Published online Jun 14, 2022. doi: 10.3748/wjg.v28.i22.2437
Peer-review started: December 8, 2021
First decision: March 11, 2022
Revised: March 24, 2022
Accepted: April 27, 2022
Article in press: April 27, 2022
Published online: June 14, 2022
Processing time: 184 Days and 2.1 Hours
Gastric carcinoma (GC) is a common gastrointestinal malignancy worldwide. Based on the cancer-related mortality, the current prevention and treatment strategies for GC still show poor clinical results. Therefore, it is important to find effective drug treatment targets.
To explore the mechanism by which 18β-glycyrrhetinic acid (18β-GRA) regulates mitochondrial ribosomal protein L35 (MRPL35) related signal proteins to inhibit the proliferation of GC cells.
Cell counting kit-8 assay was used to detect the effects of 18β-GRA on the survival rate of human normal gastric mucosal cell line GES-1 and the proliferation of GC cell lines MGC80-3 and BGC-823. The apoptosis and cell cycle were assessed by flow cytometry. Cell invasion and migration were evaluated by Transwell assay, and cell scratch test was used to detect cell migration. Furthermore, a tumor model was established by hypodermic injection of 2.5 × 106 BGC-823 cells at the selected positions of BALB/c nude mice to determine the effect of 18β-GRA on GC cell proliferation, and quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was used to detect MRPL35 expression in the engrafted tumors in mice. We used the term tandem mass tag (TMT) labeling combined with liquid chromatography–tandem mass spectrometry to screen for differentially expressed proteins (DEPs) extracted from GC cells and control cells after 18β-GRA intervention. A detailed bioinformatics analysis of these DEPs was performed, including Gene Ontology annotation and enrichment analysis, Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis, and so on. Moreover, STRING database (https://string-db.org/) was used to predict protein-protein interaction (PPI) relationships and Western blot was used to detect the expression of proteins of interest in GC cells.
The results indicated that 18β-GRA could inhibit the proliferation of GC cells in a dose- and time-dependent manner. It could induce GC cell apoptosis and arrest the cell cycle at G0/G1 phase. The proportion of cells arrested at S phase decreased with the increase of 18-GRA dose, and the migration and invasiveness of GC cells were inhibited. The results of animal experiments showed that 18β-GRA could inhibit tumor formation in BALB/c nude mice, and qRT-PCR results showed that MRPL35 expression level was significantly reduced in the engrafted tumors in mice. Using TMT technology, 609 DEPs, among which 335 were up-regulated and 274 were down-regulated, were identified in 18β-GRA intervention compared with control. We found that the intervention of 18β-GRA in GC cells involved many important biological processes and signaling pathways, such as cellular processes, biological regulation, and TP53 signaling pathway. Notably, after the drug intervention, MRPL35 expression was significantly down-regulated (P = 0.000247), TP53 expression was up-regulated (P = 0.02676), and BCL2L1 was down-regulated (P = 0.01699). Combined with the Retrieval of Interacting Genes/Proteins database, we analyzed the relationship between MRPL35, TP53, and BCL2L1 signaling proteins, and we found that COPS5, BAX, and BAD proteins can form a PPI network with MRPL35, TP53, and BCL2L1. Western blot analysis confirmed the intervention effect of 18β-GRA on GC cells, MRPL35, TP53, and BCL2L1 showed dose-dependent up/down-regulation, and the expression of COPS5, BAX, and BAD also increased/decreased with the change of 18β-GRA concentration.
18β-GRA can inhibit the proliferation of GC cells by regulating MRPL35, COPS5, TP53, BCL2L1, BAX, and BAD.
Core Tip: 18β-glycyrrhetinic acid (18β-GRA) is a pentacyclic triterpene derivative extracted from the natural medicine licorice. Our results showed that 18β-GRA could inhibit gastric carcinoma (GC) cell proliferation, migration, invasion, and tumor formation, induce GC cell apoptosis, and arrest the cell cycle. Tandem mass tag analysis revealed that the expression of mitochondrial ribosomal protein L35 (MRPL35) was significantly decreased after 18β-GRA intervention in GC cells, which was confirmed by Western blot results. These data indicate that 18β-GRA inhibits the proliferation/migration and promotes apoptosis of GC cells by down-regulating MRPL35 expression, suggesting that MRPL35 is a therapeutic target for GC.
- Citation: Yuan L, Yang Y, Li X, Zhou X, Du YH, Liu WJ, Zhang L, Yu L, Ma TT, Li JX, Chen Y, Nan Y. 18β-glycyrrhetinic acid regulates mitochondrial ribosomal protein L35-associated apoptosis signaling pathways to inhibit proliferation of gastric carcinoma cells. World J Gastroenterol 2022; 28(22): 2437-2456
- URL: https://www.wjgnet.com/1007-9327/full/v28/i22/2437.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i22.2437
Gastric carcinoma (GC) is among the most common cancers as well as one of the most common causes of cancer-related mortality worldwide. Based on the Global Cancer Observatory (GLOBOCAN) database, GC ranks fifth in the incidence and fourth in the mortality among various types of cancers worldwide, after lung cancer, colorectal cancer, and liver cancer, and there would be more than one million new cases per year since 2020 by the World Health Organization predicting[1]. The current treatment of GC is mainly based on surgery, chemotherapy, and radiotherapy[2]. Most patients with early and advanced GC can be treated by surgery, but the treatment effect is very poor for patients with advanced GC and metastasis[3]. Although chemotherapy and radiotherapy are effective, they have severe side effects, such as hair loss and gastrointestinal reactions, leading to low compliance. Therefore, it is critical to find an effective therapeutic target for GC.
18β-glycyrrhetinic acid (18β-GRA) is a pentacyclic triterpene derivative extracted from the natural medicine licorice (Figure 1). It has significant anti-inflammatory[4], anti-oxidant, anti-bacterial, and anti-proliferative effects[5]. Recent studies have shown that 18β-GRA has reliable anticancer effects on human malignant tumors, such as lung cancer[6], breast cancer[7], colon cancer, and GC. Huang et al[6] found that 18β-GRA inhibited the extracellular signal-regulated kinase (ERK)/cyclic adenosine monophosphate response element binding protein (CREB) pathway by inhibiting thromboxane synthase (TxAS), thereby reducing the cell proliferation of non-small cell lung cancer (NSCLC) cells[6]. 18β-GRA showed potent inhibitory effects on human breast carcinoma MCF-7 cell proliferation in a concentration- and time-dependent manner, without affecting immortalized normal mammary epithelial cell line. 18β-GRA induced apoptosis in MCF-7 cells via caspase activation and modulation of the protein kinase B (Akt)/phosphorylated forkhead box O3 (FOXO3a) pathway[7]. In vivo and in vitro, 18β-GRA inhibited the proliferation, invasion, and migration of colorectal cancer cells, and had no obvious inhibitory effect on normal cells. It prolonged the survival of tumor-bearing mice by promoting apoptosis of colorectal cancer cells. Moreover, 18β-GRA inhibited the initiation and progression of gastric tumors by ameliorating the inflammatory microenvironment through down-regulation of cyclooxygenase-2 (COX-2) expression and by inhibiting Wnt-1 expression through the up-regulation of tumor suppressor miR-149-3p[8]. 18β-GRA inhibited the migration and invasion of SGC-7901 cells via the reactive oxygen species (ROS)/protein kinase C-α/ERK signaling pathway, and suppressed MMP-2 and 9 activities in SGC-7901 cells in a dose-dependent manner[9]. Toll-like receptor 2 (TLR2) mRNA and protein expression levels were elevated in the GC cell lines and human GC tissues. Overexpression of TLR2 was correlated with high histological grade. However, the expression of TLR2 was found to be down-regulated by 18β-GRA through a dynamic process of DNA methylation regulation. GRA pretreatment inhibited TLR2-activated GC cell proliferation, energy metabolism, and carcinogenesis[10]. In general, 18β-GRA showed an excellent inhibitory effect on GC, and its pharmacological mechanism is still worth exploring.
The human mitochondrial ribosomal protein (MRP) gene family consists of 30 members encoding small mitochondrial ribosomal subunits and 50 members encoding large subunits. All MRPs encoded by nuclear genes are important for the mitochondrial function and protein synthesis, which are involved in ribosomal protein transcription and translation, and mitochondrial oxidative phosphorylation. MRPs can regulate cellular functions outside mitochondria, such as cell proliferation, apoptosis, protein biosynthesis, and signal transduction. In NSCLC, overexpression of MRPL15 was associated with a poor prognosis[11]. MRPL15 plays a role in ovarian cancer through cell cycle, DNA repair, and the mTOR1 signaling pathway, suggesting that MRPL15 may be a prognostic indicator and therapeutic target for ovarian cancer[12]. Hao et al[13] found that MRPL42 expression was tested to be up-regulated in glioma tissues compared with normal tissues. The proliferation of glioma cells was largely blunted by silencing MRPL42. The results suggested that MRPL42 silencing resulted in increased distribution of cell cycle at G and G/M phases, while the S-phase cells decreased. Taken together, MRPL42 is a novel oncogene in glioma[13]. In conclusion, these findings suggest that MRPs should play a crucial role in regulating the development of various malignant tumors.
Mitochondrial ribosomal protein L35 (MRPL35) is a 25 ku protein encoded by the MRPL35 gene located on chromosome 2p11.2, which is a mitotic component of the central protuberance of the mitochondria. It is a member of the large subunit family of MRPs[14] and plays an important role in the assembly of cytochrome C oxidase complex (COX) as the specific component of mitochondrial ribosomes[15]. The expression of MRPL35 rises in the tissues of colorectal carcinoma (CRC), compared with the matched cancer-adjacent tissues. In vitro, down-regulation of MRPL35 resulted in increased production of ROS along with DNA damage, reduction of cell proliferation, G/M arrest, apoptosis, and autophagy induction[16]. Silencing MRPL35 suppresses the proliferation of esophageal cancer TE-1 cells, down-regulates MMP-2, and promotes cell apoptosis, after transfection of human esophageal cancer cells with lentivirus[17]. Our previous study found that the expression of MRPL35 was significantly up-regulated in GC, and the survival analysis showed that the up-regulated MRPL35 was associated with a poor survival rate in GC. Knockdown of MRPL35 was showed to inhibit proliferation of GC cells by regulating BCL-XL, PICK1, and AGR2 which are apoptosis-related proteins[18].
In order to more significantly explore the mechanism of 18β-GRA, we detected the expression abundance of MRPL35 in AGS, SGC-7901, MGC80-3, and BGC-823 cell lines by quantitative reverse transcription-polymerase chain reaction (qRT-PCR), and the results showed that MRPL35 expression levels in MGC80-3 and BGC-823 cell lines were similar, so we did experiment on these two cell lines (Supplementary Figure 1).
