Published online Oct 7, 2021. doi: 10.3748/wjg.v27.i37.6306
Peer-review started: March 22, 2021
First decision: June 14, 2021
Revised: June 28, 2021
Accepted: September 2, 2021
Article in press: September 2, 2021
Published online: October 7, 2021
Processing time: 191 Days and 0.3 Hours
Gluten is a complex mixture of proteins with immunogenic peptide sequences triggering the autoimmune activity in patients with celiac disease (CeD). Gluten immunogenic peptides (GIP) are resistant to gastrointestinal digestion and are then excreted via the stool and urine. Most common detection methods applied in the follow-up visits for CeD patients such as serology tests, dietetic interviews, questionnaires, and duodenal biopsy have been proved to be inefficient, invasive, or inaccurate for evaluating gluten-free diet (GFD) compliance. Determination of excreted GIP in stool and urine has been developed as a non-invasive, direct, and specific test for GFD monitoring.
To summarize published literature about the clinical utility of GIP determination in comparison to the tools employed for GFD monitoring.
PubMed and Web of Science searches were performed using the keywords “gluten immunogenic peptides” or “gluten immunogenic peptide” and a combination of the previous terms with “feces”, “stools”, “urine”, “celiac disease”, “gluten-free diet”, and “adherence” to identify relevant clinical studies published in English and Spanish between 2012 to January 2021. Reference lists from the articles were reviewed to identify additional pertinent articles. Published articles and abstracts reporting the clinical use of GIP determination in stool and/or urine for the follow-up of patients with CeD in comparison with other tools in use were included. Case reports, commentaries, reviews, conference papers, letters, and publications that did not focus on the aims of this review were excluded.
Total of 15 publications were found that involved the use of GIP determination in stool and/or urine to monitor the adherence to the GFD in comparison to other tools. Studies included both children and adults diagnosed with CeD and healthy volunteers. Overall, these preliminary studies indicated that this novel technique was highly sensitive for the detection of GFD transgressions and therefore could facilitate the follow-up of patients with CeD. Tools identified in this work included the CeD-specific serology, dietetic questionnaires, symptomatology, and the duodenal biopsy. Review of the literature revealed that the rates of GFD adherence may vary between 30%-93% using either stool or urine GIP determination, 49%-96% by the serology, 59%-94% using the dietetic questionnaires, 56%-95% by the reported symptoms and 44%-76% with the duodenal biopsy. In addition, the association between the different methods and histological abnormalities (Marsh II-III) was found to be 33%-100% for GIP determination (stool and urine), 25%-39% for CeD-specific serology, 3%-50% for dietetic questionnaires, and 22%-28% for the symptomatology.
Excreted GIP detection is the precise approach for determining voluntary or involuntary gluten consumption in CeD patients preventing future complications arising from gluten exposure.
Core Tip: A strict gluten-free diet (GFD) is the only available treatment for celiac disease. However, treatment adherence is difficult due to the ubiquitous nature of gluten, hurting patients’ quality of life. Despite several tests to evaluate GFD compliance, it has been proven to be invasive or inefficient. The determination of gluten immunogenic peptides (GIP) in stool and urine has been developed as a non-invasive, direct, and specific test for GFD monitoring. We herein summarized the current available literature meeting the clinical utility of GIP determination compared to the available tools in use.
- Citation: Coto L, Mendia I, Sousa C, Bai JC, Cebolla A. Determination of gluten immunogenic peptides for the management of the treatment adherence of celiac disease: A systematic review. World J Gastroenterol 2021; 27(37): 6306-6321
- URL: https://www.wjgnet.com/1007-9327/full/v27/i37/6306.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i37.6306
Gluten is a heterologous polymorphic mixture of proteins called prolamins. Wheat prolamins are termed gliadins and glutenins. Prolamins provide food products with special functional properties such as elasticity as well as extensibility and are characterized by high proline and glutamine content, allowing for the more efficiently packing of proteins but also complicating the enzyme-mediated hydrolysis of their tight structures[1]. Consequently, many of these proteins are insufficiently degraded by gastric and pancreatic enzymes in the gastrointestinal tract. Therefore, after the ingestion of gluten-containing foods, some gluten peptides can enter the intestinal epithelium and trigger an immune response in genetically predisposed individuals suffering from celiac disease (CeD)[2-4].
Evidence suggests that gluten can cross the intestinal barrier via the transcellular pathway perpetuating intestinal inflammation in the context of gluten intolerance[5,6]. The α-gliadin 33-mer peptide has been described as one of the most immuno
CeD is a systemic disease, involving well-known key immune factors, including the human leukocyte antigen (HLA-DQ2 and HLA-DQ8), the anti-tissue trans-glutaminase (anti-tTG) antibodies, and gluten[10]. As a systemic disease, there are intestinal and extraintestinal symptoms that can be presented individually or in combination. In addition, patients may also be completely asymptomatic[11]. Intestinal presentation of CeD is common in both the pediatric and adult patient population and is characterized by diarrhea, loss of appetite, abdominal distention, bloating, pain, constipation, or weight loss[12,13]. The most common extraintestinal symptoms are iron deficiency anemia, osteopenia or osteoporosis, hypertransaminasemia, neuro
Following CeD diagnosis, patients must follow a strict, life-long gluten-free diet (GFD), the only treatment currently available, which not only reduces disease symptoms but also allows for the healing of the intestinal epithelia and prevents long-term complications[15-17]. However, following a strict GFD is challenging and requires substantial daily effort, hurting the quality of life in addition to psychological problems and fear of involuntary gluten intake, even in patients considering themselves to be strictly adherent[15,18-20]. Gluten-free food availability, inadequate food labeling regulations, cost, and safety are the main barriers related to GFD[18,21,22]. Therefore, the frequency of voluntary and involuntary transgressions is high. It was reported that at least 50% of adult patients are not fully adherent to the GFD in a daily or weekly period of observation[8,9,23,24]. In addition, 36%-55% of patients who expressed their complete adherence to a GFD did not achieve histological remission, potentially from inadvertent lapses in their daily gluten intake[24-27]. Inadvertent gluten ingestion was suggested to be more frequent than intentional intake, not only when eating out, but also at home[28-30]. A systematic review recently reported adherence rates ranging from 23% to 98% in the pediatric population, determined by using all available methods for evaluating adherence[31].
Although continuous patient monitoring is expected to improve GFD adherence, there is no consensus on how frequent and which tools to use in the patient follow-up[32-35]. All available guidelines recommended clinical and dietary evaluation as well as serology tests at least once a year or every two years to confirm the GFD adherence in addition to detecting possible complications[14]. However, monitoring approaches have not been comprehensively assessed with a clear lack of a gold standard for comparisons[36-38]. Thus, various questions often arise among caregivers and patients regarding the best approach for detecting gluten exposure. GFD follow-up through the detection of excreted GIP has the benefits of being non-invasive, objective, and specific, highlighting its potential as a complementary technique for monitoring CeD treatment[8,9].
Despite the increasing number of studies on the use of GIP excretion determination for the assessment of GFD compliance, there are limited reference guidelines on the detection of GIP in stool and urine for monitoring the treatment in patients diagnosed with CeD. The purpose of this systematic review is to compile insights from studies that tackled the practical issues related to the clinical utility of the available methods for GIP determination compared to current GFD adherence monitoring methods.
PubMed and Web of Science searches were performed using the search terms “gluten immunogenic peptides” or “gluten immunogenic peptide” and a combination of the previous terms with “feces”, “stools”, “urine”, “celiac disease”, “gluten-free diet”, and “adherence” using the Boolean AND operator, which allows the establishments of logical relations among concepts. References of included full-text articles were scrutinized for additional studies.
