Published online Oct 28, 2021. doi: 10.3748/wjg.v27.i40.6908
Peer-review started: April 30, 2021
First decision: July 2, 2021
Revised: July 19, 2021
Accepted: September 8, 2021
Article in press: September 8, 2021
Published online: October 28, 2021
Processing time: 180 Days and 3.8 Hours
Hepatic stellate cells (HSCs) are the key effector cells mediating the occurrence and development of liver fibrosis, while aerobic glycolysis is an important metabolic characteristic of HSC activation. Transforming growth factor-β1 (TGF-β1) induces aerobic glycolysis and is a driving factor for metabolic reprogramming. The occurrence of glycolysis depends on a high glucose uptake level. Glucose transporter 1 (GLUT1) is the most widely distributed glucose transporter in the body and mainly participates in the regulation of carbohydrate metabolism, thus affecting cell proliferation and growth. However, little is known about the relationship between TGF-β1 and GLUT1 in the process of liver fibrosis and the molecular mechanism underlying the promotion of aerobic glycolysis in HSCs.
To investigate the mechanisms of action of GLUT1, TGF-β1 and aerobic glycolysis in the process of HSC activation during liver fibrosis.
Immunohistochemical staining and immunofluorescence assays were used to examine GLUT1 expression in fibrotic liver tissue. A Seahorse extracellular flux (XF) analyzer was used to examine changes in aerobic glycolytic flux, lactate production levels and glucose consumption levels in HSCs upon TGF-β1 stimulation. The mechanism by which TGF-β1 induces GLUT1 protein expression in HSCs was further explored by inhibiting/promoting the TGF-β1/mothers-against-decapentaplegic-homolog 2/3 (Smad2/3) signaling pathway and inhibiting the p38 and phosphoinositide 3-kinase (PI3K)/AKT signaling pathways. In addition, GLUT1 expression was silenced to observe changes in the growth and proliferation of HSCs. Finally, a GLUT1 inhibitor was used to verify the in vivo effects of GLUT1 on a mouse model of liver fibrosis.
GLUT1 protein expression was increased in both mouse and human fibrotic liver tissues. In addition, immunofluorescence staining revealed colocalization of GLUT1 and alpha-smooth muscle actin proteins, indicating that GLUT1 expression was related to the development of liver fibrosis. TGF-β1 caused an increase in aerobic glycolysis in HSCs and induced GLUT1 expression in HSCs by activating the Smad, p38 MAPK and P13K/AKT signaling pathways. The p38 MAPK and Smad pathways synergistically affected the induction of GLUT1 expression. GLUT1 inhibition eliminated the effect of TGF-β1 on HSC proliferation and migration. A GLUT1 inhibitor was administered in a mouse model of liver fibrosis, and GLUT1 inhibition reduced the degree of liver inflammation and liver fibrosis.
TGF-β1 induces GLUT1 expression in HSCs, a process related to liver fibrosis progression. In vitro experiments revealed that TGF-β1-induced GLUT1 expression might be one of the mechanisms mediating the metabolic reprogramming of HSCs. In addition, in vivo experiments also indicated that the GLUT1 protein promotes the occurrence and development of liver fibrosis.
Core Tip: Liver fibrosis is a repair response of the liver to various chronic injuries. However, fibrosis may eventually evolve into liver cirrhosis or even liver cancer if it progresses. Hepatic stellate cell activation is the initiating factor for liver fibrosis. Transforming growth factor-β1 (TGF-β1) is a pleiotropic cytokine that induces aerobic glycolysis. Glucose transporter 1 (GLUT1) regulates glucose metabolism. This study examined the effects of TGF-β1-mediated pathways on GLUT1 expression in vivo and in vitro, explored the relationship between GLUT1 and TGF-β1 and further investigated the potential underlying mechanisms.
- Citation: Zhou MY, Cheng ML, Huang T, Hu RH, Zou GL, Li H, Zhang BF, Zhu JJ, Liu YM, Liu Y, Zhao XK. Transforming growth factor beta-1 upregulates glucose transporter 1 and glycolysis through canonical and noncanonical pathways in hepatic stellate cells. World J Gastroenterol 2021; 27(40): 6908-6926
- URL: https://www.wjgnet.com/1007-9327/full/v27/i40/6908.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i40.6908
Liver fibrosis is the inevitable result of chronic liver inflammation caused by various etiologies. With progressive destruction of liver parenchymal cells, liver fibrosis eventually develops into liver cirrhosis and even liver cancer[1,2]. Although liver cirrhosis and liver cancer are irreversible, liver fibrosis can be reversed. Therefore, the mechanism of and clinical studies on liver fibrosis have always been the focus of liver disease research. The main pathological feature of liver fibrosis is the excessive deposition of extracellular matrix (ECM), while the key initiating factors are activation of quiescent hepatic stellate cells (HSCs) and transformation of their phenotypes and functions[3]. The transforming growth factor-β1 (TGF-β1) pathway is the key fibrogenic pathway that drives HSC activation and induces ECM production. HSC activation requires metabolic reprogramming and a continuous energy supply[4,5]. Aerobic glycolysis is an important metabolic characteristic of the transdifferentiation of quiescent stellate cells, a process similar to the Warburg effect in tumor cells, and the core metabolic changes include a transition from oxidative phosphorylation to aerobic glycolysis[6]. Dysregulated glycolysis has been implicated in experimental models of lung and liver fibrosis, and inhibition of glycolysis reduces ECM accumulation[7]. In view of the mechanisms involved, targeting and inhibiting the metabolic reprogramming of activated HSCs during liver fibrosis may be a promising anti-liver fibrosis strategy.
TGF-β1 is a multifunctional cytokine and a major profibrotic cytokine that regulates cell differentiation, cell proliferation and ECM production and directly regulates multiple cellular signal transduction networks[8]. In the canonical TGF-β1/mothers-against-decapentaplegic-homolog 2/3 (Smad2/3) pathway, ligands induce the assembly of the TGF-β1 receptor I (TβRI)/TGF-β1 receptor II (TβRII) heterocomplex, which targets Smad4 via Smad2 and Smad3 proteins to form the Smad complex, leading to phosphorylation and nuclear translocation of Smad2/3; this R-Smad/Co-Smad4 complex translocates to the nucleus where it binds to DNA either directly or in association with other DNA-binding proteins[9-11]. Phosphorylated Smad2/3 binds to specific Smad binding elements (SBEs) in gene promoter regions to activate/suppress the expression of target genes[12,13]. In addition to Smads, TGF-β1 also triggers other protein-mediated signaling pathways, e.g., p38, mitogen-activated protein kinases (MAPKs) and phosphoinositide 3-kinase (PI3K). Some functions of TGF-β1 have been studied in depth, such as the mediation of cell differentiation and proliferation. However, TGF-β1 has recently been reported to induce aerobic glycolysis and is considered a driving factor in metabolic reprogramming[14]. TGF-β is also a strong activator of glycolysis in mesenchymal cells[15]. Extracellular accumulation of lactic acid induces epithelial-mesenchymal transition (EMT) by directly reconstituting the ECM and releasing activated TGF-β1. EMT induced by TGF-β in hepatocellular carcinoma cells reprograms lipid metabolism to sustain the elevated energy requirements associated with this process[16]. Research on the mechanism of idiopathic pulmonary fibrosis has shown that TGF-β1-induced aerobic glycolysis causes lactic acid accumulation and changes the cellular microenvironment, thereby activating latent TGF-β1 in the ECM and eventually forming a positive feedback loop to promote the effects of TGF-β1[17].
Glucose transporter 1 (GLUT1) is a member of the GLUT transporter family, the most conserved and most widely distributed glucose transporter in mammals and the main transporter regulating glucose uptake[18]. An increasing number of studies have found that GLUT1 plays an important role in accelerated metabolism. Research on the mechanism of neurodegenerative diseases has revealed that GLUT1 controls the activation of microglia by promoting aerobic glycolysis[19]. GLUT1 enhances the stimulating effect of TGF-β1 on mesangial cells, breast cancer cells and pancreatic cancer cells. As glucose uptake increases during TGF-β1-induced EMT of breast cancer cells, GLUT1 expression also increases and is correlated with EMT markers (including E-cadherin and vimentin). GLUT1 is the key mediator of the aerobic glycolysis phenotype in ovarian cancer and is required to maintain a high level of basic aerobic glycolysis. In models of bleomycin-induced pulmonary fibrosis, GLUT1-dependent aerobic glycolysis has been reported to be essential for pulmonary parenchymal fibrosis[20-23]. Certain signaling molecules (such as cAMP, p53, PI3K and AKT) reduce alpha-smooth muscle actin (α-SMA) protein expression in primary mouse fibroblasts by inhibiting GLUT1 expression. Exosomes secreted by activated HSCs affect the metabolic switch of liver nonparenchymal cells through delivery of the glycolysis-related proteins GLUT1 and PKM2; GLUT1 is involved in metabolic reprogramming of HSCs[24]. TGF-β1 and GLUT1 play important regulatory roles in metabolic reprogramming. To date, however, researchers have not explored whether the increases in TGF-β1 and GLUT1 Levels during HSC activation are related. Therefore, this study investigated the effect of the TGF-β1 signaling pathway on the regulation of GLUT1 and aerobic glycolysis. We hypothesized that TGF-β1 drives HSC activation and aerobic glycolysis by inducing GLUT1 expression, thereby promoting liver fibrosis progression. As shown in the present study, GLUT1 expression was significantly increased in mouse and human fibrotic liver tissue samples. Further in vitro experiments showed that the aerobic glycolysis capacity of HSCs was enhanced and GLUT1 expression increased with increasing TGF-β1 Levels. Inhibition/ promotion of the Smad2/3 signaling pathway and inhibition of the p38 and PI3K/AKT signaling pathways confirmed that TGF-β1 induced GLUT1 expression by targeting the pSmad2/3, p38 and PI3K/AKT pathways, thus promoting HSC activation. Finally, administration of a specific GLUT1 inhibitor in a mouse model of liver fibrosis resulted in a significant reduction in liver fibrosis. Based on these findings, the TGF-β1 signaling pathway enhances aerobic glycolysis by promoting GLUT1 expression, thereby promoting the development of liver fibrosis.