In this study, we focused on examining the effects of 18β-GRA intervention on GC cell function in vitro and tumor formation in nude mice in vivo. Tandem mass tag (TMT) analysis and STRING database prediction were used to investigate the molecular mechanism, and Western blot was used to verify the expression changes of related proteins. An important conclusion is that 18β-GRA inhibits GC cell proliferation/migration and promotes apoptosis by interfering with the expression of MRPL35-related signaling proteins, which may contribute to the development of effective therapies. These results suggest that MRPL35 may be a potential therapeutic target for the inhibition of GC by 18β-GRA.
Human gastric mucosal epithelial cells GES-1 (Cat. No. BNCC353464, BNCC, China) were cultured in DMEM (Cat. No. C11995500BT, Gibco, United States) containing 100 mL/L fetal bovine serum (FBS; Cat. No. SH30256.01, Gibco) and penicillin and streptomycin (Cat. No. p1400-100, Gibco). GC cell lines BGC-823 (Cat. No. GCC-ST0008CS, GeneChem, China) and MGC80-3 (Cat. No. GCC-ST0004CS, GeneChem) were cultured in RPMI-1640 (Cat. No. C11875500BT, Gibco) containing 100 mL/L FBS and penicillin and streptomycin. The culture flasks were placed in an incubator at a constant temperature of 37 ℃, with 50 mL/L CO2 and saturated humidity. BALB/c nude mice (male, weighing 18-22 g) of specific pathogen-free (SPF) grade were purchased from the Animal Experiment Center of Ningxia Medical University. All animals were fed standard laboratory feed and water in 12 h light/dark cycle environment. The animal protocols (IACUC-NYLAC-2019-083) were approved by the Institutional Animal Care and Use Committee of Ningxia Medical University.
Normal gastric mucosal epithelial GSE-1 cells were cultured in an incubator at 37 ℃ and 50 mL/L CO2, collected, and inoculated in a 96-well plate at a density of 4 × 104 cells/mL. On the next day, the cells were treated with different concentrations of 18β-GRA (purity > 97%; Cat. No. G10105-10G, Sigma, United States) for 24 h, 48 h, and 72 h. Thereafter, 10 μL of cell counting kit-8 (CCK-8) (Cat. No. KGA317, KeyGEN, China) was added to each well and incubated for 2 h. Optical density (OD) was detected at 450 nm. All experiments were repeated three times.
MGC80-3 and BGC-823 cells were cultured in an incubator at 37 ℃ and 50 mL/L CO2. Cells in the logarithmic growth phase were inoculated in 96-well plates at an adjusted concentration of 4 × 104 cells/mL. After 24 h, cells were treated with different concentrations of 18β-GRA for 24 h, 48 h, and 72 h. Thereafter, 10 μL of CCK-8 was added to each well and incubated for 2 h. OD was detected at 450 nm. All experiments were independently repeated at least three times.
MGC80-3 and BGC-823 cells were inoculated in 6-well plates. The cells were treated with 18-GRA for 48 h on the second day. Then the cells were collected and centrifuged at 3000 r/min for 5 min. Annexin V-FITC apoptosis detection kit (Cat. No. KGA107, KeyGEN) was used for staining and apoptosis was detected by flow cytometry after 15 min at room temperature in dark. Cells from each group were collected, washed with pre-cooled PBS, and fixed overnight in 750 mL/L ethanol. The cells were stained with cell cycle kit and incubated at room temperature for 30-60 min in the dark. The cell cycle was detected by flow cytometry. All experiments were repeated three times.
First, 8 μmol/L pore size Transwell (Cat. No. Costar.3422, Corning, United States) with and without Matrigel were placed into a 24-well plate. MGC80-3 and BGC-823 cells were inoculated into the upper chambers at 2 × 105 cells/mL. After adding 750 μL complete medium in the lower chambers and culturing at 37 ℃ for 24 h, the medium was replaced with medium containing different concentrations of 18β-GRA and cultured for 48 h. The cells were gently wiped off with cotton swabs and fixed with 4% paraformaldehyde for 5 min, stained with crystal violet for 5 min, and washed with PBS. Thereafter, the cells were observed with an inverted microscope and photographed to count and compare the differences in the cell invasion between groups. The number of migrated cells was counted, and the average value was used for statistical analysis. All experiments were repeated three times.
GC cells MGC80-3 and BGC-823 were inoculated on 6-well plates. After the cells were attached, a scratch was made with a 200 μL pipette tip. The cells were washed with PBS. Then medium containing 18-GRA was added, and the cells were cultured in a 50 mL/L CO2 incubator at 37 ℃ for 0 h and 48 h, and photographed with a microscope to calculate the cell migration rate of each group. All experiments were repeated three times.
Four-week-old male BALB/c nude mice (18-22 g) of SPF grade were purchased from the Animal Experiment Center of Ningxia Medical University. All animals were housed in polypropylene cages at a temperature of 22 ± 1 ℃ and 50 ± 5% humidity, in a 12:12 h light/dark cycle, and provided free access to standard laboratory chow and water. There were six BALB/c nude mice in each group. A microsyringe was used to extract 200 μL of BGC-823 cell suspension in logarithmic growth phase at a concentration of 2.5 × 106 cells/mL, which was subcutaneously injected into the right back of each nude mouse. The 18β-GRA group was intragastrically administered with 18β-GRA at 50 mg/kg[10,19] every other day, and the control group was similarly administered with equal volume of normal saline. Intragastric gavage administration was carried out in conscious animals. The tumor volume was measured daily using the formula 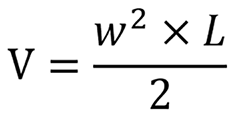 (V, volume; W, width; L, length). All animals were euthanized by exposure to 1000 mL/L CO2 for 5 min for tissue collection. The animal protocols were approved by the Institutional Animal Care and Use Committee of Ningxia Medical University.
(V, volume; W, width; L, length). All animals were euthanized by exposure to 1000 mL/L CO2 for 5 min for tissue collection. The animal protocols were approved by the Institutional Animal Care and Use Committee of Ningxia Medical University.
Total RNA of the transplanted tumor tissue from mice was extracted using Trizol reagent (Cat. No. DP424, Tiagen Biochemical Technology, China), and PrimeScript RT reagent Kit with gDNA Eraser was used to synthesize first-strand cDNA from total RNA. qRT-PCR was used to detect by using BlazeTaq SYBR Green qPCR Mix 2.0 (Cat. No. QP031-S, GeneCopoeia, United States) according to the manufacturer's instructions. The primers used are as follows: MRPL35: Forward, 5’-TTGGCATCTTCAACCTACCGC-3’ and reverse, 5’-GGAGGAAACAACTGGTGTCTGA-3’; GAPDH: Forward, 5’-TGACTTCAACAGCGACACCCA-3’ and reverse, 5’-CACCCTGTTGCTGTAGCCAAA-3’. All experiments were repeated three times.
SDT buffer was added into the control and 18β-GRA-treated GC cells. After being boiled for 15 min, the protein content was quantified with the BCA Protein Assay Kit (Cat. No. P0012, Beyotime, China), 20 μg proteins were taken, and 6 × loading buffer (Cat. No. P0015F, Beyotime) was added. After being boiled for 5 min, the proteins were separated by 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and protein bands were visualized by Coomassie Blue R-250 staining.
The method of filter-aided sample preparation digestion was performed by adding dithiothreitol (DTT; Cat. No. 43819-5G, Sigma) to a 200 μg protein solution to a final concentration of 100 mmol/L, boiling reduced protein lysate for 5 min, and alkylation with 100 μL iodoacetamide buffer (IAA) for 15 min at room temperature protected from light. Then, 100 μL of 0.1 M tetraethyl ammonium bromide (TEAB; Cat. No. SE252676/90114, Thermo Fisher, United States) solution was added, and the mixture was centrifuged for 15 min. After 40 μL of trypsin buffer (4 μg trypsin in 40 μL of 0.1 M TEAB solution) was added, the filtrate was collected overnight at 37 ℃ to obtain peptide segments. One hundred microliters of peptide mixture was then labeled using TMT 6plex Isobaric Mass Tag Labeling kit (Thermo Fisher, United States) according to the manufacturer’s instructions.
The labeled peptides of the two groups were fractionated with Agilent 1260 Infinity HPLC system. The peptide mixture was diluted with buffer A (10 mmol/L HCOONH4, 5%/85% ACN, pH 10.0). Liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis was performed on a Q Exactive Plus mass spectrometer (Thermo Fisher, United States), and peptides were detected on MS at a resolution of 70000 with a scan range of 350-1800 m/z and with automatic gain control of 5e4. All the original data files obtained were processed with Proteome Discoverer 2.2 (Thermo Fisher, United States) software. Proteins that met the expression fold change > 1.2 and P value (Student’s t test) < 0.05 were considered to be differentially expressed proteins (DEPs).
Blast2GO and InterProScan software are used to analyze the cellular processes of DEPs, the biological processes involved, and the corresponding molecular functions. The Fisher’s exact test was employed to test Gene Ontology (GO) annotation/Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway/InterPro domain enrichment analysis. Fisher’s exact test was used to test enrichment pathways; InterPro was used to determine enrichment domains. We used WoLFPSOR to locate and predict DEPs. Matplotlib was used to classify samples and protein expression levels at the same time to form hierarchical cluster heat maps. A protein-protein interaction (PPI) network incorporating identified target proteins was prepared with the STRING database, using a score > 0.4 as the significance threshold for identified interactions.
The cells were collected after drug intervention. Whole cell lysates were obtained by gentle lysis (Cat. No. KGP250, KeyGEN). Protein quantitation was performed by the BCA method (Cat. No. KGPBCA, KeyGEN). The protein was separated by SDS-PAGE, and the protein was transferred to PVDF membranes by wet-transfer method. Thereafter, the membrane was incubated with 50 mL/L skim milk powder at room temperature for 1 h. Later on, the membranes were incubated with the primary antibodies, including anti-MRPL35 (Cat. No. YT5669, Immunoway; 1:2000), anti-TP53 (Cat. No. YM3052, Immunoway; 1:2000), anti-BCL2L1 (Cat. No. YT0477, Immunoway; 1:2000), anti-BAX (Cat. No. YM3619, Immunoway; 1:500), anti-COPS5 (Cat. No. ab124720, Abcam; 1:2000), anti-BAD (Cat. No. 9292, Cell Signaling Technology; 1:1000), and anti-β-tubulin (Cat. No. YM3030, Immunoway; 1:10000). Then, the membranes were washed with TBST and incubated with anti-mouse/rabbit IgG antibody (Cat. No. S001 S004, TDYBio; 1:10000) at 37 ℃ for 1 h. After washing, the proteins were detected with an ECL detection system. ImageJ software was used to measure the band intensity.
The statistical methods of this study were reviewed by Li-Qun Wang from Department of Epidemiology, Department of Medical Statistics, Institute of Public Health and Management, Ningxia Medical University. All analyses were performed using GraphPad Prism 7.0. All the data are expressed as the mean ± SE. Statistical significance was analyzed using one-way ANOVA. P < 0.05 was considered statistically significant.
