Published online Jul 28, 2021. doi: 10.3748/wjg.v27.i28.4722
Peer-review started: January 26, 2021
First decision: February 27, 2021
Revised: March 7, 2021
Accepted: July 5, 2021
Article in press: July 5, 2021
Published online: July 28, 2021
Processing time: 180 Days and 22 Hours
Dysbacteriosis may be a crucial environmental factor for ulcerative colitis (UC). Further study is required on microbiota alterations in the gastrointestinal tract of patients with UC for better clinical management and treatment.
To analyze the relationship between different clinical features and the intestinal microbiota, including bacteria and fungi, in Chinese patients with UC.
Eligible inpatients were enrolled from January 1, 2018 to June 30, 2019, and stool and mucosa samples were collected. UC was diagnosed by endoscopy, pathology, Mayo Score, and Montreal classification. Gene amplicon sequencing of 16S rRNA gene and fungal internal transcribed spacer gene was used to detect the intestinal microbiota composition. Alpha diversity, principal component analysis, similarity analysis, and Metastats analysis were employed to evaluate differences among groups.
A total of 89 patients with UC and 33 non-inflammatory bowel disease (IBD) controls were enrolled. For bacterial analysis, 72 stool and 48 mucosa samples were obtained from patients with UC and 21 stool and 12 mucosa samples were obtained from the controls. For fungal analysis, stool samples were obtained from 43 patients with UC and 15 controls. A significant difference existed between the fecal and mucosal bacteria of patients with UC. The α-diversity of intestinal bacteria and the relative abundance of some families, such as Lachnospiraceae and Ruminococcaceae, decreased with the increasing severity of bowel inflammation, while Escherichia-Shigella showed the opposite trend. More intermicrobial correlations in UC in remission than in active patients were observed. The bacteria-fungi correlations became single and uneven in patients with UC.
The intestinal bacteria flora of patients with UC differs significantly in terms of various sample types and disease activities. The intermicrobial correlations change in patients with UC compared with non-IBD controls.
Core Tip: Further study is required on gut microbiota alterations in patients with ulcerative colitis (UC). This study aimed to analyze the relationship between clinical features and the intestinal microbiota, including bacteria and fungi, in Chinese patients with UC. Gene amplicon sequencing of 16S rRNA gene and fungal internal transcribed spacer gene was applied. A total of 89 UC patients and 33 controls were enrolled. Results showed that the intestinal bacteria flora of patients with UC differed significantly in terms of various sample types and disease activities. The intermicrobial correlations changed in patients with UC compared with controls.
- Citation: He XX, Li YH, Yan PG, Meng XC, Chen CY, Li KM, Li JN. Relationship between clinical features and intestinal microbiota in Chinese patients with ulcerative colitis. World J Gastroenterol 2021; 27(28): 4722-4737
- URL: https://www.wjgnet.com/1007-9327/full/v27/i28/4722.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i28.4722
Inflammatory bowel disease (IBD) is a group of chronic inflammatory disorders of the gastrointestinal tract that are divided into ulcerative colitis (UC), Crohn’s disease, and IBD-unclassified[1]. The worldwide incidence rate of IBD varies greatly[2]. Among countries in the Asia-Pacific area, where the rate of IBD has rapidly increased, China has the highest incidence, with UC being the most common type[3]. The etiology of UC is still unclear, and it may be related to an abnormal epithelial barrier function and immune response driven by environmental factors in individuals with genetic susceptibility[1].
Disordered intestinal microbiota, or dysbacteriosis, is considered one of the crucial environmental factors for IBD[1]. Animal experiments have indicated the association between microbiota and UC. Some animal models, including mice with an immune deficiency, spontaneously develop chronic colitis resembling human UC[4]. However, under germ-free conditions or durative antibiotic therapy, the development of spontaneous colitis in these mice was prevented[4]. Dozens of studies found that bacterial strains derived from mice with colitis or patients with IBD could induce intestinal inflammation in mice with an immune deficiency and even in wild-type mice[4].
