Published online May 7, 2022. doi: 10.3748/wjg.v28.i17.1781
Peer-review started: October 18, 2021
First decision: December 12, 2021
Revised: December 31, 2021
Accepted: March 27, 2022
Article in press: March 27, 2022
Published online: May 7, 2022
Processing time: 192 Days and 21.7 Hours
Colorectal cancer (CRC) is an extremely malignant tumor with a high mortality rate. Little is known about the mechanism by which forkhead Box q1 (FOXQ1) causes CRC invasion and metastasis through the epidermal growth factor receptor (EGFR) pathway.
To illuminate the mechanism by which FOXQ1 promotes the invasion and metastasis of CRC by activating the heparin binding epidermal growth factor (HB-EGF)/EGFR pathway.
We investigated the differential expression and prognosis of FOXQ1 and HB-EGF in CRC using the Gene Expression Profiling Interactive Analysis (GEPIA) website (http://gepia.cancer-pku.cn/index.html). Quantitative real-time polymerase chain reaction (qRT-PCR) and Western blotting were used to detect the expression of FOXQ1 and HB-EGF in cell lines and tissues, and we constructed a stable low-expressing FOXQ1 cell line and verified it with the above method. The expression changes of membrane-bound HB-EGF (proHB-EGF) and soluble HB-EGF (sHB-EGF) in the low-expressing FOXQ1 cell line were detected by flow cytometry and ELISA. Western blotting was used to detect changes in the expression levels of HB-EGF and EGFR pathway-related down
GEPIA showed that the expression of FOXQ1 in CRC tissues was relatively high and was related to a lower overall survival rate. PCR array results showed that FOXQ1 is related to the HB-EGF and EGFR pathways. Knockdown of FOXQ1 suppressed the expression of HB-EGF, and led to a decrease in EGFR and its downstream genes AKT, RAF, KRAS expression levels. After knockdown of FOXQ1 in CRC cell lines, cell proliferation, migration and invasion were attenuated. Adding HB-EGF restored the migration and invasion ability of CRC, but not the cell proliferation ability. Kaplan–Meier survival analysis results showed that the combination of FOXQ1 and HB-EGF may serve to predict CRC survival.
Based on these collective data, we propose that FOXQ1 promotes the invasion and metastasis of CRC via the HB-EGF/EGFR pathway.
Core Tip: Invasion and metastasis play important roles in tumorigenesis, resulting in the death of most colorectal cancer (CRC) patients. Forkhead Box q1 (FOXQ1) is a well-established oncogene in multiple tumors, including CRC. Our previous study suggested that FOXQ1 positively regulates the expression of heparin-binding epidermal growth factor (HB-EGF) and triggers the activation of the epidermal growth factor receptor (EGFR) pathway in CRC. However, the role and mechanism of how FOXQ1 promotes tumorigenesis in CRC by activating the HB-EGF/EGFR pathway remain unexplored. In the present study, our findings demonstrated that the essential role of FOXQ1-induced invasion and metastasis in CRC was related to activation of the HB-EGF/EGFR pathway.
- Citation: Zhang JJ, Cao CX, Wan LL, Zhang W, Liu ZJ, Wang JL, Guo Q, Tang H. Forkhead Box q1 promotes invasion and metastasis in colorectal cancer by activating the epidermal growth factor receptor pathway. World J Gastroenterol 2022; 28(17): 1781-1797
- URL: https://www.wjgnet.com/1007-9327/full/v28/i17/1781.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i17.1781
Colorectal cancer (CRC) is the third most common cancer in the world and the fourth most common cause of cancer death in the world[1]. Among all cancerous processes involved, local invasion and metastasis are the main factors related to cancer. The metastatic dissemination of primary tumors is directly related to the survival rate of patients, accounting for approximately 90% of all colon cancer deaths[2]. The median survival of metastatic CRC is less than 2 years[3]. Therefore, clarifying the mechanism of invasion and metastasis is the key to improving the survival rate of patients with CRC.
Forkhead Box q1 (FOXQ1) is a member of the forkhead transcription factor family[4], and it promotes tumorigenesis by activating cell proliferation, invasion and apoptosis[5]. Kaneda et al[6] found that compared to adjacent tissues, FOXQ1 is overexpressed in CRC. Overexpression of FOXQ1 reduces cell proliferation but increases cell tumorigenicity and tumor growth, and it inhibits apoptosis and promotes angiogenesis, thereby promoting CRC tumorigenesis. Liu et al[7] also demonstrated that the expression of FOXQ1 in either CRC tissue samples or cancer cell lines is higher than that in normal colorectal tissues and cell lines, and they reported that FOXQ1 promotes cancer metastasis by regulating PI3K/AKT signaling. Weng et al[8] verified that FOXQ1 can be used as an independent indicator of the prognosis of CRC patients.
The epidermal growth factor receptor (EGFR) signaling pathway plays an important role in physiological processes, such as cell growth, proliferation, and differentiation. The expression level of EGFR gradually increases from normal mucosa, adenomas with low-grade dysplasia, and adenomas with high-grade dysplasia to CRC, confirming that EGFR plays an important role in CRC[9]. Heparin-binding epidermal growth factor (HB-EGF) is one of the seven major ligands of EGFR. HB-EGF was originally identified as a secreted product from human macrophage U937 cells, and it induces cell proliferation and differentiation[10,11]. Soluble HB-EGF (sHB-EGF) is the main stimulator of cell proliferation. The affinity of sHB-EGF binding to target cells to promote proliferation of the target cells and activation of EGFR tyrosine kinase activity is 20-40 times higher than that of EGF. Thus, sHB-EGF is the most effective EGFR signaling pathway activator[12,13]. Membrane-bound HB-EGF (proHB-EGF) is affected by a variety of proteins, and it can be shed into active sHB-EGF to promote cell proliferation[14-16].
There is evidence that poor prognosis and low survival rates for CRC are associated with abnormally activated signaling pathways, including the EGFR signaling pathway[17]. In advanced CRC the most commonly used targeted therapies are the monoclonal antibodies cetuximab and panitumab, which block EGFR activation[18]. Activation of EGFR signaling leads to resistance to chemotherapy in CRC cells and promotes cell survival, while inhibition of EGFR signaling significantly reduces proliferation in CRC cells[19]. In nasopharyngeal carcinoma, Luo et al[20] reported that FOXQ1 induces vasculogenic mimicry through the EGFR signaling pathway, thereby promoting the metastasis of nasopharyngeal carcinoma cells. Our previous study suggested that FOXQ1 positively regulates the expression of HB-EGF and triggers the activation of the EGFR pathway in CRC[21]. However, the role and mechanism of how FOXQ1 promotes tumorigenesis in CRC by activating the HB-EGF/EGFR pathway remain largely unknown. Therefore, we analyzed the correlation between FOXQ1 and the HB-EGF/EGFR pathway by constructing FOXQ1 knockdown cells and using tissue microarrays, cell function experiments, quantitative real-time polymerase chain reaction (qRT-PCR), and western blotting. Our findings elucidated the critical role of the FOXQ1 and HB-EGF/EGFR pathways in CRC, providing theoretical support for the clinical application of targeted FOXQ1 in the treatment of CRC.
The human CRC cell lines, DLD1 and SW480, as well as the human embryonic kidney 293 (HEK293) cell line were purchased and authenticated from the Cell Bank of the Chinese Academy of Science in Shanghai, China. DLD1 cells were cultured in RPMI 1640. SW480 and HEK293 cells were cultured in DMEM. All media were supplemented with 10% heat-inactivated fetal bovine serum (Life Technologies BRL), and the cells were maintained in a 5% CO2-humidified atmosphere at 37 °C.
Three siRNAs targeting the human FOXQ1 sequence (NM_033260.3) were designed using siRNA Target Finder (InvivoGen, San Diego, CA, United States), and one scrambled siRNA was designed as a negative control, refer to our previous research[21]. The pSPAX2 packaging system from Addgene was used to construct a lentiviral PLKO.1 vector (PLKO.1-puro-shFOXQ1). After each group of recombinant plasmids was confirmed by sequencing, lentiviral vectors and packing vectors (pRSV-rev, pMDlg-pRRE and pCMV-VSV-G) were cotransfected into HEK293 cells using Lipofectamine® 2000 transfection reagent (Life Technologies). Lentivirus was collected to infect DLD1 and SW480 cells. Stable cells were generated after selection with puromycin (Solarbio, Beijing, China) (0.4 ng/μL for DLD1 cells and 0.1 ng/μL for SW480 cells) for 7-14 d after infection. The most effective knockdown cells were designated DLD1-shFOXQ1 and SW480-shFOXQ1, and the corresponding controls were named DLD1-shControl and SW480-shControl, respectively.