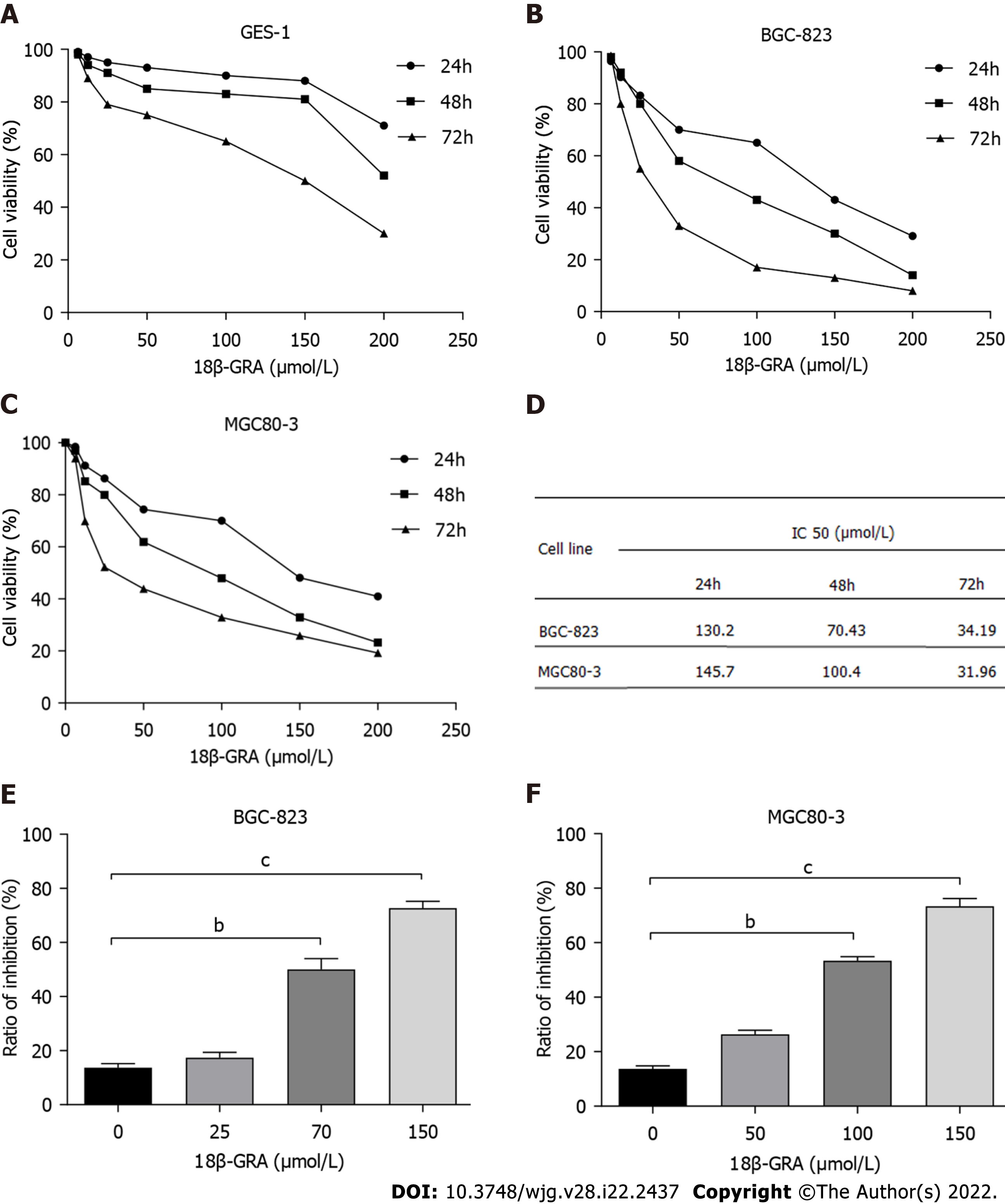
CCK-8 assay was used to evaluate the effect of 18-GRA on the survival rate of normal gastric mucosal GES-1 cells. The results showed that 12.5-150 μmol/L of 18β-GRA had little effect on the survival rate of GES-1 cells at 24 h and 48 h. However, when the concentration of 18β-GRA was greater than 150 μmol/L, the survival rate of GES-1 cells was significantly decreased (Figure 2A).
CCK-8 results showed that 6.25-200 μmol/L of 18β-GRA inhibited the proliferation of BGC-823 and MGC80-3 cells in a time- and dose-dependent manner (Figure 2B and C). After the intervention of 18-GRA in BGC-823 and MGC80-3 cells for 24 h, 48 h, and 72 h, the 50% inhibitory concentration (IC50) of cells was calculated and is shown in Figure 2D. Based on the above results, 25 μmol/L (low), 70 μmol/L (medium), and 150 μmol/L (high) were determined for BGC-823 cells, and 50 μmol/L (low), 100 μmol/L (medium), and 150 μmol/L (high) were determined for MGC80-3 cells. Compared with the 0 μmol/L (control), the different concentrations of 18-GRA inhibited the proliferation of BGC-823 and MGC80-3 cells (P < 0.01) (Figure 2E and F).
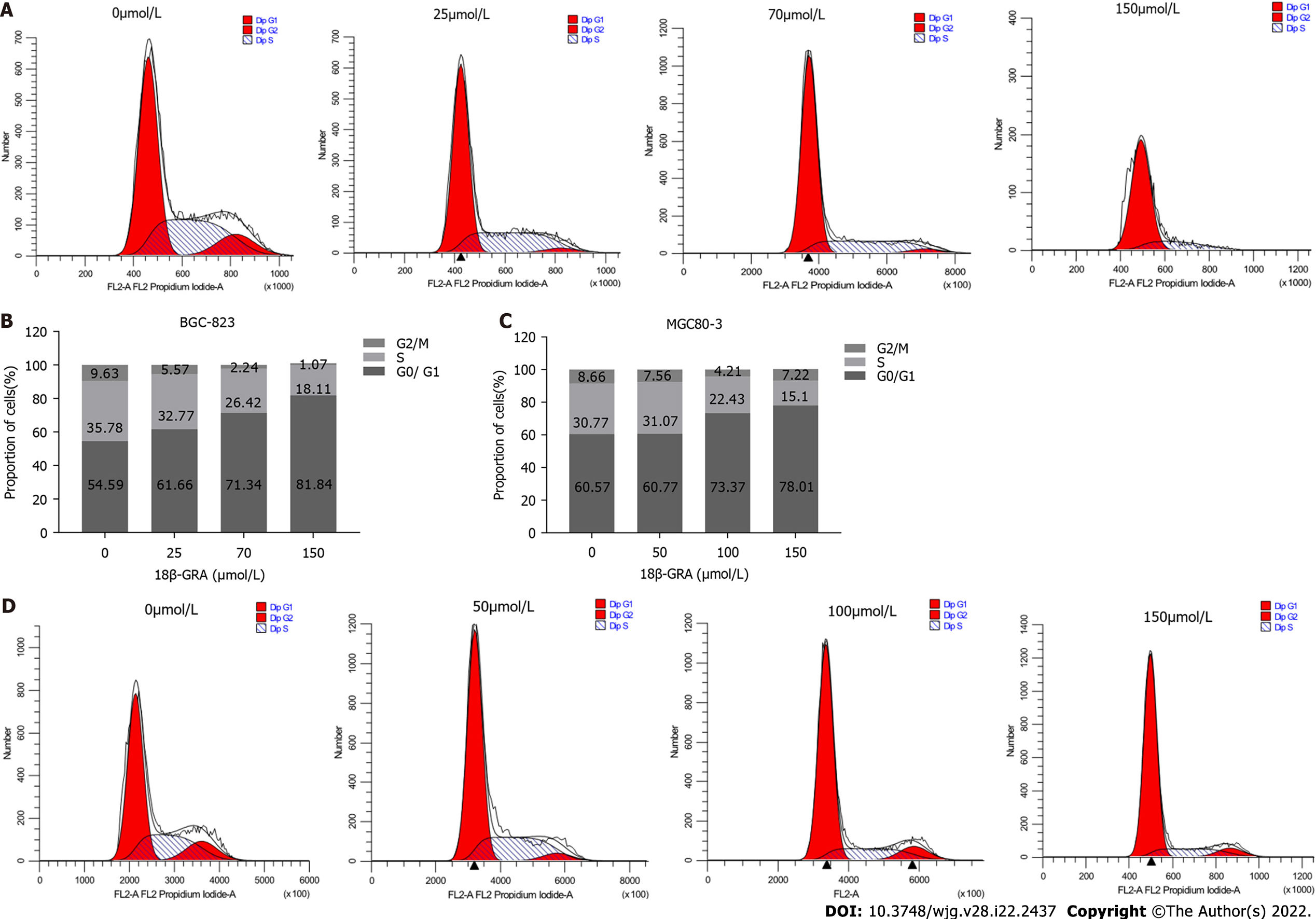
In order to study the effect of 18β-GRA on GC cell cycle, BGC-823 and MGC80-3 cells were treated with different concentrations of 18β-GRA, and the cell cycle of BGC-823 was arrested at G0/G1 phase; the proportions of cells at G0/G1 phase were 81.84% (150 μmol/L), 71.34% (70 μmol/L), and 61.66% (25 μmol/L) (Figure 3A and B). The cell cycle of MGC80-3 cells was arrested at G0/G1 phase; the proportions of cells at G0/G1 phase were 78.01% (150 μmol/L) and 73.37% (100 μmol/L) and higher than 60.57% (control). And with the increase of 18-GRA concentration, the proportion of cells in S-phase decreased (Figure 3C and D).
