the genes encoding ANO1 and ANO3 has been associated with several cancer types, specifically gastrointestinal stromal tumors, breast cancer, and squamous cell carcinoma[12,13]. Furthermore, ANO6 has been strongly implicated in the metastatic potential of breast cancer[14]. The expression levels of other members of the ANO family are also associated with cell proliferation and cancer development[15-17].
In our previous studies, we identified a crucial role for several chloride ion channels and transporters in patients with gastric cancer (GC); intracellular chloride regulates proliferation and cell cycle progression[18,19], whereas furosemide, a potent inhibitor of the Na+/K+/2Cl- cotransporter, induces G0/G1 arrest[20]. Furthermore, leucine rich repeat containing 8 VRAC subunit A regulate the proliferation, apoptosis, migration, and invasion of GC cells[21].
ANO5 has recently been implicated in various cancers, such as thyroid[22] and pancreatic cancer[23]; however, limited information is presently available on its involvement in tumor progression in patients with GC or the clinical significance of its expression. Therefore, in the present study, we investigated whether ANO5 contributes to the regulation of cancer growth and evaluated its clinicopathological significance in GC.
MKN7, MKN45, MKN74, HGC27, and NUGC4 human GC cell lines were purchased from the Riken Cell Bank (Tsukuba, Japan). Cells were cultured in RPMI-1640 (Nacalai Tesque, Kyoto, Japan) containing 100 μg/mL of streptomycin, 100 U/mL penicillin, and 10% FBS at 37 °C in a 5% CO2 incubator. Rabbit polyclonal anti-ANO5 antibody was obtained from Funakoshi (GTX81161) for immunohistochemical (IHC) analysis and western blotting. Mouse monoclonal anti-β-actin antibody was provided by Sigma-Aldrich (St. Louis, MO, United States) and HRP-conjugated anti-rabbit and mouse secondary antibodies by Cell Signaling Technology (Beverly, MA, United States).
RNA was extracted from cancer cells using an RNeasy kit (Qiagen, Valencia, CA, United States). The Step One plusTM Real-Time polymerase chain reaction (PCR) System (Applied Biosystems, Foster City, CA, United States) and TaqMan Gene Expression Assays (Applied Biosystems) were employed for reverse transcription-quantitative PCR analysis using the following PCR thermocycling conditions: initial denaturation step at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. The expression levels of the following genes were assessed: ANO5 (Hs01381106_m1), CDKN1A (Hs00355782_m1), CDK2 (Hs00608082_m1), CDK4 (Hs00175935_m1), CDK6 (Hs00608037_m1), cyclin E2 (CCNE2; Hs00180319_m1), and E2F1 (Hs00153451_m1) (all from Applied Biosystems). The expression of each gene was normalized using the housekeeping gene β-actin (Hs01060665_g1; Applied Biosystems). All assays were performed in triplicates.
The cells were washed twice with ice-cold PBS and harvested in M-PER lysis buffer (Pierce, Rockford, IL, United States) supplemented with protease inhibitors (Pierce Biotechnology). Protein concentrations were measured using a modified Bradford assay (Bio-Rad, Hercules, CA, United States). Cell lysates containing equal amounts of total protein (10 mg/lane) were resolved using 10% SDS-PAGE and subsequently transferred to polyvinylidene fluoride membranes (GE Healthcare, Piscataway, NJ, United States). Membranes were incubated with antibodies for 24 h at 4 °C. Band densities were quantified using ImageJ (version 1.52; National Institutes of Health).
All small interfering RNA (siRNA) reverse transfection procedures were performed using Lipofectamine® RNAiMAX reagent (Invitrogen, Carlsbad, CA, United States) with a final siRNA concentration of 20 nmol/L, according to the manufacturer’s instructions. ANO5 siRNA (Stealth RNAi siRNA; HSS137119, Stealth RNAi siRNA; HSS137120) and control siRNA (Stealth RNAiTM siRNA Negative Control) were obtained from Invitrogen.
Control-HaloTagR plasmid (Promega, G6591) and ANO5-HaloTagR plasmid (pFN21AE5809) were transfected using P3000TM (Invitrogen) and lipofectamine 3000 (Invitrogen) following the manufacturer’s instructions. After passaging, ANO5-expressing cells were used for the cell proliferation assay.
NUGC4 and MKN45 cells were seeded at densities of 1.0 and 2.0 × 105 cells/well, respectively, on six-well plates and incubated at 37 °C in a 5% CO2 incubator. The siRNA was transfected 24 h after seeding. The cells were detached from the plates with trypsin-EDTA 48 h and 72 h after siRNA transfection and counted using a hemocytometer.
Cell proliferation activity was measured using the water-soluble tetrazolium salts-8 assay with Cell Count Reagent SF (Nacalai Tesque). NUGC4, MKN45, and MKN7 cells were seeded at a density of 1.0 × 104, 1.0 × 104, and 1.5 × 104 cells/well, respectively, in 24-well plates and were incubated at 37 °C in a 5% CO2 incubator. The siRNA was transfected 24 h after seeding. Cell proliferation was evaluated every 24 h by measuring the absorbance at 450 nm using a Thermo Scientific Multiskan FC (Thermo Fisher Scientific).
Cell cycle progression was assessed 48 h after siRNA transfection by flow cytometry. Cells were detached from the plates using trypsin-EDTA and subsequently treated with 0.2% Triton X-100 and stained with propidium iodide with RNase staining buffer (BD Biosciences, San Jose, CA, United States). Flow cytometry data were acquired using a BD Accuri C6 plus flow cytometer (BD Biosciences) to assess DNA content in at least 10000 cells.
Cells were evaluated 72 h after transfection and stained using the ANNEXIN V-FITC Kit (Beckman Coulter, Brea, CA, United States). The frequencies of early and late apoptotic cells among at least 10000 cells were assessed using a BD Accuri C6 plus flow cytometer.
Migration assays were performed using 24-well cell culture inserts with 8-μm pores (BD Biosciences), whereas invasion assays were performed using Biocoat Matrigel® (BD Biosciences). At 48 h post-transfection, NUGC4 and MKN45 cells were seeded at a density of 3.0 × 105 cells/well in serum-free RPMI-1640 in the upper chamber, whereas the lower chamber contained RPMI-1640 with 10% FBS. Matrigel and the cells remaining in the upper chamber after a 48-h incubation were removed. Diff-Quick staining reagents (Sysmex) were used to stain migrated or invaded cells, which were counted in four independent fields of view. Both assays were conducted thrice.
NUGC4 and MKN45 cells were transfected with either control or ANO5 siRNA. At 48 h after siRNA transfection, total RNA was extracted using RNeasy kit. Cyanine 3 (Cy3)-labeled cRNA was prepared from 0.1 μg total RNA using the Low Input Quick Amp Labeling Kit (Agilent Technologies, CA, United States) and then subjected to RNeasy column purification (Qiagen). Dye incorporation and cRNA yields were assessed using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific). Subsequently, 0.6 μg Cy3-labeled cRNA was fragmented in a 25 μL reaction volume containing 1 × Agilent fragmentation buffer and 2 × Agilent blocking agent at 60 °C for 30 min. The 2 × Agilent hybridization buffer (25 μL) was then added, and hybridization to SurePrint G3 Human GE 8 × 60K Microarray Ver3.0 (Agilent Technologies) was conducted at 65 °C for 17 h in a rotating Agilent hybridization oven. The microarrays were then washed with GE Wash Buffer 1 (Agilent Technologies) at room temperature for 1 min, followed by GE Wash buffer 2 (Agilent Technologies) at 37 °C for 1 min.
Slides were scanned using the Agilent SureScan Microarray Scanner (G2600D) with the one color scan setting for 8 × 60k array slides. The scanned images were analyzed with Feature Extraction Software (Agilent Technologies) using the default parameters to obtain background-subtracted and spatially detrended processed signal intensities. Microarray data were analyzed using ingenuity pathway analysis software (Ingenuity Systems, Redwood City, CA).
Histologically proven primary GC tumor samples were obtained from 195 consecutive patients who underwent curative gastrectomy between 2011 and 2013 at Kyoto Prefectural University of Medicine, Japan. For mRNA analysis, frozen tissue samples of normal stomach and tumors were collected from surgical specimens and stored at -80 °C. Written informed consent was obtained from all patients prior to enrollment. Patients with noncurative resection or preoperative chemotherapy were excluded from the study. Tumor staging was conducted according to the International Union Against Cancer/TNM Classification of Malignant Tumors (8th edition)[24]. The present study was approved by the Institutional Review Board of the Kyoto Prefectural University of Medicine (ERB-C-1195).