Cell surface HB-EGF was detected using APC-conjugated anti-human HB-EGF (eBioscience, San Diego, CA, United States). APC-conjugated mouse IgG2ak isotype was used as a control (eBioscience, San Diego, CA, United States) according to the manufacturer’s directions. Briefly, CRC cells were harvested and blocked with blocking buffer (PBS containing 2% BSA) for 10 min at 4 °C and then stained with 5 μL of monoclonal HB-EGF antibody (eBioscience, San Diego, CA, United States) for 30 min at 4 °C. After two washes, cells were resuspended in 100 μL of PBS. Samples were analyzed using a MoFlo flow cytometer (Beckman Coulter) and FlowJo software (Becton, Dickinson and Company).
Protein extracts were isolated from 106 treated cells using mammalian cell lysis reagent containing protease inhibitor cocktail (Sigma–Aldrich, St. Louis, MO, United States) and phosphatase inhibitors (Roche, United States) following the manufacturer’s directions. Equal amounts of protein (30 μg) were resolved on a 10% sodium dodecyl sulfate (SDS)-precast polyacrylamide gel (Bio–Rad Laboratories) and transferred to an Immobilon-polyvinylidene difluoride membrane (Millipore, Billerica, MA, United States). The membranes were blocked and incubated with the following primary antibodies: FOXQ1 (Abcam, Cambridge, MA, United States), HB-EGF (Abcam, Cambridge, MA, United States), phosphor-PI3K (Cell Signaling Technology, Cold Spring Harbor, NY, United States), PI3K (Proteintech, Wuhan, China), Akt (Proteintech, Wuhan, China), phosphor-Akt (Proteintech, Wuhan, China), phosphor-MAPK (Cell Signaling Technology, Cold Spring Harbor, NY, United States), MAPK (Proteintech, Wuhan, China), EGFR (Proteintech, Wuhan, China), KRAS (Proteintech, Wuhan, China), RAF (Proteintech, Wuhan, China), E-cadherin, N-cadherin, vimentin and β-actin (Proteintech, Wuhan, China). Blots were then incubated with the appropriate peroxidase-conjugated secondary antibody as follows: HRP-Rb-anti-goat (Cell Signaling Technology, Cold Spring Harbor, NY, United States) or HRP-goat-anti-mouse (Proteintech, Wuhan, China), respectively. The proteins were detected using an ECL system (Millipore, Braunschweig, Germany) and visualized with a ChemiDoc XRS system (Bio–Rad, Hercules, CA, United States).
Total RNA was isolated using TRIzol (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s protocol. Total RNA (1 μg) was reverse transcribed into cDNA using PrimeScript RT reagent (TakaraBio, Japan), and qRT–PCR was performed using a LightCycler 480 (Roche, United States) with SYBR Premix Ex Taq (Takara, China). Each sample was analyzed in triplicate, and Glyceraldehyde-3-phosphate dehydrogenase(GAPDH) was used as the reference. Quantitative results were calculated using the 2-∆∆CT method. Primers used for qRT–PCR were designed and synthesized by Takara (Dalian, China) (Table 1).
| Gene name | Forward (5'→3') | Reverse (5'→3') |
| FOXQ1 | TGACTTCAACAGCGACACCCA | CACCCTGTTGCTGTAGCCAAA |
| EGFR | AGACGCAGATAGTCGCCCAAAG | TCCATCAGGGCACGGTAGAAG |
| HB-EGF | CCATGTCTTCGGAAATACAAGGA | CCAGGATGGTTGTGTGGTCA |
| Akt | AGACGCAGATAGTCGCCCAAAG | TCCATCAGGGCACGGTAGAAG |
| RAF1 | AGACGCAGATAGTCGCCCAAAG | TCCATCAGGGCACGGTAGAAG |
| KRAS | GTGGACGAATATGATCCAACAATAG | TGCTGTGTCGAGAATATCCAAGAG |
| GAPDH | GGCAACGGGCTACAGCTTTA | GGCACCCCACATACATAATCAA |
The human epidermal growth factor (EGF)/platelet-derived growth factor (PDGF) Signaling RT² Profiler™ PCR Array (SABiosciences), which profiles the expression of 84 genes related to EGF/PDGF-mediated signal transduction, five housekeeping genes and three controls, was used to analyze the effect of FOXQ1 on EGF/PDGF signaling-related gene expression (Table 2). Total RNA was extracted with TRIzol reagent according to the manufacturer’s manual. DNase treatment was performed by amplification grade I DNase I (Sigma–Aldrich, St. Louis, MO, United States) according to the manufacturer’s instructions. Each total RNA preparation (5 μg) was digested with 1 μL of DNase I (1 unit/μL) and 1 μL of 10 reaction buffer in a volume of 10 μL. After incubation and addition of Stop Solution, DNase I was denatured by incubation at 70 °C for 10 min. The RNA samples were kept on ice for another 5 min and then converted into cDNA with the RT2 PCR Array First Strand Kit (SuperArray) according to the manufacturer’s protocol. cDNA (20 ng) was combined with RT2 SYBR Green/ Fluorescein PCR master mix (SuperArray), and equal amounts of this mixture (25 μL) were added to each well of the RT2 Profiler PCR plate containing the predispensed gene-specific primer sets. PCR cycles were performed according to the manufacturer's instructions. The relative level of mRNA expression for each gene in each sample was first normalized to the expression of GAPDH in that sample and then normalized to the level of mRNA expression in the DLD1-shControl.
| Symbol | Gene information | Fold change (shFOXQ1/shControl) | P value | FOXQ1 binding sites (TSS: -2k_+1k) |
| PLAT | Plasminogen activator, tissue | -8.01 | 0.0375 | 3 |
| COL1A1 | Collagen, type I, alpha 1 | -5.72 | 0.0138 | 1 |
| ELK1 | ELK1, member of ETS oncogene family | -5.45 | 0.0228 | 1 |
| BCAR1 | Breast cancer anti-estrogen resistance 1 | -3.97 | 0.0479 | 2 |
| PIK3R2 | Phosphoinositide-3-kinase, regulatory subunit 2 | -3.63 | 0.0197 | 3 |
| NCK2 | NCK adaptor protein 2 | -3.51 | 0.0237 | 6 |
| JUN | Jun proto-oncogene v-jun | -3.43 | 0.0077 | 12 |
| EGR1 | Early growth response 1 | -3.36 | 0.0166 | 11 |
| LTA | Lymphotoxin alpha (TNF superfamily, member 1) | -3.18 | 0.0796 | 3 |
| PDGFB | Platelet-derived growth factor beta | -2.91 | 0.1896 | 5 |
| STAT3 | Signal transducer and activator of transcription 3 (acute-phase response factor) | -2.49 | 0.0038 | 6 |
| HB-EGF | Heparin-binding EGF-like growth factor | -2.19 | 0.0278 | 3 |
| FN1 | Fibronectin 1 | -2.10 | 0.0005 | 6 |
| FASLG | Fas ligand (TNF superfamily, member 6) | -2.08 | 0.2026 | 17 |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | -2.06 | 0.0058 | 2 |
| MAP2K1 | Mitogen-activated protein kinase kinase 1 | -2.02 | 0.0243 | 5 |
| AKT3 | V-akt murine thymoma viral oncogene homolog 3 | -2.02 | 0.1440 | 11 |
| FOS | FBJ murine osteosarcoma viral oncogene homolog | -2.01 | 0.0470 | 5 |
Exogenous recombinant HB-EGF protein at a final concentration of 50 ng/mL was added to the cell culture medium when the cell density reached 80%, and the culture medium was changed after incubation for 24 h[22]. The supernatant was collected after the cells were cultured for another 24 h. The protein concentrations of soluble HB-EGF, ADAM9, ADAM10, ADAM12 and MMP-7 in the cell culture medium were determined by ELISA detection kits against human HB-EGF, ADAM9, ADAM10, ADAM12 and MMP-7, respectively (R&D Systems, Minneapolis, United States).
DLD1-shFOXQ1 and SW480-shFOXQ1 cells (2,000 cells/well in a 96-well plate) were incubated with medium containing 10% FBS at 37 °C for 24, 48, 72 and 96 h. At the end of incubation, 10 μL of Cell Counting Kit-8 (CCK-8) solution (Beyotime Biotech, Shanghai, China) was added to each well with 100 μL of medium and incubated for another 4 h at 37 °C, and the OD at 450 nm was measured by a microplate reader (BioTek, Winooski, VT, United States). The effect of siRNA FOXQ1 on CRC cell viability was assessed as the percent of cell viability compared to vehicle-treated control cells, which were arbitrarily assigned as 100% viability.
Cell migration was analyzed using Transwell inserts with 8.0 μm membrane pores (BD, San Jose, CA, United States) according to the manufacturer’s protocol. Migration was additionally evaluated with the wound-healing assay. Briefly, DLD1-shFOXQ1/SW480-shFOXQ1 and DLD1-shControl/SW480-shControl cells were seeded in 6-well plates at a density that enabled a confluency of 80% to be attained 24 h after plating. A 10 μL filter tip was used to gently scratch the cell monolayer across the center of the well. Cells were then gently washed twice with PBS to remove the dislodged cells, and fresh medium was added. The first images of the scratch area were then acquired. Cells were cultured in serum-free medium for another 48 h, and a second set of images was then acquired to determine the extent of wound closure.