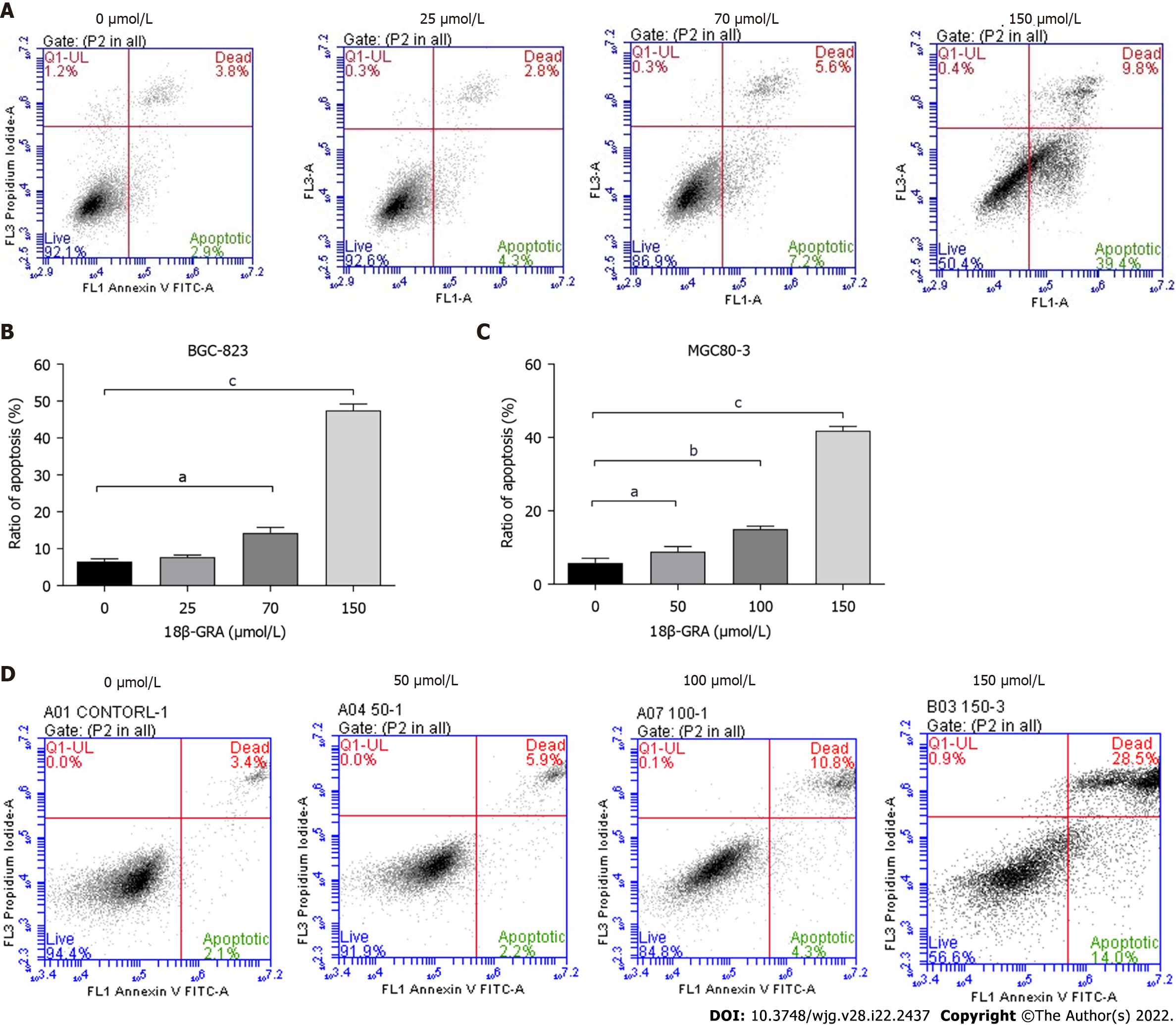
Flow cytometry was used to detect the effects of different concentrations of 18β-GRA on GC cell apoptosis. After intervention with different concentrations of 18β-GRA in BGC-823 and MGC80-3 cells for 48 h, the apoptosis rates of BGC-823 cells were 14.27% (70 μmol/L) and 47.47% (150 μmol/L), which were higher than 6.53% (control) (Figure 4A and B). The apoptosis rates of MGC80-3 cells were 8.87% (50 μmol/L), 15.03% (100 μmol/L), and 41.8% (150 μmol/L), which were all higher than 5.8% (control) (Figure 4C and D).
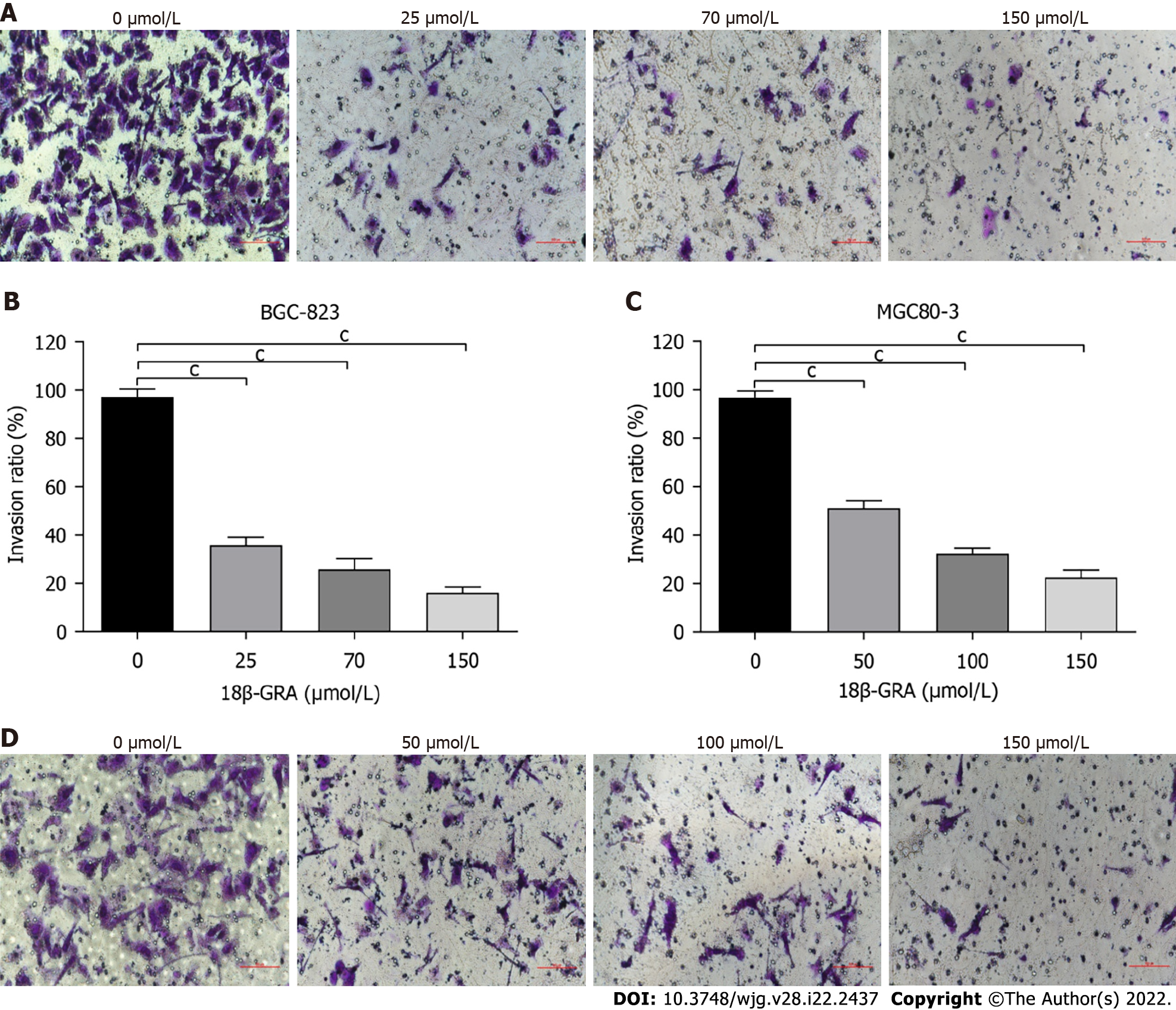
BGC-823 and MGC80-3 cells were treated with different concentrations of 18β-GRA for 48 h, and the cell invasion ability was detected. The results of Transwell assay are shown in Figure 5. Compared with control, the different concentrations of 18β-GRA inhibited the invasion of BGC-823 and MGC80-3 cells (P < 0.05).
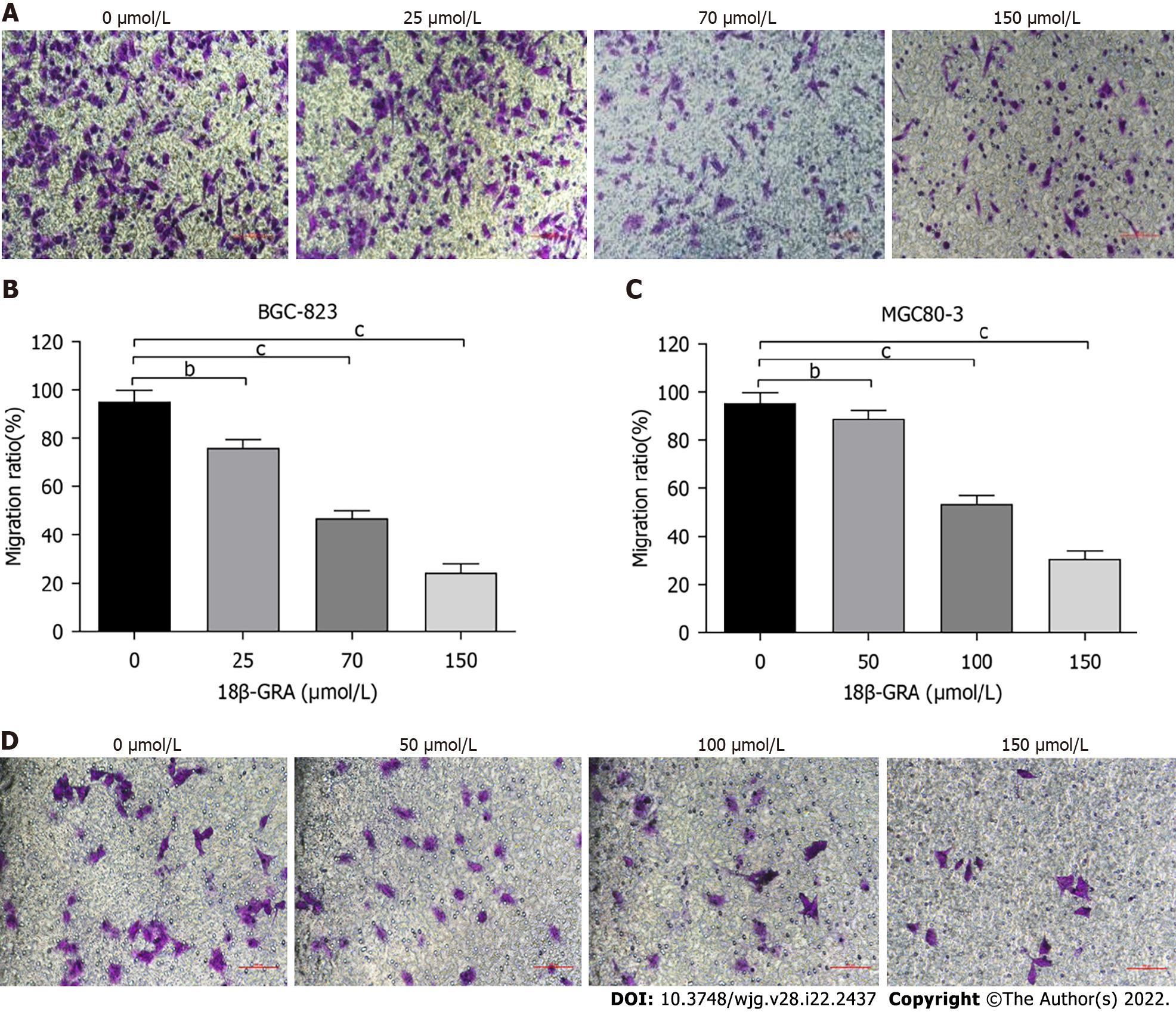
The effect of 18β-GRA on GC cell migration was evaluated in Transwell chambers without matrigel. The migration of BGC-823 and MGC80-3 cells was inhibited after 48 h of treatment with different concentrations of 18-GRA (Figure 6).