The eligibility criteria were based on the PICOS (Participants, Intervention, Comparison, Outcomes, Study design) acronym. Articles eligible to be included in this review were required to meet the following criteria: (1) The study reported both children and adult patients diagnosed with CeD, since it would be more useful for readers as the evidence available about the use of this tool is limited; (2) The inter
We excluded publications that did not focus on the aim of this review. If a full-text paper could not be obtained, but the abstract presented sufficient data, the publication was included. Case reports, commentaries, reviews, conference papers, and letters were excluded.
Retrieved manuscripts were reviewed by the authors, and the data were extracted and described.
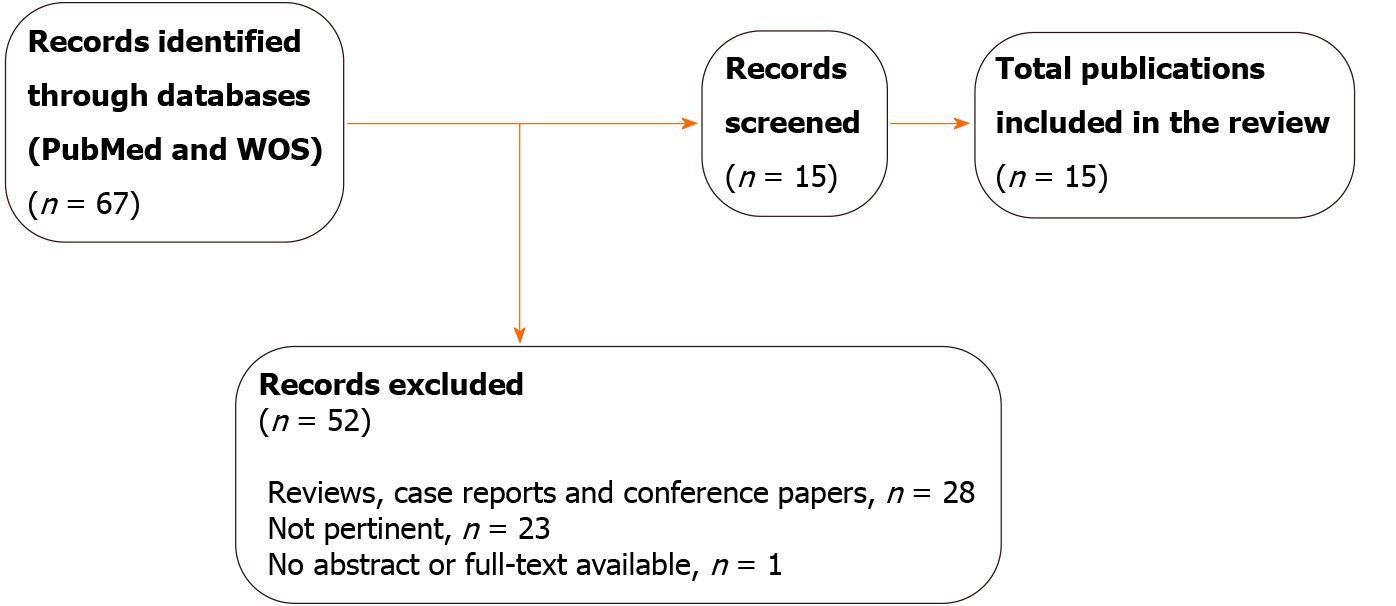
Through the literature review, our searches yielded 67 results after the removal of duplicates. In total, 52 entries were excluded for different reasons: 23 did not respond to the aim of this publication, 28 were reviews, case reports, or conference papers, and one had not abstract or full-text available. After screening, 15 publications were eligible for further study. Of these, 14 had a full text in English and one had an English abstract. Two publications reported complementary data about the same study (Figure 1).
The 15 publications included both children and adults diagnosed with CeD. Healthy adult and pediatric volunteers were also included in many of the studies. The articles were published between 2016 and 2021. The characteristics of included studies are summarized in Table 1[8,9,22-24,29,30,39-46].
| Ref. | Design | Study population | Intervention | Main results | ||
| Comino et al[8] | Prospective, multicenter, observational study | 184 adult and pediatric CeD patients | Fecal GIP ELISA, serology, questionnaires, and symptoms to evaluate adherence to the GFD | GIP-positive results were found in 12%-28% of children < 12 years-old, 30% in > 13 years-old females and 60% in > 13 years-old males. Low correlation of anti-tTG and anti-DGP markers and poor adherence to the GFD | ||
| Moreno et al[9] | Randomized controlled study | 58 adult and pediatric CeD patients and 76 healthy controls | Urine GIP LFIA test, serology, and duodenal biopsy to evaluate adherence to the GFD | About 50% CeD patients were GIP-positive. High correlation of GIP quantifiable concentration in urine with persistent villus atrophy in treated CeD patients (n = 25). No correlation between serology and mucosal damage | ||
| Gerasimidis et al[39] | Cross-sectional study cohort for a subgroup | 63 pediatric CeD patients | Fecal ELISA GIP test, serology, and questionnaires to evaluate gluten intake during diagnosis and adherence to the GFD after diagnosis | GIP-positive results in 95% of de novo patients with CeD during diagnosis. GIP-positive results were found in 17% and 27% of patients after 6 and 12 months of the beginning of the GFD, respectively. GIP-positive results were found in 16%, 16%, and 14% of patients considered compliant according to the Biagi score, tTG, and clinical assessment, respectively | ||
| Comino et al[40] | Prospective, multicenter, observational study | 64 pediatric CeD patients | Fecal GIP ELISA, serology, questionnaires, and symptoms to evaluate adherence to the GFD after diagnosis | Most children (97%) were GIP-positive at diagnosis. A decrease of GIP detection was observed on a GFD, but the rate of GIP-positive results increased from 13% at 6 months to 25% at 24 months. Anti-tTG antibody levels showed low sensitivity to identify patients with GIP-positive results. Dietitian assessment was only moderately correlated with GIP detection | ||
| Costa et al[41] | Cross-sectional study and prospective cohort | 44 adult CeD patients | Fecal GIP ELISA, stool and urine LFIA GIP tests, serology, questionnaires, and symptoms to evaluate adherence to the GFD | 25% of patients had at least one GIP-positive test, 32% in asymptomatic patients and 15.8% in symptomatic patients. Dietary assessment estimated gluten intake in only 50% of GIP-positive samples. Anti-tTG and anti-DGP positive results in 3/12 and 6/12 of GIP-positive cases, respectively | ||
| Silvester et al[29,30] | Prospective longitudinal study | 18 adult CeD patients | Monitoring GFD adherence by collection of daily food, stool, and urine samples for the analysis of GIP content, and relationship with duodenal biopsy, serology, questionnaires, and symptoms | GIP were detected in 66,7% patients. No significant correlation was found between gluten ingestion and non-invasive measures of GFD adherence. Most patients with normal anti-tTG had ≥ 1 GIP-positive sample (64%), 2/3 of these had persistent villous atrophy (Marsh 3a) and 2/3 of those with all GIP-negative samples had normal villous architecture (Marsh 0-1) but 4/6 with Marsh 0 had detectable gluten in ≥ 1 sample | ||
| Ruiz-Carnicer et al[23] | Prospective observational study | 22 newly diagnosed CeD patients, 77 CeD patients following a GFD and 13 healthy volunteers | Urine LFIA GIP test to evaluate adherence to the GFD and comparison with serology, clinical manifestations, dietary questionnaire, and histological results | Mucosal damage (Marsh II-III) was found in 24% of CeD patients, 94%of these had ≥ 1 GIP urine sample. 60-80% of these were asymptomatic, had negative serologic results and were compliant with treatment regarding the dietary questionnaire. GIP-negative results were found in 97% of the patients without mucosal damage | ||
| Fernandez-Miaja et al[22] | Cross-sectional study | 80 pediatric CeD patients | Relationship of fecal LFIA GIP for GFD monitoring GFD with CDAT, serology and sociodemographic and clinical data | Acceptable agreement was found between GIP detection and CDAT questionnaire (92.5% and 86.3% adherence rate, respectively). Most patients (83.3%) with GIP-positive results had negative anti-tTG antibodies | ||
| Porcelli et al[42] | Cross-sectional study | 25 CeD patients | Assessment of compliance with the GFD using Fecal GIP ELISA testing, the Biagi questionnaire, evaluation of symptoms and serology | GIP-positive results were found in 4 patients, 2 of these complied with the GFD according to the Biagi questionnaire. All GIP-negative patients were asymptomatic. Levels of anti-tTG antibodies were significantly higher in GIP-positive patients than in GIP-negative patients | ||
| Roca et al[43] | Prospective, cross-sectional study | 43 pediatric CeD patients at follow-up (Group 1) and 18 at diagnosis (Group 2) | Fecal GIP ELISA and LFIA analysis to monitor in real life the adherence to GFD Comparison to food record questionnaire and serology | Group 1: GIP-positive results were found in of 34.9% patients by ELISA (46,7% also by LFIA). 48.8% of patients had positive anti-tTG antibodies (4 reported symptoms) and 10 of these had GIP-positive results by ELISA (70% also by LFIA) (2 reported symptoms). All the transgressions detected by food record were also detected with GIP | ||