Tissue microarrays containing a total of 90 pairs of colorectal tumor tissues and matched adjacent normal tissues, together with pathological staging data in accordance with TNM classification of the American Joint Committee on Cancer (2010) and follow-up survival time after surgery, were obtained from Shanghai Biochip Co. Ltd., Shanghai, China (HCol-Ade180Sur-06). FOXQ1 (ab51340) and HB-EGF (ab192545) antibodies were purchased from Abcam (Cambridge, MA, United States). Tissue microarray analysis was performed using a standard immunohistochemistry (IHC) protocol. The median value of the immunoreactivity score (IRS) was selected as the cutoff for high and low protein expression levels based on a measure of heterogeneity according to the log-rank test with respect to disease-specific survival (DSS) as described previously. Cutoff values for the scoring system were assigned as follows: High expression of FOXQ1 and HB-EGF were defined as an IRS of ≥ 4 (4, 6, 8, 9 and 12); and low expression was defined as an IRS of < 4 (0, 1, 2 and 3)[23]. Immunostained sections were scanned using a microscope (Axiovert 200, Carl Zeiss, Gttingen, Germany). Data for 25 patients were excluded because the dots were off the chips during the experiment. In total, data for 65 patients with CRC were included in the final analysis (Tables 3 and 4).
| Clinicopathological variables | Cohort tumor HB-EGF expression | P value | ||
| Negative (n = 26) | Positive (n = 39) | |||
| Age (yr) | 65.72 (10.37) | 69.43 (7.98) | ||
| Gender | Female | 11 | 23 | > 0.05 |
| Male | 14 | 16 | ||
| Size of tumor (mm) | ≤ 10 | 5 | 7 | > 0.05 |
| > 10 | 21 | 32 | ||
| Lymphatic metastasis | Absent | 19 | 19 | > 0.05 |
| Present | 7 | 20 | ||
| Tumor differentiation | I-II | 12 | 24 | > 0.05 |
| III-IV | 14 | 15 | ||
| TNM stage | I-II | 11 | 8 | > 0.05 |
| III-IV | 15 | 31 | ||
| AJCC clinical stage according to 7th issue | 1 and 2A | 17 | 24 | > 0.05 |
| 3 and 3B | 9 | 15 | ||
| FOXQ1, negative (n = 31) | FOXQ1, positive (n = 34) | P value | |||
| HB-EGF | Negative (n = 26) | 20 | 6 | 4.116 | < 0.05 |
| Positive (n = 39) | 11 | 28 |
Statistical analyses were performed using GraphPad Prism 8 and SPSS v.19. An unpaired two-tailed Student’s t-test was performed for two-group comparisons, and one-way analysis of variance (ANOVA) was performed for multiple group comparisons. Survival curves were calculated using the Kaplan–Meier algorithm and log-rank test. P < 0.05 was considered to indicate a statistically significant difference.
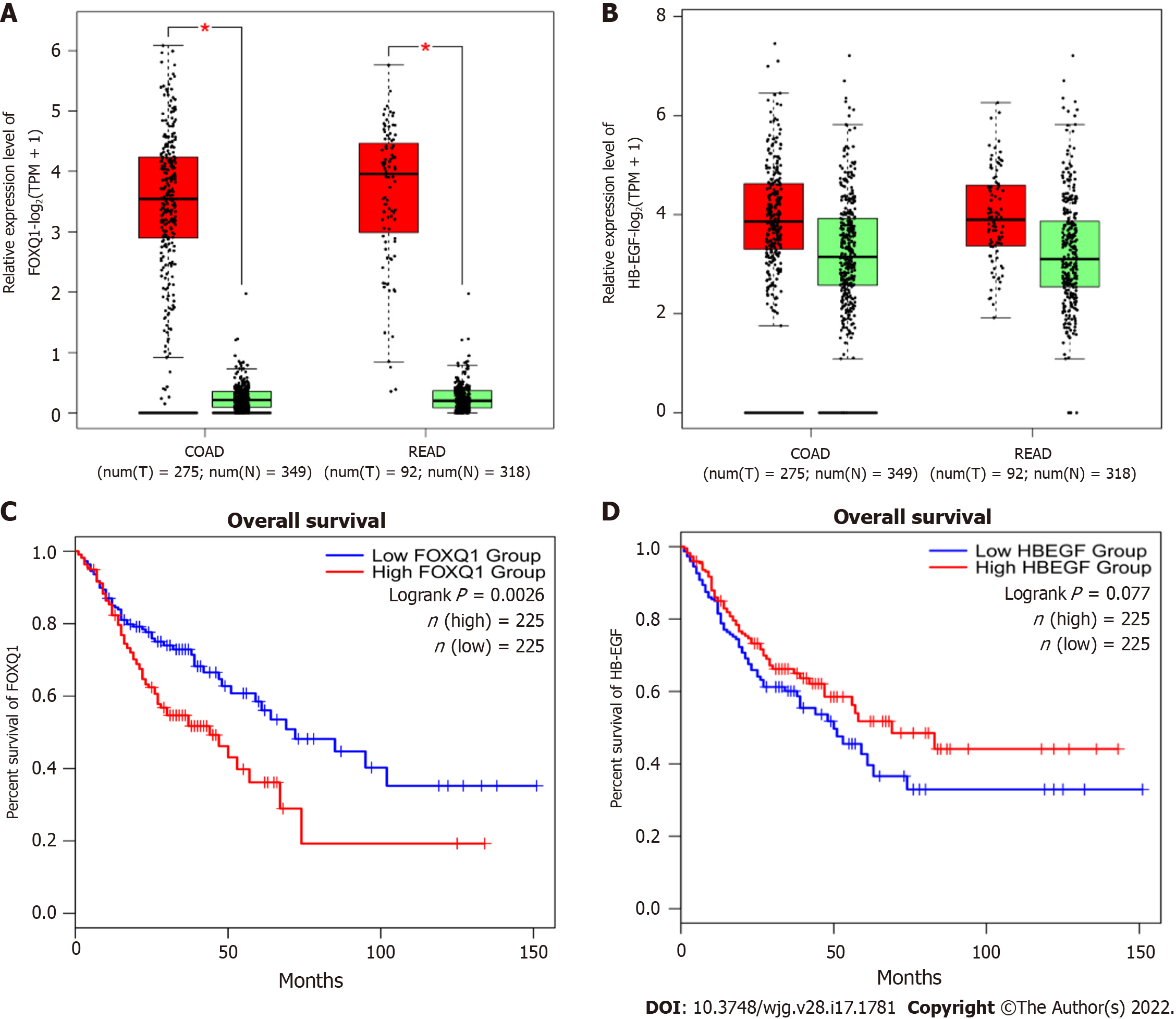
To determine the expression levels of FOXQ1 and HB-EGF in CRC, we investigated the expression and prognosis of FOXQ1 and HB-EGF in CRC in the Gene Expression Profiling Interactive Analysis (GEPIA) online database (Figure 1). The results showed that FOXQ1 was upregulated in CRC compared to normal samples according to GEPIA (Figure 1A). Increased expression of FOXQ1 was also associated with worse overall survival (Figure 1C). The expression of HB-EGF in CRC was not significantly different from that in normal colorectal tissues (Figure 1B), and its expression had no significant effect on the overall survival of patients with CRC (Figure 1D).