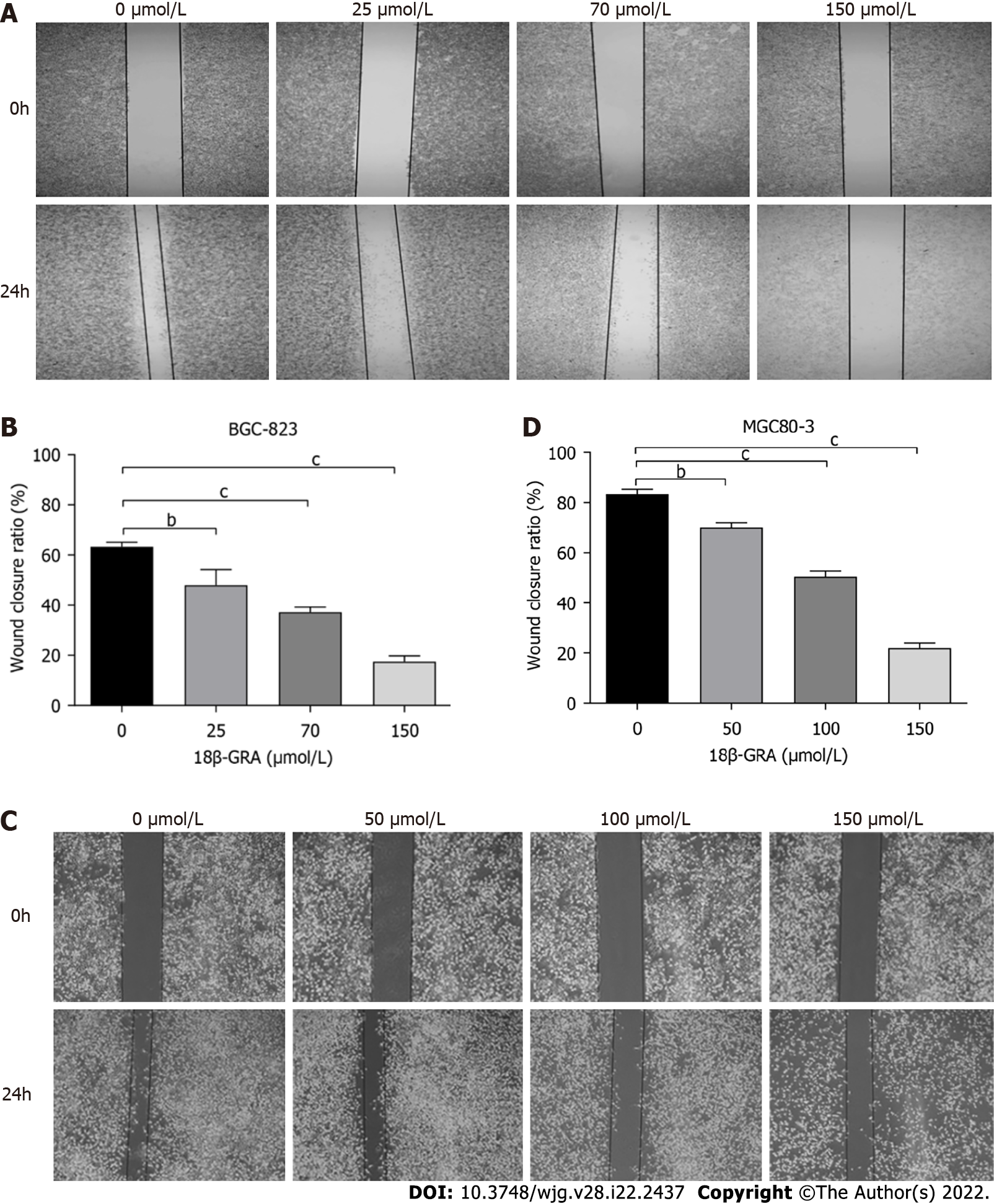
We also evaluated the effect of 18β-GRA on GC cell migration by wound scratch assays. The migration of MGC80-3 and BGC-823 cells was inhibited by different concentrations of 18β-GRA for 48 h (Figure 7). With the increase of 18β-GRA concentration, the inhibitory effect was gradually enhanced (P < 0.05).
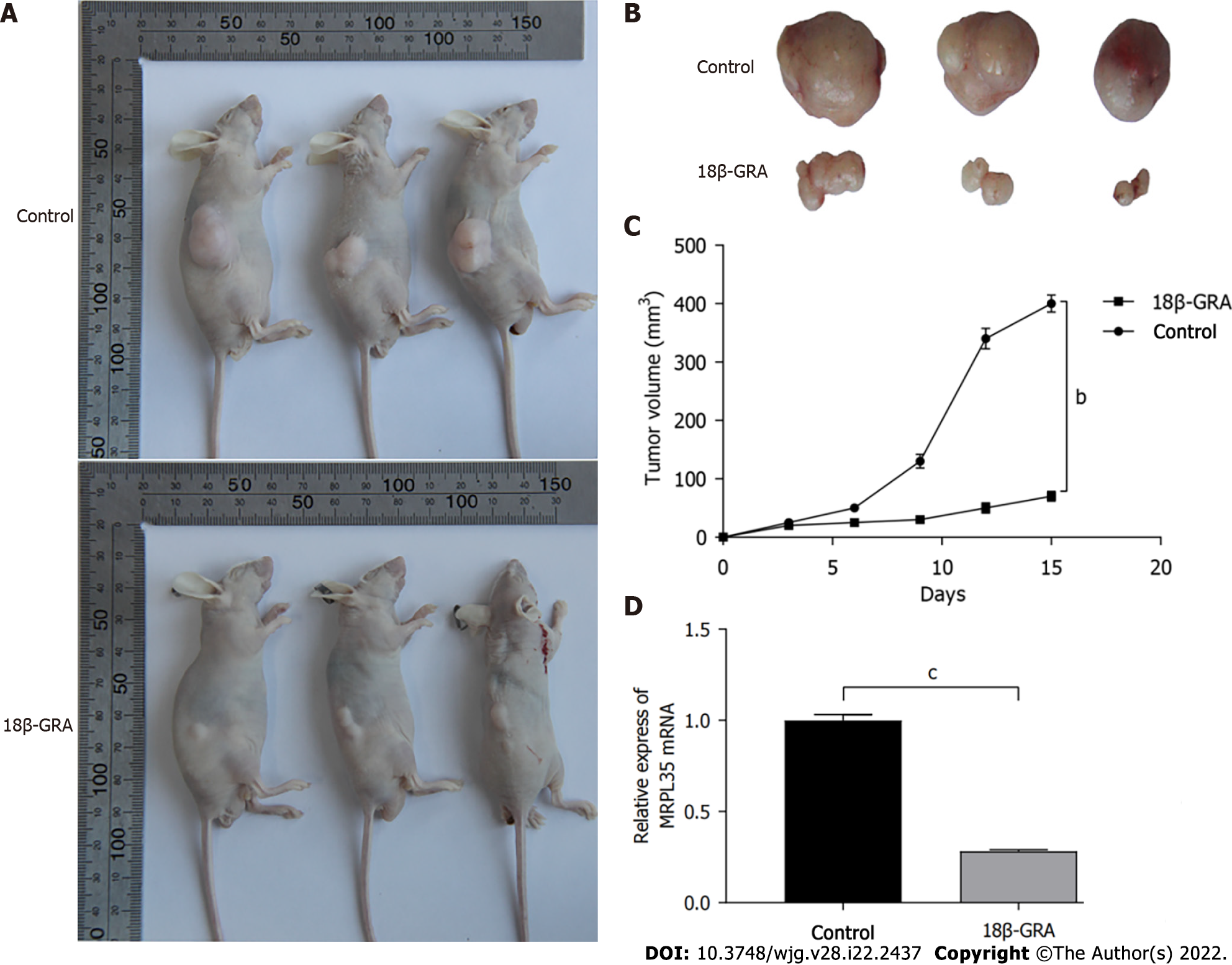
The results of tumor formation experiments in nude mice showed that after 2 wk of continuous intragastric administration, the tumor volume in the 18β-GRA group was significantly smaller than that in the control (Figure 8A and B). The tumor volume was recorded within 2 wk, and the tumor growth curve was drawn (Figure 8C). The tumor growth rate of the 18β-GRA group was lower than that of the control (P < 0.01).
qRT-PCR was used to detect the expression level of MRPL35 in the transplanted tumor tissues of mice. The results showed that the expression level of MRPL35 in the transplanted tumor tissues of mice in the 18β-GRA group was significantly lower than that in the control (P < 0.001) (Figure 8D).
During the experiment, we also monitored the influence on body weight, food, and water intake in mice, and found that there was little toxic effect of 18β-GRA on them (Supplementary Figures 2-4).