The Vectastain avidin-biotinylated peroxidase complex Elite Kit (Vector Laboratories, Burlingame, CA, United States) was employed for IHC staining using the avidin-biotinylated peroxidase complex method. After deparaffinization in xylene, sections were rehydrated in a graded series of ethanol solutions. The sections were then incubated in 0.3% H2O2 for 30 min to block endogenous peroxidase activity. Endogenous biotin, biotin receptors, and avidin-binding sites were also blocked using an Avidin/Biotin Blocking Kit (Vector laboratories). Sections were incubated with ANO5 antibody diluted 1:100 at 37 °C for 1 h and then at 4 °C overnight. Cells were visualized using the standard avidin-biotinylated peroxidase complex method, with hematoxylin as the counterstain.
ANO5 expression levels in immunohistochemically stained samples were semi-quantitatively graded based on the staining intensity and proportion of cytoplasm in the stained cancer cells. The staining intensity was scored as 0 (no staining), 1 (weak staining), 2 (moderate staining), or 3 (strong staining). The proportion of stained tumor cells as a percentage of the stained area in the cancer area was scored from 0 to 1.0. IHC scores were calculated as the maximum multiplied product of intensity and proportion scores (0-3.0). IHC diagnosis was based on tumor ANO5 expression assessment, and other IHC parameters were performed by at least two physicians, including an experienced pathologist.
MQAE reagent, a chloride-sensitive fluorescence probe (Dojindo Laboratories, Kumamoto, Japan) was used to assess intracellular chloride concentrations. NUGC4 and MKN45 cells were seeded in 24-well plates at a density of 3.0 × 104 cells/well and then incubated in normal medium at 37 °C with 5% CO2. The medium was then replaced with standard and low-chloride medium in which MQAE was dissolved, and cells were incubated at 37 °C in a CO2 incubator for a further 12 h. Following washing with PBS five times, the fluorescence intensity of MQAE was evaluated under a fluorescence microscope (BZ-X800; Keyence, Osaka, Japan). Three fields of view were analyzed per sample at × 100 magnification. Quantification was performed using a BZ-X800 analyzer and accompanying software (BZ-H4C, v.1.1.1.8; Keyence).
A low chloride stimulation experiment was conducted to examine the effects of changes in intracellular chloride concentrations on GC cells. A low-chloride medium supplemented with 10% FBS was prepared in chloride-free RPMI-1640 (chloride replaced with NO3-) (Nacalai Tesque).
To block the c-Jun N-terminal kinase (JNK) signaling pathway, NUGC4 and MKN45 cells were incubated with the JNK inhibitor SP600125 (10µm, ab120065, Abcam) according to manufacturer’s instructions. The cells were divided into 3 groups: control, ANO5 siRNA, and JNK inhibitor (ANO5 siRNA + SP600125). Cell proliferation was detected every 24 h after ANO5 silencing.
Statistical analysis was performed using the Mann-Whitney U test for two-group comparisons. Categorical data were analyzed using Fisher’s exact test. The Kaplan–Meier method was used to construct survival curves, and differences in survival were examined using the log-rank test for equality. Prognostic factors were identified using the Cox proportional hazard model. These analyses were performed using the JMP statistical software (version 15; SAS Institute, Cary, NC, United States). Data are presented in the graphs as the mean ± standard error of the mean. P < 0.05 was considered a statistically significant difference.
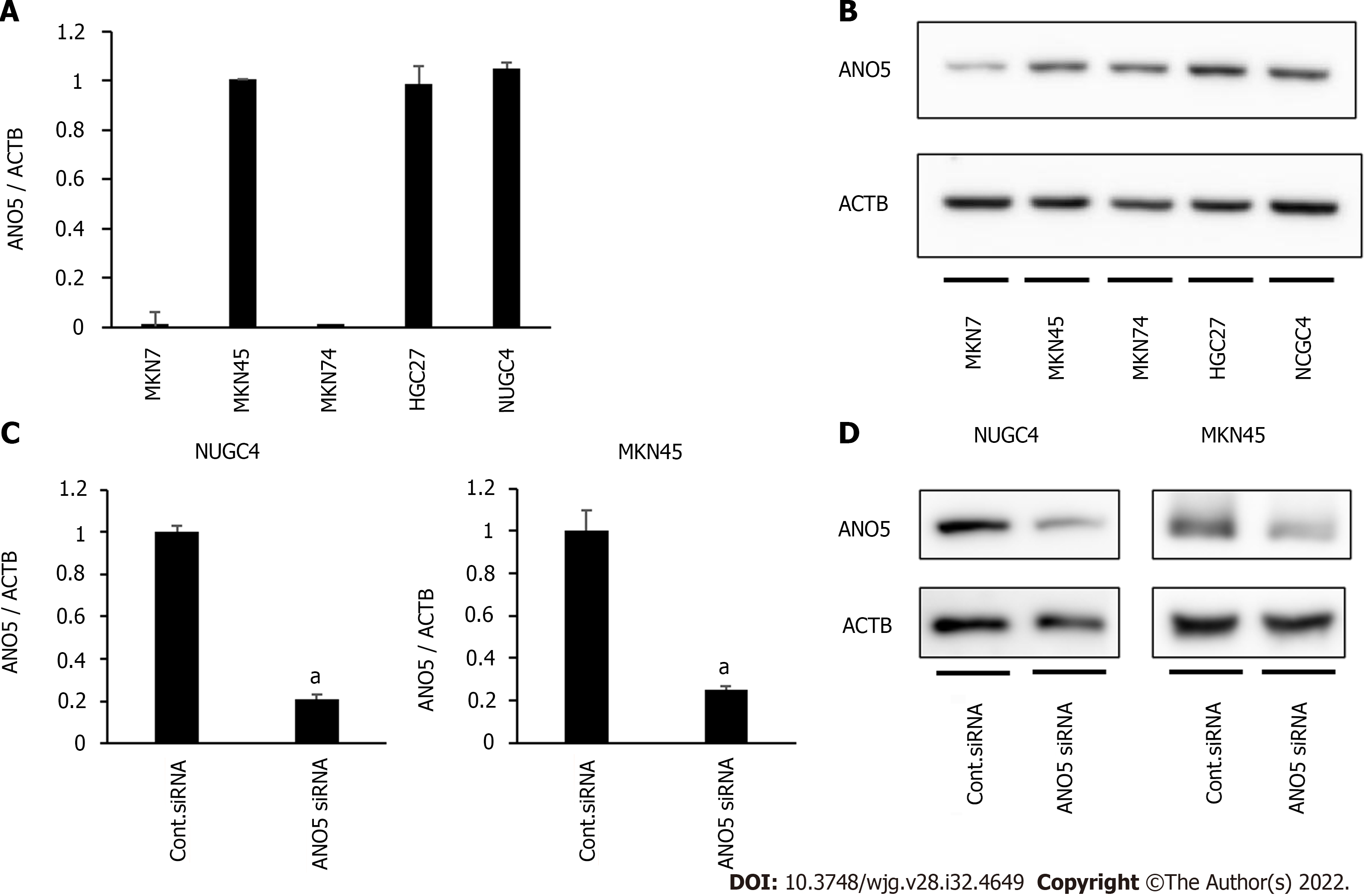
ANO5 gene and protein expression were first examined in five human GC cell lines, MKN7, MKN45, MKN74, HGC27, and NUGC4, by reverse transcription-quantitative PCR and western blotting. ANO5 expression was detected in several cells in the five GC cell lines (Figure 1A and B). Compared to paired adjacent normal tissue, ANO5 expression was significantly upregulated in GC tissue (P = 0.004; n = 12; Supplementary Figure 1).
ANO5 expression was knocked down using siRNA in NUGC4 and MKN45 cells, and its effects on tumor progression were assessed. ANO5 mRNA (Figure 1C) and protein levels (Figure 1D) were downregulated in NUGC4 and MKN45 cells. We also conducted an overexpression study in MKN7 cells. The ANO5 plasmid increased ANO5 mRNA levels (Supplementary Figure 2A, left panel).