Existing evidence from animal experiments has stressed the necessity of a thorough understanding of the constitution and changes of the intestinal microbiota in patients with UC to elucidate the possible pathogenesis and investigate potential treatments. Numerous studies have focused on the variations in bacterial diversity and composition. Compared with non-IBD controls, there is a trend of decreased microbial α-diversity in patients with UC[5-7], although the total bacterial count decreased or increased in different studies[8,9]. Altered bacterial composition in patients with UC was discovered in almost all the related studies. The intestinal bacterial microbiota in the feces was different from that in the colon mucosa[10,11]. Differences were also found between patients with UC and non-IBD controls[6,7,12-16], among patients with UC of various severity[9,17,18] or disease types[11,19], between inflamed and noninflamed biopsies[6,16,19,20], among patients who accepted different therapies[10,11], and between patients who responded or did not respond to certain therapies[13]. In recent years, studies have also focused on changes in intestinal fungi in IBD patients[5,17]. However, existing studies concerning the above areas have arrived at various conclusions, which may be attributed to the limited sample size, imprecise stratification of patients, or other factors. Therefore, further study is required to obtain more information about microbiota alterations in the gastrointestinal tract of patients with UC for better clinical management and treatment.
In this study, our main purpose was to characterize the intestinal microbiota of patients with UC by 16S rRNA gene amplicon sequencing and compare the differences between the fecal and mucosa samples of the same UC patient, among patients with active UC, those with UC in remission, and non-IBD controls, among patients with different degrees of disease activity or with different disease types, and between biopsy samples collected from inflamed and noninflamed sites. In addition, the fecal fungal microbiota in the patients and controls was also analyzed using internal transcribed spacer (ITS) rRNA gene amplicon sequencing. Finally, we compared the intermicrobial correlations in patients with UC and non-IBD controls.
Patients with UC and non-IBD controls were recruited from patients admitted to Peking Union Medical College Hospital in Beijing, China from January 1, 2018 to June 30, 2019. The diagnosis of UC was determined by endoscopic and pathological findings. Only patients with regular follow-ups (more than 3 times in 1 year, outpatient and/or inpatient) were included, and all these patients orally took 1 g of mesalazine per time, 3-4 times per day. Patients with any history of colectomy, antibiotic use or probiotic application within 3 mo before sample collection, or other types of gastroenteric complications (including gastrointestinal tumor, gastroduodenal ulcer, and irritable bowel syndrome) were excluded. The clinical activity of disease was determined by the Mayo score/Disease Activity Index[21], and the patients were classified into UC in remission or active UC groups, with the latter further divided into mild, moderate, and severe groups. The patients were also classified into proctitis (type E1), distal colitis (type E2), or pancolitis (type E3) groups based on their disease types according to the Montreal classification[22]. The inflammatory status of the biopsy was confirmed by pathological evidence. The non-IBD controls were asymptomatic patients with colonic polyps.
This work was been approved by the ethical committees of Peking Union Medical College Hospital (No. JS-1488). All the patients included have signed an informed consent form.
Fecal samples were collected 1-2 d before bowel preparation and endoscopic examination, and were stored in stool tubes (Allwegene Company, Beijing) at -80 °C within 2 h after collection. Two biopsies were taken from each biopsy site during endoscopy. One of the biopsies was stored in formalin for standard histological staining, and the other was snap-frozen and then stored at -80 °C for sequencing.