The TGF-β1 antibody was purchased from R&D Systems (Minneapolis, MN, United States); antibodies against GLUT1, p-Smad2/3, Smad2/3, p-P38, p-AKT and desmin were purchased from Abcam (Cambridge, MA, United States), and the tubulin antibody was purchased from Research Diagnostics (Flanders, NJ, United States). The anti-α-SMA antibody, carbon tetrachloride (CCl4), corn oil, OptiPrep and other chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO, United States) and Fisher Scientific (Waltham, MA, United States). A TβRI/II inhibitor (APExBIO Technology, United States) was used at 2 μmol/L. Inhibitors of p38 MAPK and PI3K, namely, SB203580 and LY294002, respectively, were purchased from Abcam (Cambridge, MA, United States). The Smad3 inhibitors SIS3 and phloretin were purchased from Abcam (Cambridge, MA, United States).
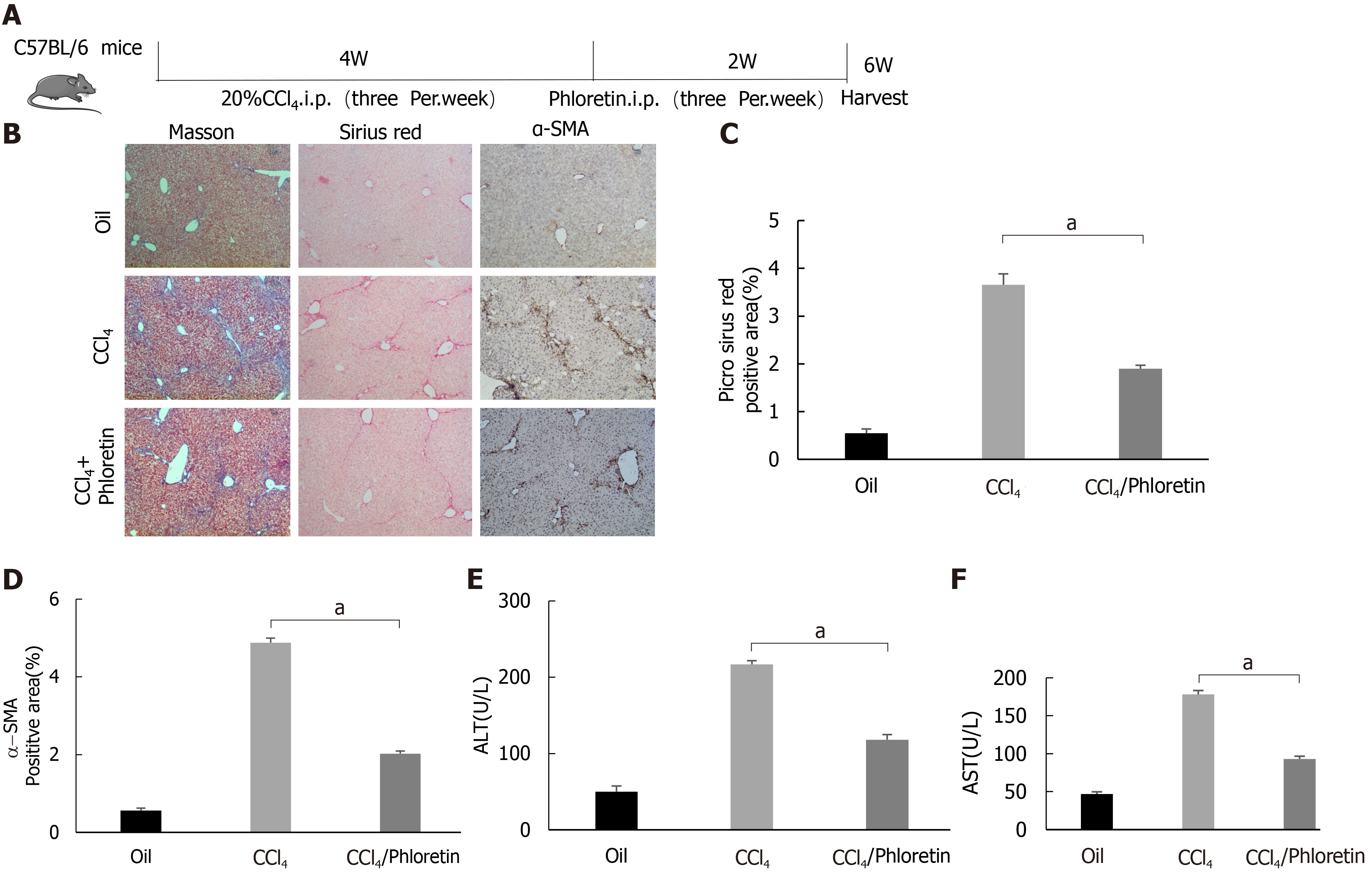
The animal protocol was designed to minimize pain or discomfort to the animals. The animals were acclimated to laboratory conditions (22 °C, 12-h/12-h light/dark cycle, 50% humidity, ad libitum access to food and water) for 1 wk prior to experimentation. The methods and experimental procedures were carried out in accordance with the relevant guidelines and regulations. Mice (C57BL6, eight to ten weeks old) were housed in standard conditions, and sex-matched mice were treated with 2.0 μL/g body weight CCl4 [diluted 1:10 (v/v) with corn oil] or corn oil as a control by intraperitoneal (i.p.) injections three times per week for 4 wk[25]. Mice were challenged with CCl4 or corn oil (control), followed by an i.p. injection of phloretin (10 mg/kg, three times per week for 2 wk) or 0.9% saline (vehicle). Mice were sacrificed at 48 h after the experiment ended, and tissues were harvested.
Normal and liver fibrosis tissue samples were obtained from patients treated at the Department of Hepatobiliary Surgery of the Affiliated Hospital of Guizhou Medical University (Guiyang, China). Written informed consent was obtained from the patients.
Immunoblotting was performed using whole-liver tissue lysates or whole-cell lysates prepared in buffer containing 1% NP-40 as described previously[26]. Total proteins were extracted and quantified using Bradford protein quantification kits. Protein samples (40 μg each) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were transferred onto polyvinylidene fluoride (PVDF) membranes and incubated with primary antibodies overnight at 4 °C. On the next day, signals were developed with an electrochemiluminescence detection kit after incubation with the appropriate secondary antibodies.
Primary mouse HSCs were isolated and cultured as described previously[26]. Briefly, cells were isolated from livers through in situ liver perfusion with pronase, Liberase and collagenase followed by density gradient centrifugation. The dispersed cell suspension was filtered and gradiently centrifuged for 2 min to remove hepatocytes. The remaining cell fraction was washed and resuspended in 11.5% OptiPrep and then gently transferred to a tube containing 15% OptiPrep at the bottom, followed by PBS addition as the top layer. The cell fraction was then centrifuged at 1400 rpm/min for 20 min. The HSC fraction layer was obtained at the interface between the top and intermediate layers. The purity of the HSC fraction was estimated based on autofluorescence 1 d after isolation and was always greater than 97%. Flow cytometry was used to identify the purity of primary cells. In brief, the cells were digested with trypsin, centrifuged at 1000 rpm/min for 10 min, washed twice with PBS, resuspended in EP tubes (100 μL/tube) and centrifuged at 2000 rpm/min for 6 min. Then, the supernatant was discarded, 100 μL of PBS was added, and the cells were resuspended and dispersed. A mouse monoclonal antibody against desmin (desmin is a typical molecular marker of HSCs) was added, and the cells were incubated at 1:100 for 1.5 h and centrifuged at 2000 rpm/min for 6 min. Centrifugation was repeated twice. The cells were resuspended and dispersed by adding 100 μL of PBS. Fluorescence-labeled anti-mouse secondary antibody (1:1000) was added, followed by incubation at 4 °C for 30 min in the dark. Next, 1 mL of PBS was added to each tube, followed by centrifugation at 2000 rpm/min for 6 min, which was repeated twice. Finally, 0.5 mL of PBS was added to resuspend the cells, and then the cells were subjected to flow cytometry measurements. HSCs were also confirmed to lack E-cadherin expression. Cell viability was also examined, and HSCs were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and antibiotics as described previously[26]. Cells were starved or treated on day 2 after isolation, and the duration of starvation or treatment is described in each figure legend. The duration of the whole experiment was 5-7 d after HSC isolation, and cells at passages 1-2 were used (as cells were passaged from regular culture flasks to experimental cell culture wells for some experiments).
Liver samples were fixed with formalin, embedded in paraffin, sectioned and processed routinely for Masson’s trichrome and Sirius red staining. Antibodies used for immunohistochemical (IHC) staining of GLUT1 and α-SMA were purchased from Abcam Technology, United States.