| Porcelli et al[44] | Cross-sectional study | 55 CeD patients: 27 adults and 28 children | Assessment of compliance with the GFD using Fecal GIP ELISA, the Biagi questionnaire, evaluation of symptoms and serology | GIP-positive results were found in 8 patients, 71.4% of these were asymptomatic and 37.5% had raised anti-tTG antibodies. A significant association was found between the Biagi score and GIP-positive results but according to the Biagi score, 57.1% of GIP-positive patients followed the diet strictly and 5.4% of GIP-negative subjects did not comply with the diet | ||
| Laserna-Mendieta et al[45] | Prospective observational study | 97 adolescent and adult CeD patients | Evaluation of the sensitivity and specificity of fecal GIP LFIA test to detect duodenal lesions in CeD patients on a GFD and comparison to serology and questionnaires | Compared to the duodenal histology, GIP LFIA test showed similar sensitivity (33%) and specificity (81%) to anti-tTG antibodies. No relationship was found between GIP and questionnaires but an association between GIP and patients’ self-reported gluten consumption was seen | ||
| Stefanolo et al[24] | Prospective observational study | 53 adult CeD patients | Fecal GIP ELISA and urine LFIA GIP test, anti-tTG, anti-DGP, and questionnaires to evaluate adherence to the GFD in symptomatic and asymptomatic patients | At least one GIP-positive result in 88.7% of patients for the 4 wk period. Patients who had symptoms had elevated GIP levels for more weeks than patients who did not have these symptoms (P < 0.05). Correlation was found between GIP and anti-DGP antibodies but not with levels of anti-tTG antibodies | ||
| Fernández-Bañares et al[46] | Multicenter prospective observational study | 76 adult CeD patients | Fecal GIP ELISA, anti-tTG, questionnaires and symptomatology to evaluate villous atrophy persistence after 2 years on a GFD | Persistent villous atrophy was present in 53% of patients at follow-up, 72% of these were asymptomatic and 75% had negative anti-tTG antibodies. Most patients were adherent to the GFD according to the dietary evaluation. In contrast, GIP-positive results were found in ≥ 1 fecal sample of 77% of patients with villous atrophy and in 60% of patients with mucosal recovery | ||
Analytical methods require a standard analyte for quantitative determination. The main issue is the selection of a standard gluten peptide in the huge heterogeneity of the gluten proteins and the countless number of gluten hydrolyzed fragments. The a-gliadin 33-mer peptide (LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF) is resistant to hydrolysis via gastric, pancreatic, and intestinal brush-border membrane proteases, which allows for its detection in human excretions. This peptide is considered one of the most immunodominant gluten peptides for CeD patients, as it contains three overlapping T cell epitopes, repetitions of p57-68[2,47,48]. Monoclonal antibodies A1 and G12 were generated against the α-2-gliadin 33-mer peptide specifically and sensitively detecting significant amounts of excreted GIP in stool and urine[8,9]. The reactive epitope profile may thus play an important role in the detection of excreted GIP. Even though there are a variety of antibodies on the market that allow for gluten detection in food, such as R5, 401.21, α20, 14G11, and 13F6, only G12 and/or A1 are useful for gluten peptide sensitive detection in stool and urine[8,9,49-52].
Urine samples are highly heterogeneous matrices with low protein content, making complicating the development of immunoassays for biomarkers detection. Urine contains organic molecules such as urea, creatinine, and uric acid, inorganic ions such as K+, Na+, Cl-, and Ca2+, cells, as well as peptides of more than 1500 proteins[53]. The concentration of these compounds and the pH usually exhibit considerable variability not only among individuals but also between different urine samples taken from the same individual[54]. The complex composition of these samples and its variability, in addition to the high frequency of matrix interferences, complicate the reproducibility and robustness of urine immunoassays. Chatziharalambous et al[53] evaluated 11 commercial ELISA assays for the detection of urine biomarkers, reporting that only three of them met the requirements of FDA validation guidelines[55].
The currently available tests for GIP detection in stool and/or urine (Table 2) are immunoassays adapted from those used for gluten detection in foods to maximize sensitivity. Lateral flow immunoassay (LFIA) tests can detect GIP from concentrations of 0.15 µg GIP/g in stool and 2.2 ng GIP/mL in urine after less than 30 minutes, showing both tests a high sensitivity (98.5% and 97%, respectively) and specificity of 100%[9,56]. While these tests provide qualitative data, semiquantitative results could also be obtained in urine samples using the LFIA coupled with a lateral flow reader[9]. A quantitative G12-based sandwich ELISA test for stools was developed to increase the sensitivity and quantitative determinations. The analytical sensitivity of the assay was 0.16 µg GIP/g stool (limit of quantification), and it was validated in a multicenter clinical study, showing a diagnostic sensitivity and specificity of 98.5% and 100%, respectively[8].
Although a significant correlation between gluten intake and excreted GIP concentration was reported, high variability is usually observed among subjects[9,29,30,43]. Interindividual diversity (weight, sex, age, gut microbiota, etc.), the type of gluten-containing food (beer, pasta, bread, cookies, etc.), the amount of daily liquid intake, and the accompanying diet may have a considerable impact on the resulting GIP concentration as well as the excretion time in urine and stool samples, especially when the gluten exposure is not regular[9,29,30,43].
It appears difficult to predict the specific time and amount of gluten intake with high accuracy based on excreted GIP concentration and even considering the time of sample collection. However, some constraints regarding the expected time for GIP detection and the limits of detection could have been determined in different studies (Table 3).
| Specifications for the determination of GIP excretion after gluten intake | Sample | Time ranges (h) | Gluten source (amount) | Method |
| Shortest time to detect GIP | Urine | 3-9 | GCD (> 2 g) | LFIA [9,29,30] |
| Stool→ | < 24 | GCD (> 2 g) | LFIA; ELISA[43] | |
| Longest time to detect GIP | Urine | 36 | GCD (> 2 g) | LFIA[9,29,30] |
| Stool | > 72 | GCD (> 2 g) | LFIA; ELISA[29,30,43] | |
| Minimal gluten intake to detect excreted GIP | Urine | > 40-500 mg/d; | LFIA; SPE + LFIA[9,41] | |
| 25-50 mg | ||||
| Stool | > 40 mg/d | ELISA, LFIA[41,43] |
It was observed that after consuming normal gluten containing diet, a negative result in both urine and stool samples was rarely observed in the following 6-12 h and 3-5 days, respectively, after the last gluten-containing meal[9,43]. Moreover, in individuals with multiple regular transgressions, the interval between gluten ingestion and urine sample collection for maximal sensitivity of detection was generally consistent between 4 and 24 h[29,30].