DNA was extracted from samples using the DNeasy PowerSoil Kit (QIAGEN, Dusseldorf, GER) according to the manufacturer’s protocol. The purity and quality of the genomic DNA were checked on 0.8% agarose gels. The V3-4 hypervariable region of the bacterial 16S rRNA gene was amplified with the primers 338F (ACTCCTACGGGAGGCAGCAG) and 806R (GGACTACHVGGGTWTCTAAT)[23]. The fungal ITS gene was amplified with the primers ITS3-F (GCATCGATGAAGAACGCAGC) and ITS4-R (TCCTCCGCTTATTGATATGC)[24]. For each mucosa or fecal sample, a 10-digit barcode sequence was added to the 5’ end of the forward and reverse primers. Polymerase chain reaction (PCR) was carried out on a Mastercycler Gradient (Eppendorf, Germany) using a 25 μL reaction volume containing 12.5 μL of 2 × Taq PCR MasterMix, 3 μL of BSA (2 ng/μL), primers (5 μM), 2 μL of template DNA, and 5.5 μL of ddH2O. The cycling parameters were 95 °C for 5 min, followed by 32 cycles of 95 °C for 45 s, 55 °C for 50 s, and 72 °C for 45 s with a final extension at 72 °C for 10 min. Three PCR products per sample were pooled to mitigate reaction-level PCR biases. The PCR products were purified using a QIAquick Gel Extraction Kit (QIAGEN, Germany), quantified using real-time PCR, and sequenced by Allwegene Company (Beijing, China).
Deep sequencing was performed on the Miseq platform at Allwegene Company (Beijing). After the run, image analysis, base calling, and error estimation were performed using Illumina Analysis Pipeline Version 2.6. The raw data were first screened, and sequences were removed if they were shorter than 150 bp or longer than 500 bp, had a low-quality score (≤ 20), contained ambiguous bases, or did not exactly match the primer sequences and barcode tags. After screening, the qualified reads were separated using the sample-specific barcode sequences and trimmed with Illumina Analysis Pipeline Version 2.6. Then, the dataset was analyzed using Quantitative Insights Into Microbial Ecology (QIIME, Ver2.1, Rob Knight laboratory and Greg Caporaso laboratory, United States). After 16S rRNA amplicon sequencing and screening, an average of 59307 ± 4058 [mean ± standard error of mean (SEM)] qualified reads were obtained from the fecal samples, and an average of 54201 ± 3834 qualified reads were obtained from the mucosa samples. After ITS sequencing and screening, an average of 47051 ± 4675 qualified reads were obtained from the fecal samples. In the subsequent subgroup analysis, singletons were further removed from the qualified sequences and then the latter was subsampled and clustered into operational taxonomic units (OTUs) at a similarity level of 97% to generate rarefaction curves and to calculate the richness and diversity indices (the number of subsampled sequences and the corresponding OTUs for each subgroup analysis are described in Supplementary Table 1. The Ribosomal Database Project Classifier tool was used to classify all sequences into different taxonomic groups. Finally, the bacterial taxonomic groups were matched to specific genera based on Silva database (Ver. 128), and the fungal groups were based on the Unite database (Ver. 7.1).
A t-test was used to detect the difference in age among groups. The chi-square test was used to compare the distributions of sex, disease type, and disease activity among groups. The alpha diversity, principal component analysis (PCA), and analysis of similarities (ANOSIM) were performed based on the OTUs and the abundance acquired from the subsampled sequences of samples involved in the subgroup analysis. Alpha diversity was estimated using Shannon index metric, and one-way ANOVA (SPSS 19.0) was used to measure the difference among Shannon indices. To examine the similarity of the microbiota structure between different samples, PCA was performed using R software. PCA was used to reduce the high dimensionality of the intestinal microbiota structure, which contained numerous original variables, and to observe the relationship among samples in a rectangular coordinate. To further examine the similarity between different samples and to determine whether the between-group difference was greater than the inter-group difference, ANOSIM was performed in the R. The Metastats analysis using Mothur was employed to detect interindividual differences and the differences in microbiota composition among groups at the phylum and genus levels were measured. Only bacteria or fungi with relative abundances higher than 1% in either group were displayed in this study and were discussed with emphasis. Generally, the numerical value is shown as the mean ± SD. Under the condition of multiple comparisons, the P values were corrected to control for the false discovery rate by Benjamini-Hochberg method (α = 0.05). Differences with a P < 0.05 were considered statistically significant. Moreover, Spearman correlation coefficient analysis was applied and inter-microbial correlations at the genus level with a P value less than 0.05 and an R value greater than 0.6 were selected and put into a correlation network using Cytoscape v3.7.0. The statistical methods of this study were reviewed by Dr. Tao-Tao Han from Chinese Academy of Medical Sciences and Peking Union Medical College.