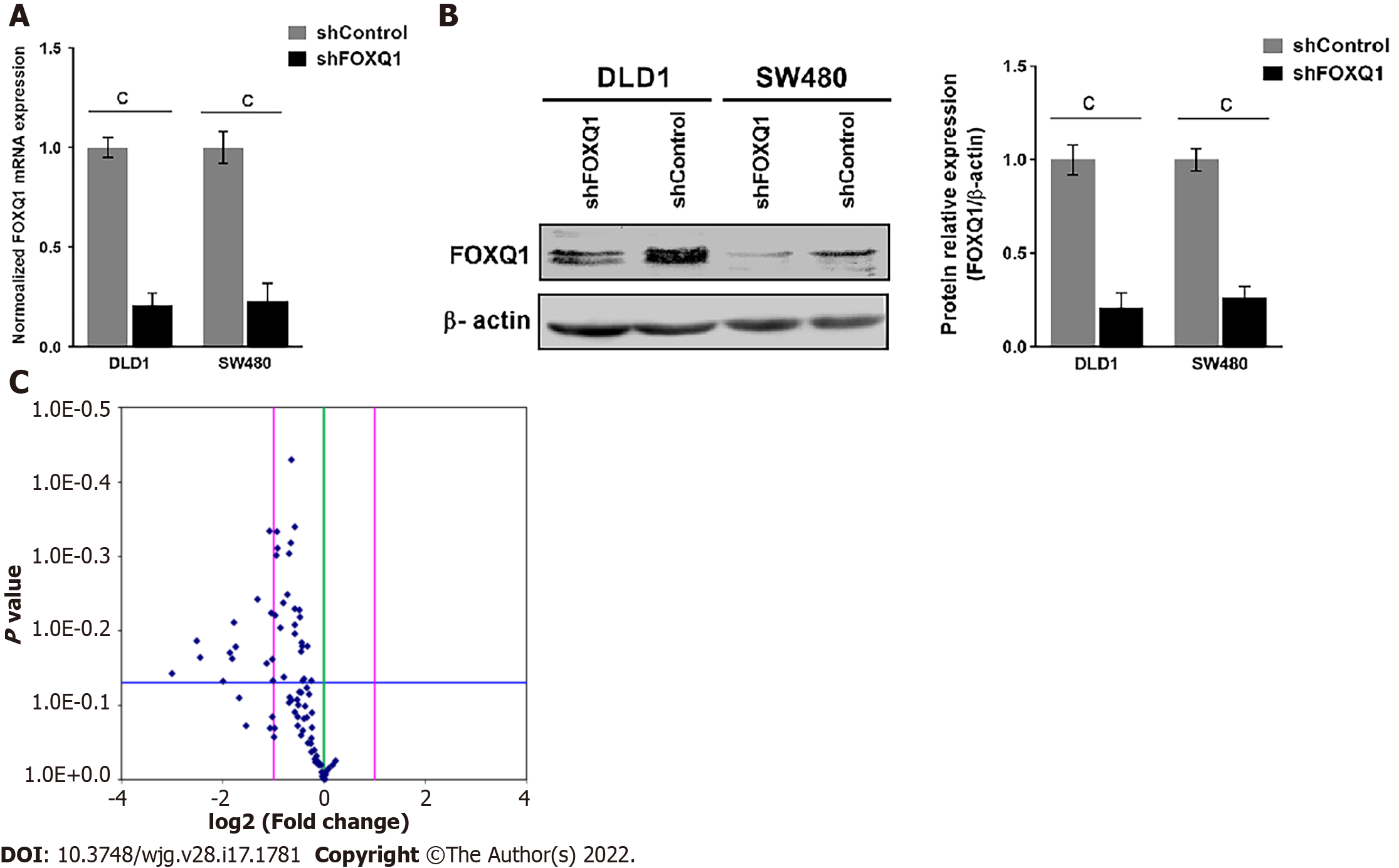
To elucidate the functional roles of FOXQ1 in CRC, we generated CRC lines with stable FOXQ1 knockdown. Among the CRC lines we tested, DLD1 and SW480 cells had relatively high endogenous FOXQ1 expression as described previously[21]. Because the high expression of FOXQ1 in these two cells lines has also been confirmed in several other independent studies[3,4,8,9,24], we selected these two cell lines for knockdown studies. qRT–PCR assays verified significant FOXQ1 knockdown in DLD1-shFOXQ1 and SW480-shFOXQ1 cells. FOXQ1 mRNA expression in DLD1-shFOXQ1 and SW480-shFOXQ1 cells was significantly reduced compared to that in the control DLD1-shControl and SW480-shControl cells (Figure 2A). Western blot analysis also verified that the expression of FOXQ1 was significantly downregulated (Figure 2B).
A panel of PCR arrays consisting of 84 representative genes related to the EGF/PDGF signaling pathways was used to detect the transcriptional signatures of DLD1-shFOXQ1 and DLD1-shControl cells. Differentially expressed genes with statistical significance were identified by volcano plot filtering. The results showed that 18 genes had expression changes with a fold-change ≥ 2.0 (P < 0.05)(Figure 2C). Among these 18 genes, 12 genes were associated with EGFR signaling pathways, including HB-EGF (FC = -2.19), and there were several novel genes of the three main EGF/PDGF downstream signaling pathways (MAPK/ERK1/2, PI3K/AKT and JAK/STAT3). These novel genes included RAS, PI3K and STAT3 as well as several intracellular transcription factors activated by these signaling pathways, such as c-JUN and c-FOS (Table 2).
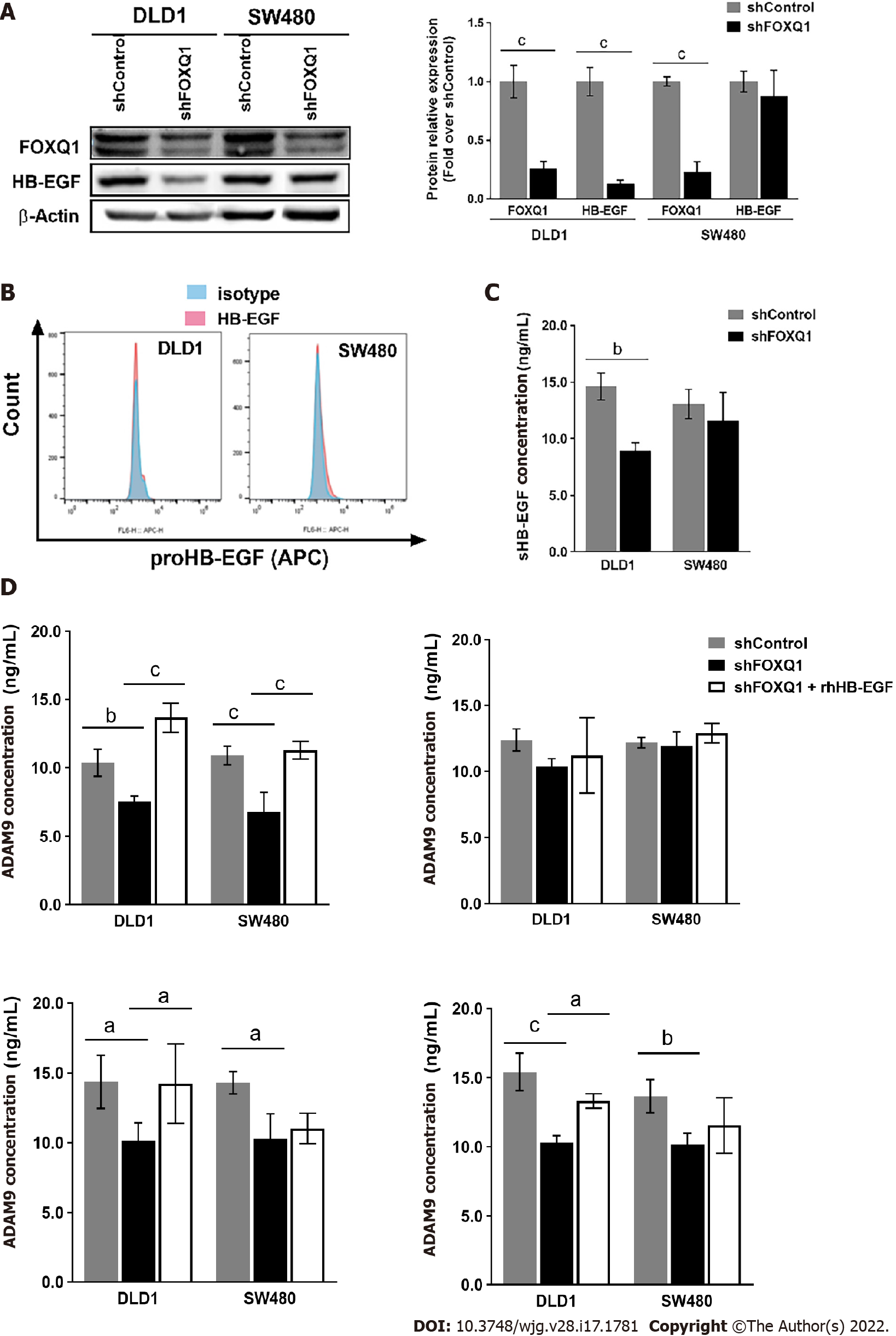
Western blot analysis confirmed that knockdown of FOXQ1 in CRC cells resulted in a significant decrease in the expression of HB-EGF in DLD1 cells (Figure 3A), but no significant changes in HB-EGF were observed in SW480 cells (Figure 3A). Flow cytometry analysis confirmed that knockdown of FOXQ1 expression in CRC cells did not affect the expression level of proHB-EGF (Figure 3B). ELISA results indicated that knockdown of FOXQ1 significantly reduced the shedding of soluble HB-EGF (sHB-EGF) in DLD1 cells but did not affect the shedding of HB-EGF in SW480 cells (Figure 3C). Among the four proteins that affect the release of the extracellular domain of proHB-EGF, three (ADAM9, ADAM12 and ADAM7) were decreased significantly in both DLD1-shFOXQ1 and SW480-shFOXQ1 cells, while ADAM10 was not changed. The feedback regulation of ADAM9, ADAM12 and MMP7 secretion by HB-EGF from CRC cells was studied by adding exogenous recombinant human HB-EGF (rhHB-EGF). The results showed that rhHB-EGF reversed the decline in the expression of ADAM9, ADAM12 and MMP7 in DLD-shFOXQ1 cells. However, only the decreased expression of ADAM9 was reversed by rhHB-EGF in SW480-shFOXQ1 cells (Figure 3D).