Despite the variability of GIP concentrations found among individuals in most studies, a significantly low GIP content was typically observed in patients with CeD compared to healthy individuals with no diet restrictions[8,9].
Fecal immunoassays were first suggested as a novel method for the detection of gluten intake during diagnosis and GFD monitoring[57]. Two different methods could be used to estimate the amount of ingested gluten depending on the situation: LFIA test for either clinical laboratories or point-of-care settings due to its simplicity, while ELISA would be more suitable when a quantitative analysis and/or high throughput are convenient[58,59]. In a prospective, nonrandomized, partially-blinded, multicenter study GFD compliance of CeD children and adults was examined by measuring fecal GIP (determined by ELISA), a dietary questionnaire, celiac serology, and clinical response, as markers of diet adherence[8]. Their results revealed detectable amounts of GIP in the stools of about 30% of the analyzed patients on a GFD for at least one year in comparison to the 18% found when assessing by dietary questionnaire or by determination of anti-tTG antibodies in the serum alone. Indeed, less GIP-positive results were found in those declared non-compliant by the food questionnaire, while 70% of patients who did not declare any gluten intake had positive levels of GIP in stools[8].
Then, Moreno et al[9] demonstrated that urine samples could be used to monitor GFD compliance via an LFIA test. These tests revealed a high level of GFD infringement in patients on long-term treatment (48% and 45% in adults and children, respectively)[9]. Thus, several studies have compared GIP detection results in stool and urine for GFD monitoring, with information obtained via CeD-specific serology, dietary questionnaires, symptomatology, and duodenal biopsies, revealing the limitations of all these methods used to evaluate GFD transgressions and the concordance between them (Table 4).
| Ref. | Stool GIP+ | Urine GIP+ | anti-tTG+ | anti-DGP+ | Questionnaires1 | Symptoms | Duodenal biopsy (Marsh II/III) |
| Comino et al[8] | 56 (30) | - | 32 (18) | 11 (6) | 25 (18) | 9 (5) | - |
| Moreno et al[9] | - | 12 (48) | 4 (16) | - | - | 7 (28) | |
| Gerasimidis et al[39] | 11 (19) | - | 12 (20) | - | 4 (6) | - | - |
| Comino et al[40] | 6 (25) | - | 7 (20) | 0 (0) | - | - | - |
| Costa et al[41] | 11 (25) | 3 (7) | 9 (21) | 18 (45) | 18 (41) | 19 (43) | - |
| Silvester et al[29,30] | 5 (28) | 8 (44) | 7 (39) | - | 4 (22) | 8 (44) | 10 (56) |
| Ruiz-Carnicer et al[23] | - | 44 (58) | 9 (12) | - | 14 (23) | 18 (23) | 18 (24) |
| Fernández-Miaja et al[22] | 6 (8) | - | 3 (4) | - | 10 (13) | - | - |
| Roca et al[43] | 15 (35) | - | 22 (51) | - | 4 (9) | 4 (9) | - |
| Porcelli et al[44] | 8 (15) | - | 3 (6) | - | 5 (11) | 16 (34) | - |
| Laserna-Mendieta et al[45] | 22 (23) | - | 11 (12) | - | 17 (18) | - | 6 (28) |
| Stefanolo et al[24] | 33 (62) | 37 (70) | 22 (42) | 25 (47) | - | 18 (34) | - |
| Fernández-Bañares et al[46] | 53 (70) | - | 17 (22) | - | 6 (8) | 15 (20) | 40 (53) |
A branch of the first study with newly diagnosed pediatric CeD patients (n = 64) showed that the percentage of diet adherence decreased on follow-up at 6, 12, and 24 mo, as the rate of GIP-positive stools increased from 13% to 25%[40]. Meanwhile, anti-deamidated gliadin peptide (anti-DGP) antibodies normalized by 24 mo, and anti-tTG antibodies were elevated in 20% of the patients. Some children, particularly the older ones, were reported to have a propensity for GFD non-compliance, and 46% of non-adherent participants had at least two GIP-positive stools during follow-up[40]. Similar data were reported by other groups[22,44]. Patients with higher levels of gluten exposure exhibited prolonged ascension of anti-tTG antibodies (P < 0.05) than those with GIP-negative results.
In agreement with these data, Gerasimidis et al[39] found that most patients following a GFD had GIP-negative results, while 18% exhibited recent gluten exposure, which was not detected by using anti-tTG antibodies and the Biagi score. These findings confirmed the limitations of dietary evaluation and serology in adult patients with CeD on a GFD. A quarter of patients considered adherent by those methods had detectable GIP, using both LFIA and ELISA tests, in at least one of two independent tests during the 2 wk of the study. A 65,9% of concordance was observed between dietary reports and GIP results. Only four patients had high serum antibody values and two of them confirmed dietary non-compliance, in agreement with GIP results[41].
Recent research demonstrated that both ELISA and LFIA methods confirmed suspected and unsuspected dietary exposure in stool samples of CeD children and adolescent patients on a long-term GFD. However, no significant association was found between longer GFD duration, and the amount of GIP recovered in stool[43]. GIP-positive results were obtained from 35% of patients by ELISA (47% of these were also confirmed by LFIA). However, based on the dietary questionnaire, 90.7% of patients were compliant with the treatment. All the patients revealed as non-adherent by the questionnaire had GIP-positive stools. Furthermore, anti-tTG antibodies were detected in five patients, three of whom were also GIP-positive. However, the authors suggested that these elevated levels may be related to the short length of treatment (< 12 mo) and a lack of GFD adherence, as a patient with negative serology and GIP-negative stools was identified.
Differences in diet compliance rates based on a GIP LIFA test in stools and a validated adherence questionnaire (CDAT) (92.5% vs 86.3%, respectively) were observed among 80 CeD children and adolescent patients[22]. Nevertheless, the methods exhibited acceptable concordance (Kappa: 0.31, P = 0.004). Of those patients with good adherence using CDAT (n = 66), three had GIP-positive results. Porcelli et al[44] also found a significant association between strict diet adherence estimated by the Biagi score and fecal GIP detection. According to the Biagi score, 94.6% of GIP-negative patients exhibited good adherence. However, the questionnaire failed in the identification of 57.1% of GIP-positive patients, while GIP detection did not recognize 5.4% of gluten exposures declared via the questionnaire. In this study, 62.5% of GIP-positive patients exhibited negative anti-tTG antibodies levels.
The results from the DOGGIE BAG study[29,30] also confirmed that diet transgressions in patients with CeD were frequent despite efforts made to strictly follow the GFD. In this prospective longitudinal study, the food consumed by patients, in addition to their urine and stool samples, were analyzed for gluten intake and GIP excretion, respectively, throughout 10 days before a biopsy at 24 mo after the diagnosis. Gluten was found in the food of 9/18 participants at concentrations as high as 200 ppm, and GIP-positive results were obtained in 12/18 participants reporting good adherence to the GFD (8 of these had positive urine samples, and five had positive stool samples). No correlation was observed between gluten exposure and commonly used non-invasive measures of GFD adherence. Most of the participants (73%) who suspected a gluten exposure had at least one positive food, stool, or urine sample. Among the remaining participants who did not suspect any gluten intake, four of them resulted positive for GIP[29,30]. Other authors also found a significant association between patient self-reported gluten consumption and GIP detection. GIP was detected in 52.9% of patients who suspected gluten intake in the last month in comparison to 16.3% for those who were not aware of any gluten exposure[45].