Eighty-nine patients with UC and 33 non-IBD controls were included in the present study. Among the 89 patients with UC, 31 provided both stool and mucosa samples and 19 patients with UC at active stage provided mucosa samples from inflamed and noninflamed sites, and finally 72 stool samples and 67 mucosa samples were obtained. Twenty-one stool samples and 12 mucosa samples were obtained from non-IBD controls diagnosed with tubular adenoma, tubulovillous adenoma, or a hyperplastic polyp according to histological evidence. The basic clinical characteristics of the subjects among the groups under different stratifications were compared (Supplementary Tables 2-4), including age, sex proportion, disease activity, and disease type distribution in patients with UC. Most of the characteristics showed little intergroup difference, except for the mean age of the non-IBD controls and patients with UC and the disease type distribution of active UC and UC in remission. Among the stool samples, the compositions of fungal flora in 15 non-IBD samples and 42 UC samples were detected by ITS sequencing. The rest of the samples (6 non-IBD controls and 30 UC patients) failed to undergo ITS sequencing due to the insufficiency of tissues. Although a difference existed in the results of OTUs clustering in different subgroup analysis, in both the fecal and the mucosa sample, Firmicutes, Bacteroidetes, Proteo -bacteria, and Actinobacteria were always the four chief components among the identified phyla, and the identified fecal fungi mainly consisted of Basidiomycota and Ascomycota.
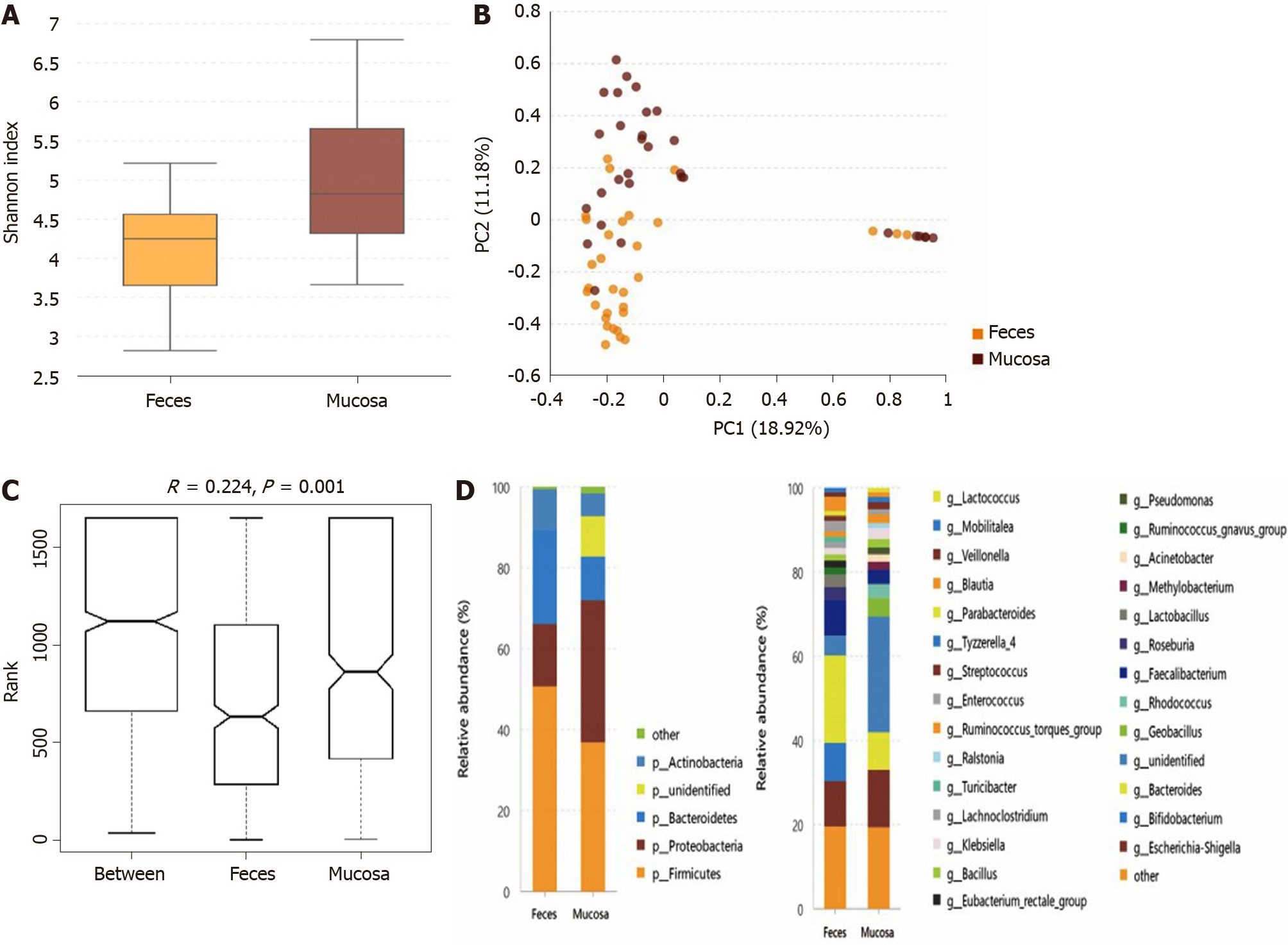