Cells were transfected with small interfering RNAs (siRNAs) using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s recommendations. The siRNAs used were an siRNA mix targeting sequences in Smad2 and Smad3 purchased from Santa Cruz Biotechnology (siRNA Smad2/3) and siRNAs targeting four different sequences of Smad4 purchased from Santa Cruz Biotechnology. GLUT1 siRNA was purchased from Cell Signaling Technology. The sequences of the siRNAs used are shown in Table 1.
| Forward | Reverse | |
| siGlut1-1 | 5′-CACCGGGAGTGACAAAGACTTTGTTCAAGCA-3′ | 5′-GATCCAAAAAAGGGAGTGACAAAGACTTCTC-3′ |
| Negative control | 5′-ATCCGACTTCATAAGGCGCATGCT-3′ | 5′-AGTATTCCGCGTACGAAGTTCTGC-3′ |
| siRNA Smad4 targeting four different sequences1 | ||
| GCAAUUGAAAGUUUGGUAA, CCCACAACCUUUAGACUGA, GAAUCCAUAUCACUACGAA and GUACAGAGUUACUACUUAG | ||
| siRNA Smad2/31 | Sense | Antisense |
| sc-37239A | CUUGCUGGAUUGAACUUCAtt | UGAAGUUCAAUCCAGCAAGtt |
| sc-37239B | CCGUCGUAGUAUUCAUGUAtt | UACAUGAAUACUACGACGGtt |
| sc-37239C | CUGACUCCUUGUUUAAUGAtt | UCAUUAAACAAGGAGUCAGtt |
| sc-37239D | GGAAGCUGAGAGUUAUAGAtt | UCUAUAACUCUCAGCUUCCtt |
Primary mouse HSCs were plated on XF96 cell culture microplates. The extracellular acidification rate (ECAR), a glycolytic flux parameter, was measured with a Seahorse XF96 bioanalyzer using the XF Glycolysis Stress Test kit according to the manufacturer’s instructions (102194-100, Seahorse Bioscience). Lactate levels were measured spectrophotometrically in 700 μL of supernatants from cells receiving the corresponding treatment using standard enzymatic methods.
Primary mouse HSCs were seeded in a 96-well plate at a density of 3000 cells per well. CCK-8 reagent was added to each well every 24 h, and the plates were incubated for an additional 1 h at 37 °C and measured by recording the absorbance at 450 nm with an Elx800™ spectrophotometer (BioTek, Winooski, VT, United States).
Alanine aminotransferase (ALT) and aspartate transaminase (AST) levels in mouse serum samples and the supernatant of cell culture medium were detected using an automatic biochemical analyzer (Siemens Advia 1650; Siemens, Bensheim, Germany).
GLUT1, hexokinase 2 (HK-2), pyruvate kinase 2 (PKM-2) and α-SMA mRNA levels were determined using real-time polymerase chain reaction (RT-PCR) with a SYBR Green Master Mix Kit (Roche, Indianapolis, IN, United States). The primer sequences used are shown in Table 2.
| Gene | Forward sequence | Reverse sequence |
| HK2 | 5’-GGGTAGCCACGGAGTACAAA-3’ | 5’-TGGATTGAAAGCCAACTTCC-3’ |
| GLUT1 | 5’-GCTTCTCCAACTGGACCTC-3’ | 5’-AAGAAGAGCACGAGGAGCAC-3’ |
| PKM2 | 5’-TGGGATGGAAACTGTGAAGAG-3’ | 5’-CGGAGTTCCTCGAATAGCTG-3’ |
| α-SMA | 5’-AAGAGCATCCGACACTGCTGAC-3’ | 5’-AGCACAGCCTGAATAGCCACATAC-3’ |
The migratory properties of HSCs were assessed using a Transwell assay. Cells were seeded at a density of 4 × 105 cells/well in the upper compartment of Transwell chambers with serum-free medium, and the lower compartment contained 700 μL of 5% glucose-containing medium per well. Migration was subsequently observed and measured.
Tissue sections were placed at room temperature for 10 min and deparaffinized in water for further antigen retrieval. After the sections were dried slightly, a histochemical pen was used to draw circles around the tissue, and 3%-5% BSA was added dropwise inside the circle for blocking, followed by incubation for 30 min. The primary antibody was added dropwise at the recommended ratio to the sections, and the sections were placed in a refrigerator (4 °C) and incubated overnight. After 3 washes, the sections were incubated with a FITC (CK-18)-labeled secondary antibody at room temperature for 45 min, and nuclei were stained with DAPI (300 nmol/L) for 1-5 min. After 3 washes, an antifluorescence quencher was added, and the sections were sealed with resin. Photographs of random fields were taken under an upright fluorescence microscope (ZEISS Axiovert).
Data were analyzed using Student’s t test (SigmaPlot, SPSS 17.0, United States) to determine differences between two groups and are presented as the mean ± SE. For comparisons between multiple groups, three-way analysis of variance was performed, followed by t tests with Bonferroni correction using SAS 9.3 software (SAS Institute Inc., Cary, NC, United States). In addition, a log-rank test was used for survival analysis. All experiments were repeated at least three times. Differences were considered statistically significant at P < 0.05 (aP < 0.05, bP < 0.01).
The classic mouse model of CCl4-induced liver fibrosis was used to first clarify whether GLUT1 is related to liver fibrosis. Successful establishment of the liver fibrosis model was confirmed by Sirius red staining and α-SMA IHC staining (Figure 1A-C). Notably, a significant increase in GLUT1 expression was detected in the liver tissue specimens from the model group (Figure 1A and D). Subsequently, tissue immunofluorescence staining was performed, and the results showed significantly increased GLUT1 expression in liver tissue samples from the CCl4 liver fibrosis model. More importantly, GLUT1 colocalized with α-SMA, indicating a correlation between GLUT1 and liver fibrosis (Figure 1E). IHC staining for GLUT1 and α-SMA was performed using human liver fibrosis specimens and liver specimens from a healthy control group; as expected, GLUT1 expression was significantly higher in the human liver fibrosis specimens (Figure 1F). Finally, whole-liver lysates were prepared from human liver tissue specimens and specimens from the mouse liver fibrosis model, and GLUT1 protein expression was analyzed. The results were consistent with the IHC data (Figure 1G-H). In summary, these results indicate that GLUT1 expression is related to liver fibrosis progression.