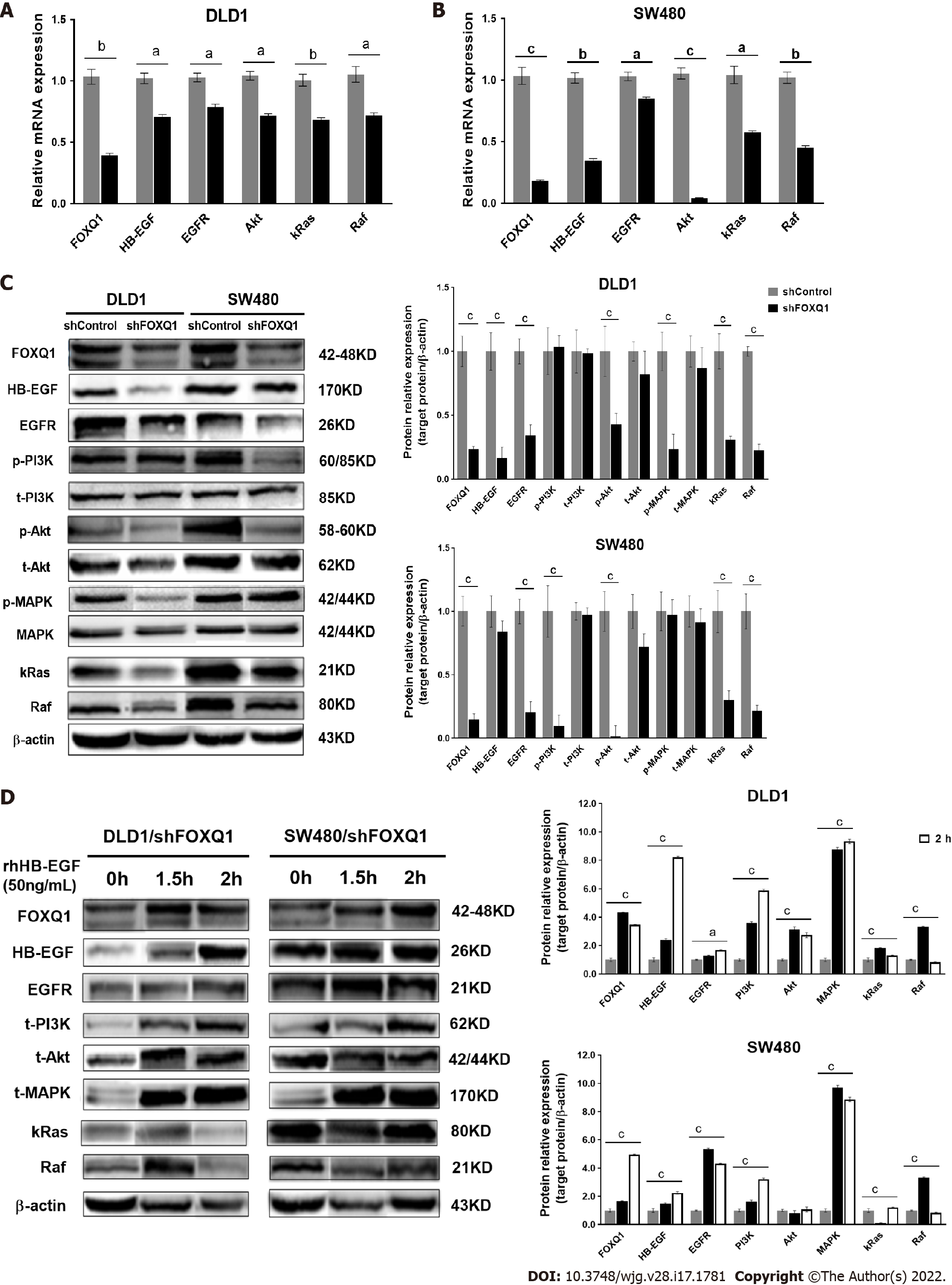
To verify the role of FOXQ1 in activating the HB-EGF/EGFR signaling pathway, we performed qRT–PCR analysis in DLD1-shFOXQ1 and SW480-shFOXQ1 cells. The results confirmed that FOXQ1 knockdown resulted in a significant decrease in the mRNA expression of HB-EGF, EGFR and downstream genes (AKT, RAF and KRAS) in DLD1-shFOXQ1 and SW480-shFOXQ1 cells (Figure 4A and B). In addition, the Western blot analysis showed that knockdown of FOXQ1 resulted in a significant decrease in HB-EGF and EGFR expression as well as decreased AKT and MAPK phosphorylation in DLD1-shFOXQ1 cells compared to DLD1-shControl cells. In SW480-shFOXQ1 cells, EGFR and HB-EGF expression was significantly decreased, and AKT and PI3K phosphorylation was inhibited compared to that in SW480-shControl cells (Figure 4C). Furthermore, Western blot analysis confirmed that the decreased expression of important downstream genes was rescued by rhHB-EGF protein in either DLD1-shFOXQ or SW480-shFOXQ1 cells (Figure 4D).
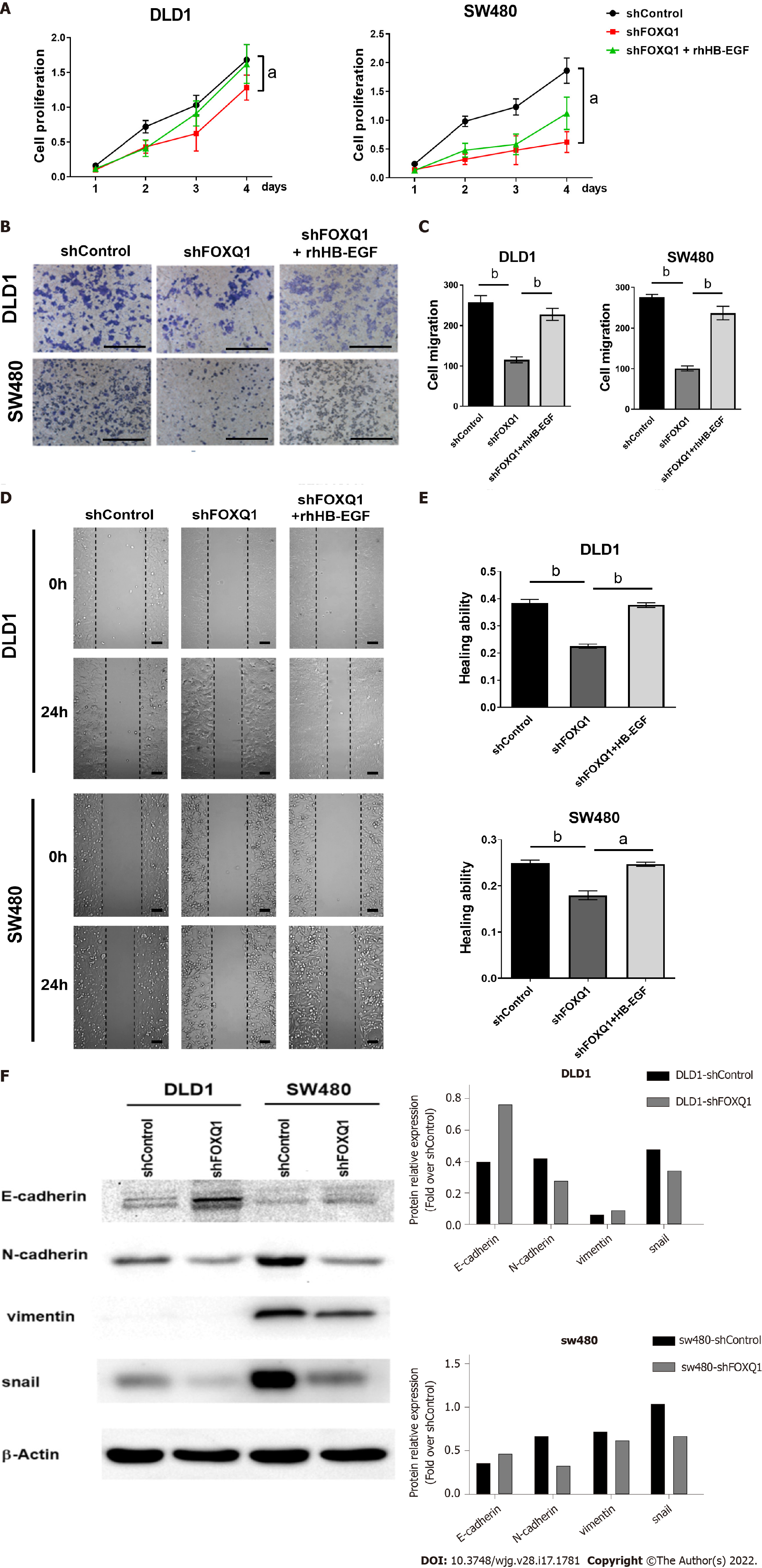
CCK-8 results confirmed that FOXQ1 knockdown significantly inhibited the proliferation of DLD1-shFOXQ1 and SW480-shFOXQ1 cells, and the inhibitory effect was partially reversed by exogenous rhHB-EGF (Figure 5A). The results of the Transwell migration assay confirmed that FOXQ1 knockdown also reduced the migration of DLD1 and SW480 cells, and the inhibitory effect was also reversed to a large extent by rhHB-EGF (Figure 5B and C). Scratch experiment results confirmed that FOXQ1 knockdown reduced the wound-healing ability of DLD1 and SW480 cells, which was also reversed to a large extent by rhHB-EGF (Figure 5D and E). We next analyzed the protein expression during cell invasion and metastasis (Ecadherin, N-cadherin, Vimentin and Snail), Western blot analysis indicated that FOXQ1 knockdown reduced the expression in DLD1-shFOXQ1 and SW480-shFOXQ1 cells (Figure 5F).
