Published online Sep 28, 2021. doi: 10.3748/wjg.v27.i36.6064
Peer-review started: March 3, 2021
First decision: June 3, 2021
Revised: June 4, 2021
Accepted: August 9, 2021
Article in press: August 9, 2021
Published online: September 28, 2021
Processing time: 203 Days and 23.4 Hours
Colorectal cancer (CRC) is the third most common cancer and the second most common cause of cancer-related death worldwide. The 5-year survival rate of patients with early-stage CRC could reach 90%, but it is very low in patients with advanced-stage CRC. Recent studies have shown that circular RNAs play important roles in regulating the migration and invasion of CRC cells.
To elucidate the role of circRNA_0084927 (circ_0084927) in the migration and invasion of CRC cells and its underlying mechanism.
Clinical tissue samples and cells were collected, and the expression of circ_0084927 was detected by quantitative polymerase chain reaction (qPCR). The diagnostic performance of circ_0084927 was assessed by receiver operating characteristic curve analysis. The role of circ_0084927 in CRC cell proliferation, migration, and invasion was determined using cell counting kit-8 assay, wound healing assay, and transwell assay, respectively. The regulatory relationship among circ_0084927, miRNA-20b-3p (miR-20b-3p), and glutathione S-transferase mu 5 (GSTM5) was identified using databases, luciferase reporter assay, qPCR, and Western blot analysis. AKT-mTOR signaling was also verified after circ_0084927 knockdown or miR-20b-3p mimic treatment.
The expression of circ_0084927 was significantly increased in CRC tissues and cells, and it was higher in advanced-stage CRC compared with early-stage CRC. The area under the curve (AUC) of circ_0084927 was 0.806 [95% confidence interval (CI): 0.683-0.896]. In addition, the AUC was 0.874 (95%CI: 0.738-0.956) in patients with advanced-stage CRC and 0.713 (95%CI: 0.555-0.840) in those with early-stage CRC. Knockdown of circ_0084927 inhibited the migration and invasion of HCT116 cells. Moreover, circ_0084927 was found to act as a sponge of miR-20b-3p. MiR-20b-3p activation reduced the circ_0084927 level, whereas miR-20b-3p inhibition increased the circ_0084927 level. But the effect was not found after circ_0084927 mutation. In addition, miR-20b-3p expression in CRC patients was also reduced and negatively correlated with circ_0084927 expression. The function of circ_0084927 in HCT116 cells with circ_0084927 knockdown was rescued by miR-20b-3p. Moreover, GSTM5 expression was significantly decreased after overexpressing miR-20b-3p or inhibiting circ_0084927, but its expression was rescued when circ_0084927 and miR-20b-3p were both inhibited. Finally, AKT-mTOR signaling was markedly regulated by circ_0084927 and miR-20b-3p.
The expression of circ_0084927 is significantly increased in CRC and higher in advanced-stage CRC than in early-stage CRC. Moreover, circ_0084927 potentially regulates CRC cell migration and invasion via the miR-20b-3p/GSTM5/ AKT/mTOR pathway.
Core Tip: The 5-year survival rate of patients with early-stage colorectal cancer (CRC) could reach 90%, but it is very low in patients with advanced-stage CRC. Recent studies have shown that circular RNAs play important roles in regulating the migration and invasion of CRC cells. To elucidate the role of circRNA_0084927 (circ_0084927) in the migration and invasion of CRC cells and the underlying mechanism, this study was performed. The expression of circ_0084927 was significantly increased in CRC tissues and cells, and it was markedly higher in advanced-stage CRC compared with early-stage CRC. Knockdown of circ_0084927 inhibited the migration and invasion of HCT116 cells. Moreover, circ_0084927 potentially regulates CRC migration and invasion via the miRNA-20b-3p/glutathione S-transferase mu 5 pathway.
- Citation: Liu F, Xiao XL, Liu YJ, Xu RH, Zhou WJ, Xu HC, Zhao AG, Xu YX, Dang YQ, Ji G. CircRNA_0084927 promotes colorectal cancer progression by regulating miRNA-20b-3p/glutathione S-transferase mu 5 axis. World J Gastroenterol 2021; 27(36): 6064-6078
- URL: https://www.wjgnet.com/1007-9327/full/v27/i36/6064.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i36.6064
Colorectal cancer (CRC) is the third most common cancer and the second most common cause of cancer-related death worldwide, and the numbers of new cases and deaths in 2018 were 1.8 million and 881000, respectively[1]. With the development of diagnostic and therapeutic technologies, the 5-year survival rate for CRC patients has reached 65%[2] and can reach 90% in early-stage CRC patients. However, the 5-year survival rate of patients with advanced-stage CRC is very low due to tumor metastasis and other complications. Although colonoscopy is the gold standard of CRC screening, approximately 60% of CRC patients are diagnosed at an advanced stage given the rate of missed lesions and incomplete colonoscopy coverage[3-5]. Therefore, it is urgent to elucidate the pathogenesis and molecular mechanism of CRC with metastasis.
Circular RNAs (circRNAs), as important regulatory noncoding RNAs, have become a new research hotspot following microRNAs (miRNAs) and long noncoding RNAs. Researchers have demonstrated that circRNAs play important roles in tumors, including CRC[6-9], and their functions are completed in a variety of ways, such as acting as sponges of miRNAs[6,7], acting as transcriptional regulators[10,11], and translating proteins by combining with RNA binding proteins[12-15]. However, to date, the functions of circRNAs in CRC remain largely unknown, especially in CRC with metastasis. Further study to elucidate the function of circRNAs in CRC with metastasis is needed.
In this study, we first verified the differential expression of circRNA_0084927 (circ_0084927) according to previous circRNA sequencing data[16] and further found that circ_0084927 was associated with the pathological stage of CRC. Then, we demonstrated that knockdown of circ_0084927 markedly inhibited HCT116 cell migration and invasion. Regarding the regulatory mechanism of circ_0084927, it acts as a sponge of miRNA-20b-3p (miR-20b-3p), regulating glutathione S-transferase mu 5 (GSTM5) and the AKT-mTOR pathway. Therefore, our findings elucidated the role of circ_0084927 and provided a new treatment strategy for CRC with metastasis.
Thirty pairs of CRC tissues and adjacent normal tissues (normal) were obtained during surgery at the Longhua Hospital Affiliated to Shanghai University of Traditional Chinese Medicine (Shanghai, China). The diagnosis of CRC was confirmed based on pathological evidence. Tissues were snap-frozen in liquid nitrogen and stored at -80 °C before detection. The study was approved by the Ethics Committee of Longhua Hospital (No. 2019LCSY020), and informed consent was obtained from all participants.