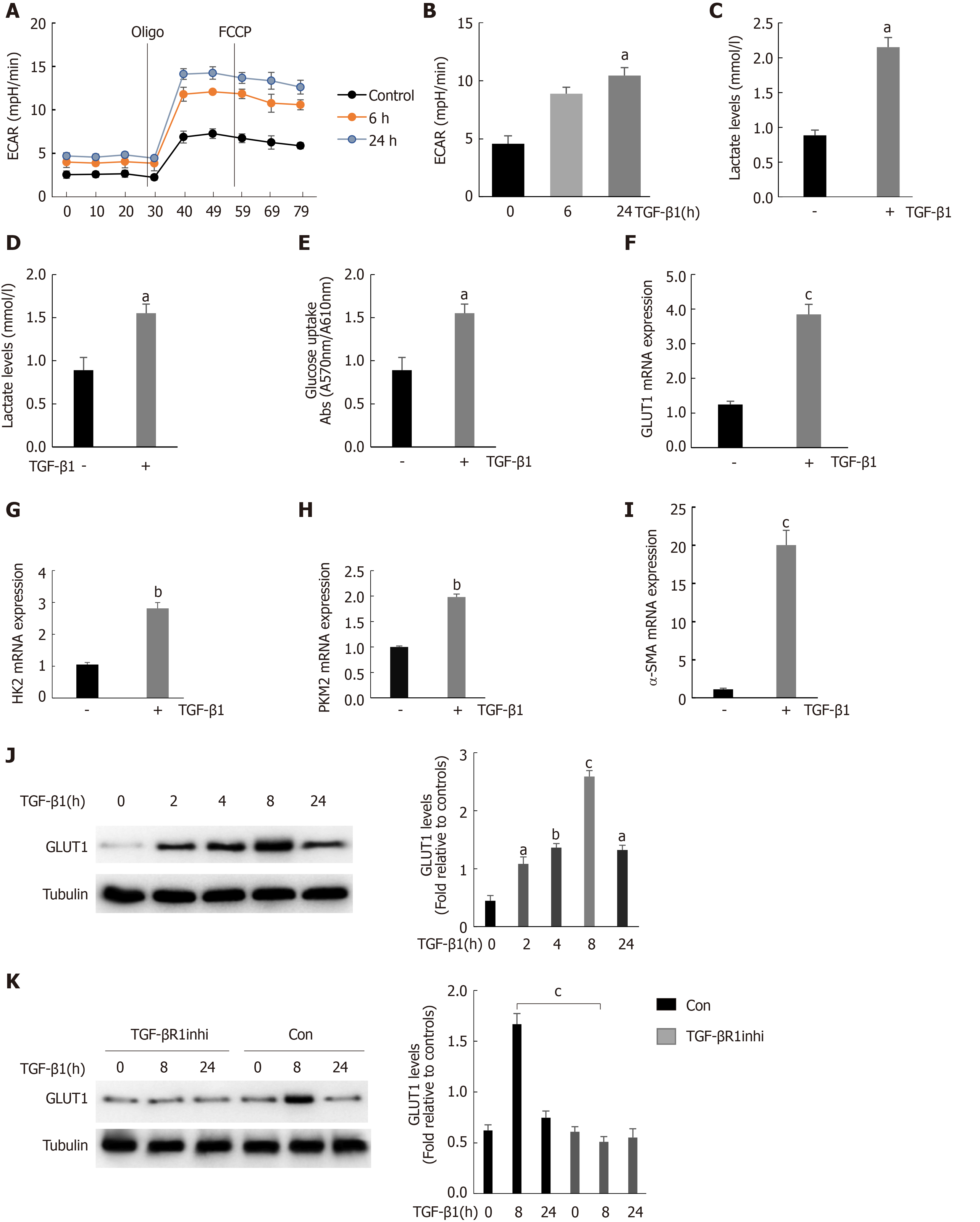
We found that GLUT1 colocalized with α-SMA, indicating that GLUT1 expression was increased mainly in activated HSCs because α-SMA is a major marker of HSC transdifferentiation. TGF-β1 is a very important profibrotic factor and regulates metabolic reprogramming in pulmonary fibrosis[27], as reported in some studies. Therefore, we questioned whether the increase in GLUT1 expression in liver fibrosis and TGF-β1 are related. This study first determined the effect of TGF-β1 on glycolysis in HSCs. To assess the dose responses to TGF-β1, we incubated HSCs with different TGF-β1 concentrations ranging from 3 ng/mL to 5 ng/mL, and the results showed similar effects on GLUT1 protein levels (Supplementary Figure 1). Therefore, a TGF-β1 concentration of 3 ng/mL was used for subsequent experiments. Mouse primary HSCs were isolated and cultured, and the cells were stimulated with TGF-β1 for 24 h prior to the experiments. TGF-β1 stimulation led to an early and continuous increase in the ECAR (an indicator of extracellular acid production) in HSCs, indicating that glycolysis was enhanced in these cells (Figure 2A and B). In addition, the intracellular and extracellular levels (in the medium) of lactic acid were examined to further confirm the glycolytic changes in HSCs. Both intracellular and extracellular lactic acid levels were significantly increased. Consistent with the increase in glycolysis, the level of glucose consumption also increased in these cells (Figure 2C and D). Therefore, TGF-β1 induces glycolysis during the process of HSC transdifferentiation. The expression levels of key glycolytic enzymes in these cells were evaluated; the expression levels of HK-2, PKM-2 and GLUT1 were upregulated, and the increase in GLUT1 expression was particularly significant (Figure 2F-H). Based on these findings, the increase in glycolysis during HSC transdifferentiation is related to the upregulation of key glycolysis enzymes. Similarly, the expression of α-SMA, a marker of transdifferentiation, also increased during the TGF-β1-mediated activation of HSCs (Figure 2I). GLUT1 protein expression was examined at various time points after stimulating HSCs with TGF-β1 (3 ng/mL) to determine whether TGF-β1 stimulates GLUT1 expression in a time-dependent manner, and the results showed that GLUT1 expression increased 2 h after stimulation with TGF-β1 and peaked at 8 h. These results indicate a time-dependent relationship between the increase in GLUT1 expression and TGF-β1 stimulation, which is consistent with the early increase in aerobic glycolysis in HSCs (Figure 2J). Finally, the addition of an inhibitor of the type 1 TGF-β1 receptor inhibited TGF-β1-induced GLUT1 expression in HSCs, suggesting that GLUT1 induction was mediated by TGF-β1 (Figure 2K). Based on these data, TGF-β1 is involved in glycolysis during HSC transdifferentiation and mediates GLUT1 expression, thereby promoting HSC transdifferentiation.
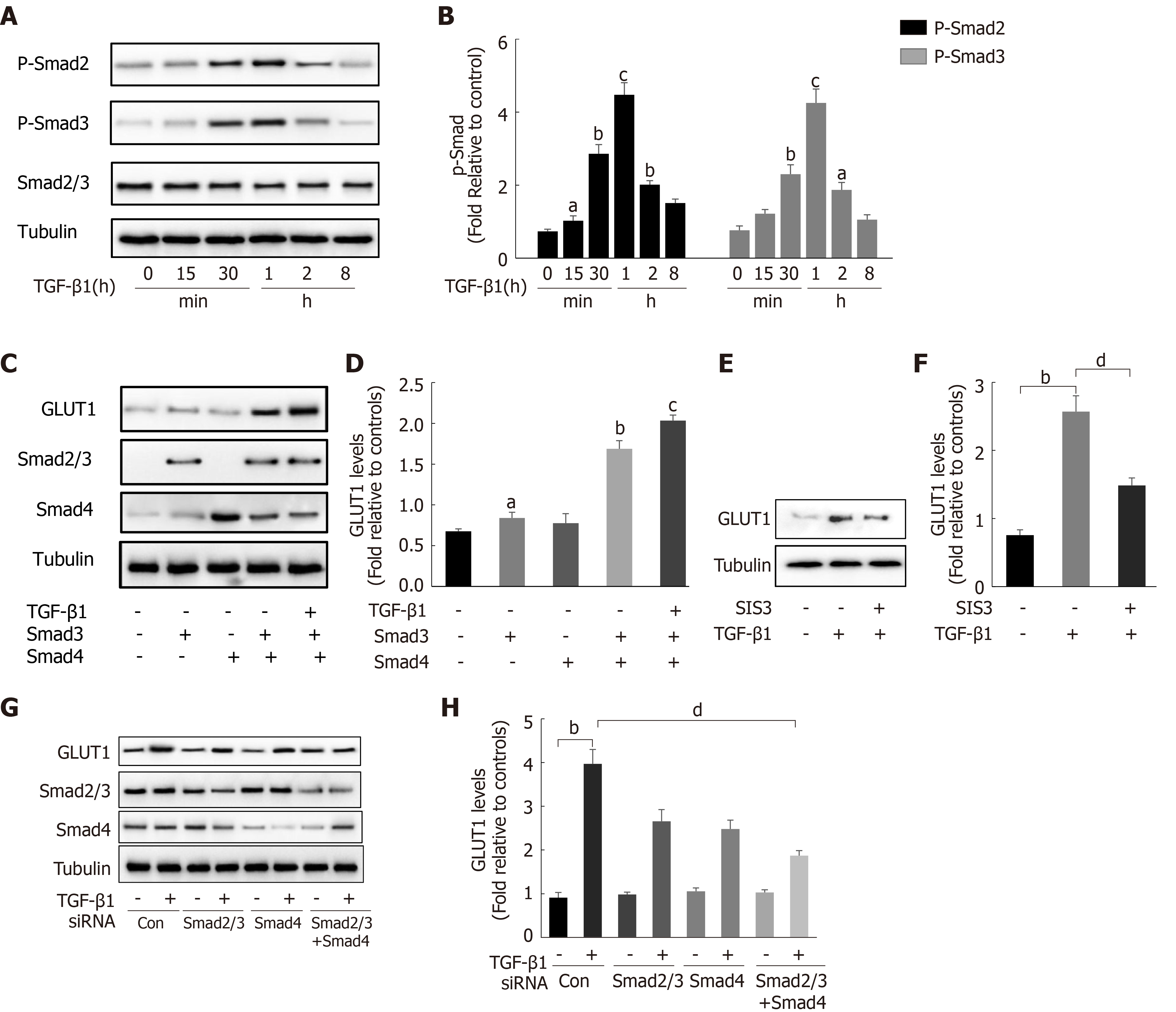
After finding that TGF-β1 stimulation induces GLUT1 expression, the specific mechanism by which TGF-β1 induces GLUT1 expression was further explored. Changes in the expression levels of Smad proteins in the canonical pathway activated by TGF-β1 stimulation were first examined. Western blot analysis revealed a time-dependent relationship between Smad2/Smad3 phosphorylation and TGF-β1 stimulation (Figure 3A and B), and phosphorylation occurred at time points close to when GLUT1 expression increased. Next, the direct role of Smads in GLUT1 induction was explored. Smad3 or Smad4 overexpression plasmids were first transiently transfected into HSCs, and then certain groups of HSCs were induced with TGF-β1 for 4 h. Smad3 or Smad4 overexpression promoted GLUT1 expression, and TGF-β1 addition amplified these effects, resulting in a further increase in GLUT1 expression (Figure 3C and D). Smad2/3 and/or Smad4 siRNAs were used to silence their expression levels and to better understand the roles of Smads in the relationship between TGF-β1 and GLUT1 expression, and the analysis performed at 48 h after the transfection of Smad2/3 and/or Smad4 siRNAs showed that TGF-β1-mediated GLUT1 expression was significantly reduced. This change was more significant and the decrease in GLUT1 expression was more substantial when the cells were transfected simultaneously with both siRNAs (Figure 3G and H). Finally, HSCs were sequentially treated with the Smad inhibitor SIS3 for 2 h and then with TGF-β1 for 4 h, resulting in a significant decrease in GLUT1 protein expression (Figure 3E and F). These results preliminarily indicate the important regulatory role of Smad proteins in TGF-β1-mediated GLUT1 expression and suggest that Smad proteins directly participate in the regulation of GLUT1 by TGF-β1.