On the other hand, several studies examined the association between the available tools for GFD monitoring and the duodenal biopsy, currently considered the gold standard (Table 5). Moreno et al[9] assessed the correlation between duodenal biopsies and GIP concentration in urine samples. Analysis of duodenal biopsies revealed that all the adult patients with small intestine damage (Marsh II/III) had GIP-positive urine samples (n = 7). In addition, there was a significant correlation between the absence of GIP in urine and the absence of villus atrophy in the gut intestinal epithelium. In agreement with other publications[60-62], this study confirmed the poor correlation of serological tests with mucosal healing as well as the limitations of dietary history questionnaires in the assessment of GFD adherence.
| Ref. | GIP+ (Stool and/or urine) | Serology+ | Questionnaires1 | Symptoms |
| Duodenal biopsy (Marsh II/III) | ||||
| Moreno et al[9] | 7 (100) | 2 (29) | - | - |
| Silvester et al[29,30] | 8 (80) | - | - | - |
| Ruiz-Carnicer et al[23] | 17 (94) | 7 (39) | 6 (43) | 4 (22) |
| Laserna-Mendieta et al[45] | 2 (33) | 2 (33) | 3 (50) | - |
| Fernández-Bañares et al[46] | 31 (78) | 10 (25) | 1 (3) | 11 (28) |
| Duodenal biopsy (Marsh 0/I) | ||||
| Moreno et al[9] | 5 (28) | 2 (11) | - | - |
| Silvester et al[29,30] | 4 (50) | - | - | - |
| Ruiz-Carnicer et al[23] | 27 (47) | 2 (3) | 8 (17) | 14 (24) |
| Laserna-Mendieta et al[45] | 20 (22) | 9 (10) | 77 (85) | - |
| Fernández-Bañares et al[46] | 22 (61) | 7 (19) | 5 (14) | 4 (11) |
In addition, a comprehensive study with a cohort of 77 participants under treatment with a GFD for ≥ 24 mo revealed that 58% had detectable GIP in their urine in at least one sample of three collected during the week[23]. Among the patients with GIP-negative results, 97% of them did not present histological abnormalities (Marsh 0–I), while among patients with GIP-positive results, 17 of 44 (39%) had histological damage in the intestinal epithelial (Marsh II-III), with only 16% presenting positive serological results. Significant differences were found in GIP concentrations between participants with Marsh II–III and Marsh 0–I. The highest sensitivity was observed when at least one of three urine samples tested GIP-positive (94.4%). However, this combination exhibited low specificity (53.4%). In contrast, the optimal specificity was obtained when a single urine sample was collected on the visit day (84.2%). Then, it was expected that one GIP-positive urine sample on the day of the visit would reveal a regular habit of GFD non-compliance not-restrained by the medical supervision. The authors did not observe concordance between gluten exposure measured by GIP and serology, or through a CDAT questionnaire and symptomatology, all of which exhibited low sensitivity for mucosal damage (38.9%, 22.2%, and 42.9%, respectively).
About the association of duodenal biopsy and GIP in fecal samples, Fernández-Bañares et al[46] found that most of CeD patients (77%) with persistent villous atrophy obtained a GIP-positive result in at least 1 stool sample after 2 years under GFD. In contrast, dietary evaluation failed to detect most of these gluten exposures, con
Other authors reported a weaker association between evident gluten exposure, as measured by GIP detection in urine and stool, and persistent villous atrophy[29,30,45]. In the DOGGIE BAG study referred to above, at least one GIP-positive sample was found in 7 of 11 patients with normal serology, and 2/3 of them had persistent villous atrophy (Marsh IIIa) after 24 mo of follow-up. Further, 2/3 of patients with GIP-negative results for all samples were Marsh 0-I, showing significant mucosal recovery[29,30]. A possible explanation for the discordance described before is that occasional or low gluten exposure may be sufficient for GIP detection but not enough to induce mucosal damage in some patients with CeD[23,29,30,45]. A delay of intestinal mucosal recovery[29,30,45] and improved compliance of participants during the study was also considered[29,30]. Besides, Laserna-Mendieta et al[45] reported a low sensitivity but acceptable specificity of GIP for the detection of mucosal damage, however for the calculation they included in the same group Marsh 1 patients (n = 21), who do not present mucosal damage but lymphocytic infiltration, and Marsh 2 and 3 patients (n = 6). Moreover, in this study, weak GIP-positive results were discarded for analysis and duodenal biopsies were performed in two hospitals with different pathologists, entailing a risk of interobserver variability[63,64]. Hence, with these results, the introduction of GIP detection as a non-invasive and sensitive assessment approach for GFD adherence may reduce the need for endoscopy and could identify potential intestinal mucosal damage not detectable via serological tests or dietary questionnaires. In addition, intestinal mucosal recovery could be predicted based on recurring negative GIP tests.
Regarding the relationship between symptoms and GIP excretion, only a small percentage of CeD patients reported symptoms in the studies with the highest number of celiac volunteers, although a score system for symptomatology was not used[8]. Roca et al[43] detected GIP in samples from 22.7% of asymptomatic pediatric patients negative for anti-tTG antibodies. In contrast, Costa et al[41] reported that most adult patients with gluten transgressions determined via GIP detection were asymptomatic, although the difference was not statistically significant due to the low number of cases enrolled. Similar data were published by Porcelli et al[44] wherein the rate of asymptomatic patients with GIP-positive results was 71.4%.
A recent publication[24] studied the relationship of stool and urine GIP results with gastrointestinal symptoms in CeD adult patients for 4 wk to represent a real-life scenario. 62% of the patients were found to have at least one GIP-positive stool test, and 69.9% had positive urine samples. The results suggested that symptoms in CeD patients under a GFD are the consequence of gluten exposure. A significantly higher rate of GIP-positive stool samples was observed in symptomatic compared to asymptomatic patients (77.8% vs 54.3%, respectively). However, a group of asym
Considering that avoiding symptoms is the main motivation for adhering to a GFD, GIP content could be a useful indicator for the identification of asymptomatic patients with considerable immunogenic gluten exposure preventing gut mucosal recovery. Furthermore, GIP detection may help in symptomatic patients with CeD to determine whether non-responsive celiac disease or refractory celiac disease occurs due to recurrent gluten exposure or due to additional factors inducing persistent symptoms, such as the consumption of FODMAPs or intestinal dysbiosis[40,41,65,66].
Excreted GIP detection in either stool or urine is a precise approach for determining voluntary or involuntary gluten consumption. However, isolated GIP measurements might not identify intermittent compliance, unless punctual transgressions took place close to the moment of sample collection. It appears that the use of multiple samples (at least two) contributes to higher sensitivity and specificity of GIP detection[23,24]. In a considerable number of studies, biopsies and recurrent excretion of GIP indicated that serological tests, symptoms, and dietary questionnaires were not sufficient indicators of GFD adherence[9,23,29-31]. However, anti-tTG antibodies and clinical dietary assessment have been recently suggested as complementary tools in the evaluation of GFD adherence[44,45]. In this context, Porcelli et al[44] proposed the use of the Biagi score in combination with fecal GIP tests using a binary logistic regression model. In contrast, other groups did not find anti-tTG levels to be a relevant marker of adherence even though anti-DGP antibodies levels have been shown to significantly correlate with the quantity and frequency of GIP excretion in stools[24,41].
Regarding which method is more appropriate, Roca et al[43,67] proposed GIP testing of fecal samples as a non-invasive method that allows patient empowerment for self-managing the disease. ELISA is to be used for the laboratory quantification of GIP, while LFIA strips should be used for patient self-control following suspected involuntary infringements. Even though GIP evaluation may represent an extra cost for CeD patients monitoring, its inclusion in the pursuing of strict GFD adherence would reduce health expenditure by preventing future complications arising from gluten exposure in overlooked asymptomatic patients and would improve the self-esteem or “peace of mind” of patients adhering to the GFD, decreasing the need for additional assessment and endoscopy in non-responsive celiac disease or refractory celiac disease, especially in children.