Normal colon cells (FHC) and CRC cells (HCT116, HT29, SW480, and SW620) (Shanghai Cell Bank, shanghai, China) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and penicillin/streptomycin (100 U/mL) (Gibco, Carlsbad, United States) in an incubator with 5% CO2 at 37 °C. In addition, human embryonic kidney 293T (HEK-293T) cells obtained from American Type Culture Collection (ATCC) (Manassas, United States) were cultured in Roswell Park Memorial Institute 1640 medium with 10% fetal bovine serum and peni
After transfection, HCT116 cells were seeded into 96-well plates at a concentration of 1 × 104 cells and cultured for 0 h, 24 h, 48 h, and 72 h. Then, 10 μL of cell counting kit-8 reagent was added to each well. After incubation at 37 °C for 1 h, the absorbance value was detected at 450 nm.
After transfection, HCT116 cells were seeded in a 6-well dish with a culture insert (Ibidi, Germany) at a concentration of 3 × 104 cells. After 24 h, the culture insert was removed, and the cells were washed twice with polybutylene succinate. Two milliliters of serum-free medium were added to each dish for 48 h. Images were captured, and the wound area was measured using Image J software (National Institutes of Health, United States).
Six-well plates with 8-μm chambers (Corning, United States) were used to assess cellular migration (without Matrigel) or invasion (with Matrigel). Briefly, transfected HCT116 cells were seeded in 6-well plates at a concentration of 1 × 105 cells. Two hundred microliters of serum-free medium was added to the upper chamber, and six hundred microliters of medium with 30% fetal bovine serum was added to the lower chamber for 48 h. Then, the cells were fixed with 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet solution for 15 min. Four fields were randomly selected to calculate the number of migrating or invading cells and to evaluate the ability of cell migration or invasion.
Total RNA was extracted using TRIzol reagent (Ambion, United States). For circRNA and mRNA analysis, cDNA was synthesized using an EVM-MLV reverse transcription kit (Aikeri Biotech, Hunan, China). For miRNA analysis, cDNA was synthesized using a miRNA cDNA synthesis kit (Sangon Biotech, Shanghai, China). The amplification reaction was performed using the SYBR-Green quantitative polymerase chain reaction (qPCR) kit (Thermo Fisher Scientific, MA, United States) or the miRNA fluorescence quantitative PCR kit (Sangon Biotech). Gene expression was normalized using β-actin or U6. The primers are listed in Table 1.
| Gene name | Forward (5'→3') | Reverse (5'→3') |
| β-actin | GGCTGTATTCCCCTCCATCG | CCAGTTGGTAACAATGCCATGT |
| circRNA_0084927 | AGCACTACAGAGGCACAAACATC | GTGCCCTGACTACGGTGTTATC |
| circRNA_0138996 | CTGATCCCAATGGATTGCATC | GTCCTCCCGTTCCTCTTCG |
| circRNA_0110477 | CGAATCAAAGCAGCCTATCAAG | TGCCACATAGAATTTGGGTGTC |
| circRNA_0133544 | CATGAGGTTAGGCAGTTGTATCG | TGTTTTTAGCCTTTTCTCCATCTC |
| circRNA_0002867 | AACCAGAGCACATTAGCCAAAG | CAAACTCGGCGTGTTCTTCTC |
| GSTM5 | GAAGATGGGAGGGAGGAG | CCTTGGGGAAGAGAAGAGA |
| U6 | AGAGAAGATTAGCATGGCCCCTG | ATCCAGTGCAGGGTCCGAGG |
| miRNA-20b-3p | ACTGTAGTATGGGCACTTCCAG | ATCCAGTGCAGGGTCCGAGG |
Cells were collected and lysed. Protein concentration was determined using a protein assay kit (Beyotime, China). The protein was separated and transferred to a polyvinylidene fluoride membrane followed by incubation with 5% milk at room temperature for 1 h. The membrane was incubated at 4 °C overnight with the following antibodies: GSTM5 (GTX108776, Genetex, United States), PI3 kinase p85 (PI3K, 4292S, CST, United States), phospho-PI3K (BS-3332R, Bioss, China), Akt (4685S, CST), phospho-Akt (4060S, CST), mTOR (2983S, CST), phospho-mTOR (5536S, CST), and β-actin (4970s, CST). Then, the secondary antibody was added and incubated at room temperature for 2 h, and protein expression was observed using a chemiluminescence gel imaging system (Tanon 5200, China).
HEK-293T cells were seeded in 24-well plates and transfected using FuGene HD transfection reagent (Promega, United States) according to previous studies[17,18]. Briefly, HEK-293T cells were transfected using circ_0084927 wild type (circ_0084927-WT; Genomeditech, China) or circ_0084927 mutant (circ_0084927-Mut; Genomeditech) plasmid with or without miR-20b-3p mimic/inhibitor. After transfection for 22 h, luciferase activity was detected using the dual-luciferase reporter system (Promega).
Statistical analyses were conducted using SPSS 24.0 software. Data were assessed using Student’s t-test or one-way ANOVA. Pearson correlation was used to analyze the correlation between circRNA and miRNA. Receiver operating characteristic (ROC) curve analysis was performed to evaluate the diagnostic value using MedCalc software. P < 0.05 was considered statistically significant. Dr. Ming Yang from the Good Clinical Practice Office, Longhua Hospital, Shanghai University of Traditional Chinese Medicine reviewed the statistical methods of this study before submission.