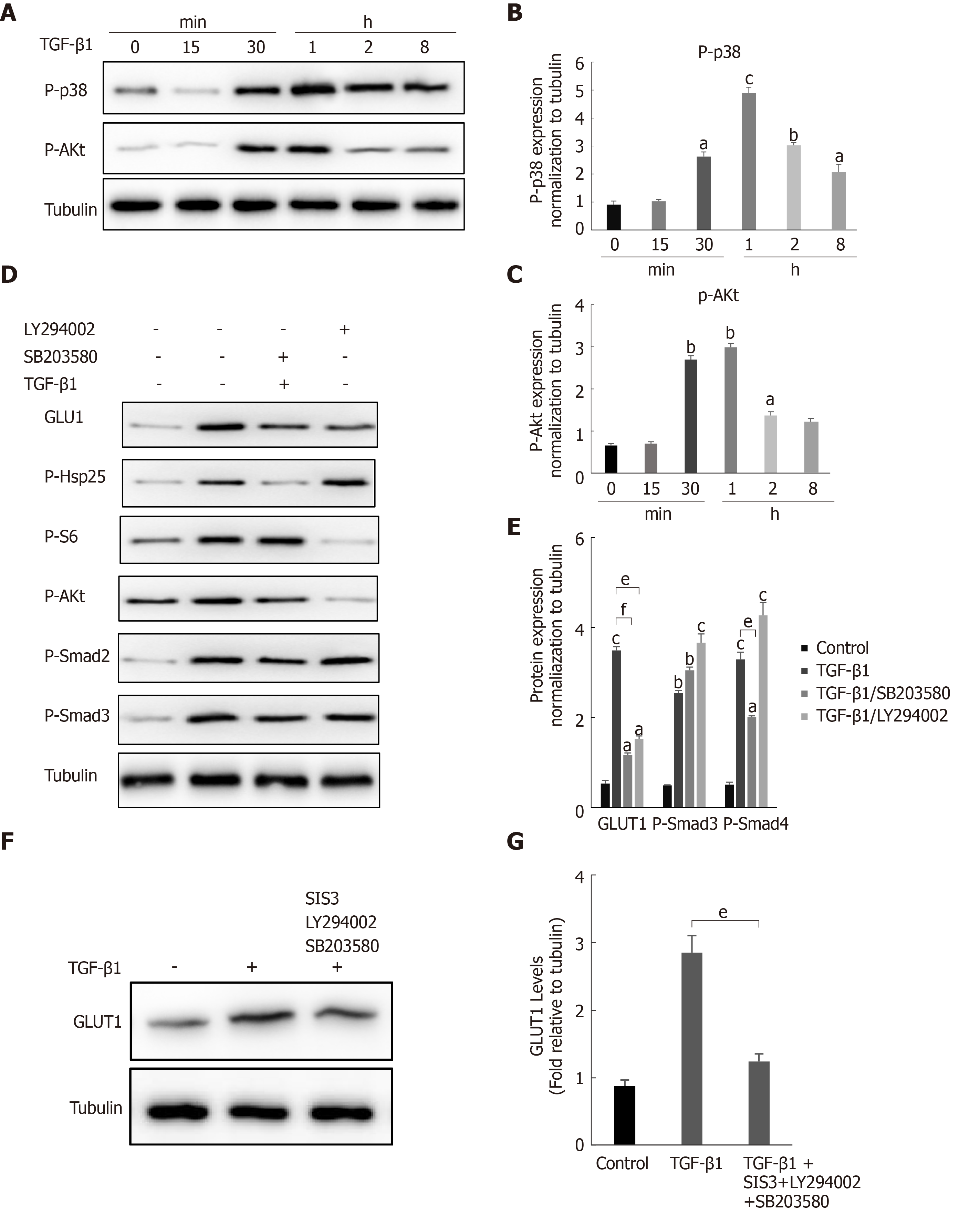
During fibrosis development, TGF-β1 activates not only the canonical Smad pathway but also noncanonical pathways (such as the PI3K/AKT and p38 MAPK signaling pathways). In addition, the Smad pathway and non-Smad pathways are mutually dependent[28]. Therefore, we questioned whether a non-Smad pathway is involved in the TGF-β1-mediated induction of GLUT1. Changes in the phosphorylation levels of p38 and AKT in HSCs after TGF-β1 stimulation were examined to answer this question. Western blot analyses showed increased levels of phosphorylated p38 and AKT in HSCs after TGF-β1 treatment (Figure 4A-C). HSCs were pretreated with the specific p38 MAPK inhibitor SB203580 and the PI3K inhibitor LY294002 for 1 h and then induced with TGF-β1 to understand the bridging role of p38 MAPK and AKT in TGF-β1-mediated GLUT1 expression. Western blot analyses showed that p-AKT activity was significantly inhibited and that GLUT1 protein expression was significantly reduced (Figure 4D). S6 ribosomal protein and heat shock protein 25 (Hsp25) are downstream proteins in the PI3K/AKT and p38 MAPK pathways, and their phosphorylation was also inhibited. Addition of the p38 inhibitor reduced the phosphorylation of the S6 protein, and the phosphorylation level of Smad2 was also affected; however, Smad3 was not significantly affected (Figure 4D and E). The above results indicate that (1) GLUT1 expression in HSCs did not rely solely on the TGF-β1-mediated Smad pathway, i.e., the p38 MAPK and PI3K/AKT signaling pathways were also involved in TGF-β1-mediated GLUT1 expression, and (2) TGF-β1-mediated pathways did not act independently, as mutual restrictions and interactions between the pathways were observed. Based on the results described above, the effects of inhibiting the Smad3, p38 MAPK and PI3K/AKT pathways on GLUT1 expression were analyzed, and the simultaneous addition of inhibitors of the Smad3, p38 MAPK and PI3K/AKT pathways significantly reduced TGF-β1-mediated GLUT1 expression (Figure 4F and G). In summary, TGF-β1 requires the participation of non-Smad pathways to induce GLUT1 expression during HSC activation.
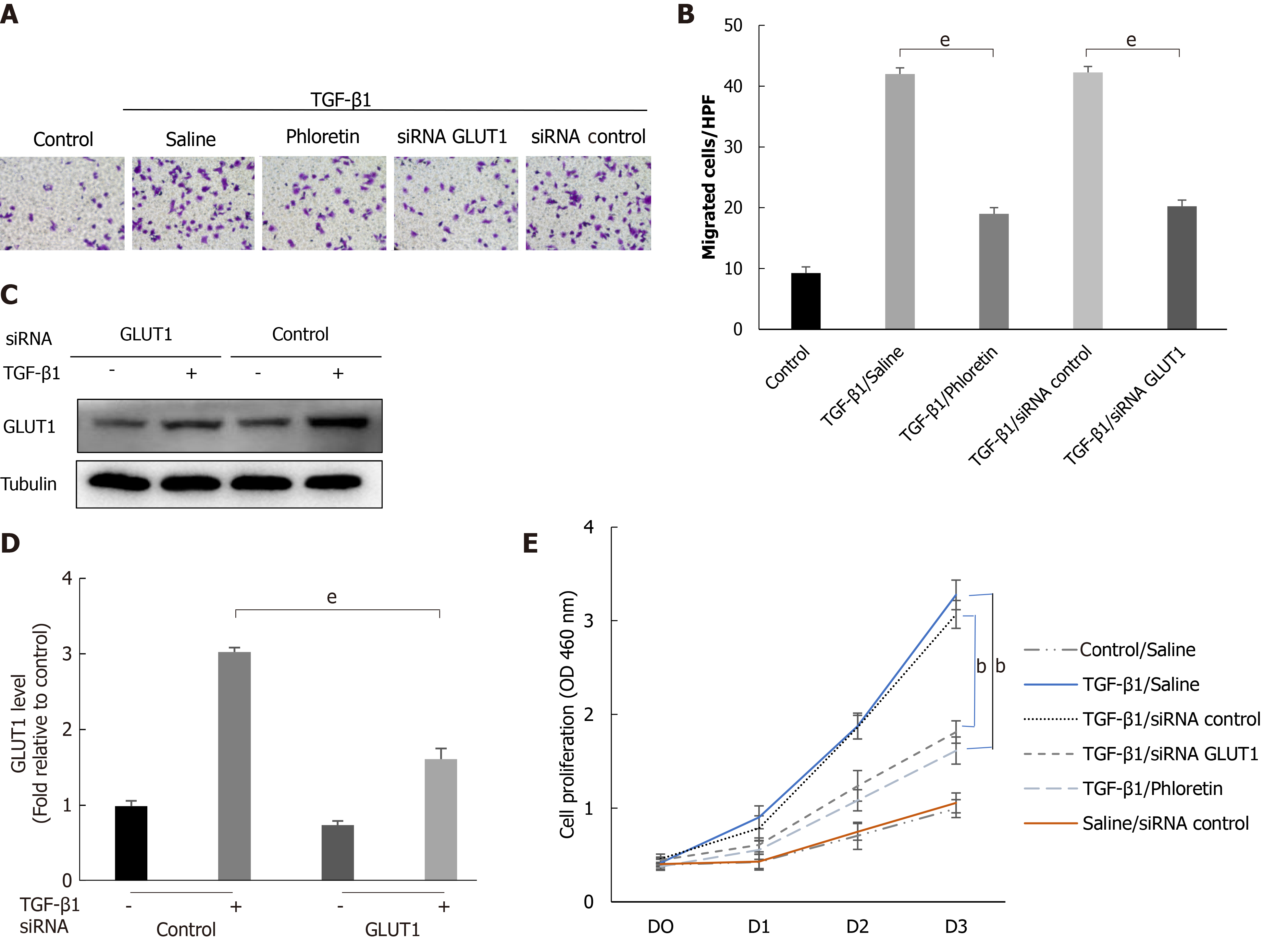
Cells were first treated with phloretin (a specific inhibitor of GLUT1) for 30 min or transfected with an siRNA targeting GLUT1, followed by treatment with TGF-β1 for 4 h. The targeted inhibition of GLUT1 by phloretin and the siRNA suppressed the effect of TGF-β1 on the migration and proliferation of HSCs (Figure 5A, B and E). Western blot analyses also showed that siRNA transfection effectively inhibited TGF-β1-induced GLUT1 expression (Figure 5C and D). Therefore, inhibition of GLUT1 expression reverses the effect of TGF-β1 on the migration and proliferation of HSCs and delays the process of HSC transdifferentiation into myofibroblasts. No noticeable effect of the control siRNA on proliferation was identified between TGF-β1-treated cells and TGF-β1/siRNA-control-treated cells or between control/saline-treated cells and saline/siRNA-control-treated cells (Figure 5E). In addition, no obvious effect of siRNA interference on cell viability (Supplementary Figure 2) or the expression of TGF-β receptors was found (Supplementary Figure 3).