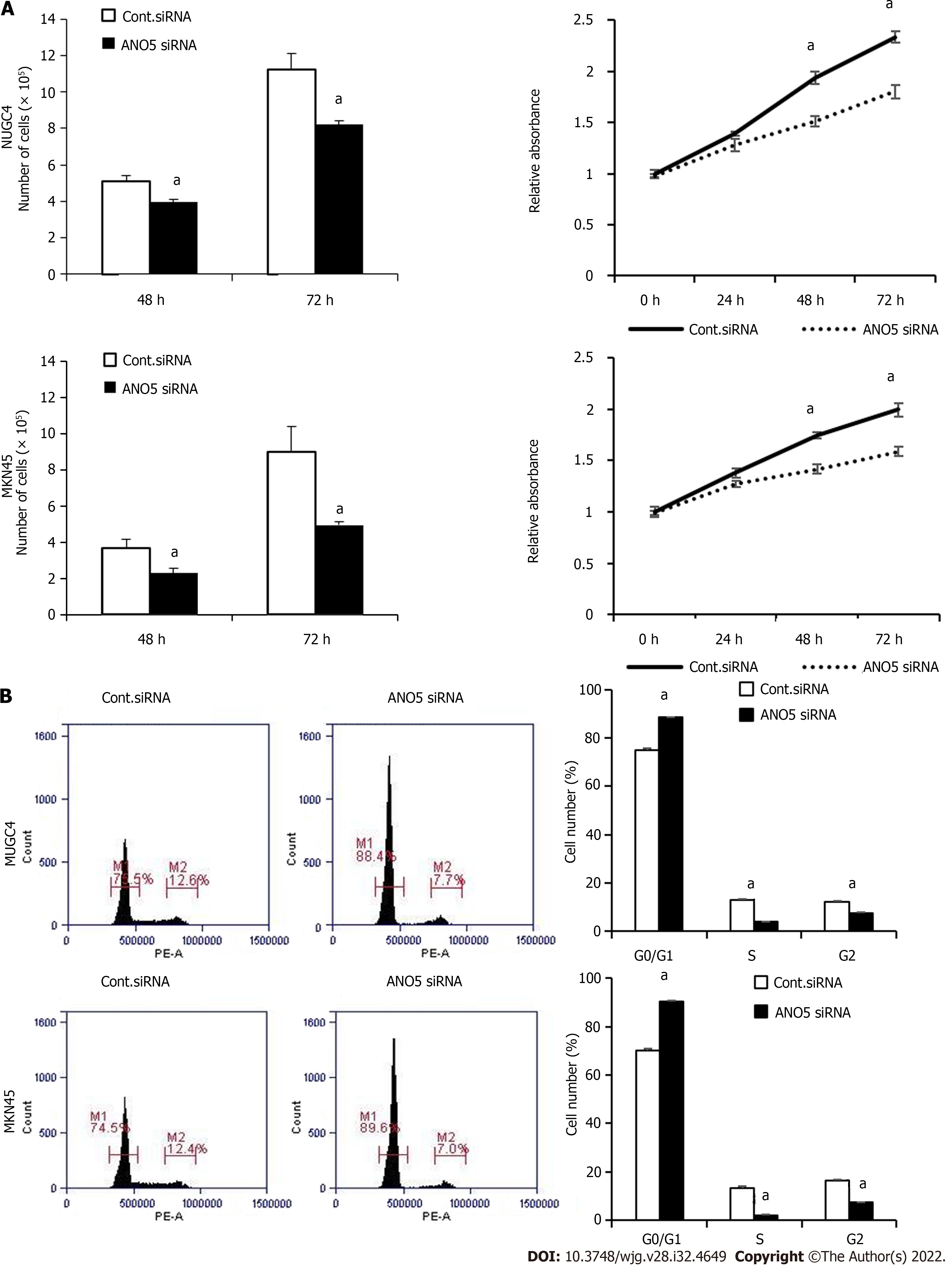
The effect of ANO5 siRNA transfection on the proliferation and cell cycle progression of NUGC4 and MKN45 cells were subsequently examined. Compared with the control siRNA, the number of NUGC4 and MKN45 cells was significantly reduced at 48 h and 72 h after the transfection with ANO5 siRNA (Figure 2A, left panel). The results of the cell proliferation assay showed that the relative absorbance of GC cells transfected with the control siRNA (NUGC4 and MKN45) was significantly lower than that of GC cells transfected with ANO5 siRNA (HSS137119) (NUGC4 and MKN45) (Figure 2A, right panel). Whereas, ANO5 plasmid increased the relative absorbance of MKN7 cell (Supplementary Figure 2B). Moreover, ANO5 silencing increased the numbers of NUGC4 and MKN45 cells in the G0/G1 phase (Figure 2B). These results indicated that ANO5 regulated the proliferation and cell cycle of GC cells.
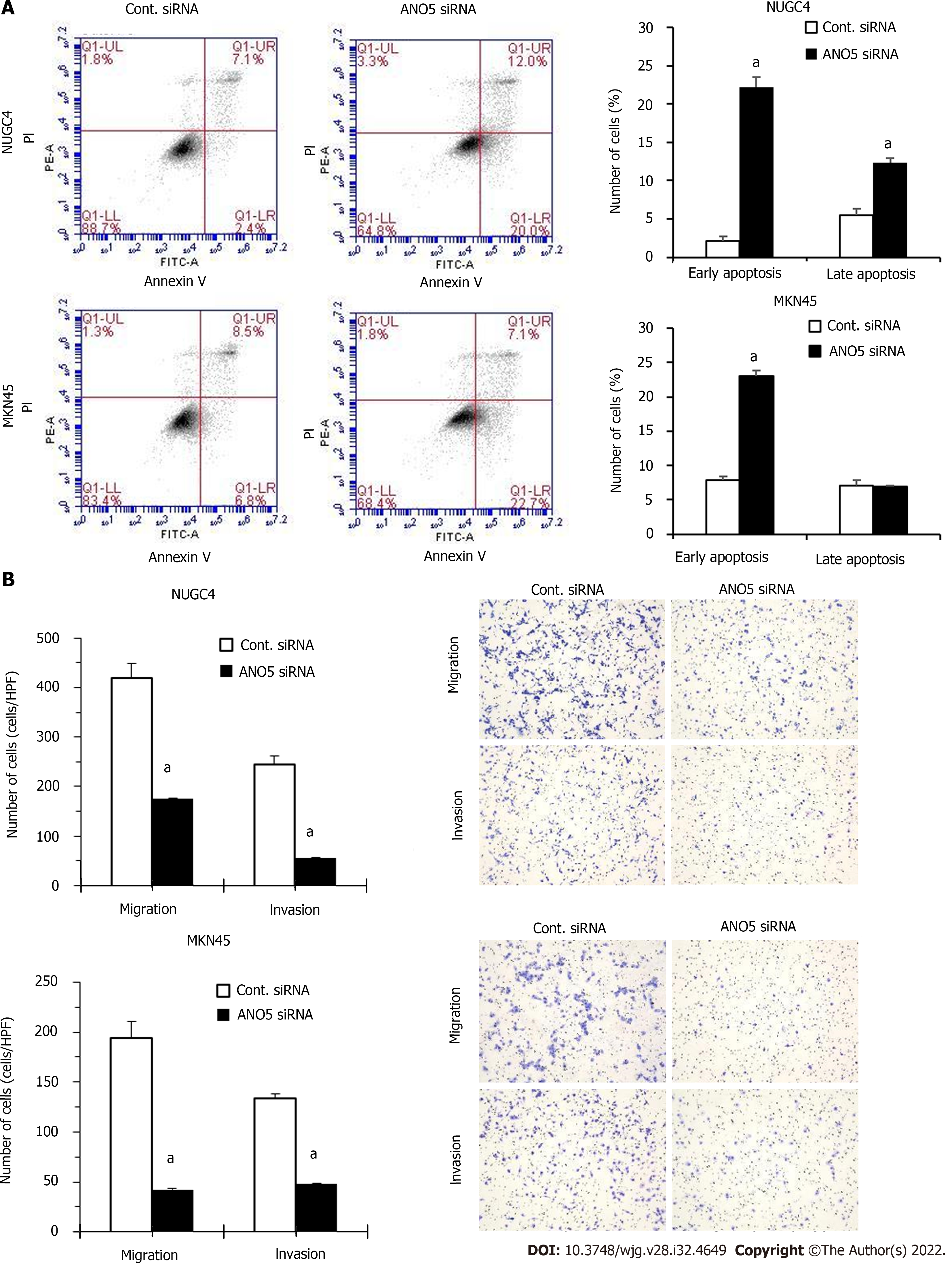
To further clarify the role of ANO5, apoptosis assays were performed in NUGC4 and MKN45 cells. ANO5 silencing significantly increased the frequency of early and late apoptotic NUGC4 cells and the frequency of early apoptotic MKN45 cells 72 h after siRNA transfection (Figure 3A). These results indicated that the apoptosis in NUGC4 and MKN45 cells was regulated by ANO5 expression. Another ANO5 siRNA (HSS137120) was used to assess its impact on cell growth and survival, with results similar to those of HSS137119 (Supplementary Figure 3).
The effects of the ANO5 silencing on NUGC4 and MKN45 cell migration and invasion were examined using a Boyden chamber assay. The results demonstrated that ANO5 knockdown in NUGC4 and MKN45 cells significantly reduced their migration and invasion (Figure 3B).