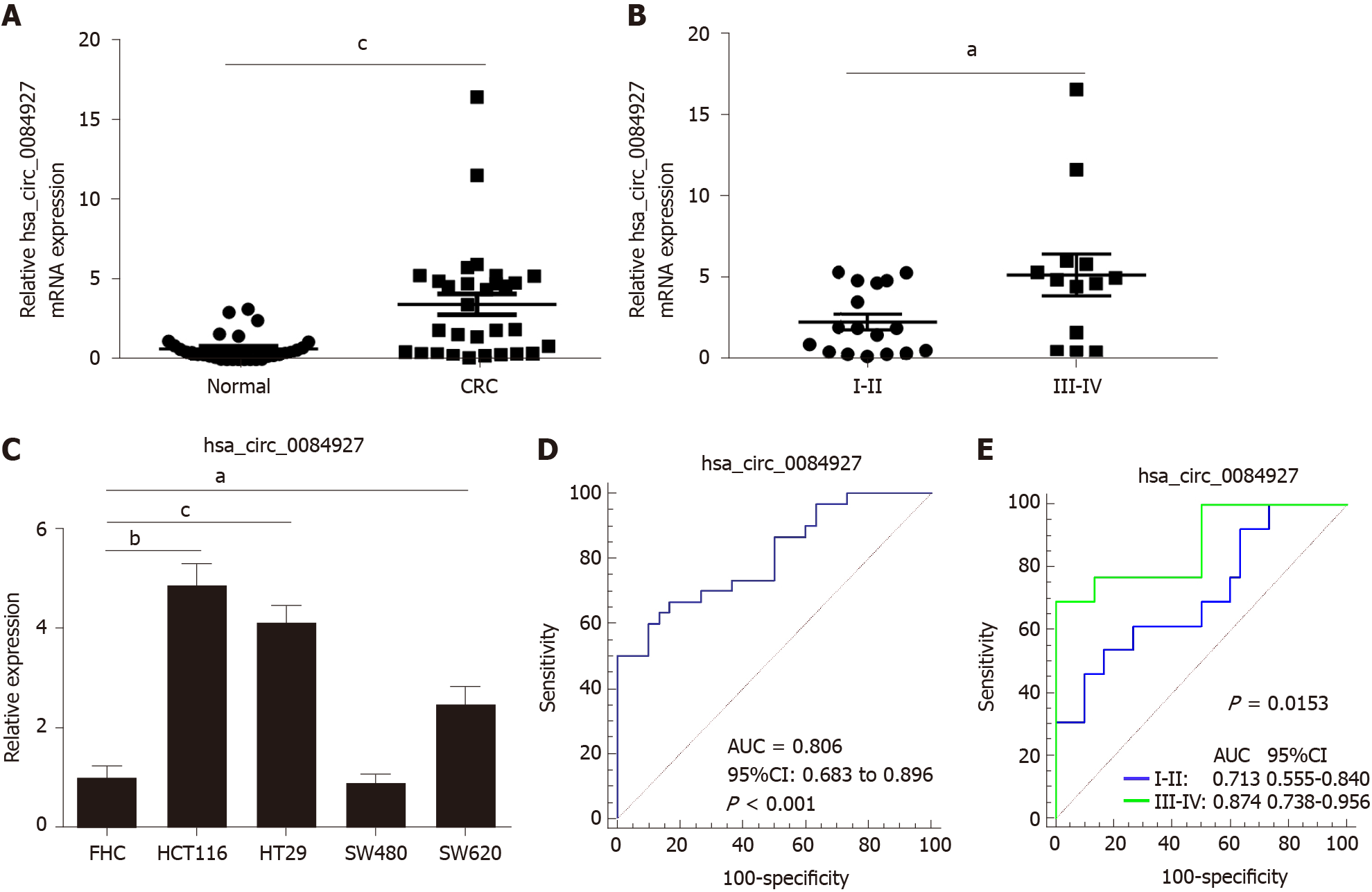
Our previous study has proved that circRNAs play important roles in the transition of adenoma into CRC[16], but the function of circRNAs in advanced-stage CRC remains largely undetermined. According to previous circRNA sequencing data[16], five differentially expressed circRNAs were chosen and verified. The results showed that circ_0084927 expression was significantly increased in the CRC group compared with the normal group and was higher in advanced-stage CRC compared with early-stage CRC (Figure 1A and B). The other four circRNAs were not significantly different (Supplementary Figure 1). In addition, circ_0084927 expression in CRC cells (HCT116, HT29, and SW620) was also increased (Figure 1C). ROC curve analysis was used to evaluate the diagnostic performance of circ_0084927 in CRC. The area under the curve (AUC) of circ_0084927 was 0.806 (95%CI: 0.683 to 0.896) (P < 0.001). In addition, the AUC was 0.874 (95%CI: 0.738-0.956) in patients with advanced-stage CRC and 0.713 (95%CI: 0.555-0.840) in those with early-stage CRC, indicating that circ_0084927 had higher diagnostic performance in advanced-stage CRC (P = 0.0153) (Figure 1D and E).
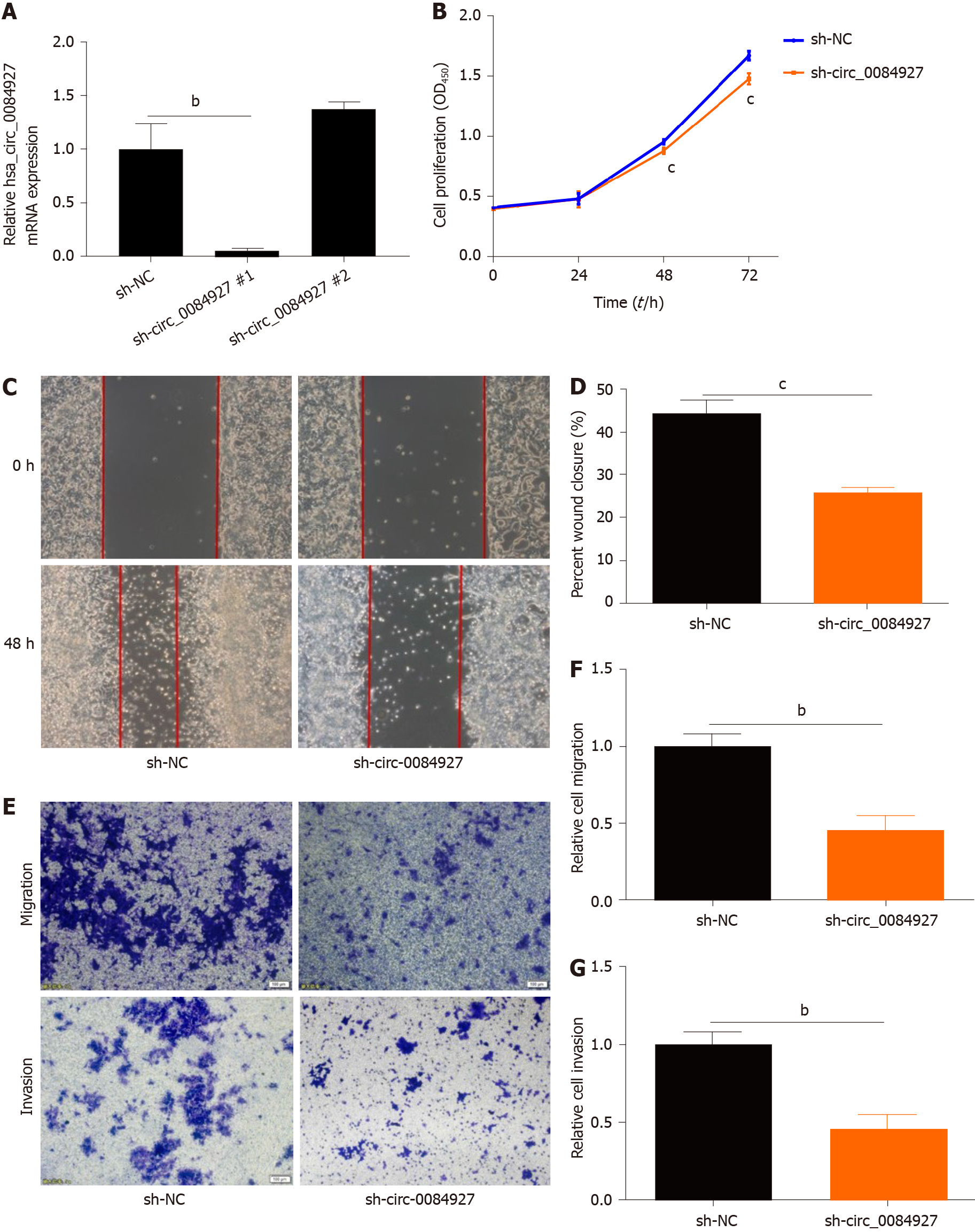
To explore the function of circ_0084927 in CRC, circ_0084927 expression was inhibited in HCT116 cells using shRNA plasma (Figure 2A), and circ_0084927 knockdown markedly inhibited HCT116 cell proliferation at 48 h and 72 h (Figure 2B). In addition, circ_0084927 knockdown also significantly inhibited HCT116 cell migration and invasion (Figure 2C-G). These results demonstrated that circ_0084927 is an oncogene that promotes CRC function.