The microbiota in the stool and mucosa samples of the same patient with UC were compared if collected within less than 3 d, and 31 pairs of samples were included. Compared with the mucosal sample (Shannon index, 5.04 ± 1.14), the α-diversity of the microbiota in the fecal sample (Shannon index, 4.08 ± 0.89) was significantly lower (P = 0.001) (Figure 1A). As directly assessed by PCA (Figure 1B), the flora structure of the mucosa sample and that of the stool sample showed obvious differences and ANOSIM indicated significant intergroup differences (Figure 1C). The microbiota compositions at the phylum and genus levels of the mucosa and fecal samples are shown in Figure 1D. At the phylum level, the relative abundance of mucosal Proteobacteria and unidentified phyla was higher than that of fecal Proteobacteria, while the relative abundances of Bacteroidetes and Firmicutes were lower in the mucosa sample. At the genus level, 16 types of bacterial genera differed significantly between the mucosa and stool samples (the relative abundance and the P value of each bacterium are described in Supplementary Table 5).
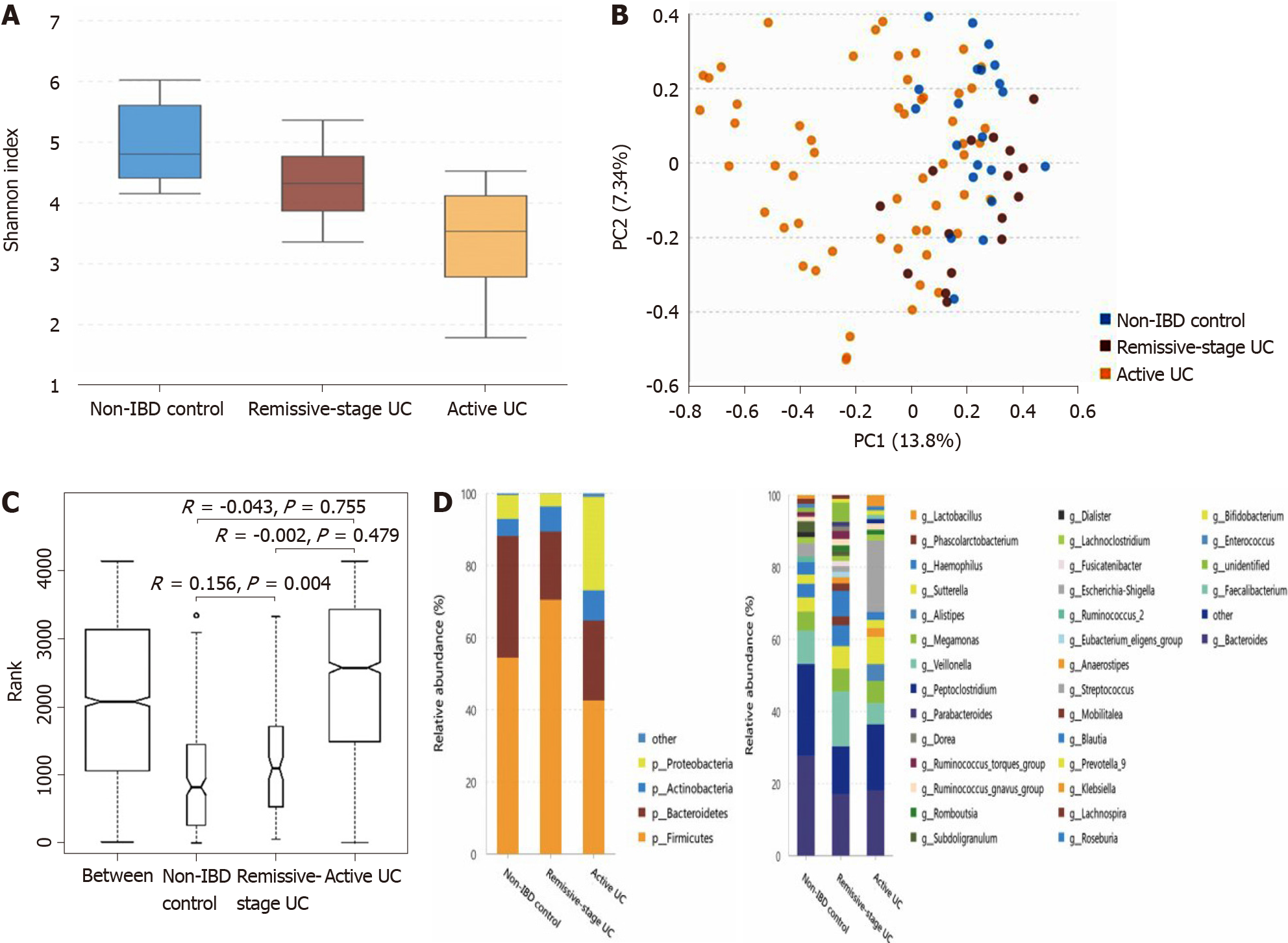
The α-diversity of intestinal bacteria decreases in patients with UC and the microbiota composition differs among UC patients in remission, patients with active UC, and non-IBD controls: As shown in Figure 2A, compared with the non-IBD controls (Shannon index, 4.71 ± 0.64), the α-diversity of the intestinal microbiota evaluated by the Shannon index significantly decreased in patients with UC in remission (Shannon index, 4.11 ± 0.74; P = 0.013), as well as in patients