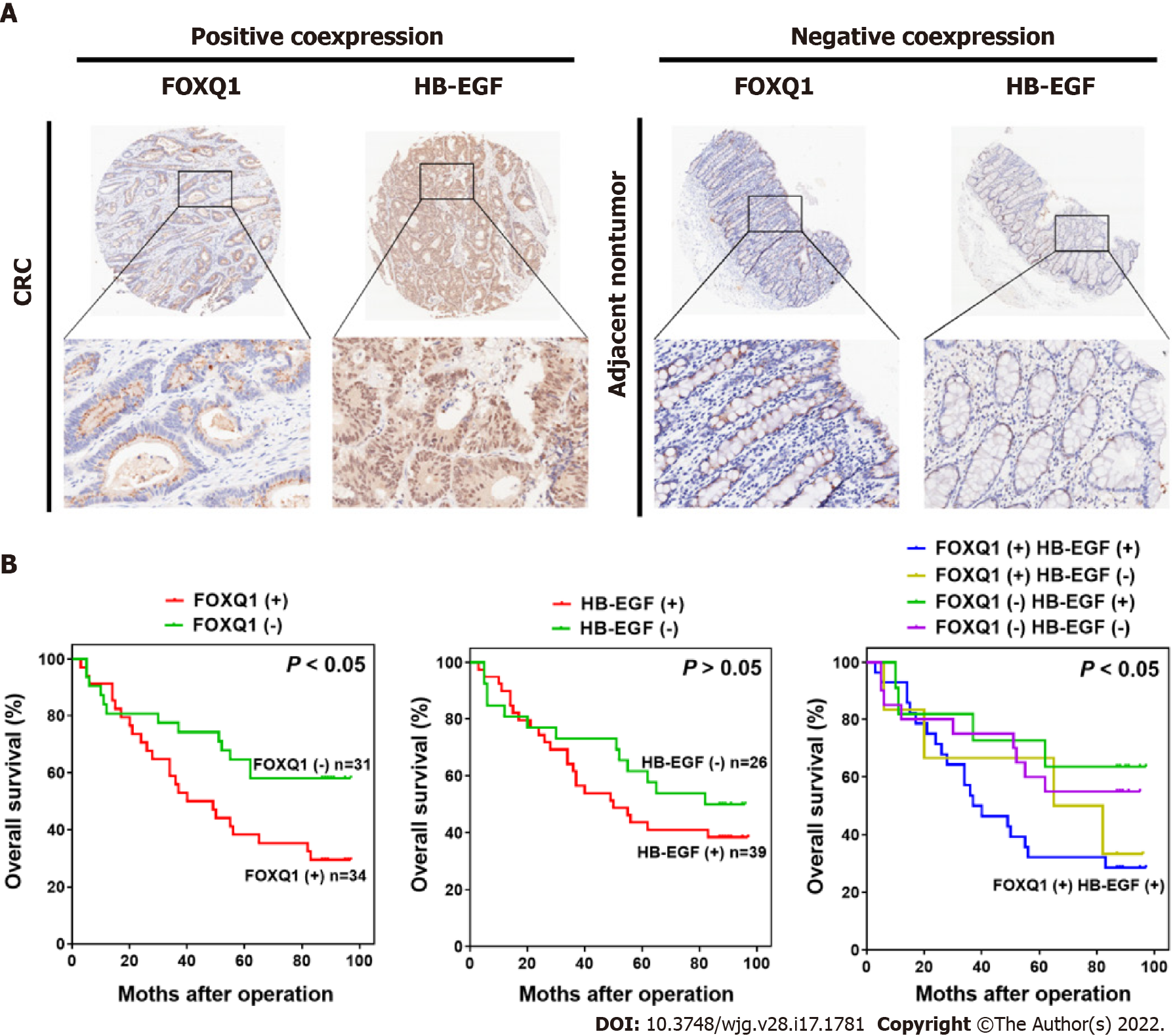
To verify the clinical relevance of our findings, we evaluated the expression of FOXQ1 and HB-EGF in human CRC tissue biopsies (cohort, n = 65) (Tables 3 and 4). IHC analysis showed that FOXQ1 was significantly upregulated in CRC tissues compared to adjacent nontumorous tissues and that HB-EGF was moderately upregulated (Figure 6A). Although the overexpression of HB-EGF had no significant correlation with any clinicopathological characteristics of CRCs (Table 3), further analysis verified the positive correlation between FOXQ1 and HB-EGF (Table 4). In a cohort of 65 CRC patients, Kaplan–Meier survival analysis results showed that CRC patients with positive expression of FOXQ1 had shorter overall survival than those with negative expression of FOXQ1. Furthermore, Kaplan–Meier survival analysis of CRC patients with positive coexpression of FOXQ1 and HB-EGF had the shortest overall survival times compared to the corresponding single-negative or double-negative groups in the cohort of 65 CRC patients (Figure 6B).
Studies have indicated that FOXQ1 is an oncogene in multiple tumors, including CRC[24], breast cancer[25], lung cancer[26], gastric cancer[27], liver cancer[28], pancreatic cancer[29], ovarian cancer[30], and neuroglioma[31]. In CRC, FOXQ1 promotes tumor invasion and metastasis through the Wnt signaling pathway, and it affects the prognosis of patients[24]. EGFR is overexpressed in a variety of cancers, including CRC. EGFR overexpression and activation have a positive effect on the cell growth and metastasis of a variety of solid tumors, including CRC[17]. Metastasis is a multistep process in which tumor cells spread from the primary site to distant sites and form secondary tumors[32], and is the leading cause of death in CRC patients. Many downstream targets of HB-EGF and EGFR include MAPK, p-MAPK, RAS, RAF, PI3K, p-PI3K, AKT, p-AKT, and AKT protein kinases, leading to many processes related to tumor progression, including cell growth[33], epithelial-mesenchymal transition (EMT)[34], metastasis[35], and angiogenesis[36]. FOXQ1 can regulate FAK, PI3K, AKT, and many other key proteins in the PI3K/AKT signaling pathway and promotes the phosphorylation of the above proteins to maintain the activation of PI3K/AKT signaling[37]. FOXQ1 can also combine with VEGFR2 and VE-cadherin to promote angiogenesis and endothelial cell migration and rearrangement[24].
In our previous studies, we found that the mRNA and protein expression levels of FOXQ1 gradually increase with the pathological development of colorectal adenoma to colorectal adenocarcinoma, and the increased expression of FOXQ is not only involved in the process of colorectal adenoma carcinogenesis, but is also closely related to invasion and metastasis of CRC[38]. The differential expression of genes that are involved in the process of colorectal adenoma carcinogenesis screened by the gene chip has been analyzed by signal pathway analysis, suggesting that the abnormally high expression of FOXQ1 is closely related to the activation of the EGFR signaling pathway[39]. Studies have shown that the abnormal activation of the EGFR pathway plays an important role in malignant growth, invasion, and metastasis of colorectal tumors. When sHB-EGF binds to EGFR, the tyrosine kinase activity of EGFR is activated, mainly by activating the MAPK/ERK1/2, PI3K/AKT, and JAK/STAT3 signaling pathways, leading to the proliferation, invasion, metastasis, and apoptosis of tumor cells[40]. The results of this study also proved that knockout of FOXQ1 caused a decrease in gene expression in these three signaling pathways.
Combined with the analysis in Figure 3B-D, these results suggested that FOXQ1 may regulate the EGFR pathway by promoting the separation of ProHB-EGF into sHB-EGF. AMAD7, AMAD9, and AMAD12 are also factors that affect the separation of ProHB-EGF[41]. In this study, decreased expression levels of AMAD7, AMAD9, and AMAD12 were also observed in DLD1-shFOXQ1 and SW480-shFOXQ1 cells, but this indirect regulation should be further verified in other CRC cell lines.