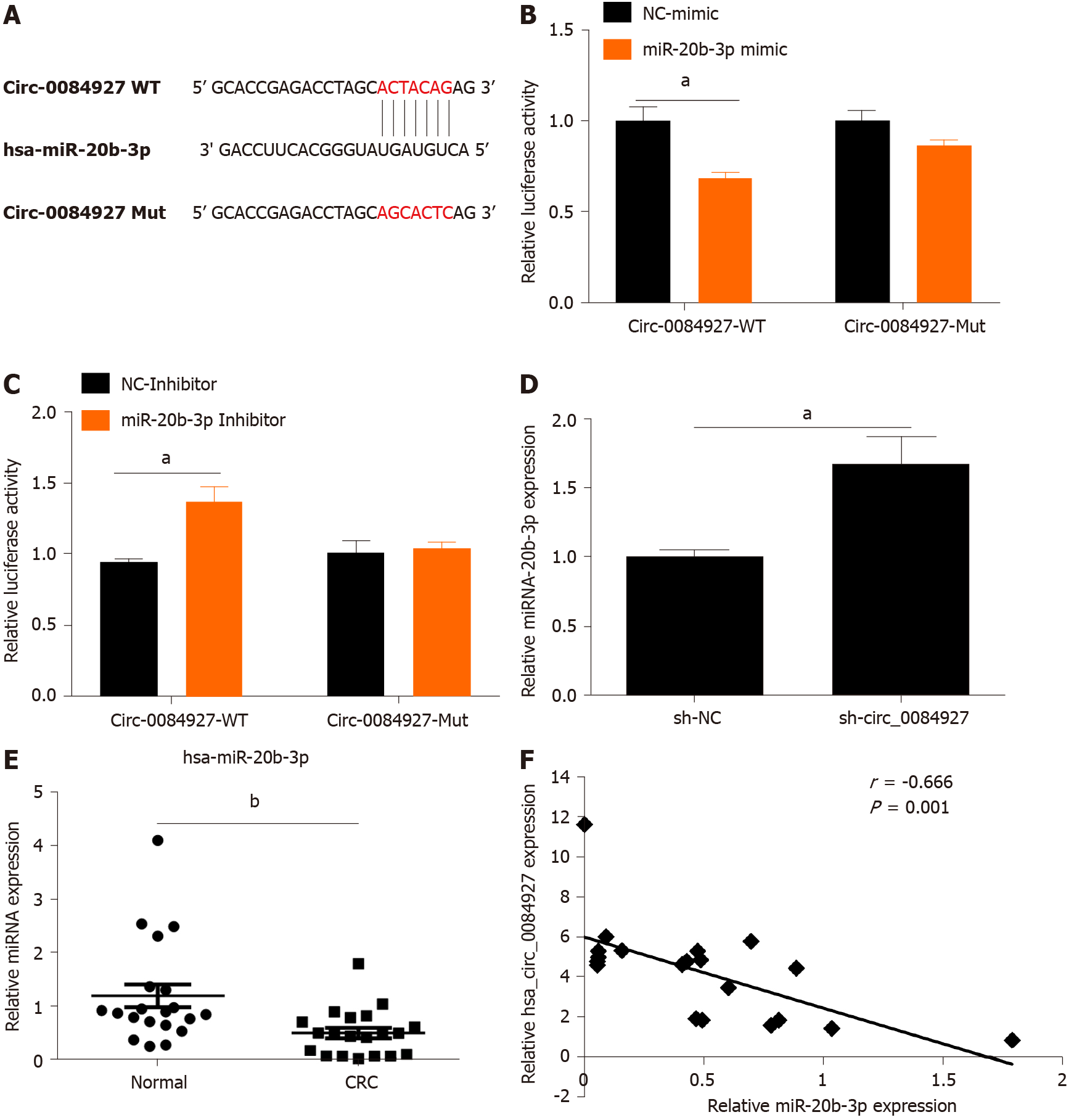
The biological function of circRNAs mainly involves acting as a sponge of miRNAs[19]. In this study, circ_0084927 may act as a sponge of miR-20b-3p as demonstrated by bioinformatics analysis (Figure 3A). The miR-20b-3p mimic markedly reduced the luciferase activity of circ_0084927, whereas the miR-20b-3p inhibitor markedly increased the luciferase activity of circ_0084927, but had no effect on circ_0084927-Mut (Figure 3B and C). qPCR results also indicated that knockdown of circ_0084927 promoted the expression of miR-20b-3p (Figure 3D). We also tested the expression of miR-20b-3p in CRC tissue samples, and its expression was significantly reduced (Figure 3E). Spearman correlation coefficient analysis revealed that circ_0084927 was negatively correlated with miR-20b-3p (Figure 3F). All of the data suggested that circ_0084927 acts as a sponge of miR-20b-3p.