Some authors have already proposed algorithms for the application of the determination of GIP in stools[39] and urine[23] for the follow-up of patients with CeD. It is expected that the application of those protocols with some variations will be progressively introduced in the guidelines to assess the adherence of patients to the GFD. Future studies may also determine the utility of GIP excretion analysis for other applications, such as the confirmation that sufficient gluten has been ingested before diagnosis or to discover unknown aspects of gluten protein metabolism.
A lifelong strict gluten-free diet (GFD) is the only available treatment for celiac disease (CeD), which reduces symptoms and allows the healing of the intestinal mucosa, preventing long-term complications. However, total exclusion of gluten is difficult to achieve in practice and voluntary and involuntary transgressions are highly frequent.
Gluten immunogenic peptides (GIP) detection in stool and urine is becoming increasingly apparent as a noninvasive and reliable marker for close and efficient GFD monitoring in patients with CeD.
The authors aimed to summarize published data regarding the performance of GIP determination in stool and urine in comparison to other available tools in assessing GFD adherence in patients with CeD.
The authors conducted a systematic review searching in PubMed and Web of Science clinical studies that reported the performance of GIP determination in stool and/or urine with other biomarkers for the evaluation of treatment adherence in adult and pediatric patients with CeD.
The authors screened 67 articles and 15 articles were included for full-text analysis. In the selected publications GIP determination in stool and/or urine were compared with at least one of the following markers: levels of CeD-specific serology, results from CeD-specific non-specific questionnaires, symptomatology, and duodenal biopsy. Fecal determination by ELISA was the most investigated GIP marker, followed by urine rapid test and stool rapid test. Variability was seen in the concordance between the different tools among the studies reviewed due to the differences found in the study design, the target population, and the methods used. One of the main outcome measures reviewed was the diagnostic accuracy of these tools in assessing mucosal healing. An association of mucosal status and GIP detection was observed in some publications, where serology, questionnaires, and symptomatology showed lower sensitivity. Furthermore, GIP detection may help in symptomatic patients to determine whether non-responsive CeD or refractory CeD occurs due to recurrent gluten exposure or due to additional factors.
The introduction of GIP detection in stool and/or urine as a non-invasive marker for GFD monitoring in patients with CeD may reduce health expenditure by preventing future complications arising from gluten exposure not detectable by other tools in use. Stool and urine rapid tests may be an option for disease self-managing by the patient, while fecal ELISA test could be used for GIP quantification in the laboratory.
The introduction of algorithms for GIP determination within the follow-up of patients with CeD in guidelines will allow its routine use in clinical practice. In addition, future studies may also determine their utility for other applications such as the confirmation of gluten consumption during the diagnosis process or the investigation of gluten metabolism.
This paper will be part of Laura Coto’s doctorate that is being carried out within the context of “Human Nutrition Program” at the University of Granada.
| 1. | Shewry PR, Halford NG, Belton PS, Tatham AS. The structure and properties of gluten: an elastic protein from wheat grain. Philos Trans R Soc Lond B Biol Sci. 2002;357:133-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 382] [Cited by in RCA: 344] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 2. | Shan L, Molberg Ø, Parrot I, Hausch F, Filiz F, Gray GM, Sollid LM, Khosla C. Structural basis for gluten intolerance in celiac sprue. Science. 2002;297:2275-2279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1153] [Cited by in RCA: 1113] [Article Influence: 46.4] [Reference Citation Analysis (1)] |
| 3. | Fasano A, Catassi C. Clinical practice. Celiac disease. N Engl J Med. 2012;367:2419-2426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 433] [Cited by in RCA: 477] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 4. | Abadie V, Sollid LM, Barreiro LB, Jabri B. Integration of genetic and immunological insights into a model of celiac disease pathogenesis. Annu Rev Immunol. 2011;29:493-525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 357] [Cited by in RCA: 363] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 5. | Loeff T, Araya M, Pérez-Bravo F. Frequency of MYO9B polymorphisms in celiac patients and controls. Rev Esp Enferm Dig. 2012;104:566-571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 6. | Wapenaar MC, Monsuur AJ, van Bodegraven AA, Weersma RK, Bevova MR, Linskens RK, Howdle P, Holmes G, Mulder CJ, Dijkstra G, van Heel DA, Wijmenga C. Associations with tight junction genes PARD3 and MAGI2 in Dutch patients point to a common barrier defect for coeliac disease and ulcerative colitis. Gut. 2008;57:463-467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 127] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 7. | Tye-Din JA, Stewart JA, Dromey JA, Beissbarth T, van Heel DA, Tatham A, Henderson K, Mannering SI, Gianfrani C, Jewell DP, Hill AV, McCluskey J, Rossjohn J, Anderson RP. Comprehensive, quantitative mapping of T cell epitopes in gluten in celiac disease. Sci Transl Med. 2010;2:41ra51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 327] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 8. | Comino I, Fernández-Bañares F, Esteve M, Ortigosa L, Castillejo G, Fambuena B, Ribes-Koninckx C, Sierra C, Rodríguez-Herrera A, Salazar JC, Caunedo Á, Marugán-Miguelsanz JM, Garrote JA, Vivas S, Lo Iacono O, Nuñez A, Vaquero L, Vegas AM, Crespo L, Fernández-Salazar L, Arranz E, Jiménez-García VA, Antonio Montes-Cano M, Espín B, Galera A, Valverde J, Girón FJ, Bolonio M, Millán A, Cerezo FM, Guajardo C, Alberto JR, Rosinach M, Segura V, León F, Marinich J, Muñoz-Suano A, Romero-Gómez M, Cebolla Á, Sousa C. Fecal Gluten Peptides Reveal Limitations of Serological Tests and Food Questionnaires for Monitoring Gluten-Free Diet in Celiac Disease Patients. Am J Gastroenterol. 2016;111:1456-1465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 158] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 9. | Moreno ML, Cebolla Á, Muñoz-Suano A, Carrillo-Carrion C, Comino I, Pizarro Á, León F, Rodríguez-Herrera A, Sousa C. Detection of gluten immunogenic peptides in the urine of patients with coeliac disease reveals transgressions in the gluten-free diet and incomplete mucosal healing. Gut. 2017;66:250-257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 181] [Cited by in RCA: 244] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 10. | Caio G, Volta U, Sapone A, Leffler DA, De Giorgio R, Catassi C, Fasano A. Celiac disease: a comprehensive current review. BMC Med. 2019;17:142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 564] [Cited by in RCA: 616] [Article Influence: 88.0] [Reference Citation Analysis (1)] |
| 11. | Leonard MM, Sapone A, Catassi C, Fasano A. Celiac Disease and Nonceliac Gluten Sensitivity: A Review. JAMA. 2017;318:647-656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 247] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 12. | Vivas S, Ruiz de Morales JM, Fernandez M, Hernando M, Herrero B, Casqueiro J, Gutierrez S. Age-related clinical, serological, and histopathological features of celiac disease. Am J Gastroenterol. 2008;103:2360-2365; quiz 2366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 98] [Article Influence: 5.4] [Reference Citation Analysis (1)] |
| 13. | Reilly NR, Aguilar K, Hassid BG, Cheng J, Defelice AR, Kazlow P, Bhagat G, Green PH. Celiac disease in normal-weight and overweight children: clinical features and growth outcomes following a gluten-free diet. J Pediatr Gastroenterol Nutr. 2011;53:528-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 95] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 14. | Lindfors K, Ciacci C, Kurppa K, Lundin KEA, Makharia GK, Mearin ML, Murray JA, Verdu EF, Kaukinen K. Coeliac disease. Nat Rev Dis Primers. 2019;5:3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 291] [Article Influence: 41.6] [Reference Citation Analysis (0)] |