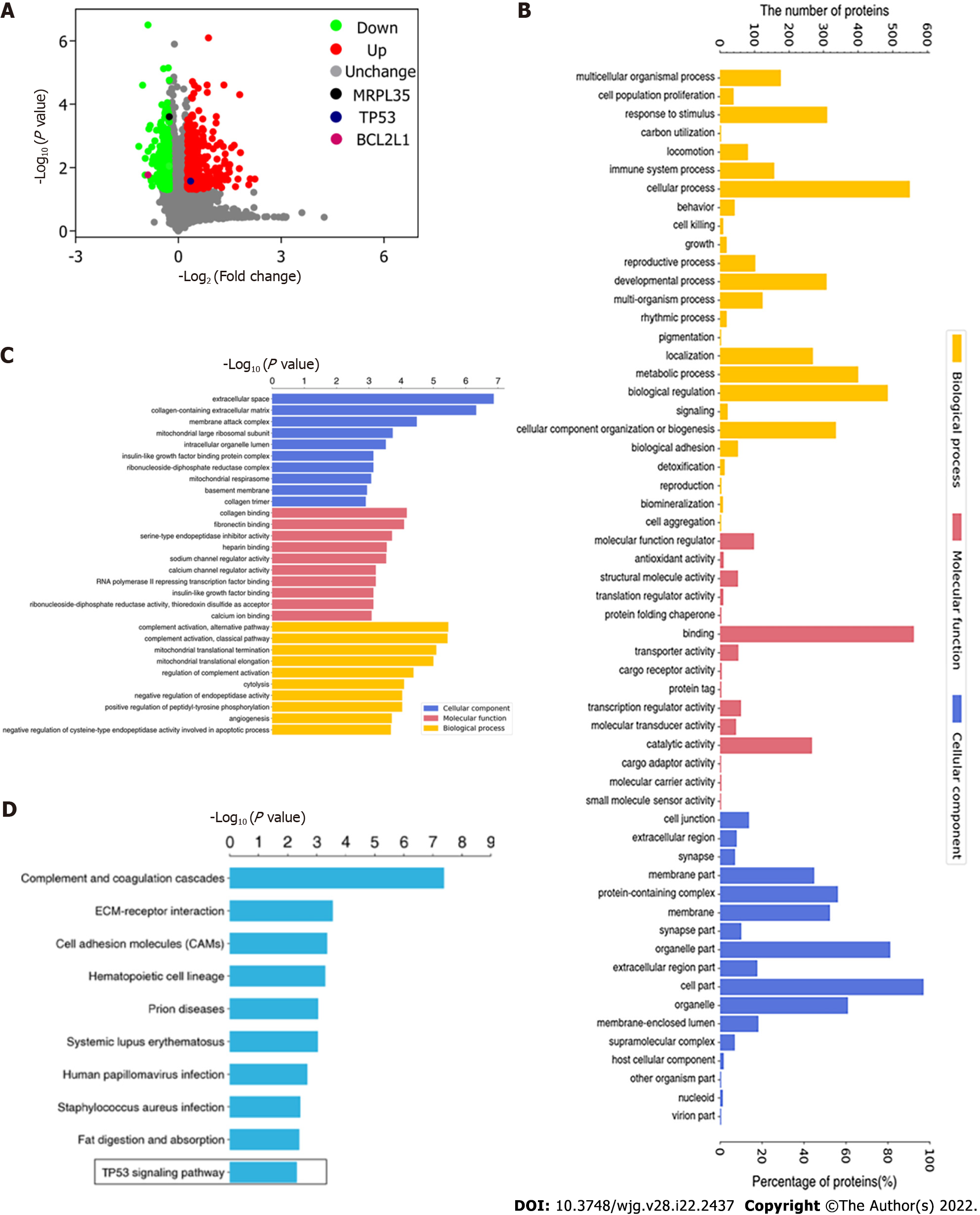
Proteomic analysis based on TMT labeling-based was performed on control and GC cells treated with 18β-GRA. A total of 6275 proteins were identified by proteomic analysis, and 609 DEPs were screened using the expression difference ratio > 1.2 and P < 0.05 as the standard, among which 335 were up-regulated and 274 were down-regulated. To better visualize the DEPs, we drew the volcano plot. The red dots indicate significantly up-regulated proteins and blue dots indicate significantly down-regulated proteins (Figure 9A). Notably, after 18β-GRA intervention, MRPL35 expression was significantly down-regulated (P = 0.000247), TP53 expression was up-regulated (P = 0.02676), and BCL2L1 was down-regulated (P = 0.01699). The GO annotation and enrichment analysis was used to analyze the DEPs and to determine their biological processes (BP), molecular functions (MF), and cellular components (CC) (Figure 9B and C). In the BP analysis, DEPs were mainly involved in cellular processes, biological regulation, and molecular metabolism. It is mainly enriched in complement activation, alternative pathway and classical pathway, and mitochondrial translational termination. With respect to MF, the DEPs are involved in the binding, catalytic activity, molecular function regular, and so on. It is mainly enriched in the binding of collagen and fibronectin, and serine-type endopeptidase inhibitor activity. In the CC analysis, we found that the DEPs were highly localized in the cell, organelle, and protein-containing complex part. Those DEPs are mainly enriched in extracellular space, collagen-containing extracellular matrix, membrane attack complex, and mitochondrial large ribosomal subunit. The DEPs were classified by KEGG pathway enrichment analysis (Figure 9D). KEGG pathway enrichment analyses revealed these DEPs to be enriched in complement and coagulation cascades, ECM-receptor interaction, and cell adhesion molecules. It is worth noting that these DEPs are also involved in the TP53 signaling pathway.
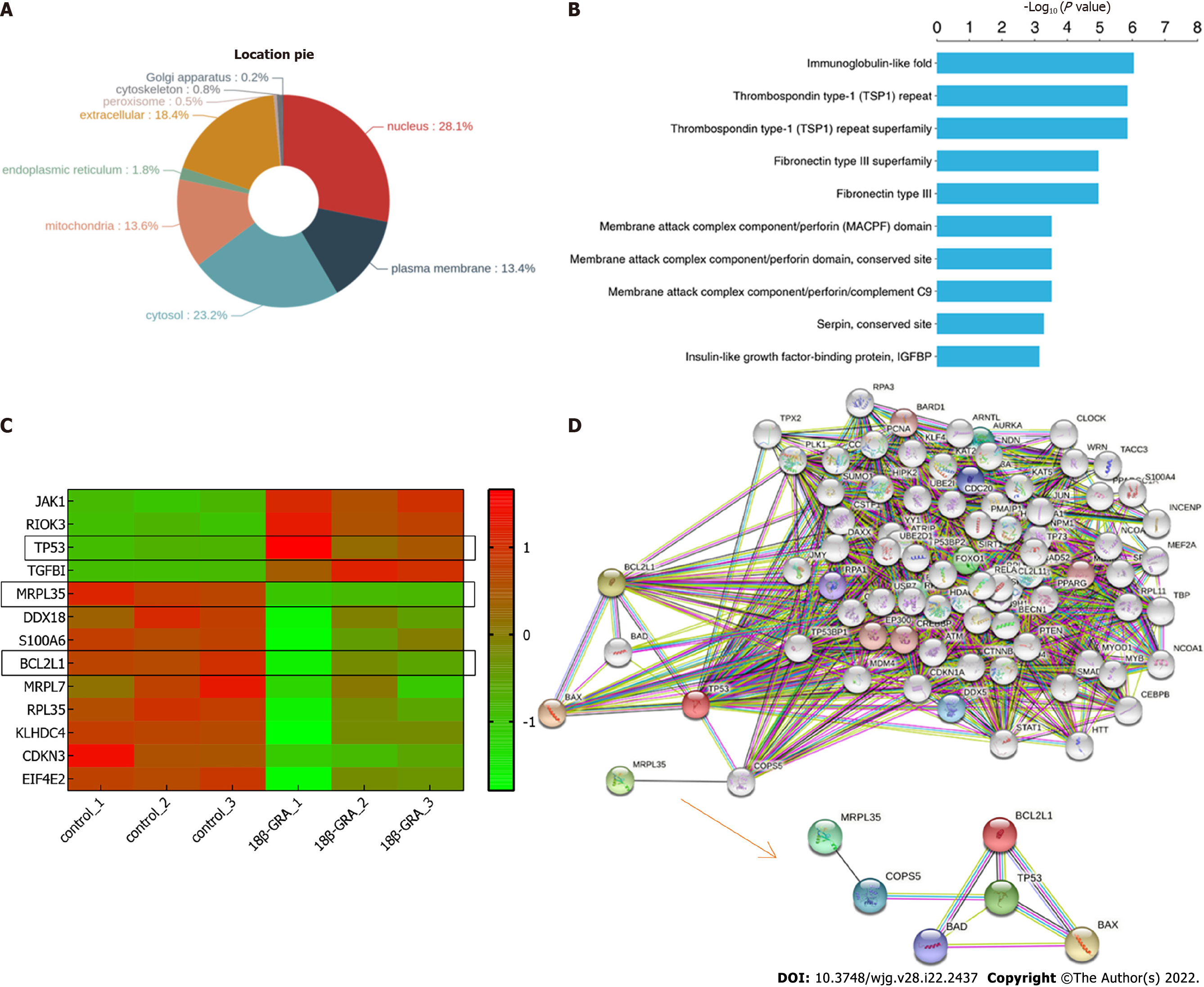
Wolfpsort was used to predict the subcellular localization of these DEPs, which were primarily localized to the nucleus (28.1%), cytosol (23.2%), extracellular space (18.4%), mitochondria (13.6%), and so on (Figure 10A). Further protein domain enrichment analysis showed that DEPs were significantly enriched in the immunoglobulin-like fold domain, thrombospondin type-1 (TSP1) repeat and superfamily, and fibronectin type III and superfamily protein domain (Figure 10B). Cluster analysis was performed on DEPs and drawn as a heat map (Figure 10C). Each row in the figure represents a DEP, red represents up-regulated DEP expression and green for down-regulated DEP expression; the darker the color is, the more significant the difference is.
Finally, in order to further explore the function of MRPL35, TP53, and BCL2L1, the STRING database was used to construct a PPI network, and the results are shown in Figure 10D. And we found that COPS5 is the protein connecting MRPL35 and TP53. In addition, we also found that TP53 and BCL2L1 are closely related to the pro-apoptotic proteins BAX and BAD.