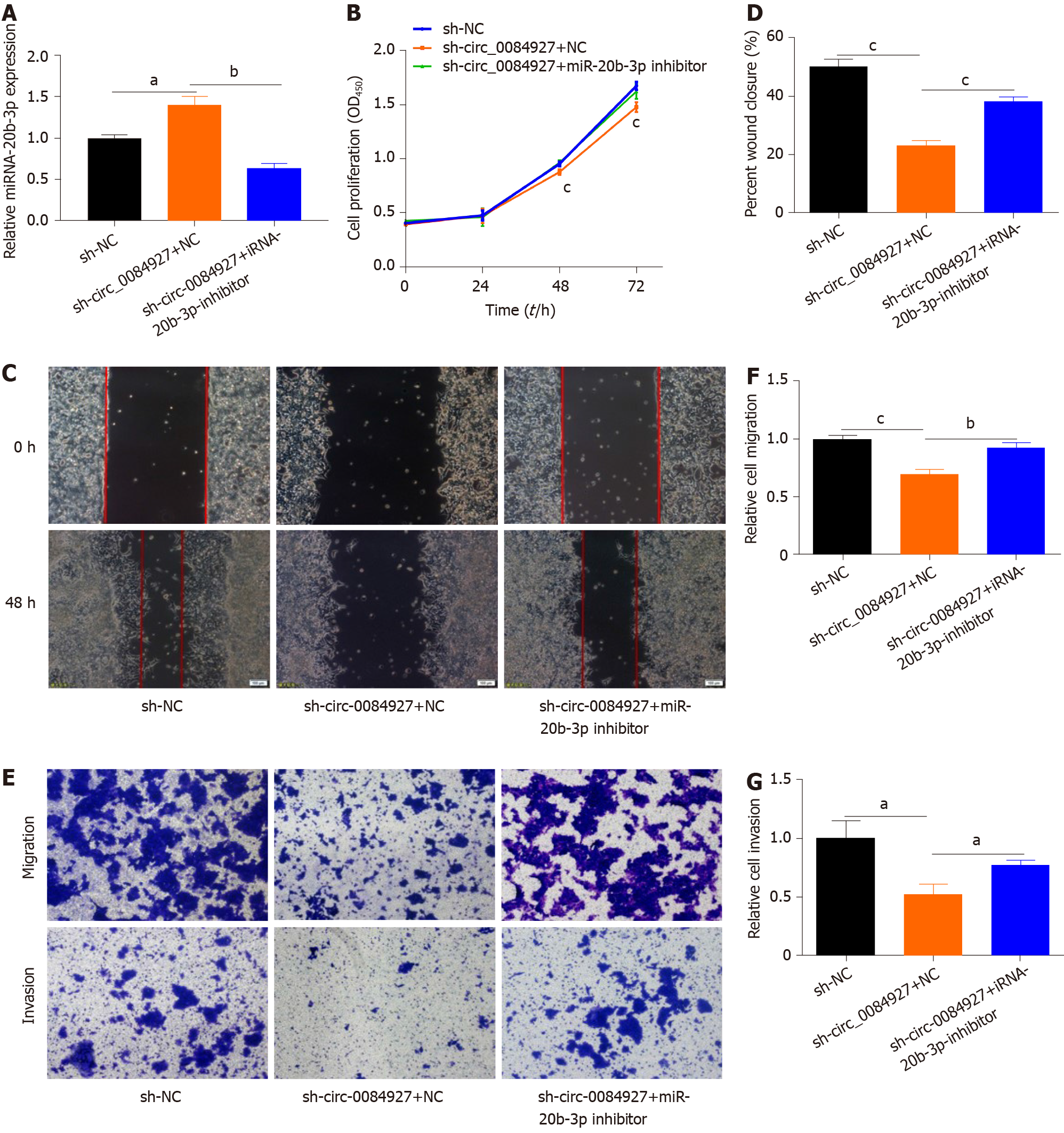
To assess whether the function of circ_0084927 in HCT116 cells is mediated by miR-20b-3p, HCT116 cells with circ_0084927 knockdown was transfected with or without miR-20b-3p inhibitor (Figure 4A). The CCK-8 assay showed that circ_0084927 knockdown inhibited the proliferation of HCT116 cells, while the effect was rescued by the miR-20b-3p inhibitor (Figure 4B). Wound healing and transwell assays showed that circ_0084927 knockdown inhibited the migration and invasion of HCT116 cells, whereas miR-20b-3p inhibition rescued this effect (Figure 4C-G).
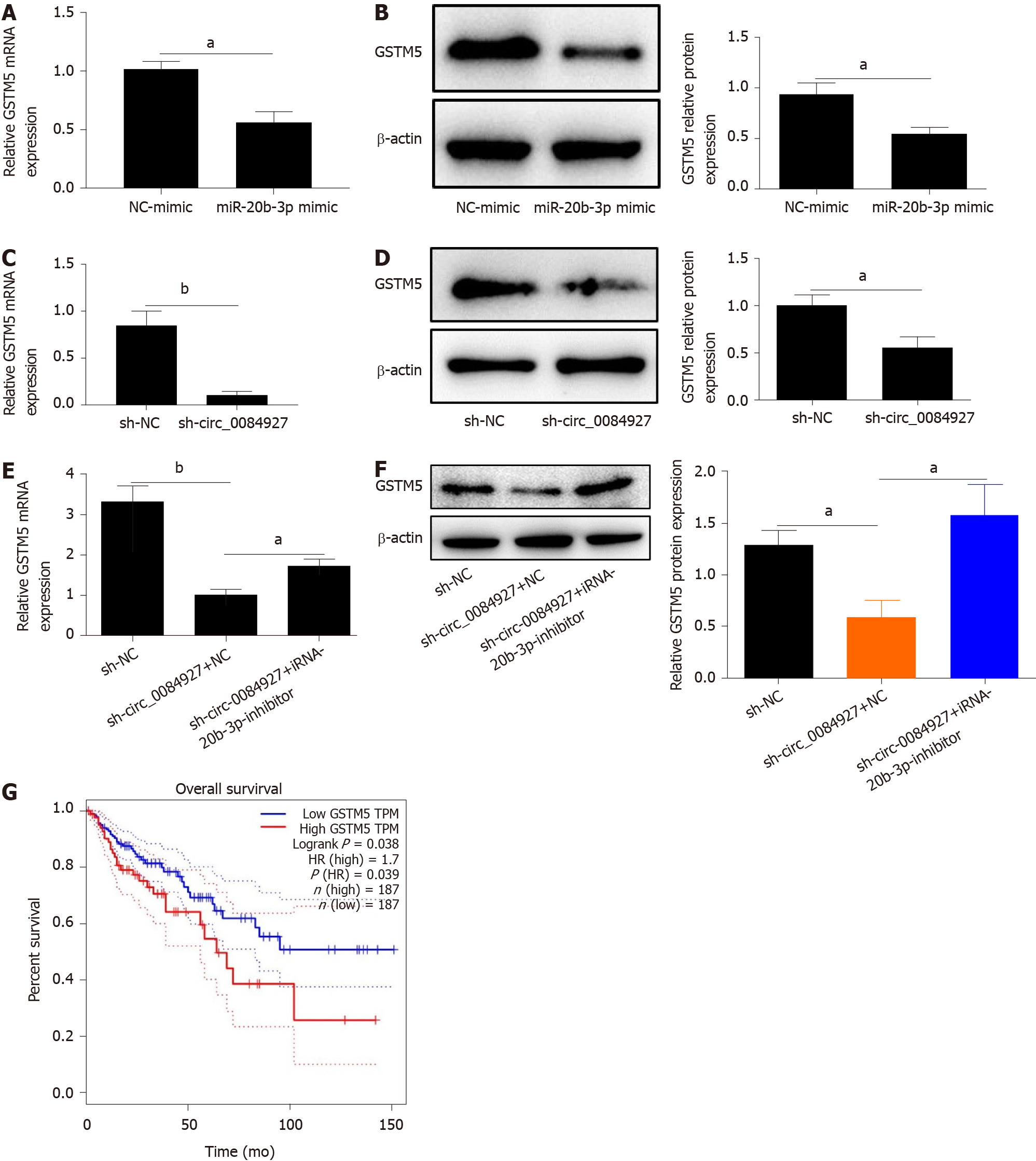
According to bioinformatics analysis, we found that GSTM5 is a target of miR-20b-3p. We then assessed GSTM5 expression after overexpressing miR-20b-3p. The results showed that GSTM5 mRNA expression was significantly reduced after overexpressing miR-20b-3p (Figure 5A). GSTM5 protein level was consistent with its mRNA level (Figure 5B). Moreover, GSTM5 mRNA and protein expression was also markedly reduced after silencing circ_0084927 (Figure 5C and D). After transfecting the miR-20b-3p inhibitor into HCT116 cells with circ_0084927 knockdown, GSTM5 mRNA and protein expression was rescued (Figure 5E and F). The survival curve results using The Cancer Genome Atlas (TCGA) data also showed that patients with high GSTM5 levels had a poor survival, indicating that GSTM5 was negatively correlated with CRC patient survival (Figure 5G).