| 15. | Lebwohl B, Granath F, Ekbom A, Smedby KE, Murray JA, Neugut AI, Green PH, Ludvigsson JF. Mucosal healing and risk for lymphoproliferative malignancy in celiac disease: a population-based cohort study. Ann Intern Med. 2013;159:169-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 185] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 16. | Elli L, Ferretti F, Orlando S, Vecchi M, Monguzzi E, Roncoroni L, Schuppan D. Management of celiac disease in daily clinical practice. Eur J Intern Med. 2019;61:15-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 17. | Therrien A, Kelly CP, Silvester JA. Celiac Disease: Extraintestinal Manifestations and Associated Conditions. J Clin Gastroenterol. 2020;54:8-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 108] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 18. | See JA, Kaukinen K, Makharia GK, Gibson PR, Murray JA. Practical insights into gluten-free diets. Nat Rev Gastroenterol Hepatol. 2015;12:580-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 126] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 19. | Leinonen H, Kivelä L, Lähdeaho ML, Huhtala H, Kaukinen K, Kurppa K. Daily Life Restrictions are Common and Associated with Health Concerns and Dietary Challenges in Adult Celiac Disease Patients Diagnosed in Childhood. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 20. | Troncone R, Auricchio R, Granata V. Issues related to gluten-free diet in coeliac disease. Curr Opin Clin Nutr Metab Care. 2008;11:329-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 33] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 21. | Vriezinga SL, Schweizer JJ, Koning F, Mearin ML. Coeliac disease and gluten-related disorders in childhood. Nat Rev Gastroenterol Hepatol. 2015;12:527-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 22. | Fernández Miaja M, Díaz Martín JJ, Jiménez Treviño S, Suárez González M, Bousoño García C. [Study of adherence to the gluten-free diet in coeliac patients]. An Pediatr (Barc). 2021;94:377-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 23. | Ruiz-Carnicer Á, Garzón-Benavides M, Fombuena B, Segura V, García-Fernández F, Sobrino-Rodríguez S, Gómez-Izquierdo L, Montes-Cano MA, Rodríguez-Herrera A, Millán R, Rico MC, González-Naranjo C, Bozada-García JM, Díaz J, Coronel-Rodríguez C, Espín B, Romero-Gómez M, Cebolla Á, Sousa C, Comino I, Argüelles F, Pizarro Á. Negative predictive value of the repeated absence of gluten immunogenic peptides in the urine of treated celiac patients in predicting mucosal healing: new proposals for follow-up in celiac disease. Am J Clin Nutr. 2020;112:1240-1251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 67] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 24. | Stefanolo JP, Tálamo M, Dodds S, de la Paz Temprano M, Costa AF, Moreno ML, Pinto-Sánchez MI, Smecuol E, Vázquez H, Gonzalez A, Niveloni SI, Mauriño E, Verdu EF, Bai JC. Real-World Gluten Exposure in Patients With Celiac Disease on Gluten-Free Diets, Determined From Gliadin Immunogenic Peptides in Urine and Fecal Samples. Clin Gastroenterol Hepatol. 2021;19:484-491.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 60] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 25. | Stoven S, Murray JA, Marietta E. Celiac disease: advances in treatment via gluten modification. Clin Gastroenterol Hepatol. 2012;10:859-862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 26. | Tio M, Cox MR, Eslick GD. Meta-analysis: coeliac disease and the risk of all-cause mortality, any malignancy and lymphoid malignancy. Aliment Pharmacol Ther. 2012;35:540-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 107] [Article Influence: 7.6] [Reference Citation Analysis (1)] |
| 27. | Hall NJ, Rubin GP, Charnock A. Intentional and inadvertent non-adherence in adult coeliac disease. A cross-sectional survey. Appetite. 2013;68:56-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 83] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 28. | Muhammad H, Reeves S, Jeanes YM. Identifying and improving adherence to the gluten-free diet in people with coeliac disease. Proc Nutr Soc. 2019;78:418-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 29. | Silvester JA, Comino I, Rigaux LN, Segura V, Green KH, Cebolla A, Weiten D, Dominguez R, Leffler DA, Leon F, Bernstein CN, Graff LA, Kelly CP, Sousa C, Duerksen DR. Exposure sources, amounts and time course of gluten ingestion and excretion in patients with coeliac disease on a gluten-free diet. Aliment Pharmacol Ther. 2020;52:1469-1479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 30. | Silvester JA, Comino I, Kelly CP, Sousa C, Duerksen DR; DOGGIE BAG Study Group. Most Patients With Celiac Disease on Gluten-Free Diets Consume Measurable Amounts of Gluten. Gastroenterology. 2020;158:1497-1499.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 88] [Article Influence: 14.7] [Reference Citation Analysis (1)] |
| 31. | Myléus A, Reilly NR, Green PHR. Rate, Risk Factors, and Outcomes of Nonadherence in Pediatric Patients With Celiac Disease: A Systematic Review. Clin Gastroenterol Hepatol. 2020;18:562-573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 69] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 32. | Villafuerte-Galvez J, Vanga RR, Dennis M, Hansen J, Leffler DA, Kelly CP, Mukherjee R. Factors governing long-term adherence to a gluten-free diet in adult patients with coeliac disease. Aliment Pharmacol Ther. 2015;42:753-760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 78] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 33. | Bebb JR, Lawson A, Knight T, Long RG. Long-term follow-up of coeliac disease--what do coeliac patients want? Aliment Pharmacol Ther. 2006;23:827-831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 58] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 34. | Haines ML, Anderson RP, Gibson PR. Systematic review: The evidence base for long-term management of coeliac disease. Aliment Pharmacol Ther. 2008;28:1042-1066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 148] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 35. | Herman ML, Rubio-Tapia A, Lahr BD, Larson JJ, Van Dyke CT, Murray JA. Patients with celiac disease are not followed up adequately. Clin Gastroenterol Hepatol. 2012;10:893-899.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 73] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 36. | Catassi C, Fabiani E, Iacono G, D'Agate C, Francavilla R, Biagi F, Volta U, Accomando S, Picarelli A, De Vitis I, Pianelli G, Gesuita R, Carle F, Mandolesi A, Bearzi I, Fasano A. A prospective, double-blind, placebo-controlled trial to establish a safe gluten threshold for patients with celiac disease. Am J Clin Nutr. 2007;85:160-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 422] [Cited by in RCA: 386] [Article Influence: 20.3] [Reference Citation Analysis (1)] |
| 37. | Walker MM, Murray JA. An update in the diagnosis of coeliac disease. Histopathology. 2011;59:166-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 43] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 38. | Catassi C. Protocollo per la diagnosi ed il follow-up della malattia celiaca. Encicl del Dirit. 2015;263-358. |