with active UC (Shannon index, 3.20 ± 1.04; P = 0.000). The α-diversity of patients with active UC was also significantly lower than that of UC patients in remission (P = 0.002). According to the PCA and ANOSIM results, the intergroup difference between the non-IBD controls and UC patients in remission group was significant (Figure 2B and C). The microbiota compositions of the different groups are shown in Figure 2D (the relative abundance and P value are listed in Supplementary Table 6). At the phylum level, the abundance of Proteobacteria significantly increased in patients with active UC but decreased in patients with UC in remission, while that of Firmicutes showed the opposite pattern. The abundance of Bacteroidetes significantly decreased in both the active UC and UC in remission groups. At the genus level, compared with the non-IBD controls, the relative abundances of Alistipes, Bacteroides, Dialister, and Escherichia-Shigella in patients with UC in remission significantly decreased, while that of Lachnospira increased. The relative abundances of Escherichia-Shigella, Enterococcus, and Peptoclostridium were significantly higher in the active UC group than in the non-IBD group, while those of Alistipes, Subdoligranulum, Roseburia, Ruminococcus 2, and Ruminococcus torques were significantly lower in patients with active UC. When comparing patients with active UC and UC in remission, the relative abundances of Escherichia-Shigella, Enterococcus, Haemophilus, and Klebsiella were higher in the former, while those of Lachnospira, Faecalibacterium, Roseburia, and Blautia were higher in the latter.