In vitro experiments confirmed the importance of GLUT1 in liver fibrosis. Next, we examined whether inhibition of GLUT1 expression suppressed liver fibrosis in vivo. Phloretin, a specific inhibitor of GLUT1, was used, and its effect on CCl4-induced liver fibrosis was examined. After successful establishment of the CCl4-induced model, an i.p. injection of phloretin was administered three times a week; the intervention was discontinued after 2 wk. A simple technical roadmap is shown in Figure 6A. Compared with those in the model group, the areas of collagen fiber deposition were significantly reduced in the liver tissues from mice in the drug intervention group; these findings were confirmed by Masson’s trichrome and Sirius red staining (Figure 6B-D). The degree of liver inflammation was determined by performing serological assessments of changes in ALT and AST levels, and the results indicated that the degree of inflammation was significantly reduced in the drug intervention group (Figure 6E and F). Therefore, the in vivo results revealed that GLUT1 inhibition reduces CCl4-induced liver fibrosis.
In normal liver tissues, quiescent HSCs express TGF-β1 at low levels, while TGF-β1 is immediately upregulated after acute or chronic liver injury and interacts with multiple signaling pathways to induce HSC activation and proliferation and extensive ECM production[29,30]. TGF-β1 enhances aerobic glycolysis, amino acid uptake and lactic acid production in Ras- and Myc-transformed cells. TGF-β1 contributes to the metabolic reprogramming of cancer cells and tumor-associated stromal cells[31]. When used to replace a peritoneal dialysis solution, TGF-β1 stimulates glycolysis and inhibits mitochondrial respiration of mesothelial cells, thus promoting the development of peritoneal fibrosis[32]. Preliminary yet strong evidence supporting the importance of metabolic reprogramming in the activation of fibroblasts is steadily accumulating. Research on the mechanism of organ fibrosis also shows that TGF-β1 is related to the occurrence of aerobic glycolysis and mitochondrial dysfunction. The transdifferentiation of resting HSCs into hepatic fibroblasts has been confirmed to be related to mutual transformation between glycolytic enzymes and gluconeogenic enzymes triggered by Hedgehog signaling[33]. Glycolysis is an important pathway of glucose metabolism, and GLUT1 is the most widely expressed glucose transporter in mammals; its expression is regulated by changes in metabolic status and oxidative stress. GLUT1 is also an important marker of liver carcinogenesis and metabolic liver diseases[34]. GLUT1-dependent glycolysis exacerbates lung fibrogenesis during Streptococcus pneumoniae infection via AIM2 inflammasome activation[35]. In the pathogenesis of diabetic glomerulosclerosis, TGF-β1 triggers GLUT1 activation by stretching glomerular mesangial cells. In breast cancer cells, long-term exposure to TGF-β1 restores GLUT1 expression and results in stable EMT and unlimited cell proliferation[36,37]. Therefore, we questioned whether TGF-β1 and GLUT1 are related to liver fibrosis.
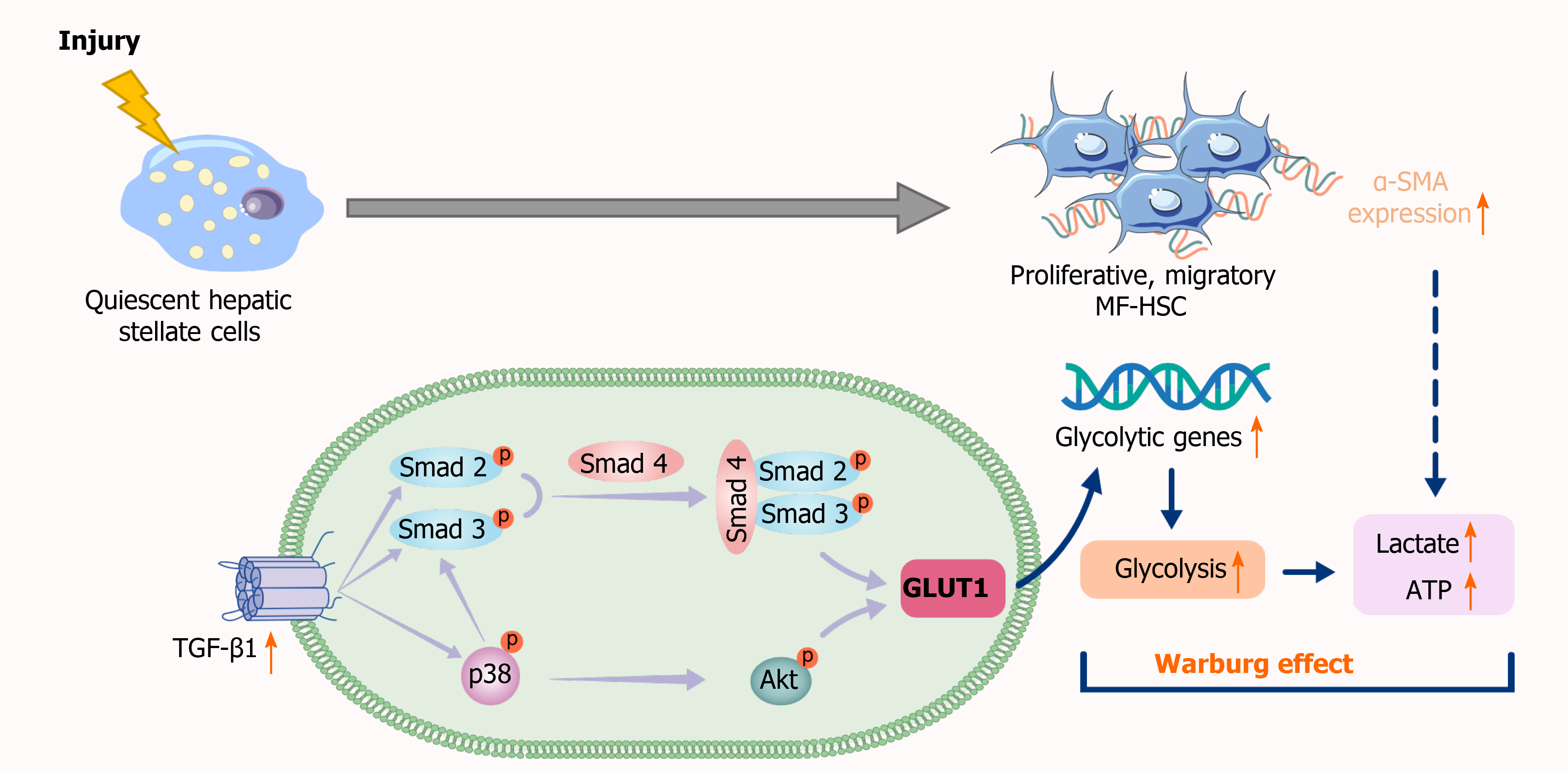
This study showed a significant increase in GLUT1 expression in human and mouse fibrotic liver tissues, which is consistent with the research results of Wan et al[24]. With the increase in TGF-β1 Levels, the gene expression levels of key enzymes, including GLUT1, in the glycolytic pathway are elevated, glucose consumption and intracellular lactate production are also increased, and glycolytic flux by HSCs is enhanced. As expected, the results of this study are consistent with those of previous studies assessing the mechanism of metabolic reprogramming of pulmonary fibrotic fibroblasts[38], indicating that TGF-β1 induces aerobic glycolysis and drives the occurrence of metabolic reprogramming during the process of stromal cell transdifferentiation. Increased GLUT1 expression also contributes to an elevated glycolytic rate, increased lactic acid production and enhanced glucose-dependent metabolic pathways in cells. In contrast, GLUT1 expression decreased significantly after the addition of a TGF-β1 receptor inhibitor, indicating that GLUT1 expression is related to TGF-β1 signaling. Experiments involving Smad overexpression, siRNA-mediated knockout and Smad inhibitors showed that the response of GLUT1 to TGF-β1 was at least partially dependent on the Smad pathway. Studies have identified a cascade of related pathways activated by TGF-β1. Therefore, this study attempted to verify whether non-Smad pathways were also involved in the induction of GLUT1 expression in HSCs. The noncanonical PI3K/AKT and p38 MAPK pathways activated by TGF-β1 were examined. In colorectal cancer (CRC) cells, silencing GLUT1 expression inactivates the TGF-β1/PI3K/AKT signaling pathway, inhibits the proliferation of CRC cells and promotes apoptosis. MAPK activation by TGF-β1 may trigger GLUT1 synthesis[39,40]. Based on the results of the present study, the simultaneous addition of specific inhibitors of the PI3K/AKT and p38 pathways, i.e., SB203580 and LY294002, respectively, reduced TGF-β1-induced GLUT1 protein expression. The addition of the p38 pathway inhibitor resulted in a decrease in Smad2 protein phosphorylation, changes in the phosphorylated AKT level and changes in the phosphorylation level of a protein downstream of PI3K/AKT signaling (namely, S6); therefore, we speculated that the p38 MAPK pathway acted as a bridge between the TGF-β1-mediated Smad and AKT pathways in HSCs and that reduced activation of the p38 MAPK pathway would inhibit the latter two pathways. In addition, the p38 MAPK pathway might limit Smad pathway-mediated GLUT1 expression to a certain extent. These results are consistent with the previously reported crosstalk between Smad and p38 MAPK in TGF-β1 signal transduction in human glioblastoma cells[41]. However, the specific mechanism underlying the interaction among TGF-β1 pathways in the induction of aerobic glycolysis in stellate cells requires further study. The significant reduction in GLUT1 protein expression was related to the simultaneous inhibition of the Smad3, p38 MAPK and PI3K signaling pathways, indicating that GLUT1 protein expression during stellate cell activation requires the activation and signaling of these three pathways. Moreover, activation of the p38 MAPK pathway might result in a certain synergistic effect with the Smad2/3 pathway (Figure 7).