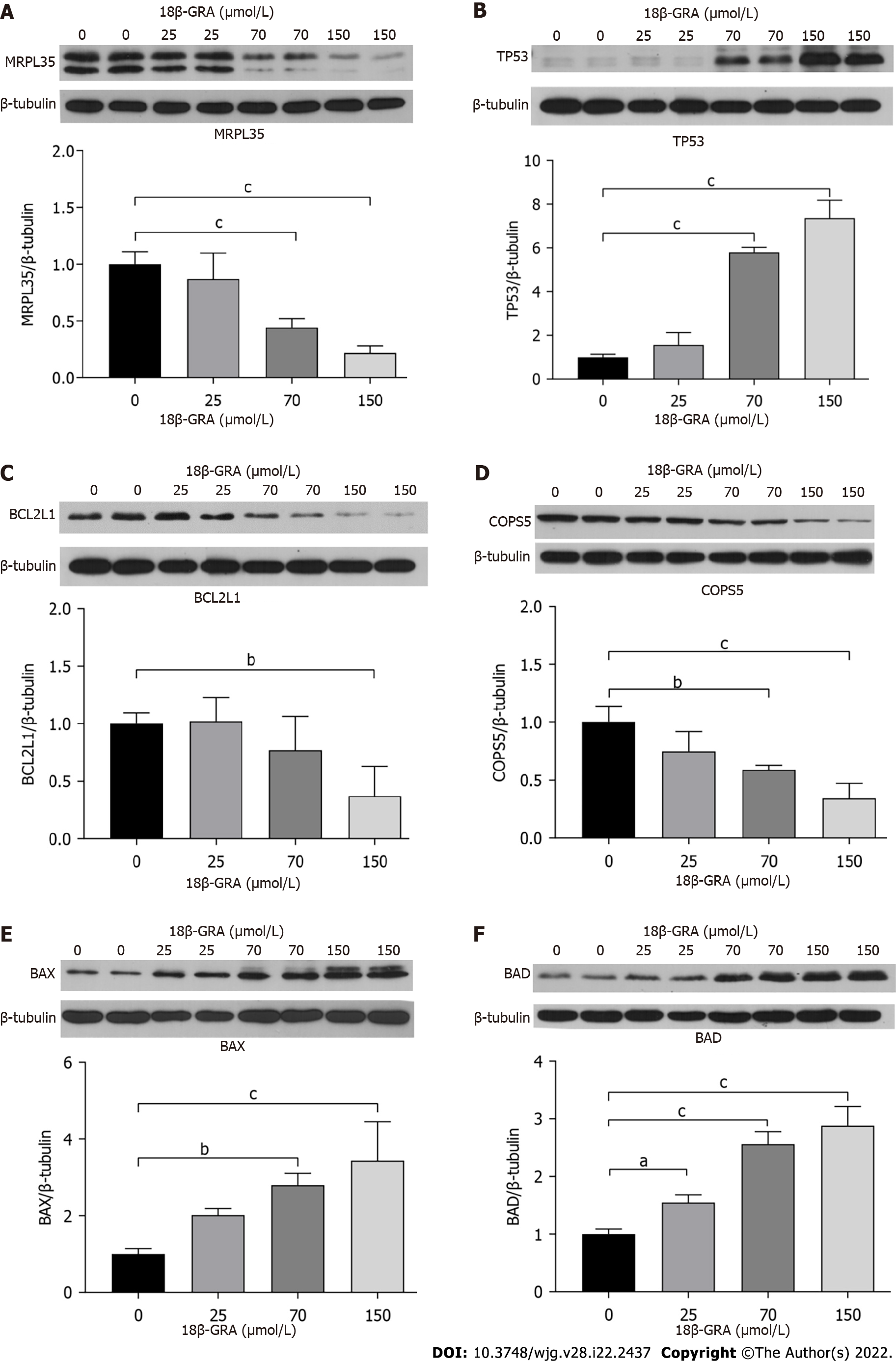
Western blot was used to detect the effect of 18β-GRA on the expression of MRPL35 related proteins in gastric cancer cells. The results showed that with the increase of 18β-GRA concentration, the expression levels of MRPL35 and BCL2L1 proteins were gradually down-regulated, while the expression of TP53 protein was up-regulated. The expression of COPS5, the protein linking MRPL35 and TP53, was down-regulated, and the expression levels of pro-apoptotic proteins BAX and BAD were up-regulated. These results confirm the results of our proteomics and bioinformatics analysis above (Figure 11).
In this study, the functional changes of GC cells after 18β-GRA intervention were tested by in vitro experiments, and the results showed that 18β-GRA had little effect on the survival rate of normal gastric epithelial GES-1 cells, and significantly inhibited the proliferation of BGC-823 and MGC80-3 cells. Next, we detected the cell cycle and found that 18β-GRA could arrest the GC cell cycle at G0/G1 phase. After drug intervention in GC cells, the cells can either exit the cell cycle reversibly, by initiating quiescence, or irreversibly, by senescence or apoptosis. The decision to exit the cell cycle solely depends on the DNA damage checkpoint[20]. In response to irreparable DNA damage, DNA damage checkpoints can initiate quiescence, senescence, or programmed cell death, primarily through TP53-dependent pathways[21,22]. Moreover, TP53 mutations are the most common mutations in cancer. However, even if cancer-related mutations prevent cell cycle exit, continuous proliferation of cells can still be prevented by blocking the entry of G1 cell cycle prior to replication, which relies on the activation of E2F-dependent transcription[23,24]. We investigated the effect of 18β-GRA on the apoptosis of two types of GC cells, and confirmed the results of cell cycle arrest. It was found that the cells chose to undergo apoptosis and exit the cell cycle, and TP53 played a crucial role in this process.
The cell invasion and migration experiments showed that cell invasion and migration were significantly suppressed after 18β-GRA intervention in GC cells. Epithelial-to-mesenchymal transition (EMT) plays an important role in the occurrence of metastasis and invasion of malignant tumors[25]. The loss of epithelial characteristics and the acquisition of mesenchymal features reduce cell adhesion and increase cell mobility. In addition, our previous experiments also showed that LINC00514 was up-regulated in GC specimens compared with non-tumor specimens, and overexpression of LINC00514 induced GC cell growth and EMT progression[26]. We used immunodeficient nude mice for tumor-forming experiments in vivo. It was found that subcutaneous tumor volume was significantly inhibited in nude mice after 2 wk of intragastric administration of 18β-GRA compared with the control group, and the expression level of MRPL35 in the transplanted tumor tissues of the 18β-GRA group was significantly decreased, which also confirmed the results of cell function in vivo.
Based on the above results, we further investigated the mechanism of 18β-GRA in inhibiting GC cell proliferation, and promoting cell apoptosis. TMT analysis showed that there were 609 DEPs in GC cells treated with 18β-GRA, among which the expression of MRPL35, TP53, and BCL2L1 was significantly altered. GO analysis found that all these DEPs were mainly involved in cellular processes, biological regulation, and molecular metabolism, and were enriched in membrane attack complexes and mitochondrial large ribosomal subunits. In addition, subcellular localization analysis of DEPs showed that these DEPs were mainly distributed in the nucleus, cytoplasm, extracellular space, and mitochondria. Our previous study showed that the high expression of MRPL35, a member of the large subunit family of MRPs, was correlated with the poor survival rate in GC, and knocking down MRPL35 could inhibit GC cell proliferation. Interestingly, KEGG results showed that these DEPs were enriched in the TP53 signaling pathway, which was ranked tenth in the analysis. TP53 is an important protein involved in the process of cell apoptosis. As a transcription factor, TP53 is mutated in most human cancers[27]. To inhibit cancer, TP53 protein regulates the transcription of many different genes, including BAX, NOXA1, and GADD45A, in response to multiple stress signals, such as DNA repair, cell cycle arrest, senescence, and apoptosis, while tumor mutations can make them resistant to apoptosis induced by the TP53 pathway. Combined with the expression changes of BCL2L1, we speculated that 18β-GRA might play a role in inhibiting the proliferation of GC by regulating TP53 apoptosis-related signaling pathways through MRPL35. Enrichment analysis of DEP domains showed that they were significantly enriched in immunoglobulin-like folding domain, TSP1 repeats and superfamily, and fibronectin type III and superfamily protein domain. Hence, we speculate that 18β-GRA might regulate the expression of MRPL35, induce the changes of apoptosis-related proteins TP53 and BCL2L1, and play a role in inhibiting the proliferation of GC cells.
Finally, we analyzed the input STRING database of these three signaling proteins, and found that COPS5 is the protein connecting MRPL35 and TP53. Therefore, we believe that MRPL35 is likely to play a pro-apoptotic role by regulating COPS5. In addition, we also found that TP53 and BCL2L1 are closely related to BAX and BAD. As a major regulator of mitochondrial involvement in the intrinsic apoptosis pathway, BAX controls a cell's commitment to the pathway by changing the physiology of resident mitochondria. BAX undergoes a series of ordered events that lead to the formation of pores in the outer membrane of mitochondria, facilitating the release of signaling molecules such as cytochrome C[28]. This event is commonly considered as the "point of no return" in cell death[29]. BAD, a ligand of the pro-survival protein 14-3-3, is an on-off protein in the mitochondria-induced apoptotic pathway[30]. When separated from 14-3-3, BAD may interact with BCL-2/BCL2L1 and release BAX, revealing its apoptotic activity. Binding of BAD to BCL2/BCL2L1 has also been reported to release the activator BH3-only molecules, such as Bim, Bid, and Puma, to directly activate BAX[31,32].
Based on the above mentioned proteomics and STRING analyses, we concluded that 18β-GRA is likely to inhibit the proliferation of GC cells by regulating the expression of MRPL35, thereby affecting the expression of TP53, BCL2L1, COPS5, BAX, and BAD proteins. Consistent with the above hypothesis, the expression of MRPL35, COPS5, apoptosis-related protein TP53, pro-apoptotic proteins BAX and BAD, and anti-apoptotic protein BCL2L1 was down-regulated after 18β-GRA intervention. The anti-tumor activity of GRA and its derivatives are associated with the mitochondrial apoptotic pathway of mitochondrial membrane potential depolarization, which leads to the activation of caspase-9 and caspase-3, and the production of ROS. Cancer cells can avoid apoptosis by destroying the balance between pro-apoptosis and pro-survival, so that the apoptotic signal of mitochondria is insensitive to cell death. Studies have shown that 18β-GRA may potentiate the heat shock protein 90 inhibition-induced apoptosis in ovarian carcinoma cell lines via the activation of the apoptosis-related proteins and the mitochondria-mediated cell death pathway, leading to activation of caspases[33].
MRPs play a critical role in protein synthesis and mitochondrial function, and also participate in the regulation of cell apoptosis, protein biosynthesis, and signal transduction. MRPL35 and its partner MRP7 play a key role in coordinating the synthesis of the COX1 subunit with its assembly into the COX enzyme[15]. The expression of MRPL35 was found to be higher in CRC tissues than in matched cancer-adjacent tissues. In vitro, down-regulation of MRPL35 led to increased production of ROS along with DNA damage, decrease in mitochondrial membrane potential, and autophagy induction, thereby inhibiting the progression of CRC[16]. Oncomine showed that the expression of MRPL35 in GC tissues was significantly higher than that in adjacent tissues. Our previous results also showed that the expression of MRPL35 was significantly up-regulated in GC, which was associated with a poor survival rate of GC patients. Moreover, the knockdown of MRPL35 could inhibit the proliferation and colony formation of GC cells and induce apoptosis[18].
This study demonstrated that 18-GRA inhibited the proliferation and promoted apoptosis of GC cells by down-regulating the expression of MRPL35, which regulated TP53 and BCL2L1 proteins, both in vivo and in vitro. Our proposed research in the future will be divided into three stages: In the first stage, the signaling pathways of MRPL35, TP53, and BCL2L1 proteins will be explored, possibly through methylation, ubiquitination, acetylation, and phosphorylation. The second stage will be to evaluate the interaction mechanism between MRPL35 and COPS5 proteins. Two modes of action will be considered: Direct action and through tool protein action. Co-immunoprecipitation (Co-IP) or yeast two-hybrid system may be used. In the third stage, MRPL35 and COPS5 protein interaction sites will be explored. After silencing the MRPL35 and COPS5 domains, GST pull-down and Co-IP will be used to verify whether the two proteins interact. The present study aimed to provide a scientific basis for the treatment of GC and identify an effective drug target for the early prevention of GC.
18β-GRA can down-regulate the expression of MRPL35 protein, suppress the expression of anti-apoptotic protein BCL2L1, and promote the pro-apoptotic proteins TP53, BAX, and BAD through COPS5, thus inhibiting the proliferation of gastric cancer cells.