To elucidate the molecular mechanisms underlying the regulation of cellular functions by ANO5, the gene expression profiles of NUGC4 cells transfected with ANO5 siRNA were investigated using microarray. The results revealed that the expression levels of 3491 genes in NUGC4 cells following ANO5 knockdown exhibited fold-changes > 1.8 compared with those in the negative control. Among these, the expression levels of 1802 genes were upregulated, whereas those of 1689 genes were downregulated in NUGC4 cells following ANO5 knockdown. The top 20 genes with significant changes in expression in the ANO5-depleted NUGC4 cells are listed in Tables 1 and 2. Ingenuity pathway analysis showed that ‘Cancer’ was the top-ranked disease and disorder, while ‘DNA Replication, Recombination, and Repair’ and ‘Cell Cycle’ were the two top-ranking molecular and cellular functions (Table 3).
| Gene symbol | Gene name | Gene ID | Fold change |
| PLCXD1 | Phosphatidylinositol-specific phospholipase C X domain containing 1 | TC0X00006433.hg.1 | 31.97 |
| NDRG4 | NDRG family member 4 | TC1600008034.hg.1 | 29.88 |
| CYP3A5 | Cytochrome P450 family 3 subfamily A member 5 | TC0700011953.hg.1 | 24.81 |
| PLCXD1 | Phosphatidylinositol-specific phospholipase C X domain containing 1 | TC0Y00006433.hg.1 | 23.44 |
| REG4 | Regenerating family member 4 | TC0100015477.hg.1 | 19.70 |
| CIDEB | Cell death-inducing DFFA-like effector b | TC1400008752.hg.1 | 17.86 |
| DHRS9 | Dehydrogenase/reductase 9 | TC0200009905.hg.1 | 17.02 |
| CIDEC | Cell death-inducing DFFA-like effector c | TC0300010217.hg.1 | 16.73 |
| SPRR1A | Small proline rich protein 1A | TC0100010017.hg.1 | 16.66 |
| APOD | Apolipoprotein D | TC0300013645.hg.1 | 16.57 |
| SEMA7A | Semaphorin 7A | TC1500010018.hg.1 | 16.09 |
| C11orf86 | Chromosome 11 open reading frame 86 | TC1100008109.hg.1 | 15.57 |
| BNIPL | BCL2 interacting protein like | TC0100009936.hg.1 | 15.52 |
| SUSD2 | Sushi domain containing 2 | TC2200006883.hg.1 | 15.25 |
| MAPRE3 | Microtubule-associated protein RP/EB family member 3 | TC0200007048.hg.1 | 14.99 |
| CYP1A1 | Cytochrome P450 family 1 subfamily A member 1 | TC1500010042.hg.1 | 14.69 |
| GOLT1A | Golgi transport 1A | TC0100017022.hg.1 | 13.79 |
| CNN2 | Calponin 2 | TC1900006507.hg.1 | 13.74 |
| ANTXR2 | ANTXR cell adhesion molecule 2 | TC0400011144.hg.1 | 13.54 |
| APOBEC1 | Apolipoprotein B mRNA editing enzyme catalytic subunit 1 | TC1200009789.hg.1 | 12.19 |
| Gene symbol | Gene name | Gene ID | Fold change |
| CDK2 | Cyclin-dependent kinase 2 | TC1200007819.hg.1 | -53.84 |
| DTL | Denticleless E3 ubiquitin ligase homolog | TC0100011512.hg.1 | -27.60 |
| RABL3 | RAB, member of RAS oncogene family like 3 | TC0300012157.hg.1 | -20.16 |
| CKS1B | CDC28 protein kinase regulatory submit 1B | TC0100010100.hg.1 | -19.34 |
| LMNB1 | Lamin B1 | TC0500008544.hg.1 | -19.18 |
| IFRD2 | Interferon-related developmental regulator 2 | TC0300013981.hg.1 | -18.61 |
| PLK1 | Polo like kinase 1 | TC1600007235.hg.1 | -17.89 |
| GINS1 | GINS complex subunit 1 | TC2000007016.hg.1 | -17.87 |
| CDK6 | Cyclin-dependent kinase 6 | TC0700011785.hg.1 | -17.08 |
| XRCC2 | X-ray repair cross complementing 2 | TC0700013119.hg.1 | -14.77 |
| POGLUT3 | Protein O-glucosyltransferase 3 | TC1100012229.hg.1 | -14.47 |
| DSG2 | Desmoglein 2 | TC1800007014.hg.1 | -14.37 |
| NEMP1 | Nuclear envelope integral membrane protein 1 | TC1200010946.hg.1 | -14.31 |
| CBX5 | Chromobox 5 | TC1200010833.hg.1 | -14.20 |
| ANKRD52 | Ankyrin repeat domain 52 | TC1200010902.hg.1 | -14.20 |
| ITGB1 | Integrin subunit beta 1 | TC1000010265.hg.1 | -14.16 |
| H2BC14 | H2B clustered histone 14 | TC0600007377.hg.1 | -13.25 |
| SCAMP2 | Secretory carrier membrane protein 2 | TC1500010047.hg.1 | -13.05 |
| CMTM7 | CKLF like MARVEL transmembrane domain containing 7 | TC0300006968.hg.1 | -13.02 |
| GINS4 | GINS complex subunit 4 | TC0800007416.hg.1 | -12.66 |
| Category | Name | Molecules | P value (range) |
| Disease and Disorders | Cancer | 3136 | 1.54 × 10-3–2.23 × 10-15 |
| Neurological Disease | 76 | 1.27 × 10-3–2.23 × 10-15 | |
| Organismal Injury and Abnormalities | 3147 | 1.54 × 10-3–2.23 × 10-15 | |
| Cardiovascular Disease | 138 | 1.43 × 10-3–4.07 × 10-14 | |
| Reproductive System Disease | 1942 | 1.54 × 10-3–1.09 × 10-10 | |
| Molecular and Cellular Functions | DNA Replication, Recombination, and Repair | 460 | 1.54 × 10-3–9.69 × 10-22 |