Further experiments found that circ_0084927 knockdown or miR-20b-3p overexpression both reduced the phosphorylation levels of AKT and mTOR (Figure 6A and B). In addition, AKT and mTOR phosphorylation levels were rescued after inhibiting miR-20b-3p in HCT116 cells with circ_0084927 knockdown (Figure 6C). These results showed that circ_0084927 regulated CRC migration and invasion via the miR-20b-3p/GSTM5/AKT-mTOR pathway.
As a malignant tumor, the incidence of CRC has recently increased. Although the 5-year survival rate of CRC patients is 65%[2], it is very low in patients with advanced-stage CRC. Therefore, it is urgent to develop techniques for the early diagnosis of CRC, which would improve patient survival. Moreover, new treatment strategies for advanced-stage CRC are also very important. Studies have indicated that circRNAs are very stable and not easily degraded due to loop properties[20] and have demonstrated the specificity of their expression in particular tissues and tumor developmental stages[20-23]. Therefore, circRNAs, such as serum exosomal hsa-circ-0004771 and circ-PNN[24,25], could act as biomarkers for the diagnosis of CRC. In addition, circRNAs could promote the progression of CRC and act as therapeutic targets[6,8,26,27]. Circ_0084927 originates from epithelial splicing regulatory protein 1 (ESRP1), which is upregulated in CRC (Supplementary Figure 2) and associated with the pathogenesis of CRC[28]. Studies have demonstrated that circ_0084927 facilitates cervical cancer advancement[29-31], but its function in CRC is unclear. In this study, we found that circ_0084927 expression was significantly increased in CRC tissues and cells, and was higher in advanced-stage CRC than in early-stage CRC. Further studies indicated that knockdown of circ_0084927 inhibited the migration and invasion of CRC cells, showing that circ_0084927 played an important role in CRC progression.
CircRNAs perform biological functions by acting as sponges of miRNAs[6,7]. In cervical cancer, miRNAs related to circ_0084927 mainly include miR-142-3p, miR-634, and miR-1179[29-31]. Our study found that circ_0084927 acts as a sponge of miR-20b-3p in CRC. In addition, miR-20b-3p activation reduced circ_0084927 level, whereas miR-20b-3p inhibition increased circ_0084927 level. The expression of circ_0084927 was not altered after the binding site with miR-20b-3p was mutated. In addition, miR-20b-3p expression in CRC patients was also reduced and was negatively correlated with circ_0084927. Studies have shown that miR-20b-3p is related to temozolomide resistance in glioblastoma[32] and diabetic retinopathy progression[33], but the function of miR-20b-3p in CRC has not been reported. In this study, we demonstrated that miR-20b-3p rescued the effect of circ_0084927 on CRC cells, indicating the role of miR-20b-3p in CRC.
Further studies found that GSTM5 is a target of miR-20b-3p. GSTM5 expression was significantly reduced after overexpressing miR-20b-3p or silencing circ_0084927. However, GSTM5 expression was rescued after silencing circ_0084927 and inhibiting miR-20b-3p. Studies have demonstrated that glutathione S-transferases are pro-carcinogenic in CRC[34]. AS a glutathione S-transferase, GSTM5 is also involved in various tumors, including breast cancer[35,36], prostate cancer[36], ovarian carcinoma[37], and CRC[38,39]. Although previous studies have demonstrated that glutathione metabolism is involved in the progression of tumors[40-42] and GSTM5 could be a prognostic and predictive marker in CRC[38,39], the regulatory mechanism is unknown. In this study, we demonstrated that circ_0084927 regulated the migration and invasion of CRC by the miR-20b-3p/GSTM5 axis and finally regulated the AKT-mTOR pathway, which plays an important role in CRC[43-45]. Therefore, our results revealed that circ_0084927 regulated the progression of CRC via the miR-20b-3p/GSTM5/AKT-mTOR pathway.
In summary, we have demonstrated that circ_0084927 expression is significantly increased in CRC and is higher in advanced-stage CRC than in early-stage CRC. Mechanistically, circ_0084927 regulates the migration and invasion of CRC cells via the miR-20b-3p/GSTM5/AKT-mTOR pathway. This is the first study to demonstrate the role of circ_0084927 in CRC, and these findings provide a new perspective for targeted therapy of CRC with metastasis.
Colorectal cancer (CRC) is the third most common cancer and the second most common cause of cancer-related death worldwide. Recent studies have shown that circular RNAs play important roles in regulating the progression of CRC. However, the biological role and underlying mechanism of circRNA_0084927 (circ_0084927) in CRC remain unclear.
Due to tumor metastasis and other complications, the 5-year survival rate of patients with advanced-stage CRC is very low. We hope to provide a new treatment strategy for CRC with metastasis.
The present study aimed to investigate the role and mechanism of circ_0084927 in regulating the progression of CRC.
Clinical tissue samples and cells were collected, and the expression of circ_0084927 was detected by quantitative polymerase chain reaction (qPCR) The diagnostic performance of circ_0084927 was assessed by receiver operating characteristic curve analysis. The role of circ_0084927 in proliferation, migration, and invasion was determined using cell counting kit-8 assay, wound healing assay, and transwell assay, respectively. The regulatory relationship among circ_0084927, miRNA-20b-3p (miR-20b-3p), and glutathione S-transferase mu 5 (GSTM5) was identified using databases, luciferase reporter assays, qPCR, and Western blot analysis. AKT-mTOR signaling was also verified after circ_0084927 knockdown or miR-20b-3p mimic treatment.