Gastric cancer (GC) is a common malignant tumor of the digestive tract in the world, with more than 1 million new cases to be expected each year since 2020. Mitochondrial ribosomal protein L35 (MRPL35) is a member of the large subunit family of mitochondrial ribosomal proteins, which plays an important role in the development of cancer.
At present, the treatment of gastric cancer is based mainly on surgery, chemotherapy, and radiotherapy, and the first-line treatment drugs are mainly harmful due to side effects.
The purpose of this study was to investigate the correlation between MRPL35 apoptosis related signaling pathway and GC.
Cell functional indexes were determined by cell counting kit-8, flow cytometry, Transwell assay, and cell scratch assay. The effect of 18β-glycyrrhetinic acid (18β-GRA) on proliferation of gastric cancer cells was observed by BALB/C tumor-forming experiments in nude mice. Tandem mass tag labeling combined with liquid chromatography–tandem mass spectrometry were used to screen for differentially expressed proteins extracted from GC cells and control cells after 18β-GRA intervention. Moreover, STRING database was used to predict protein-protein interaction (PPI) relationships and Western blot was used to detect the expression of proteins of interest in GC cells.
18β-GRA can inhibit the proliferation of gastric cancer cells, induce apoptosis, arrest the cell cycle at G0/G1 phase, and inhibit the migration and invasion of gastric cancer cells. 18β-GRA can inhibit tumor formation in BALB/c nude mice. Compared with the control group, MRPL35 and BCL2L1 expression was significantly down-regulated and TP53 expression was up-regulated after 18β-GRA intervention. STRING analysis showed that COPS5, BAX, and BAD proteins could form PPI network with MRPL35, TP53, and BCL2L1 proteins. After 18β-GRA intervention, MRPL35, TP53, and BCL2L1 were up-regulated/down-regulated in a dose-dependent manner, as were COPS5, BAX, and BAD.
18β-GRA can inhibit the proliferation of GC cells by regulating MRPL35, COPS5, TP53, BCL2L1, BAX, and BAD.
MRPL35 can be used for targeted therapy of GC, and can also be used as a new biomarker for GC.
The authors would like to acknowledge Li-Qun Wang for statistical analysis assistance, as well as Joanna Japhet Tibenda for polishing the language.
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 68647] [Article Influence: 13729.4] [Reference Citation Analysis (201)] |
| 2. | Duan R, Li X, Zeng D, Chen X, Shen B, Zhu D, Zhu L, Yu Y, Wang D. Tumor Microenvironment Status Predicts the Efficacy of Postoperative Chemotherapy or Radiochemotherapy in Resected Gastric Cancer. Front Immunol. 2020;11:609337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 3. | Sun W, Jiang C, Ji Y, Xiao C, Song H. Long Noncoding RNAs: New Regulators of Resistance to Systemic Therapies for Gastric Cancer. Biomed Res Int. 2021;2021:8853269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 4. | Wang CY, Kao TC, Lo WH, Yen GC. Glycyrrhizic acid and 18β-glycyrrhetinic acid modulate lipopolysaccharide-induced inflammatory response by suppression of NF-κB through PI3K p110δ and p110γ inhibitions. J Agric Food Chem. 2011;59:7726-7733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 157] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 5. | Huang M, Gong P, Wang Y, Xie X, Ma Z, Xu Q, Liu D, Jing Y, Zhao L. Synthesis and antitumor effects of novel 18β-glycyrrhetinic acid derivatives featuring an exocyclic α,β-unsaturated carbonyl moiety in ring A. Bioorg Chem. 2020;103:104187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 6. | Huang RY, Chu YL, Huang QC, Chen XM, Jiang ZB, Zhang X, Zeng X. 18β-Glycyrrhetinic acid suppresses cell proliferation through inhibiting thromboxane synthase in non-small cell lung cancer. PLoS One. 2014;9:e93690. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 7. | Sharma G, Kar S, Palit S, Das PK. 18β-glycyrrhetinic acid induces apoptosis through modulation of Akt/FOXO3a/Bim pathway in human breast cancer MCF-7 cells. J Cell Physiol. 2012;227:1923-1931. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 99] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 8. | Cao D, Jia Z, You L, Wu Y, Hou Z, Suo Y, Zhang H, Wen S, Tsukamoto T, Oshima M, Jiang J, Cao X. 18β-glycyrrhetinic acid suppresses gastric cancer by activation of miR-149-3p-Wnt-1 signaling. Oncotarget. 2016;7:71960-71973. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 9. | Cai H, Chen X, Zhang J, Wang J. 18β-glycyrrhetinic acid inhibits migration and invasion of human gastric cancer cells via the ROS/PKC-α/ERK pathway. J Nat Med. 2018;72:252-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 10. | Cao D, Wu Y, Jia Z, Zhao D, Zhang Y, Zhou T, Wu M, Zhang H, Tsukamoto T, Oshima M, Jiang J, Cao X. 18β-glycyrrhetinic acid inhibited mitochondrial energy metabolism and gastric carcinogenesis through methylation-regulated TLR2 signaling pathway. Carcinogenesis. 2019;40:234-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 11. | Zeng Y, Shi Y, Xu L, Zeng Y, Cui X, Wang Y, Yang N, Zhou F, Zhou Y. Prognostic Value and Related Regulatory Networks of MRPL15 in Non-Small-Cell Lung Cancer. Front Oncol. 2021;11:656172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 12. | Xu H, Zou R, Li F, Liu J, Luan N, Wang S, Zhu L. MRPL15 is a novel prognostic biomarker and therapeutic target for epithelial ovarian cancer. Cancer Med. 2021;10:3655-3673. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 13. | Hao C, Duan H, Li H, Wang H, Liu Y, Fan Y, Zhang C. Knockdown of MRPL42 suppresses glioma cell proliferation by inducing cell cycle arrest and apoptosis. Biosci Rep. 2018;38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 14. | Brown A, Amunts A, Bai XC, Sugimoto Y, Edwards PC, Murshudov G, Scheres SHW, Ramakrishnan V. Structure of the large ribosomal subunit from human mitochondria. Science. 2014;346:718-722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 260] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 15. | Box JM, Kaur J, Stuart RA. MrpL35, a mitospecific component of mitoribosomes, plays a key role in cytochrome c oxidase assembly. Mol Biol Cell. 2017;28:3489-3499. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 16. | Zhang L, Lu P, Yan L, Yang L, Wang Y, Chen J, Dai J, Li Y, Kang Z, Bai T, Xi Y, Xu J, Sun G, Yang T. MRPL35 Is Up-Regulated in Colorectal Cancer and Regulates Colorectal Cancer Cell Growth and Apoptosis. Am J Pathol. 2019;189:1105-1120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 17. | Wang AF, Zhang Q, Zhang DM, Xiu YT, Ding YM, Liu LL. The effect of silencing mitochondrial ribosomal protein L35 gene on the growth of human esophageal cancer TE-1 cells. J Jilin University. 2019;45:28-32+217. |
| 18. | Yuan L, Li JX, Yang Y, Chen Y, Ma TT, Liang S, Bu Y, Yu L, Nan Y. Depletion of MRPL35 inhibits gastric carcinoma cell proliferation by regulating downstream signaling proteins. World J Gastroenterol. 2021;27:1785-1804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 4] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 19. | Lin D, Zhong W, Li J, Zhang B, Song G, Hu T. Involvement of BID translocation in glycyrrhetinic acid and 11-deoxy glycyrrhetinic acid-induced attenuation of gastric cancer growth. Nutr Cancer. 2014;66:463-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 20. | Matthews HK, Bertoli C, de Bruin RAM. Cell cycle control in cancer. Nat Rev Mol Cell Biol. 2022;23:74-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 490] [Cited by in RCA: 958] [Article Influence: 239.5] [Reference Citation Analysis (1)] |
| 21. | Zilfou JT, Lowe SW. Tumor suppressive functions of p53. Cold Spring Harb Perspect Biol. 2009;1:a001883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 359] [Cited by in RCA: 422] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 22. | Chen J. The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression. Cold Spring Harb Perspect Med. 2016;6:a026104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 494] [Cited by in RCA: 876] [Article Influence: 87.6] [Reference Citation Analysis (0)] |
| 23. | Bertoli C, Skotheim JM, de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol. 2013;14:518-528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 844] [Cited by in RCA: 1120] [Article Influence: 86.2] [Reference Citation Analysis (0)] |
| 24. | Bertoli C, Herlihy AE, Pennycook BR, Kriston-Vizi J, de Bruin RAM. Sustained E2F-Dependent Transcription Is a Key Mechanism to Prevent Replication-Stress-Induced DNA Damage. Cell Rep. 2016;15:1412-1422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 25. | Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol. 2017;11:28-39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 348] [Cited by in RCA: 545] [Article Influence: 54.5] [Reference Citation Analysis (0)] |
| 26. | Yuan L, Li J, Yang Y, Chen Y, Bu Y, Ye M, Mao X, Ma T, Yu L, Nan Y. LINC00514 promotes gastric cancer cell growth and EMT progression via miR-204-3p/KRAS. Aging (Albany NY). 2021;13:12007-12015. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 27. | Hafner A, Bulyk ML, Jambhekar A, Lahav G. The multiple mechanisms that regulate p53 activity and cell fate. Nat Rev Mol Cell Biol. 2019;20:199-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 427] [Cited by in RCA: 808] [Article Influence: 115.4] [Reference Citation Analysis (0)] |
| 28. | Maes ME, Schlamp CL, Nickells RW. BAX to basics: How the BCL2 gene family controls the death of retinal ganglion cells. Prog Retin Eye Res. 2017;57:1-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 160] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 29. | Chang LK, Putcha GV, Deshmukh M, Johnson EM Jr. Mitochondrial involvement in the point of no return in neuronal apoptosis. Biochimie. 2002;84:223-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 91] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 30. | Chang CY, Shen CC, Su HL, Chen CJ. Gefitinib induces apoptosis in human glioma cells by targeting Bad phosphorylation. J Neurooncol. 2011;105:507-522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 31. | Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol Rev. 2007;87:99-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2548] [Cited by in RCA: 2748] [Article Influence: 144.6] [Reference Citation Analysis (3)] |
| 32. | Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L). Cell. 1996;87:619-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1984] [Cited by in RCA: 2002] [Article Influence: 66.7] [Reference Citation Analysis (0)] |
| 33. | Yang JC, Myung SC, Kim W, Lee CS. 18β-glycyrrhetinic acid potentiates Hsp90 inhibition-induced apoptosis in human epithelial ovarian carcinoma cells via activation of death receptor and mitochondrial pathway. Mol Cell Biochem. 2012;370:209-219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Matsuoka T, Japan; Nakajima N, Japan; Vorobjova T, Estonia S-Editor: Fan JR L-Editor: Wang TQ P-Editor: Yu HG