| Cell Cycle | 650 | 1.54 × 10-3–1.98 × 10-20 | |
| Cellular Assembly and Organization | 533 | 1.57 × 10-3–9.22 × 10-16 | |
| Cellular Development | 691 | 1.59 × 10-3–2.66 × 10-14 | |
| Cellular Growth and Proliferation | 677 | 1.40 × 10-3–2.66 × 10-14 | |
| Physiological System Development and Function | Connective Tissue Development and Function | 60 | 1.14 × 10-3–8.83 × 10-6 |
| Tissue Development | 164 | 1.57 × 10-3–1.11 × 10-4 | |
| Embryonic Development | 80 | 1.21 × 10-3–2.07 × 10-4 | |
| Hair and Skin Development and Function | 55 | 6.37 × 10-4–2.07 × 10-4 | |
| Organ Development | 69 | 6.37 × 10-4–2.07 × 10-4 |
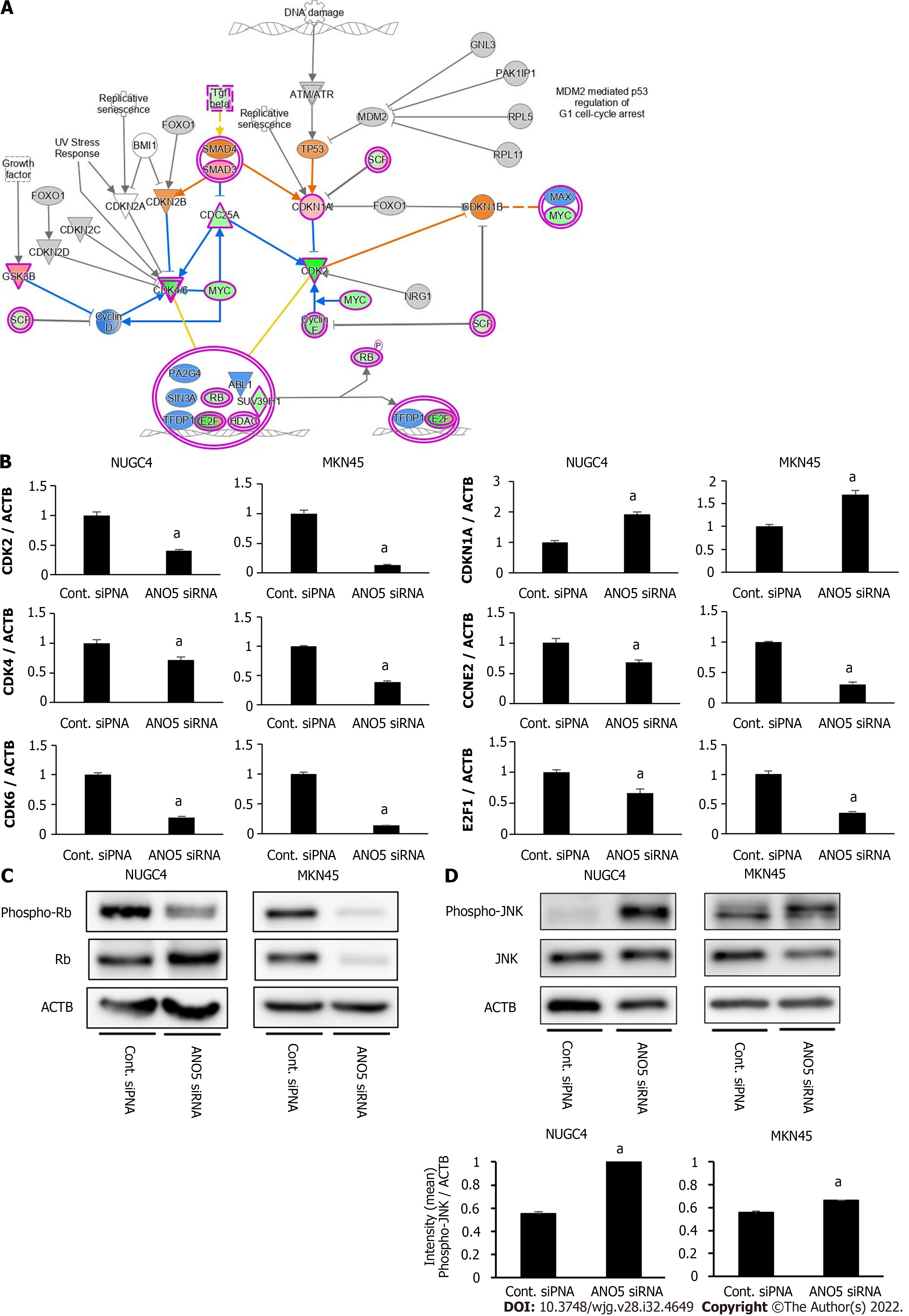
The microarray analysis identified ‘Cell Cycle: G1/S Checkpoint Regulation’ as one of the top-ranking canonical pathways in ANO5-depleted NUGC4 cells (Figure 4A). To confirm these results, seven genes were selected (CDK2, CDK4, CDK6, CDKN1A/p21, CCNE2, E2F1, and Rb). These genes were included in ‘Cell Cycle: G1/S Checkpoint Regulation,’ and CDK2 and CDK6 were the two top-ranking downregulated genes in NUGC4 cells following ANO5 knockdown (Table 2). Reverse transcription-quantitative PCR was used to confirm the expression levels of six genes. NUGC4 and MKN45 cells transfected with ANO5 siRNA had significantly lower CDK2, CDK4, CDK6, CDKN1A, CCNE2, and E2F1 expression levels and significantly higher CDKN1A expression levels than cells transfected with the control siRNA (Figure 4B). Furthermore, ANO5 plasmid decreased CDKN1A/p21 mRNA levels (Supplemen
In addition, the gene expression profile of MKN45 cells transfected with ANO5 siRNA was investigated by microarray. Changes in gene expression in ANO5-depleted NUGC4 and MKN45 cells are depicted in Supplementary Figure 4. Among the 21440 genes, 7246 genes were upregulated and 6622 genes were downregulated in both cell lines, for a total of 13868 genes (64.7%) with identical expression direction in NUGC4 and MKN45 cells. The direction of gene expression changes of gene related to ‘Cell Cycle: G1/S Checkpoint Regulation’ was consistent in both cell lines (Supplementary Table 1). Furthermore, all 40 genes displayed in Tables 1 and 2 showed the identical expression patterns in ANO5-depleted MKN45 cells (Supplementary Table 2). These results supported that ANO5 affected the cell cycle through similar mechanisms in both NUGC4 and MKN45 cell lines.
Activation of the JNK and p38 MAPK classes of protein kinases mediates cellular responses such as apoptosis and the maturation of some cell types. JNK stabilizes p21 protein through phosphorylation[25]. To elucidate the regulatory role of ANO5 in the JNK signaling pathway in GC cells, we examined the phosphorylation of the JNK protein. ANO5 silencing significantly increased JNK phosphorylation levels in NUGC4 and MKN45 cells (Figure 4D). Furthermore, the increase of CDKN1A/p21 mRNA expression induced by ANO5 silencing in NUGC4 and MKN45 cells was suppressed by JNK inhibition (Supplementary Figure 5, lower panel). Whereas, treatment with JNK inhibitors did not affect ANO5 mRNA expression (Supplementary Figure 5, upper panel). These results indicated that ANO5 expression regulated the cell cycle by upregulating p21 via JNK cascade activation in GC cells.
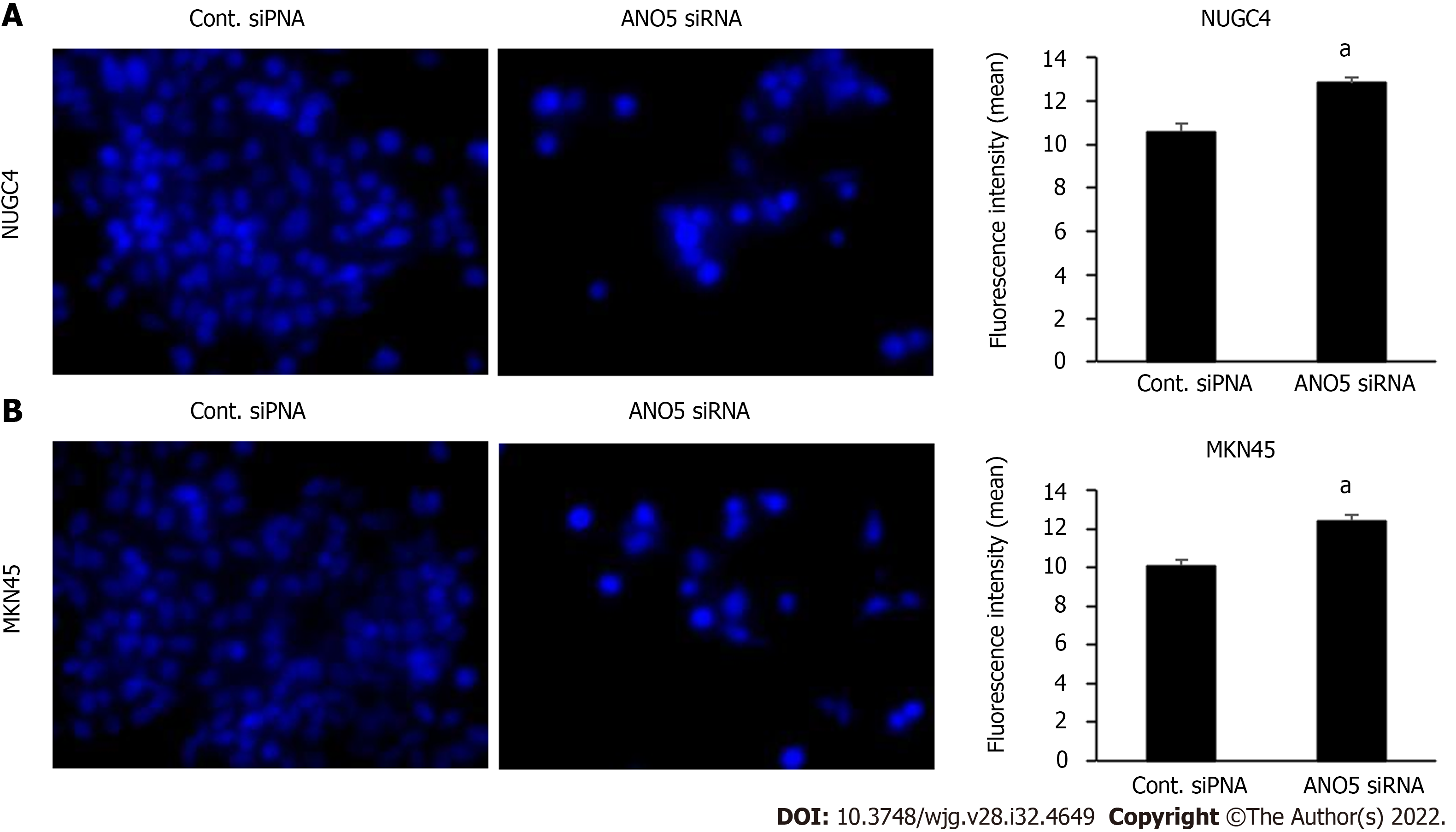
To elucidate the molecular mechanisms through which ANO5 affects the cell cycle transition from G1 to S phase, changes in the intracellular ion environment were examined. A previous study has reported that intracellular chloride affects cancer growth through the phosphorylation of several key molecules in signal transduction pathways[20]. We previously reported that the culturing in a Cl--replaced medium (replacement of Cl- by NO3-) decreased the intracellular chloride concentration [(Cl-)i] and inhibited cell growth in GC cells[19]. Our previous study also demonstrated that JNK activation under low-chloride conditions inhibited GC cell growth by upregulating p21 expression[18]. Intracellular chloride concentrations in the cells were measured based on the fluorescence intensity of MQAE, a chloride-sensitive fluorescence probe. The results revealed an increases in the fluorescence intensity of MQAE in NUGC4 and MKN45 cells following ANO5 knockdown (Figure 5). Therefore, ANO5 knockdown altered intracellular chloride concentrations in GC cells. Furthermore, low-chloride conditions effectively increased JNK phosphorylation and reduced Rb phosphorylation (Supplemen