In this study, FOXQ1 was determined to be upregulated in CRC cell lines. The results showed that FOXQ1 knockdown inhibited the proliferation, migration and repair capabilities of CRC, which was consistent with our previous results[21,42]. When rhHB-EGF protein was added, the proliferation ability of the cells was not completely restored, but the migration and repair ability of CRC cells was partially restored. These results indicated that the effect of FOXQ1 in promoting the proliferation of CRC cells is not directly mediated by HB-EGF; however, the regulation of the invasion and metastasis of CRC cells by FOXQ1 is partly mediated by HB-EGF. HB-EGF is related to the abnormal proliferation of skin and mucosal cells, and high expression of HB-EGF is closely related to the occurrence and development of a variety of tumors; the expression of the HB-EGF gene is significantly increased in various human cancers and cancer-derived cell lines, indicating that HB-EGF plays an important role in tumor invasion and metastasis[43-46].
Studies have shown that FOXQ1 is related to the poor prognosis of CRC[47]. In this study, we conducted pathological and survival analyses on 65 CRC patients. These findings suggested that the expression of FOXQ1 and its coregulatory protein HB-EGF may have a prognostic correlation with CRC. Thus, FOXQ1 may serve as a therapeutic target for CRC treatment by blocking the HB-EGF/EGFR pathway. Our research suggests that FOXQ1 activates the EGFR signaling pathway by regulating the expression of HB-EGF, thereby affecting the invasion and metastasis of CRC. Regarding the limitations of this study, it is necessary to construct a FOXQ1 high-expressing cell line and combine it with the dual luciferase reporter gene system to further verify whether FOXQ1 is directly involved in the transcriptional regulation of HB-EGF. Next, we will further explore the regulation of FOXQ1 on EGFR and its downstream signaling pathways through in vivo and in vitro studies. We will conduct a more comprehensive study on the role of HB-EGF in the invasion and metastasis of CRC to provide more possibilities for the treatment of CRC.
In conclusion, we have demonstrated that decreased FOXQ1 expression was also associated with a lower ability to invade and metastasize in CRC. FOXQ1 promotes the invasion and metastasis of CRC by activating the HB-EGF/EGFR pathway. These data indicated that FOXQ1 and HB-EGF may be potential biomarkers to improve the accuracy of CRC diagnosis and treatment.
Invasion and metastasis play important roles in tumorigenesis, resulting in the death of most colorectal cancer (CRC) patients. Forkhead Box q1 (FOXQ1) is a well-established oncogene in multiple tumors, including CRC. However, the role and mechanism of how FOXQ1 promotes tumorigenesis in CRC by activating the heparin-binding epidermal growth factor (HB-EGF)/epidermal growth factor receptor (EGFR) pathway remain largely unknown.
Our study aims to elucidate the critical role of FOXQ1 and HB-EGF/EGFR pathways in CRC, and to provide theoretical support for the clinical application of targeted FOXQ1 in the treatment of CRC.
To determine the role of FOXQ1-induced invasion and metastasis, which are related to activating the HB-EGF/EGFR pathway, and to explore the mechanism by which FOXQ1 promotes tumorigenesis by activating the HB-EGF/EGFR pathway in CRC.
We analyzed the correlation between FOXQ1 and the HB-EGF/EGFR pathway by constructing FOXQ1 knockdown cells, tissue microarray, cell function experiments, quantitative real-time polymerase chain reaction (qRT–PCR), flow cytometry, ELISA, western blot, and the Gene Expression Profiling Interactive Analysis (GEPIA) website.
GEPIA showed that the expression of FOXQ1 in CRC tissues was relatively high and was related to a lower overall survival rate. PCR array results showed that FOXQ1 is related to the HB-EGF and EGFR pathways. Knockdown of FOXQ1 suppressed the expression of HB-EGF and led to a decrease in EGFR and its downstream genes AKT, RAF, KRAS expression levels. After knockdown of FOXQ1 in CRC cell lines, cell proliferation, migration, and invasion were attenuated. Adding HB-EGF restored the migration and invasion ability of CRC, but the cell proliferation ability was not restored. Kaplan–Meier survival analysis results showed that the combination of FOXQ1 and HB-EGF may serve to predict CRC survival.
FOXQ1 promotes the invasion and metastasis of CRC by activating the HB-EGF/EGFR pathway.
In this study, our results indicated that FOXQ1 and HB-EGF may be potential biomarkers to improve the accuracy of CRC diagnosis and treatment.
| 1. | Gu MJ, Huang QC, Bao CZ, Li YJ, Li XQ, Ye D, Ye ZH, Chen K, Wang JB. Attributable causes of colorectal cancer in China. BMC Cancer. 2018;18:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 117] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 2. | Christofori G. New signals from the invasive front. Nature. 2006;441:444-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 719] [Cited by in RCA: 766] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 3. | Arlt F, Stein U. Colon cancer metastasis: MACC1 and Met as metastatic pacemakers. Int J Biochem Cell Biol. 2009;41:2356-2359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 99] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 4. | Lai E, Prezioso VR, Smith E, Litvin O, Costa RH, Darnell JE Jr. HNF-3A, a hepatocyte-enriched transcription factor of novel structure is regulated transcriptionally. Genes Dev. 1990;4:1427-1436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 343] [Cited by in RCA: 386] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 5. | Gao M, Shih IeM, Wang TL. The role of forkhead box Q1 transcription factor in ovarian epithelial carcinomas. Int J Mol Sci. 2012;13:13881-13893. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 6. | Kaneda H, Arao T, Tanaka K, Tamura D, Aomatsu K, Kudo K, Sakai K, De Velasco MA, Matsumoto K, Fujita Y, Yamada Y, Tsurutani J, Okamoto I, Nakagawa K, Nishio K. FOXQ1 is overexpressed in colorectal cancer and enhances tumorigenicity and tumor growth. Cancer Res. 2010;70:2053-2063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 151] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 7. | Liu JY, Wu XY, Wu GN, Liu FK, Yao XQ. FOXQ1 promotes cancer metastasis by PI3K/AKT signaling regulation in colorectal carcinoma. Am J Transl Res. 2017;9:2207-2218. [PubMed] |
| 8. | Weng W, Okugawa Y, Toden S, Toiyama Y, Kusunoki M, Goel A. FOXM1 and FOXQ1 Are Promising Prognostic Biomarkers and Novel Targets of Tumor-Suppressive miR-342 in Human Colorectal Cancer. Clin Cancer Res. 2016;22:4947-4957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 68] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 9. | Fujii H, Iihara H, Suzuki A, Kobayashi R, Matsuhashi N, Takahashi T, Yoshida K, Itoh Y. Hypomagnesemia is a reliable predictor for efficacy of anti-EGFR monoclonal antibody used in combination with first-line chemotherapy for metastatic colorectal cancer. Cancer Chemother Pharmacol. 2016;77:1209-1215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 10. | Higashiyama S, Abraham JA, Miller J, Fiddes JC, Klagsbrun M. A heparin-binding growth factor secreted by macrophage-like cells that is related to EGF. Science. 1991;251:936-939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 846] [Cited by in RCA: 863] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 11. | Taylor SR, Markesbery MG, Harding PA. Heparin-binding epidermal growth factor-like growth factor (HB-EGF) and proteolytic processing by a disintegrin and metalloproteinases (ADAM): a regulator of several pathways. Semin Cell Dev Biol. 2014;28:22-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 12. | Zhou Z, Harding PA. Amino-terminal deletion of heparin-binding epidermal growth factor-like growth factor4-127 stimulates cell proliferation but lacks insulin-like activity. Cell Prolif. 2007;40:213-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 13. | Nanba D, Mammoto A, Hashimoto K, Higashiyama S. Proteolytic release of the carboxy-terminal fragment of proHB-EGF causes nuclear export of PLZF. J Cell Biol. 2003;163:489-502. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 121] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 14. | Suzuki M, Raab G, Moses MA, Fernandez CA, Klagsbrun M. Matrix metalloproteinase-3 releases active heparin-binding EGF-like growth factor by cleavage at a specific juxtamembrane site. J Biol Chem. 1997;272:31730-31737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 228] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 15. | Yan Y, Shirakabe K, Werb Z. The metalloprotease Kuzbanian (ADAM10) mediates the transactivation of EGF receptor by G protein-coupled receptors. J Cell Biol. 2002;158:221-226. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 243] [Cited by in RCA: 233] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 16. | Sunnarborg SW, Hinkle CL, Stevenson M, Russell WE, Raska CS, Peschon JJ, Castner BJ, Gerhart MJ, Paxton RJ, Black RA, Lee DC. Tumor necrosis factor-alpha converting enzyme (TACE) regulates epidermal growth factor receptor ligand availability. J Biol Chem. 2002;277:12838-12845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 340] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 17. | Shi YH, Zhao DM, Wang YF, Li X, Ji MR, Jiang DN, Xu BP, Zhou L, Lu CZ, Wang B. The association of three promoter polymorphisms in interleukin-10 gene with the risk for colorectal cancer and hepatocellular carcinoma: A meta-analysis. Sci Rep. 2016;6:30809. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 18. | Wang J, Chen Y, Dai C, Shang Y, Xie J. Ginsenoside Rh2 alleviates tumor-associated depression in a mouse model of colorectal carcinoma. Am J Transl Res. 2016;8:2189-2195. [PubMed] |