The expression of circ_0084927 was significantly increased in CRC tissues and cells, and was increased in advanced-stage CRC compared with in early-stage CRC. The area under the curve (AUC) of circ_0084927 was 0.806 (95%CI: 0.683 to 0.896). In addition, the AUC was 0.874 (95%CI: 0.738-0.956) in patients with advanced-stage CRC and 0.713 (95%CI: 0.555-0.840) in those with early-stage CRC. Knockdown of circ_0084927 inhibited the migration and invasion of HCT116 cells. Moreover, circ_0084927 was found to act as a sponge of miR-20b-3p. MiR-20b-3p activation reduced the circ_0084927 level, whereas miR-20b-3p inhibition increased the circ_0084927 level. But the effect was not found after circ_0084927 mutation. In addition, miR-20b-3p expression in CRC patients was also reduced and negatively correlated with circ_0084927 expression. The function of circ_0084927 in HCT116 cells with circ_0084927 knockdown was rescued by miR-20b-3p. Moreover, GSTM5 expression was significantly decreased after overexpressing miR-20b-3p or inhibiting circ_0084927, but its expression was rescued when circ_0084927 and miR-20b-3p were both inhibited. Finally, AKT-mTOR signaling was markedly regulated by circ_0084927 and miR-20b-3p.
The expression of circ_0084927 is significantly increased in CRC and higher in advanced-stage CRC than in early-stage CRC. Moreover, circ_0084927 potentially regulates CRC migration and invasion via the miR-20b-3p/GSTM5/AKT/mTOR pathway.
Circ_0084927 could promote CRC progression and provide a new treatment strategy for CRC with metastasis.
Manuscript source: Unsolicited manuscript
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Lomperta K S-Editor: Wu YXJ L-Editor: Wang TQ P-Editor: Liu JH
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 56561] [Article Influence: 7070.1] [Reference Citation Analysis (134)] |
| 2. | Burgers K, Moore C, Bednash L. Care of the Colorectal Cancer Survivor. Am Fam Physician. 2018;97:331-336. [PubMed] |
| 3. | Zauber AG, Winawer SJ, O'Brien MJ, Lansdorp-Vogelaar I, van Ballegooijen M, Hankey BF, Shi W, Bond JH, Schapiro M, Panish JF, Stewart ET, Waye JD. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med. 2012;366:687-696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1952] [Cited by in RCA: 2371] [Article Influence: 169.4] [Reference Citation Analysis (2)] |
| 4. | Zhao S, Wang S, Pan P, Xia T, Chang X, Yang X, Guo L, Meng Q, Yang F, Qian W, Xu Z, Wang Y, Wang Z, Gu L, Wang R, Jia F, Yao J, Li Z, Bai Y. Magnitude, Risk Factors, and Factors Associated With Adenoma Miss Rate of Tandem Colonoscopy: A Systematic Review and Meta-analysis. Gastroenterology. 2019;156:1661-1674.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 422] [Article Influence: 60.3] [Reference Citation Analysis (1)] |
| 5. | Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL, Siegel RL. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69:363-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2417] [Cited by in RCA: 3117] [Article Influence: 445.3] [Reference Citation Analysis (0)] |
| 6. | Jian X, He H, Zhu J, Zhang Q, Zheng Z, Liang X, Chen L, Yang M, Peng K, Zhang Z, Liu T, Ye Y, Jiao H, Wang S, Zhou W, Ding Y, Li T. Hsa_circ_001680 affects the proliferation and migration of CRC and mediates its chemoresistance by regulating BMI1 through miR-340. Mol Cancer. 2020;19:20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 152] [Article Influence: 25.3] [Reference Citation Analysis (3)] |
| 7. | Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, Chen C, Chang W, Ping Y, Ji P, Wu J, Quan W, Yao Y, Zhou Y, Sun Z, Li D. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-β1 axis. Mol Cancer. 2020;19:117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 353] [Article Influence: 58.8] [Reference Citation Analysis (0)] |
| 8. | Li XN, Wang ZJ, Ye CX, Zhao BC, Li ZL, Yang Y. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res. 2018;37:325. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 193] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 9. | Yang H, Li X, Meng Q, Sun H, Wu S, Hu W, Liu G, Yang Y, Chen R. CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol Cancer. 2020;19:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 138] [Cited by in RCA: 177] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 10. | Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1766] [Cited by in RCA: 2423] [Article Influence: 201.9] [Reference Citation Analysis (0)] |
| 11. | Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1619] [Cited by in RCA: 2297] [Article Influence: 208.8] [Reference Citation Analysis (0)] |
| 12. | Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 280] [Cited by in RCA: 402] [Article Influence: 57.4] [Reference Citation Analysis (0)] |
| 13. | Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H, Huang S, Xie B, Zhang N. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst. 2018;110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 858] [Cited by in RCA: 853] [Article Influence: 106.6] [Reference Citation Analysis (0)] |
| 14. | Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, Chen W, Gao X, Zhao K, Zhou H, Li Z, Ming L, Xie B, Zhang N. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805-1814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 572] [Article Influence: 71.5] [Reference Citation Analysis (0)] |
| 15. | Wesselhoeft RA, Kowalski PS, Anderson DG. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat Commun. 2018;9:2629. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 220] [Cited by in RCA: 574] [Article Influence: 71.8] [Reference Citation Analysis (0)] |
| 16. | Zhu M, Dang Y, Yang Z, Liu Y, Zhang L, Xu Y, Zhou W, Ji G. Comprehensive RNA Sequencing in Adenoma-Cancer Transition Identified Predictive Biomarkers and Therapeutic Targets of Human CRC. Mol Ther Nucleic Acids. 2020;20:25-33. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 17. | Dang Y, Hu D, Xu J, Li C, Tang Y, Yang Z, Liu Y, Zhou W, Zhang L, Xu H, Xu Y, Ji G. Comprehensive analysis of 5-hydroxymethylcytosine in zw10 kinetochore protein as a promising biomarker for screening and diagnosis of early colorectal cancer. Clin Transl Med. 2020;10:e125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 18. | Dang Y, Xu J, Zhu M, Zhou W, Zhang L, Ji G. Gan-Jiang-Ling-Zhu decoction alleviates hepatic steatosis in rats by the miR-138-5p/CPT1B axis. Biomed Pharmacother. 2020;127:110127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 19. | Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4631] [Cited by in RCA: 6198] [Article Influence: 476.8] [Reference Citation Analysis (0)] |
| 20. | Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6253] [Cited by in RCA: 6203] [Article Influence: 477.2] [Reference Citation Analysis (0)] |
| 21. | Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell. 2018;71:428-442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 988] [Cited by in RCA: 1547] [Article Influence: 193.4] [Reference Citation Analysis (0)] |