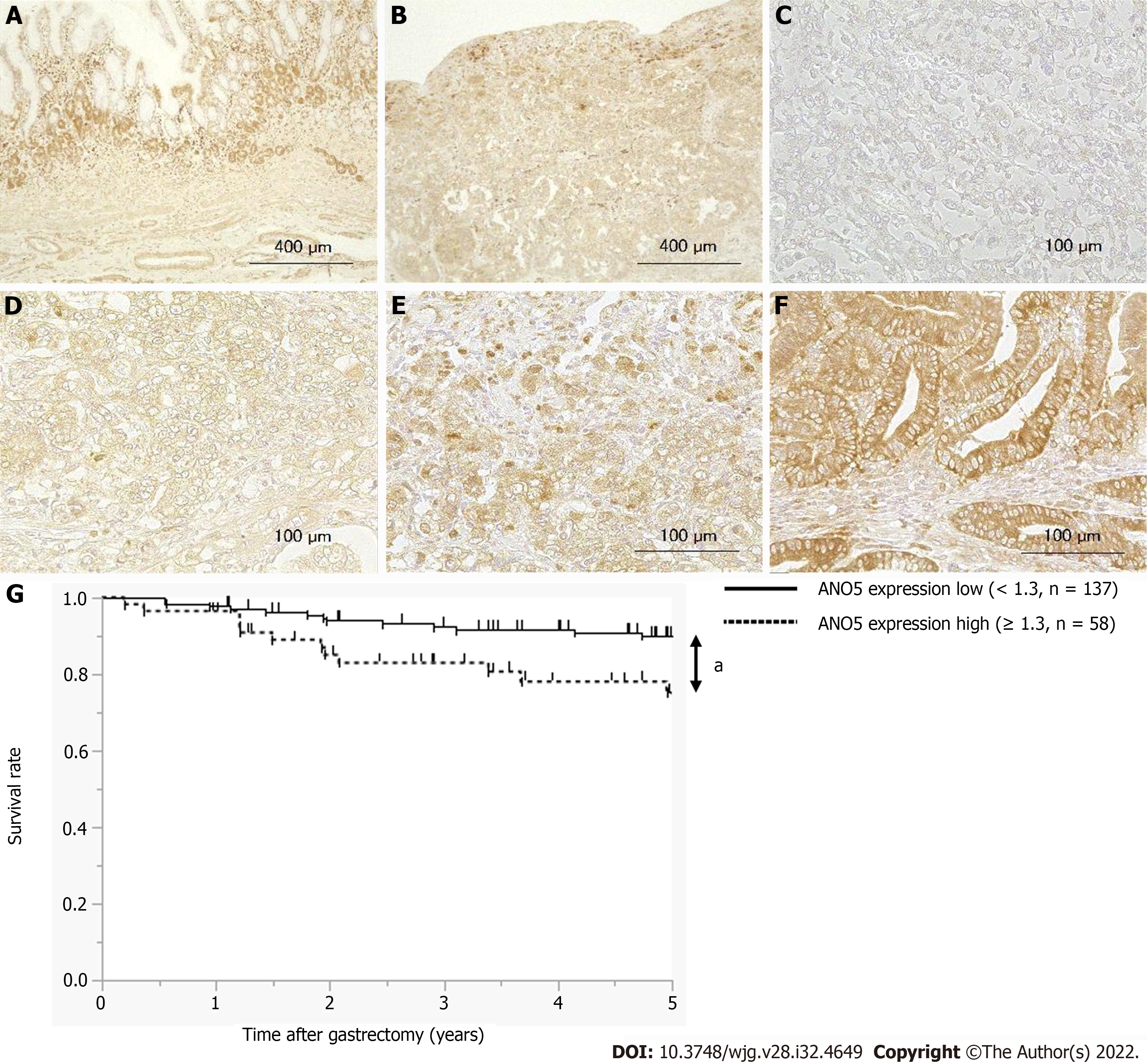
IHC detected the expression of ANO5 in non-cancerous gastric (Figure 6A) and cancerous epithelia (Figure 6B). ANO5 was observed to be expressed in the cell membranes and cytoplasm of GC tissue. The criteria for the staining intensity score were defined as 0 (no staining; Figure 6C), 1 (weak staining; Figure 6D), 2 (moderate staining; Figure 6E), or 3 (strong staining; Figure 6F). The median and mean scores for ANO5 expression were 0.9 (range, 0-2.1) and 0.97 (standard deviation = 0.53), respectively. A cut-off value of 1.3 was used to obtain the smallest P value in comparison of 5-year overall survival (OS) rates[26]. The 5-year OS rates for each cut-off value are presented in Table 4.
Patients with GC were divided into low- (ANO5 scores < 1.3, n = 137) and high-ANO5 (ANO5 scores ≥ 1.3, n = 58) expression groups based on a cut-off value of 1.3 (Figure 6G). Analysis of the clinicopathological features revealed that ANO5 expression levels were not associated with any of the variables (Table 5). To evaluate the prognostic significance of ANO5 after surgery, the following ten variables were compared: sex, age, tumor location, tumor length, histological type, lymphatic invasion, venous invasion, the pathological T stage, pathological N stage, and ANO5 IHC scores. Univariate analysis showed that patient prognosis was correlated with tumor length, lymphatic invasion, venous invasion, pathological T stage, pathological N stage, and ANO5 IHC scores (P = 0.0020, 0.0002, 0.0126, < 0.0001, < 0.0001, and 0.0104, respectively). Multivariate analysis identified high ANO5 expression (≥ 1.3) as an independent prognostic factor (P = 0.0457) (Table 6). Furthermore, the 5-year OS rate was significantly lower in the high expression group (73.9%) than in the low expression group (89.6%). Data obtained from the Kaplan-Meier plotter database also indicated that high ANO5 expression correlates with poor prognosis in GC (Supplementary Figure 7), which was consistent with the present results.
| Variables | N | IHC score | P value | ||
| High group ( 1.3) | Low group (< 1.3) | ||||
| Total | 195 | 58 | 137 | ||
| Sex | Male | 128 | 35 | 93 | 0.3262 |
| Female | 67 | 23 | 44 | ||
| Age | < 65 | 77 | 25 | 52 | 0.5246 |
| 65 | 118 | 33 | 85 | ||
| Tumor location | U | 41 | 15 | 26 | 0.3367 |
| M, L | 154 | 43 | 111 | ||
| Tumor length (mm) | < 30 | 63 | 22 | 41 | 0.3158 |
| 30 | 132 | 36 | 96 | ||
| Histological type | tub1, tub2, pap | 96 | 35 | 61 | 0.0596 |
| por, sig, muc | 99 | 23 | 76 | ||
| Lymphatic invasion | Negative | 110 | 34 | 76 | 0.7529 |
| Positive | 85 | 24 | 61 | ||
| Venous invasion | Negative | 131 | 38 | 93 | 0.7419 |
| Positive | 64 | 20 | 44 | ||
| pT | pT1-2 | 137 | 37 | 100 | 0.2312 |
| pT3-4 | 58 | 21 | 37 | ||
| pN | pN0 | 139 | 40 | 99 | 0.7294 |
| pN1-3 | 56 | 18 | 38 | ||
| Variables | N | Univariate | Multivariate | ||
| 5-yr OS rate (%) | P value | HR (95%CI) | P value | ||
| Total | 195 | ||||
| Sex | |||||
| Male | 128 | 87.9 | 0.3274 | ||
| Female | 67 | 85.3 | |||
| Age | |||||
| < 65 | 77 | 89.7 | 0.4113 | ||
| 65 | 118 | 85.2 | |||
| Location | |||||
| U | 41 | 80.9 | 0.2629 | ||
| M, L | 154 | 88.6 | |||
| Tumor length (mm) | |||||
| < 30 | 63 | 97.8 | 0.0020a | 3.010 (0.347-26.12) | 0.3176 |
| 30 | 132 | 80.1 | |||
| Histological type | |||||
| tub1, tub2, pap | 96 | 88.3 | 0.4544 | ||
| por, sig, muc | 99 | 85.6 | |||
| Lymphatic invasion | |||||
| Negative | 110 | 96.1 | 0.0002a | 1.872 (0.549-6.384) | 0.3166 |
| Positive | 85 | 75.7 | |||
| Venous invasion | |||||
| Negative | 131 | 90.6 | 0.0126a | 1.085 (0.476-2.472) | 0.8469 |
| Positive | 64 | 79.3 | |||
| pT | |||||
| pT1-2 | 137 | 95.9 | < 0.0001a | 5.240 (1.807-15.20) | 0.0023c |
| pT3-4 | 58 | 65.5 | |||
| pN | |||||
| pN0 | 139 | 94.1 | < 0.0001a | 2.148 (0.695-6.643) | 0.1844 |
| pN1-3 | 56 | 67.6 | |||
| IHC score | |||||
| < 1.3 | 137 | 89.6 | 0.0104a | 2.318 (1.016-5.288) | 0.0457c |
| 1.3 | 58 | 73.9 | |||
The ANO family of membrane proteins, also known as TMEM16, play key roles in several physiological functions, including ion transport to phospholipid scrambling[27] and ion channel regulation[28]. While the roles of ANO1 (TMEM16A) and ANO2 (TMEM16B) as calcium-activated chloride channels have been firmly established[29-32], the functions of other family members remain unclear.