TGF-β1 is a pleiotropic cytokine with an important role in the occurrence of liver fibrosis. According to previous studies, TGF-β1 signaling clearly promotes cell migration, matrix synthesis and HSC differentiation toward myofibroblasts. Moreover, the effect of TGF-β1 on fibroblast migration and proliferation depends on changes in the microenvironment[42]. As shown in the present study, TGF-β1 promoted the proliferative and migratory capabilities of HSCs, functions that are hallmarks of cell transformation. The addition of a pharmacological inhibitor of GLUT1 activity (phloretin, an effective GLUT1 inhibitor capable of inhibiting bleomycin-induced pulmonary fibrosis in vivo[43]) and silencing of the GLUT1 gene eliminated TGF-β1-induced proliferation, growth and migration. Finally, a GLUT1 inhibitor was used in in vivo experiments, and the degree of mouse liver fibrosis improved, collagen fiber deposition decreased, and the degree of inflammation decreased. Given the importance of GLUT1, the experimental results revealed that GLUT1 is involved in aerobic glycolysis during HSC activation and that aerobic glycolysis is a response to TGF-β1 signaling mediated by the Smad, PI3K/AKT and p38 MAPK pathways.
In summary, TGF-β1-induced GLUT1 expression may be one of the mechanisms involved in the reprogramming of HSCs, providing an expanded basis and new insights for the mechanism of action of TGF-β1 in metabolic reprogramming during liver fibrosis. GLUT1 plays an important role in aerobic glycolysis in HSCs and in promoting cell proliferation and transformation. GLUT1 inhibition may be an alternative therapy to the current traditional treatments for liver fibrosis. However, the extent to which GLUT1 inhibition contributes to elimination of the profibrotic effect of TGF-β1 and the specific molecular mechanisms of the interaction between the two may require verification using approaches combining proteomics and single-cell sequencing, which may be an attractive research direction in the future.
Liver fibrosis is a refractory disease that develops progressively and eventually evolves into liver cirrhosis or even liver cancer. Hepatic stellate cell (HSC) activation is the initiating factor for liver fibrosis, while aerobic glycolysis is one of the main metabolic characteristics. Transforming growth factor-β1 (TGF-β1) is the most important profibrotic factor in HSCs, and TGF-β1 drives metabolic reprogramming. Glucose transporter 1 (GLUT1) is the most widely distributed glucose transporter in mammals and is related to glycolytic metabolism. However, the role of GLUT1 in liver fibrosis and the relationship between GLUT1 and TGF-β1 remain unclear and require further investigation.
The results of this study might provide a basis for the application of GLUT1 in the treatment of liver fibrosis and provide an expanded basis for understanding the mechanism of action of TGF-β1 in metabolic reprogramming during liver fibrosis.
This study examined changes in GLUT1 expression in human and mouse fibrotic liver tissues and differences in extracellular acid production and in the expression levels of key glycolytic enzymes and GLUT1 during HSC activation induced by TGF-β1-related pathways. In addition, this study further explored the relationship between TGF-β1 pathways and GLUT1 expression and the potential underlying molecular mechanisms.
IHC was employed to examine changes in GLUT1 expression in human and mouse fibrotic liver tissues. Immunofluorescence staining was performed to examine changes in GLUT1 and alpha-smooth muscle actin (α-SMA) expression in mouse fibrotic liver tissue. Primary mouse stellate cells were isolated. After activation of the cells by TGF-β1 stimulation, changes in extracellular acid production, key glycolytic enzymes and glucose consumption were examined. In addition, changes in GLUT1 expression were explored by activating/inhibiting the Smad2/3 pathway and inhibiting the expression of proteins related to the p38 and PI3K/AKT pathways. Finally, in mice with liver fibrosis, the effect of a GLUT1 inhibitor on liver fibrosis was investigated by performing Masson’s trichrome staining and Sirius red staining and analyzing serological inflammatory markers.
The expression of the GLUT1 protein was increased in both mouse and human fibrotic liver tissue.immunofluorescence staining revealed the colocalization of GLUT1 and α-SMA proteins, indicating that GLUT1 expression was related to the development of liver fibrosis. TGF-β1 induced an increase in aerobic glycolysis in HSCs and induced GLUT1 expression in HSCs by activating the canonical and noncanonical signaling pathways. The p38 MAPK pathway and the Smad pathway synergistically affected the induction of GLUT1 expression. GLUT1 inhibition eliminated the effect of TGF-β1 on the proliferation and migration of HSCs. A GLUT1 inhibitor was administered to a mouse model of liver fibrosis, and GLUT1 inhibition reduced the degree of liver inflammation.
GLUT1 expression was upregulated in liver fibrosis, and the underlying mechanism was related to activation of the Smad2/3, p38 and PI3K/AKT pathways by TGF-β1, which directly induced GLUT1 expression and promoted glycolysis. GLUT1 inhibition eliminated TGF-β1-induced HSC activation, proliferation and migration, and GLUT1 inhibition exerted an antifibrotic effect.
The results of this study reveal that the TGF-β1 pathway directly induces GLUT1 expression and aerobic glycolysis, thus promoting liver fibrosis. This study preliminarily clarified the mechanism underlying the interaction between TGF-β1 and GLUT1 in liver fibrosis, thus providing a deeper understanding of the mechanism of liver fibrosis and providing guidance for the selection of targets to treat liver fibrosis. The results from this study indicate that GLUT1 inhibitors may have certain prospective applications as therapeutic drugs for liver fibrosis.
The authors thank Dr. Ding Q for expert technical assistance.
| 1. | Ni Y, Li JM, Liu MK, Zhang TT, Wang DP, Zhou WH, Hu LZ, Lv WL. Pathological process of liver sinusoidal endothelial cells in liver diseases. World J Gastroenterol. 2017;23:7666-7677. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 49] [Cited by in RCA: 64] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 2. | Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annu Rev Pathol. 2011;6:425-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1096] [Cited by in RCA: 1428] [Article Influence: 95.2] [Reference Citation Analysis (0)] |
| 3. | Higashi T, Friedman SL, Hoshida Y. Hepatic stellate cells as key target in liver fibrosis. Adv Drug Deliv Rev. 2017;121:27-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 530] [Cited by in RCA: 1146] [Article Influence: 127.3] [Reference Citation Analysis (0)] |
| 4. | Vallée A, Lecarpentier Y, Vallée JN. Thermodynamic Aspects and Reprogramming Cellular Energy Metabolism during the Fibrosis Process. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 52] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 5. | Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029-1033. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12491] [Cited by in RCA: 12300] [Article Influence: 723.5] [Reference Citation Analysis (0)] |
| 6. | Chen Y, Choi SS, Michelotti GA, Chan IS, Swiderska-Syn M, Karaca GF, Xie G, Moylan CA, Garibaldi F, Premont R, Suliman HB, Piantadosi CA, Diehl AM. Hedgehog controls hepatic stellate cell fate by regulating metabolism. Gastroenterology. 2012;143:1319-1329.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 232] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 7. | Henderson NC, Rieder F, Wynn TA. Fibrosis: from mechanisms to medicines. Nature. 2020;587:555-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 1318] [Article Influence: 219.7] [Reference Citation Analysis (0)] |
| 8. | Syed V. TGF-β Signaling in Cancer. J Cell Biochem. 2016;117:1279-1287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 370] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 9. | Zhang YE. Non-Smad Signaling Pathways of the TGF-β Family. Cold Spring Harb Perspect Biol. 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 462] [Cited by in RCA: 575] [Article Influence: 63.9] [Reference Citation Analysis (0)] |
| 10. | Derynck R, Budi EH. Specificity, versatility, and control of TGF-β family signaling. Sci Signal. 2019;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 585] [Article Influence: 83.6] [Reference Citation Analysis (0)] |
| 11. | Heldin CH, Moustakas A. Signaling Receptors for TGF-β Family Members. Cold Spring Harb Perspect Biol. 2016;8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 340] [Cited by in RCA: 538] [Article Influence: 53.8] [Reference Citation Analysis (0)] |
| 12. | Wrana JL, Attisano L, Wieser R, Ventura F, Massagué J. Mechanism of activation of the TGF-beta receptor. Nature. 1994;370:341-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1743] [Cited by in RCA: 1777] [Article Influence: 55.5] [Reference Citation Analysis (0)] |
| 13. | Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091-3100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1449] [Cited by in RCA: 1543] [Article Influence: 55.1] [Reference Citation Analysis (8)] |
| 14. | Dewidar B, Meyer C, Dooley S, Meindl-Beinker AN. TGF-β in Hepatic Stellate Cell Activation and Liver Fibrogenesis-Updated 2019. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 214] [Cited by in RCA: 601] [Article Influence: 85.9] [Reference Citation Analysis (0)] |