| 22. | Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1007] [Cited by in RCA: 1332] [Article Influence: 133.2] [Reference Citation Analysis (0)] |
| 23. | Cui X, Wang J, Guo Z, Li M, Liu S, Liu H, Li W, Yin X, Tao J, Xu W. Emerging function and potential diagnostic value of circular RNAs in cancer. Mol Cancer. 2018;17:123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 135] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 24. | Pan B, Qin J, Liu X, He B, Wang X, Pan Y, Sun H, Xu T, Xu M, Chen X, Xu X, Zeng K, Sun L, Wang S. Identification of Serum Exosomal hsa-circ-0004771 as a Novel Diagnostic Biomarker of Colorectal Cancer. Front Genet. 2019;10:1096. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 196] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 25. | Xie Y, Li J, Li P, Li N, Zhang Y, Binang H, Zhao Y, Duan W, Chen Y, Wang Y, Du L, Wang C. RNA-Seq Profiling of Serum Exosomal Circular RNAs Reveals Circ-PNN as a Potential Biomarker for Human Colorectal Cancer. Front Oncol. 2020;10:982. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 26. | Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, Sun H, Pan Y, He B, Wang S. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:417. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 447] [Cited by in RCA: 492] [Article Influence: 61.5] [Reference Citation Analysis (0)] |
| 27. | Chen LY, Wang L, Ren YX, Pang Z, Liu Y, Sun XD, Tu J, Zhi Z, Qin Y, Sun LN, Li JM. The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1α translation. Mol Cancer. 2020;19:164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 152] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 28. | Mager LF, Koelzer VH, Stuber R, Thoo L, Keller I, Koeck I, Langenegger M, Simillion C, Pfister SP, Faderl M, Genitsch V, Tcymbarevich I, Juillerat P, Li X, Xia Y, Karamitopoulou E, Lyck R, Zlobec I, Hapfelmeier S, Bruggmann R, McCoy KD, Macpherson AJ, Müller C, Beutler B, Krebs P. The ESRP1-GPR137 axis contributes to intestinal pathogenesis. Elife. 2017;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 29. | Chen L, Zhang X, Wang S, Lin X, Xu L. Circ_0084927 Facilitates Cervical Cancer Development via Sponging miR-142-3p and Upregulating ARL2. Cancer Manag Res. 2020;12:9271-9283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 30. | Qu X, Zhu L, Song L, Liu S. circ_0084927 promotes cervical carcinogenesis by sponging miR-1179 that suppresses CDK2, a cell cycle-related gene. Cancer Cell Int. 2020;20:333. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 31. | Shi P, Zhang X, Lou C, Xue Y, Guo R, Chen S. Hsa_circ_0084927 Regulates Cervical Cancer Advancement via Regulation of the miR-634/TPD52 Axis. Cancer Manag Res. 2020;12:9435-9448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 32. | Wu P, Cai J, Chen Q, Han B, Meng X, Li Y, Li Z, Wang R, Lin L, Duan C, Kang C, Jiang C. Lnc-TALC promotes O6-methylguanine-DNA methyltransferase expression via regulating the c-Met pathway by competitively binding with miR-20b-3p. Nat Commun. 2019;10:2045. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 145] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 33. | Wang S, Du S, Lv Y, Wang W, Zhang F. Elevated microRNA-20b-3p and reduced thioredoxin-interacting protein ameliorate diabetic retinopathy progression by suppressing the NLRP3 inflammasomes. IUBMB Life. 2020;72:1433-1448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 34. | Gorukmez O, Yakut T, Gorukmez O, Sag SO, Topak A, Sahinturk S, Kanat O. Glutathione S-transferase T1, M1 and P1 Genetic Polymorphisms and Susceptibility to Colorectal Cancer in Turkey. Asian Pac J Cancer Prev. 2016;17:3855-3859. [PubMed] |
| 35. | Yu KD, Fan L, Di GH, Yuan WT, Zheng Y, Huang W, Chen AX, Yang C, Wu J, Shen ZZ, Shao ZM. Genetic variants in GSTM3 gene within GSTM4-GSTM2-GSTM1-GSTM5-GSTM3 cluster influence breast cancer susceptibility depending on GSTM1. Breast Cancer Res Treat. 2010;121:485-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 36. | Sun C, Gu Y, Chen G, Du Y. Bioinformatics Analysis of Stromal Molecular Signatures Associated with Breast and Prostate Cancer. J Comput Biol. 2019;26:1130-1139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 37. | Wang Y, Lei L, Chi YG, Liu LB, Yang BP. A comprehensive understanding of ovarian carcinoma survival prognosis by novel biomarkers. Eur Rev Med Pharmacol Sci. 2019;23:8257-8264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 38. | Sun YL, Zhang Y, Guo YC, Yang ZH, Xu YC. A Prognostic Model Based on Six Metabolism-Related Genes in Colorectal Cancer. Biomed Res Int. 2020;2020:5974350. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 39. | Kap EJ, Seibold P, Scherer D, Habermann N, Balavarca Y, Jansen L, Zucknick M, Becker N, Hoffmeister M, Ulrich A, Benner A, Ulrich CM, Burwinkel B, Brenner H, Chang-Claude J. SNPs in transporter and metabolizing genes as predictive markers for oxaliplatin treatment in colorectal cancer patients. Int J Cancer. 2016;138:2993-3001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 40. | Yu D, Liu Y, Zhou Y, Ruiz-Rodado V, Larion M, Xu G, Yang C. Triptolide suppresses IDH1-mutated malignancy via Nrf2-driven glutathione metabolism. Proc Natl Acad Sci USA. 2020;117:9964-9972. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 101] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 41. | Xiao Y, Meierhofer D. Glutathione Metabolism in Renal Cell Carcinoma Progression and Implications for Therapies. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 73] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 42. | Miess H, Dankworth B, Gouw AM, Rosenfeldt M, Schmitz W, Jiang M, Saunders B, Howell M, Downward J, Felsher DW, Peck B, Schulze A. The glutathione redox system is essential to prevent ferroptosis caused by impaired lipid metabolism in clear cell renal cell carcinoma. Oncogene. 2018;37:5435-5450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 282] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 43. | Duan S, Huang W, Liu X, Chen N, Xu Q, Hu Y, Song W, Zhou J. IMPDH2 promotes colorectal cancer progression through activation of the PI3K/AKT/mTOR and PI3K/AKT/FOXO1 signaling pathways. J Exp Clin Cancer Res. 2018;37:304. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 161] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 44. | Weng ML, Chen WK, Chen XY, Lu H, Sun ZR, Yu Q, Sun PF, Xu YJ, Zhu MM, Jiang N, Zhang J, Zhang JP, Song YL, Ma D, Zhang XP, Miao CH. Fasting inhibits aerobic glycolysis and proliferation in colorectal cancer via the Fdft1-mediated AKT/mTOR/HIF1α pathway suppression. Nat Commun. 2020;11:1869. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 201] [Cited by in RCA: 199] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 45. | Tan X, Zhang Z, Yao H, Shen L. Tim-4 promotes the growth of colorectal cancer by activating angiogenesis and recruiting tumor-associated macrophages via the PI3K/AKT/mTOR signaling pathway. Cancer Lett. 2018;436:119-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 64] [Article Influence: 8.0] [Reference Citation Analysis (0)] |