Previous studies have evaluated the expression and role of ANO5 during tumor development in various cancer types. Song et al[33] demonstrated that ANO5 (TMEM16E) was widely expressed in the epithelial cells of the human gastrointestinal tract. ANO5 is also expressed in human pancreatic cancer tissues but not in normal pancreatic tissue[23]. Chang et al[22] reported that ANO5 expression was downregulated in thyroid cancer, which promoted thyroid cancer cell migration and invasion. However, the expression of ANO5 in human GC tissue and the pathophysiological role of its expression in GC cells have not been demonstrated.
The present study revealed that ANO5 downregulation in GC cells regulates the cell cycle and induces apoptosis, while inhibiting proliferation, migration, and invasion. These results highlight the potential of ANO5 inhibitors as therapeutic agents for the treatment of GC or other cancer types with high ANO5 expression levels. The present study also indicates that ANO5 plays a key role in the proliferation of GC cells. Cell cycle analysis showed that the number of cells remaining in the G0/G1 phase was significantly increased, whereas the number of cells in the S or G2/M phase was decreased in ANO5-depleted NUGC4 and MKN45 cells, suggesting that ANO5 downregulation inhibits GC cell proliferation via cell cycle arrest at the G0/G1 phase.
The induction of p21 is dependent on the tumor suppressor protein, p53. However, chloride ions have been shown to play important roles in cell cycle progression by regulating the expression of p21 through a p53-independent pathway in GC cells[19]. It has also been demonstrated that a decrease in chloride induced G0/G1 phase arrest by downregulating CDK2 and phosphorylated Rb expression through p21 upregulation. Furthermore, p38 and JNK activation under low-chloride conditions inhibits GC cell viability by upregulating p21 expression[18].
Moreover, ANO5 expression in GC cells affects the transition from the G1 to the S phase of the cell cycle by regulating the expression of p21 and its downstream genes through the activation of JNK signaling. To the best of our knowledge, the chloride channel activity of ANO5 has not been confirmed to date. To elucidate the molecular mechanisms underlying the effects of ANO5, we evaluated the changes in the intracellular chloride ion environment in this study. A quantitative analysis of intracellular chloride ion concentrations was conducted based on the fluorescence intensity of MQAE. Immunofluorescent analysis showed that the fluorescence intensity of MQAE increased following ANO5 silencing, indicating a decrease in intracellular chloride concentration. These results suggest that the downregulation of ANO5 induces G0/G1 phase arrest by altering the expression of G1/S checkpoint-related genes through the intracellular chloride environment of GC cells. Furthermore, since ANO5 functioned as a chloride channel, it may have the potential to inhibit tumor growth by regulating intracellular chloride concentrations in therapeutic settings.
Although ANO5 was recently implicated in various cancers, its role in tumor progression in patients with GC remains unclear. To demonstrate the clinical significance of ANO5 expression, the survival rate of 195 patients who underwent curative resection for primary GC was investigated. IHC analysis revealed that high ANO5 expression levels were a poor prognostic factor in patients with GC. Under low-chloride conditions, ANO5 appeared to function as a chloride channel in GC cells. Previous findings showed that various ion transporters function as biomarkers and therapeutic targets[34,35]. Targeting ion channels that are activated in cancer cells may be an important strategy for cancer therapy. To the best of our knowledge, the present study is the first to report a relationship between ANO5 expression and the prognosis of patients with GC. Additional functional studies are needed to provide insights into the role of ANO5 in GC progression.
This study has some limitations that must be addressed. First, it was a retrospective study. Due to the limited sample size, the pathological N stage classification factors were not correlated with the 5-year OS rate. Therefore, further studies are required to confirm our results. Second, in the selection of GC cell lines, we selected five cell lines; however, only three were used in the study.
The present study revealed that ANO5 plays a significant role in cell cycle progression in human GC cells. The results of microarray analysis showed the impact of ANO5 on the expression of G1/S checkpoint-related genes. Furthermore, ANO5 expression significantly affected JNK signaling. Collectively, these results indicate that ANO5 plays an important role in cell cycle progression by regulating the expression of p21 through JNK signaling in human GC cells. The results of the IHC analysis also suggest that high ANO5 expression levels are a poor prognostic factor in patients with GC. The present study may contribute to the identification of ANO5 as a key mediator of tumor progression, with it ultimately being a promising prognostic biomarker or a novel therapeutic target for GC.
Anoctamin 5 (ANO5) is a member of a family of calcium-activated chloride channels containing 10 members, also known as transmembrane proteins, and has been reported to be associated with various cancers.
The role of ANO5 in gastric cancer (GC) remains poorly understood. In the present study, we analyzed the relationship between ANO5 expression and tumor progression in GC.
The objectives of the present study were to investigate whether ANO5 contributes to the regulation of cancer growth and to clarify its clinicopathological significance in GC.
Knockdown experiments were performed by transfecting human GC cell lines with ANO5 small interfering RNA. Gene expression was then assessed using microarray analysis. Samples from 195 patients with GC were subjected to immunohistochemistry for ANO5, and its relationship with clinicopathological factors and prognosis were examined.
ANO5 knockdown suppressed the proliferation, migration, and invasion of cells and enhanced apoptosis. Cell cycle analysis showed that ANO5 knockdown suppressed the progression of G1-S phase. The results of microarray analysis showed up- or downregulated expression of genes related to “Cell Cycle: G1/S Checkpoint Regulation” in ANO5 knockdown NUGC4 cells. Survival analysis showed significantly poorer 5-year survival in the ANO5 high expression group (high vs low; 73.9 vs 89.6%, P = 0.0104). Immunohistochemistry multivariate analysis identified the high expression of ANO5 as an independent prognostic factor for 5-year survival in GC patients (P = 0.0457).
ANO5 plays a significant role in cell cycle progression in human GC cells. The results of the immunohistochemistry analysis suggest that high ANO5 expression levels are a poor prognostic factor in patients with GC.
The present study may contribute to the identification of ANO5 as a key mediator in tumor progression, with it ultimately being a promising prognostic biomarker or a novel therapeutic target of GC.