| 15. | Zhao X, Kwan JYY, Yip K, Liu PP, Liu FF. Targeting metabolic dysregulation for fibrosis therapy. Nat Rev Drug Discov. 2020;19:57-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 394] [Article Influence: 56.3] [Reference Citation Analysis (0)] |
| 16. | Soukupova J, Malfettone A, Bertran E, Hernández-Alvarez MI, Peñuelas-Haro I, Dituri F, Giannelli G, Zorzano A, Fabregat I. Epithelial-Mesenchymal Transition (EMT) Induced by TGF-β in Hepatocellular Carcinoma Cells Reprograms Lipid Metabolism. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 17. | Kottmann RM, Kulkarni AA, Smolnycki KA, Lyda E, Dahanayake T, Salibi R, Honnons S, Jones C, Isern NG, Hu JZ, Nathan SD, Grant G, Phipps RP, Sime PJ. Lactic acid is elevated in idiopathic pulmonary fibrosis and induces myofibroblast differentiation via pH-dependent activation of transforming growth factor-β. Am J Respir Crit Care Med. 2012;186:740-751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 315] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 18. | Chen LQ, Cheung LS, Feng L, Tanner W, Frommer WB. Transport of sugars. Annu Rev Biochem. 2015;84:865-894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 362] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 19. | Wang L, Pavlou S, Du X, Bhuckory M, Xu H, Chen M. Glucose transporter 1 critically controls microglial activation through facilitating glycolysis. Mol Neurodegener. 2019;14:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 230] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 20. | Hua W, Ten Dijke P, Kostidis S, Giera M, Hornsveld M. TGFβ-induced metabolic reprogramming during epithelial-to-mesenchymal transition in cancer. Cell Mol Life Sci. 2020;77:2103-2123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 147] [Cited by in RCA: 183] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 21. | Liu M, Quek LE, Sultani G, Turner N. Epithelial-mesenchymal transition induction is associated with augmented glucose uptake and lactate production in pancreatic ductal adenocarcinoma. Cancer Metab. 2016;4:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 77] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 22. | Li W, Wei Z, Liu Y, Li H, Ren R, Tang Y. Increased 18F-FDG uptake and expression of Glut1 in the EMT transformed breast cancer cells induced by TGF-beta. Neoplasma. 2010;57:234-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 23. | Inoki K, Haneda M, Maeda S, Koya D, Kikkawa R. TGF-beta 1 stimulates glucose uptake by enhancing GLUT1 expression in mesangial cells. Kidney Int. 1999;55:1704-1712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 77] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 24. | Wan L, Xia T, Du Y, Liu J, Xie Y, Zhang Y, Guan F, Wu J, Wang X, Shi C. Exosomes from activated hepatic stellate cells contain GLUT1 and PKM2: a role for exosomes in metabolic switch of liver nonparenchymal cells. FASEB J. 2019;33:8530-8542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 99] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 25. | Zhao XK, Yu L, Cheng ML, Che P, Lu YY, Zhang Q, Mu M, Li H, Zhu LL, Zhu JJ, Hu M, Li P, Liang YD, Luo XH, Cheng YJ, Xu ZX, Ding Q. Focal Adhesion Kinase Regulates Hepatic Stellate Cell Activation and Liver Fibrosis. Sci Rep. 2017;7:4032. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 104] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 26. | Zou GL, Zuo S, Lu S, Hu RH, Lu YY, Yang J, Deng KS, Wu YT, Mu M, Zhu JJ, Zeng JZ, Zhang BF, Wu X, Zhao XK, Li HY. Bone morphogenetic protein-7 represses hepatic stellate cell activation and liver fibrosis via regulation of TGF-β/Smad signaling pathway. World J Gastroenterol. 2019;25:4222-4234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 37] [Cited by in RCA: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 27. | Morikawa M, Derynck R, Miyazono K. TGF-β and the TGF-β Family: Context-Dependent Roles in Cell and Tissue Physiology. Cold Spring Harb Perspect Biol. 2016;8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 557] [Cited by in RCA: 1032] [Article Influence: 103.2] [Reference Citation Analysis (0)] |
| 28. | Xu F, Liu C, Zhou D, Zhang L. TGF-β/SMAD Pathway and Its Regulation in Hepatic Fibrosis. J Histochem Cytochem. 2016;64:157-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 613] [Article Influence: 61.3] [Reference Citation Analysis (0)] |
| 29. | Simeone DM, Zhang L, Graziano K, Nicke B, Pham T, Schaefer C, Logsdon CD. Smad4 mediates activation of mitogen-activated protein kinases by TGF-beta in pancreatic acinar cells. Am J Physiol Cell Physiol. 2001;281:C311-C319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 49] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 30. | Fan W, Liu T, Chen W, Hammad S, Longerich T, Hausser I, Fu Y, Li N, He Y, Liu C, Zhang Y, Lian Q, Zhao X, Yan C, Li L, Yi C, Ling Z, Ma L, Xu H, Wang P, Cong M, You H, Liu Z, Wang Y, Chen J, Li D, Hui L, Dooley S, Hou J, Jia J, Sun B. ECM1 Prevents Activation of Transforming Growth Factor β, Hepatic Stellate Cells, and Fibrogenesis in Mice. Gastroenterology. 2019;157:1352-1367.e13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 98] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 31. | Olsson N, Piek E, Sundström M, ten Dijke P, Nilsson G. Transforming growth factor-beta-mediated mast cell migration depends on mitogen-activated protein kinase activity. Cell Signal. 2001;13:483-490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 45] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 32. | Si M, Wang Q, Li Y, Lin H, Luo D, Zhao W, Dou X, Liu J, Zhang H, Huang Y, Lou T, Hu Z, Peng H. Inhibition of hyperglycolysis in mesothelial cells prevents peritoneal fibrosis. Sci Transl Med. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 94] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 33. | Nigdelioglu R, Hamanaka RB, Meliton AY, O'Leary E, Witt LJ, Cho T, Sun K, Bonham C, Wu D, Woods PS, Husain AN, Wolfgeher D, Dulin NO, Chandel NS, Mutlu GM. Transforming Growth Factor (TGF)-β Promotes de Novo Serine Synthesis for Collagen Production. J Biol Chem. 2016;291:27239-27251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 135] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 34. | Cho SJ, Moon JS, Nikahira K, Yun HS, Harris R, Hong KS, Huang H, Choi AMK, Stout-Delgado H. GLUT1-dependent glycolysis regulates exacerbation of fibrosis via AIM2 inflammasome activation. Thorax. 2020;75:227-236. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 35. | Chadt A, Al-Hasani H. Glucose transporters in adipose tissue, liver, and skeletal muscle in metabolic health and disease. Pflugers Arch. 2020;472:1273-1298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 318] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 36. | Jiang L, Deberardinis R, Boothman DA. The cancer cell 'energy grid': TGF-β1 signaling coordinates metabolism for migration. Mol Cell Oncol. 2015;2:e981994. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 37. | Nilchian A, Giotopoulou N, Sun W, Fuxe J. Different Regulation of Glut1 Expression and Glucose Uptake during the Induction and Chronic Stages of TGFβ1-Induced EMT in Breast Cancer Cells. Biomolecules. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 38. | Xie N, Tan Z, Banerjee S, Cui H, Ge J, Liu RM, Bernard K, Thannickal VJ, Liu G. Glycolytic Reprogramming in Myofibroblast Differentiation and Lung Fibrosis. Am J Respir Crit Care Med. 2015;192:1462-1474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 343] [Cited by in RCA: 442] [Article Influence: 40.2] [Reference Citation Analysis (0)] |
| 39. | Fujishiro M, Gotoh Y, Katagiri H, Sakoda H, Ogihara T, Anai M, Onishi Y, Ono H, Funaki M, Inukai K, Fukushima Y, Kikuchi M, Oka Y, Asano T. MKK6/3 and p38 MAPK pathway activation is not necessary for insulin-induced glucose uptake but regulates glucose transporter expression. J Biol Chem. 2001;276:19800-19806. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 102] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 40. | Yamamoto Y, Yoshimasa Y, Koh M, Suga J, Masuzaki H, Ogawa Y, Hosoda K, Nishimura H, Watanabe Y, Inoue G, Nakao K. Constitutively active mitogen-activated protein kinase kinase increases GLUT1 expression and recruits both GLUT1 and GLUT4 at the cell surface in 3T3-L1 adipocytes. Diabetes. 2000;49:332-339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 41. | Dziembowska M, Danilkiewicz M, Wesolowska A, Zupanska A, Chouaib S, Kaminska B. Cross-talk between Smad and p38 MAPK signalling in transforming growth factor beta signal transduction in human glioblastoma cells. Biochem Biophys Res Commun. 2007;354:1101-1106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 45] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 42. | Lu Y, Jiang F, Zheng X, Katakowski M, Buller B, To SS, Chopp M. TGF-β1 promotes motility and invasiveness of glioma cells through activation of ADAM17. Oncol Rep. 2011;25:1329-1335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 30] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 43. | Cho SJ, Moon JS, Lee CM, Choi AM, Stout-Delgado HW. Glucose Transporter 1-Dependent Glycolysis Is Increased during Aging-Related Lung Fibrosis, and Phloretin Inhibits Lung Fibrosis. Am J Respir Cell Mol Biol. 2017;56:521-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 105] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/Licenses/by-nc/4.0/
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Rojas A, Vela D, Wang Z S-Editor: Gao CC L-Editor: A P-Editor: Xing YX