| 19. | Wu L, Shi B, Huang K, Fan G. MicroRNA-128 suppresses cell growth and metastasis in colorectal carcinoma by targeting IRS1. Oncol Rep. 2015;34:2797-2805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 20. | Luo Y, Wang J, Wang F, Liu X, Lu J, Yu X, Ma X, Peng X, Li X. Foxq1 promotes metastasis of nasopharyngeal carcinoma by inducing vasculogenic mimicry via the EGFR signaling pathway. Cell Death Dis. 2021;12:411. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 21. | Tang H, Zheng J, Bai X, Yue KL, Liang JH, Li DY, Wang LP, Wang JL, Guo Q. Forkhead Box Q1 Is Critical to Angiogenesis and Macrophage Recruitment of Colorectal Cancer. Front Oncol. 2020;10:564298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 22. | Princz MA, Sheardown H. Heparin-modified dendrimer crosslinked collagen matrices for the delivery of heparin-binding epidermal growth factor. J Biomed Mater Res A. 2012;100:1929-1937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 23. | Lee JH, Lee JH, Ahn BK, Paik SS, Kim H, Lee KH. Loss of ASXL1 expression is associated with lymph node metastasis in colorectal cancer. Indian J Pathol Microbiol. 2020;63:221-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 24. | Peng X, Luo Z, Kang Q, Deng D, Wang Q, Peng H, Wang S, Wei Z. FOXQ1 mediates the crosstalk between TGF-β and Wnt signaling pathways in the progression of colorectal cancer. Cancer Biol Ther. 2015;16:1099-1109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 70] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 25. | Han X, Guo X, Zhang W, Cong Q. MicroRNA-937 inhibits the malignant phenotypes of breast cancer by directly targeting and downregulating forkhead box Q1. Onco Targets Ther. 2019;12:4813-4824. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 26. | Feng J, Xu L, Ni S, Gu J, Zhu H, Wang H, Zhang S, Zhang W, Huang J. Involvement of FoxQ1 in NSCLC through regulating EMT and increasing chemosensitivity. Oncotarget. 2014;5:9689-9702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 27. | Xiang XJ, Deng J, Liu YW, Wan LY, Feng M, Chen J, Xiong JP. MiR-1271 Inhibits Cell Proliferation, Invasion and EMT in Gastric Cancer by Targeting FOXQ1. Cell Physiol Biochem. 2015;36:1382-1394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 59] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 28. | Han S, Shi Y, Sun L, Liu Z, Song T, Liu Q. MiR-4319 induced an inhibition of epithelial-mesenchymal transition and prevented cancer stemness of HCC through targeting FOXQ1. Int J Biol Sci. 2019;15:2936-2947. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 29. | Christensen J, Bentz S, Sengstag T, Shastri VP, Anderle P. FOXQ1, a novel target of the Wnt pathway and a new marker for activation of Wnt signaling in solid tumors. PLoS One. 2013;8:e60051. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 54] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 30. | Wang C, Zhang W, Xing S, Wang Z, Wang J, Qu J. MiR-342-3p inhibits cell migration and invasion through suppressing forkhead box protein Q1 in ovarian carcinoma. Anticancer Drugs. 2019;30:917-924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 31. | Sun HT, Cheng SX, Tu Y, Li XH, Zhang S. FoxQ1 promotes glioma cells proliferation and migration by regulating NRXN3 expression. PLoS One. 2013;8:e55693. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 32. | Yang B, Tan Z, Song Y. Study on the molecular regulatory mechanism of MicroRNA-195 in the invasion and metastasis of colorectal carcinoma. Int J Clin Exp Med. 2015;8:3793-3800. [PubMed] |
| 33. | Morato A, Martignani E, Miretti S, Baratta M, Accornero P. External and internal EGFR-activating signals drive mammary epithelial cells proliferation and viability. Mol Cell Endocrinol. 2021;520:111081. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 34. | Tulchinsky E, Demidov O, Kriajevska M, Barlev NA, Imyanitov E. EMT: A mechanism for escape from EGFR-targeted therapy in lung cancer. Biochim Biophys Acta Rev Cancer. 2019;1871:29-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 148] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 35. | Shen H, He M, Lin R, Zhan M, Xu S, Huang X, Xu C, Chen W, Yao Y, Mohan M, Wang J. PLEK2 promotes gallbladder cancer invasion and metastasis through EGFR/CCL2 pathway. J Exp Clin Cancer Res. 2019;38:247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 78] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 36. | Yu X, Li W, Deng Q, You S, Liu H, Peng S, Liu X, Lu J, Luo X, Yang L, Tang M, Weng X, Yi W, Liu W, Wu S, Ding Z, Feng T, Zhou J, Fan J, Bode AM, Dong Z, Liu J, Cao Y. Neoalbaconol inhibits angiogenesis and tumor growth by suppressing EGFR-mediated VEGF production. Mol Carcinog. 2017;56:1414-1426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (12)] |
| 37. | Song Y, Zhao Y, Ding X, Wang X. microRNA-532 suppresses the PI3K/Akt signaling pathway to inhibit colorectal cancer progression by directly targeting IGF-1R. Am J Cancer Res. 2018;8:435-449. [PubMed] |
| 38. | Tang H, Guo Q, Zhang C, Zhu J, Yang H, Zou YL, Yan Y, Hong D, Sou T, Yan XM. Identification of an intermediate signature that marks the initial phases of the colorectal adenoma-carcinoma transition. Int J Mol Med. 2010;26:631-641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 39. | Zhang Y, Wang X, Wang Z, Tang H, Fan H, Guo Q. miR-182 promotes cell growth and invasion by targeting forkhead box F2 transcription factor in colorectal cancer. Oncol Rep. 2015;33:2592-2598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 40. | Normanno N, De Luca A, Bianco C, Strizzi L, Mancino M, Maiello MR, Carotenuto A, De Feo G, Caponigro F, Salomon DS. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366:2-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1302] [Cited by in RCA: 1474] [Article Influence: 70.2] [Reference Citation Analysis (0)] |
| 41. | Cissé MA, Sunyach C, Lefranc-Jullien S, Postina R, Vincent B, Checler F. The disintegrin ADAM9 indirectly contributes to the physiological processing of cellular prion by modulating ADAM10 activity. J Biol Chem. 2005;280:40624-40631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 89] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 42. | Qiao Y, Jiang X, Lee ST, Karuturi RK, Hooi SC, Yu Q. FOXQ1 regulates epithelial-mesenchymal transition in human cancers. Cancer Res. 2011;71:3076-3086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 138] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 43. | Ota I, Higashiyama S, Masui T, Yane K, Hosoi H, Matsuura N. Heparin-binding EGF-like growth factor enhances the activity of invasion and metastasis in thyroid cancer cells. Oncol Rep. 2013;30:1593-1600. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 44. | Zhou ZN, Sharma VP, Beaty BT, Roh-Johnson M, Peterson EA, Van Rooijen N, Kenny PA, Wiley HS, Condeelis JS, Segall JE. Autocrine HBEGF expression promotes breast cancer intravasation, metastasis and macrophage-independent invasion in vivo. Oncogene. 2014;33:3784-3793. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 82] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 45. | Nabeshima A, Matsumoto Y, Fukushi J, Iura K, Matsunobu T, Endo M, Fujiwara T, Iida K, Fujiwara Y, Hatano M, Yokoyama N, Fukushima S, Oda Y, Iwamoto Y. Tumour-associated macrophages correlate with poor prognosis in myxoid liposarcoma and promote cell motility and invasion via the HB-EGF-EGFR-PI3K/Akt pathways. Br J Cancer. 2015;112:547-555. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 79] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 46. | Zhang L, Shimizu D, Killeen JL, Honda SA, Lu D, Stanoyevitch A, Lin F, Wang B, Monuki ES, Carbone M. Serous carcinomatous component championed by heparin-binding EGF-like growth factor (HB-EGF) predisposing to metastasis and recurrence in stage I uterine malignant mixed mullerian tumor. Hum Pathol. 2016;53:159-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 47. | Liang SH, Yan XZ, Wang BL, Jin HF, Yao LP, Li YN, Chen M, Nie YZ, Wang X, Guo XG, Wu KC, Ding J, Fan DM. Increased expression of FOXQ1 is a prognostic marker for patients with gastric cancer. Tumour Biol. 2013;34:2605-2609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A, A
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Emran TB, Bangladesh; Herold Z, Hungary; Martin MJ, Argentina S-Editor: Fan JR L-Editor: A P-Editor: Guo X