| 1. | Pang C, Yuan H, Ren S, Chen Y, An H, Zhan Y. TMEM16A/B associated CaCC: structural and functional insights. Protein Pept Lett. 2014;21:94-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 2. | Pedemonte N, Galietta LJ. Structure and function of TMEM16 proteins (anoctamins). Physiol Rev. 2014;94:419-459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 417] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 3. | Whitlock JM, Hartzell HC. Anoctamins/TMEM16 Proteins: Chloride Channels Flirting with Lipids and Extracellular Vesicles. Annu Rev Physiol. 2017;79:119-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 141] [Article Influence: 14.1] [Reference Citation Analysis (1)] |
| 4. | Schreiber R, Uliyakina I, Kongsuphol P, Warth R, Mirza M, Martins JR, Kunzelmann K. Expression and function of epithelial anoctamins. J Biol Chem. 2010;285:7838-7845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 185] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 5. | Kunzelmann K. TMEM16, LRRC8A, bestrophin: chloride channels controlled by Ca(2+) and cell volume. Trends Biochem Sci. 2015;40:535-543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 61] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 6. | Wanitchakool P, Wolf L, Koehl GE, Sirianant L, Schreiber R, Kulkarni S, Duvvuri U, Kunzelmann K. Role of anoctamins in cancer and apoptosis. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130096. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 90] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 7. | Miettinen M. Immunohistochemistry of soft tissue tumours - review with emphasis on 10 markers. Histopathology. 2014;64:101-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 88] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 8. | Lu G, Shi W, Zheng H. Inhibition of STAT6/Anoctamin-1 Activation Suppresses Proliferation and Invasion of Gastric Cancer Cells. Cancer Biother Radiopharm. 2018;33:3-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 9. | Shang L, Hao JJ, Zhao XK, He JZ, Shi ZZ, Liu HJ, Wu LF, Jiang YY, Shi F, Yang H, Zhang Y, Liu YZ, Zhang TT, Xu X, Cai Y, Jia XM, Li M, Zhan QM, Li EM, Wang LD, Wei WQ, Wang MR. ANO1 protein as a potential biomarker for esophageal cancer prognosis and precancerous lesion development prediction. Oncotarget. 2016;7:24374-24382. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 10. | Jia L, Liu W, Guan L, Lu M, Wang K. Inhibition of Calcium-Activated Chloride Channel ANO1/TMEM16A Suppresses Tumor Growth and Invasion in Human Lung Cancer. PLoS One. 2015;10:e0136584. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 102] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 11. | Bill A, Gutierrez A, Kulkarni S, Kemp C, Bonenfant D, Voshol H, Duvvuri U, Gaither LA. ANO1/TMEM16A interacts with EGFR and correlates with sensitivity to EGFR-targeting therapy in head and neck cancer. Oncotarget. 2015;6:9173-9188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 93] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 12. | Britschgi A, Bill A, Brinkhaus H, Rothwell C, Clay I, Duss S, Rebhan M, Raman P, Guy CT, Wetzel K, George E, Popa MO, Lilley S, Choudhury H, Gosling M, Wang L, Fitzgerald S, Borawski J, Baffoe J, Labow M, Gaither LA, Bentires-Alj M. Calcium-activated chloride channel ANO1 promotes breast cancer progression by activating EGFR and CAMK signaling. Proc Natl Acad Sci U S A. 2013;110:E1026-E1034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 272] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 13. | Duran C, Hartzell HC. Physiological roles and diseases of Tmem16/Anoctamin proteins: are they all chloride channels? Acta Pharmacol Sin. 2011;32:685-692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 112] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 14. | Dutertre M, Lacroix-Triki M, Driouch K, de la Grange P, Gratadou L, Beck S, Millevoi S, Tazi J, Lidereau R, Vagner S, Auboeuf D. Exon-based clustering of murine breast tumor transcriptomes reveals alternative exons whose expression is associated with metastasis. Cancer Res. 2010;70:896-905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 54] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 15. | Li Y, Wang X, Vural S, Mishra NK, Cowan KH, Guda C. Exome analysis reveals differentially mutated gene signatures of stage, grade and subtype in breast cancers. PLoS One. 2015;10:e0119383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 16. | Jun I, Park HS, Piao H, Han JW, An MJ, Yun BG, Zhang X, Cha YH, Shin YK, Yook JI, Jung J, Gee HY, Park JS, Yoon DS, Jeung HC, Lee MG. ANO9/TMEM16J promotes tumourigenesis via EGFR and is a novel therapeutic target for pancreatic cancer. Br J Cancer. 2017;117:1798-1809. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 17. | Li C, Cai S, Wang X, Jiang Z. Identification and characterization of ANO9 in stage II and III colorectal carcinoma. Oncotarget. 2015;6:29324-29334. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 18. | Shiozaki A, Otsuji E, Marunaka Y. Intracellular chloride regulates the G(1)/S cell cycle progression in gastric cancer cells. World J Gastrointest Oncol. 2011;3:119-122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 31] [Cited by in RCA: 39] [Article Influence: 2.6] [Reference Citation Analysis (2)] |
| 19. | Miyazaki H, Shiozaki A, Niisato N, Ohsawa R, Itoi H, Ueda Y, Otsuji E, Yamagishi H, Iwasaki Y, Nakano T, Nakahari T, Marunaka Y. Chloride ions control the G1/S cell-cycle checkpoint by regulating the expression of p21 through a p53-independent pathway in human gastric cancer cells. Biochem Biophys Res Commun. 2008;366:506-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 60] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 20. | Shiozaki A, Miyazaki H, Niisato N, Nakahari T, Iwasaki Y, Itoi H, Ueda Y, Yamagishi H, Marunaka Y. Furosemide, a blocker of Na+/K+/2Cl- cotransporter, diminishes proliferation of poorly differentiated human gastric cancer cells by affecting G0/G1 state. J Physiol Sci. 2006;56:401-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 58] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 21. | Kurashima K, Shiozaki A, Kudou M, Shimizu H, Arita T, Kosuga T, Konishi H, Komatsu S, Kubota T, Fujiwara H, Okamoto K, Kishimoto M, Konishi E, Otsuji E. LRRC8A influences the growth of gastric cancer cells via the p53 signaling pathway. Gastric Cancer. 2021;24:1063-1075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 22. | Chang Z, Cai C, Han D, Gao Y, Li Q, Feng L, Zhang W, Zheng J, Jin J, Zhang H, Wei Q. Anoctamin5 regulates cell migration and invasion in thyroid cancer. Int J Oncol. 2017;51:1311-1319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 23. | Song HY, Zhou L, Hou XF, Lian H. Anoctamin 5 regulates cell proliferation and migration in pancreatic cancer. Int J Clin Exp Pathol. 2019;12:4263-4270. [PubMed] |
| 24. | James DB, Mary KG, Christian W. International Union Against Cancer (UICC) TNM classification of malignant tumors. 8th edition. Wiley: New York, 2017. |
| 25. | Kim GY, Mercer SE, Ewton DZ, Yan Z, Jin K, Friedman E. The stress-activated protein kinases p38 alpha and JNK1 stabilize p21(Cip1) by phosphorylation. J Biol Chem. 2002;277:29792-29802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 188] [Article Influence: 7.8] [Reference Citation Analysis (2)] |
| 26. | Mazumdar M, Glassman JR. Categorizing a prognostic variable: review of methods, code for easy implementation and applications to decision-making about cancer treatments. Stat Med. 2000;19:113-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 27. | Suzuki J, Umeda M, Sims PJ, Nagata S. Calcium-dependent phospholipid scrambling by TMEM16F. Nature. 2010;468:834-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 731] [Cited by in RCA: 768] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 28. | Huang F, Wang X, Ostertag EM, Nuwal T, Huang B, Jan YN, Basbaum AI, Jan LY. TMEM16C facilitates Na(+)-activated K+ currents in rat sensory neurons and regulates pain processing. Nat Neurosci. 2013;16:1284-1290. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 108] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 29. | Caputo A, Caci E, Ferrera L, Pedemonte N, Barsanti C, Sondo E, Pfeffer U, Ravazzolo R, Zegarra-Moran O, Galietta LJ. TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science. 2008;322:590-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1104] [Cited by in RCA: 1049] [Article Influence: 58.3] [Reference Citation Analysis (8)] |
| 30. | Schroeder BC, Cheng T, Jan YN, Jan LY. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell. 2008;134:1019-1029. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1015] [Cited by in RCA: 986] [Article Influence: 54.8] [Reference Citation Analysis (0)] |
| 31. | Yang YD, Cho H, Koo JY, Tak MH, Cho Y, Shim WS, Park SP, Lee J, Lee B, Kim BM, Raouf R, Shin YK, Oh U. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature. 2008;455:1210-1215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 977] [Cited by in RCA: 1088] [Article Influence: 60.4] [Reference Citation Analysis (0)] |
| 32. | Terashima H, Picollo A, Accardi A. Purified TMEM16A is sufficient to form Ca2+-activated Cl- channels. Proc Natl Acad Sci U S A. 2013;110:19354-19359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 91] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 33. | Song HY, Tian YM, Zhang YM, Zhou L, Lian H, Zhu JX. A novel finding of anoctamin 5 expression in the rodent gastrointestinal tract. Biochem Biophys Res Commun. 2014;451:258-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 34. | Siveen KS, Nizamuddin PB, Uddin S, Al-Thani M, Frenneaux MP, Janahi IA, Steinhoff M, Azizi F. TRPV2: A Cancer Biomarker and Potential Therapeutic Target. Dis Markers. 2020;2020:8892312. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 35. | Xu R, Wang X, Shi C. Volume-regulated anion channel as a novel cancer therapeutic target. Int J Biol Macromol. 2020;159:570-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Japan
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gao W, China; Gao JP, China; Zhu Z, China S-Editor: Fan JR L-Editor: Filipodia P-Editor: Cai YX