| 39. | Gerasimidis K, Zafeiropoulou K, Mackinder M, Ijaz UZ, Duncan H, Buchanan E, Cardigan T, Edwards CA, McGrogan P, Russell RK. Comparison of Clinical Methods With the Faecal Gluten Immunogenic Peptide to Assess Gluten Intake in Coeliac Disease. J Pediatr Gastroenterol Nutr. 2018;67:356-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 40. | Comino I, Segura V, Ortigosa L, Espín B, Castillejo G, Garrote JA, Sierra C, Millán A, Ribes-Koninckx C, Román E, Rodríguez-Herrera A, Díaz J, Silvester JA, Cebolla Á, Sousa C. Prospective longitudinal study: use of faecal gluten immunogenic peptides to monitor children diagnosed with coeliac disease during transition to a gluten-free diet. Aliment Pharmacol Ther. 2019;49:1484-1492. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 41. | Costa AF, Sugai E, Temprano MP, Niveloni SI, Vázquez H, Moreno ML, Domínguez-Flores MR, Muñoz-Suano A, Smecuol E, Stefanolo JP, González AF, Cebolla-Ramirez A, Mauriño E, Verdú EF, Bai JC. Gluten immunogenic peptide excretion detects dietary transgressions in treated celiac disease patients. World J Gastroenterol. 2019;25:1409-1420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 58] [Cited by in RCA: 69] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 42. | Porcelli B, Ferretti F, Cinci F, Biviano I, Santini A, Grande E, Quagliarella F, Terzuoli L, Bacarelli MR, Bizzaro N, Vascotto M, Marini M. Fecal gluten immunogenic peptides as indicators of dietary compliance in celiac patients. Minerva Gastroenterol Dietol. 2020;66:201-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 43. | Roca M, Donat E, Masip E, Crespo-Escobar P, Cañada-Martínez AJ, Polo B, Ribes-Koninckx C. Analysis of gluten immunogenic peptides in feces to assess adherence to the gluten-free diet in pediatric celiac patients. Eur J Nutr. 2021;60:2131-2140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 44. | Porcelli B, Ferretti F, Biviano I, Santini A, Cinci F, Vascotto M, Grande E, Quagliarella F, Terzuoli L, Bizzaro N, Marini M, Rentini S. Testing for fecal gluten immunogenic peptides: a useful tool to evaluate compliance with gluten-free diet by celiacs. Ann Gastroenterol. 2020;33:631-637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 45. | Laserna-Mendieta EJ, Casanova MJ, Arias Á, Arias-González L, Majano P, Mate LA, Gordillo-Vélez CH, Jiménez M, Angueira T, Tébar-Romero E, Carrillo-Ramos MJ, Tejero-Bustos MÁ, Gisbert JP, Santander C, Lucendo AJ. Poor Sensitivity of Fecal Gluten Immunogenic Peptides and Serum Antibodies to Detect Duodenal Mucosal Damage in Celiac Disease Monitoring. Nutrients. 2020;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 46. | Fernández-Bañares F, Beltrán B, Salas A, Comino I, Ballester-Clau R, Ferrer C, Molina-Infante J, Rosinach M, Modolell I, Rodríguez-Moranta F, Arau B, Segura V, Fernández-Salazar L, Santolaria S, Esteve M, Sousa C; CADER study group. Persistent Villous Atrophy in De Novo Adult Patients With Celiac Disease and Strict Control of Gluten-Free Diet Adherence: A Multicenter Prospective Study (CADER Study). Am J Gastroenterol. 2021;116:1036-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 47. | Qiao SW, Bergseng E, Molberg O, Jung G, Fleckenstein B, Sollid LM. Refining the rules of gliadin T cell epitope binding to the disease-associated DQ2 molecule in celiac disease: importance of proline spacing and glutamine deamidation. J Immunol. 2005;175:254-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 127] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 48. | Camarca A, Anderson RP, Mamone G, Fierro O, Facchiano A, Costantini S, Zanzi D, Sidney J, Auricchio S, Sette A, Troncone R, Gianfrani C. Intestinal T cell responses to gluten peptides are largely heterogeneous: implications for a peptide-based therapy in celiac disease. J Immunol. 2009;182:4158-4166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 134] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 49. | Skerritt JH, Hill AS. Monoclonal antibody sandwich enzyme immunoassays for determination of gluten in foods. J Agric Food Chem. 1991;38:1771-1778. [DOI] [Full Text] |
| 50. | Valdés I, García E, Llorente M, Méndez E. Innovative approach to low-level gluten determination in foods using a novel sandwich enzyme-linked immunosorbent assay protocol. Eur J Gastroenterol Hepatol. 2003;15:465-474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 162] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 51. | van den Broeck HC, de Jong HC, Salentijn EM, Dekking L, Bosch D, Hamer RJ, Gilissen LJ, van der Meer IM, Smulders MJ. Presence of celiac disease epitopes in modern and old hexaploid wheat varieties: wheat breeding may have contributed to increased prevalence of celiac disease. Theor Appl Genet. 2010;121:1527-1539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 117] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 52. | Zhang J, Portela SB, Horrell JB, Leung A, Weitmann DR, Artiuch JB, Wilson SM, Cipriani M, Slakey LK, Burt AM, Dias Lourenco FJ, Spinali MS, Ward JR, Seit-Nebi A, Sundvor SE, Yates SN. An integrated, accurate, rapid, and economical handheld consumer gluten detector. Food Chem. 2019;275:446-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 53. | Chatziharalambous D, Lygirou V, Latosinska A, Stravodimos K, Vlahou A, Jankowski V, Zoidakis J. Analytical Performance of ELISA Assays in Urine: One More Bottleneck towards Biomarker Validation and Clinical Implementation. PLoS One. 2016;11:e0149471. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 54. | Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7:R80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 498] [Cited by in RCA: 522] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 55. | U.S. Department of Health and Human Services FDA. Bioanalytical Method Validation Guidance for Industry. 2018; 1–41. Available from: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/bioanalytical-method-validation-guidance-industry. |
| 56. | Biomedal. iVYCHECK GIP Stool. [cited 15 June 2021]. Available from: https://ivydal.biomedal.com/tests-de-uso-profesional/ivycheck-gip-stool/. |
| 57. | Auricchio S. An innovative approach to measure compliance to a gluten-free diet. Am J Clin Nutr. 2012;95:537-538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 58. | Bethune MT, Crespo-Bosque M, Bergseng E, Mazumdar K, Doyle L, Sestak K, Sollid LM, Khosla C. Noninflammatory gluten peptide analogs as biomarkers for celiac sprue. Chem Biol. 2009;16:868-881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 59. | Ehren J, Morón B, Martin E, Bethune MT, Gray GM, Khosla C. A food-grade enzyme preparation with modest gluten detoxification properties. PLoS One. 2009;4:e6313. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 75] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 60. | Sharkey LM, Corbett G, Currie E, Lee J, Sweeney N, Woodward JM. Optimising delivery of care in coeliac disease - comparison of the benefits of repeat biopsy and serological follow-up. Aliment Pharmacol Ther. 2013;38:1278-1291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 61. | Tursi A, Brandimarte G, Giorgetti GM. Lack of usefulness of anti-transglutaminase antibodies in assessing histologic recovery after gluten-free diet in celiac disease. J Clin Gastroenterol. 2003;37:387-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 67] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 62. | Rashtak S, Ettore MW, Homburger HA, Murray JA. Comparative usefulness of deamidated gliadin antibodies in the diagnosis of celiac disease. Clin Gastroenterol Hepatol. 2008;6:426-432; quiz 370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 86] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 63. | Arguelles-Grande C, Tennyson CA, Lewis SK, Green PH, Bhagat G. Variability in small bowel histopathology reporting between different pathology practice settings: impact on the diagnosis of coeliac disease. J Clin Pathol. 2012;65:242-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 101] [Article Influence: 6.7] [Reference Citation Analysis (1)] |
| 64. | Ravelli A, Villanacci V. Tricks of the trade: How to avoid histological pitfalls in celiac disease. Pathol Res Pract. 2012;208:197-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 65. | Gibson PR, Muir JG, Newnham ED. Other Dietary Confounders: FODMAPS et al. Dig Dis. 2015;33:269-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 66. | Wacklin P, Kaukinen K, Tuovinen E, Collin P, Lindfors K, Partanen J, Mäki M, Mättö J. The duodenal microbiota composition of adult celiac disease patients is associated with the clinical manifestation of the disease. Inflamm Bowel Dis. 2013;19:934-941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 148] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 67. | Roca M, Donat E, Masip E, Crespo Escobar P, Fornes-Ferrer V, Polo B, Ribes-Koninckx C. Detection and quantification of gluten immunogenic peptides in feces of infants and their relationship with diet. Rev Esp Enferm Dig. 2019;111:106-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution-NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/Licenses/by-nc/4.0/
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Argentina
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ciacci C, Stein J S-Editor: Ma YJ L-Editor: A P-Editor: Wu RR