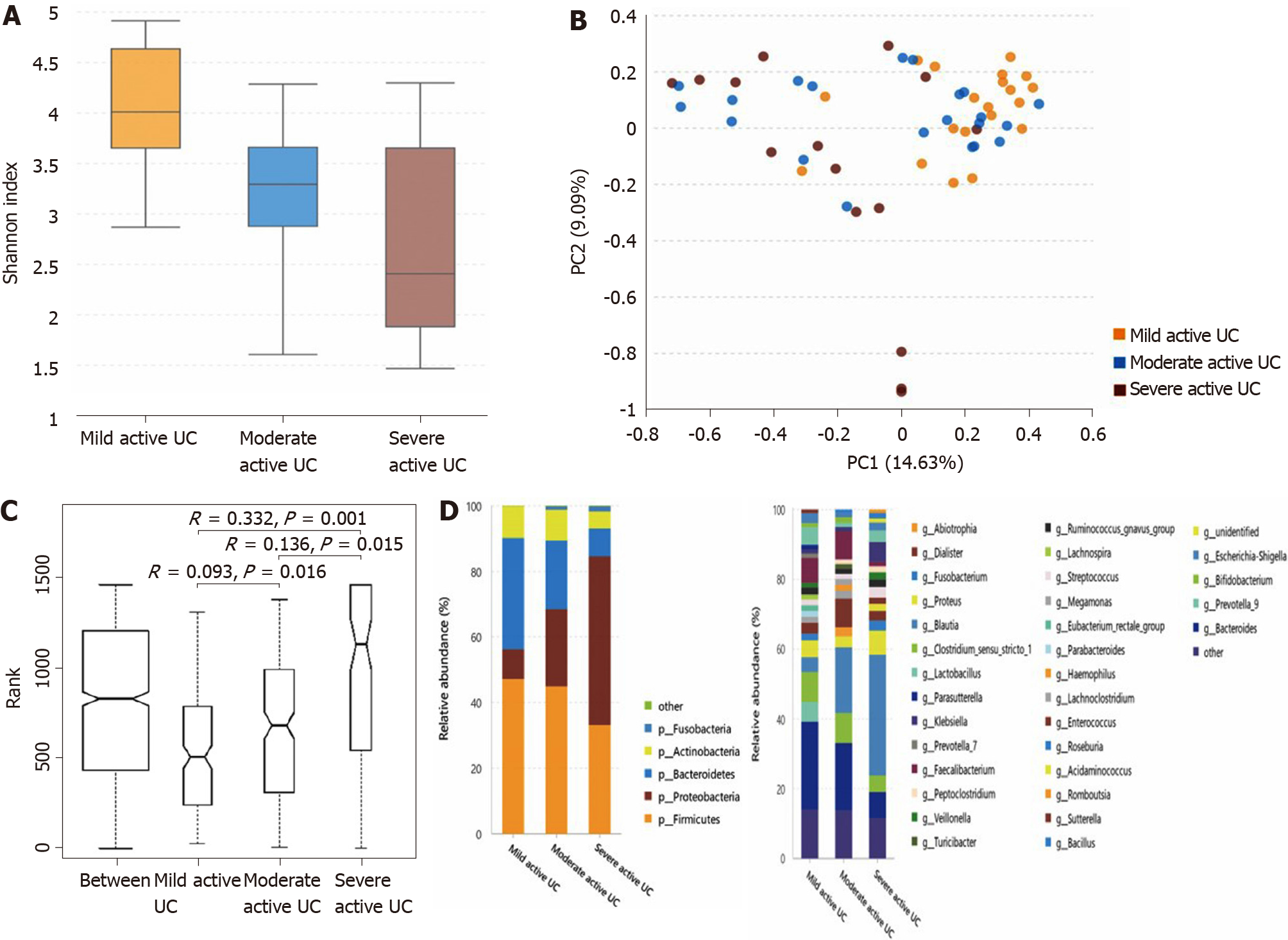
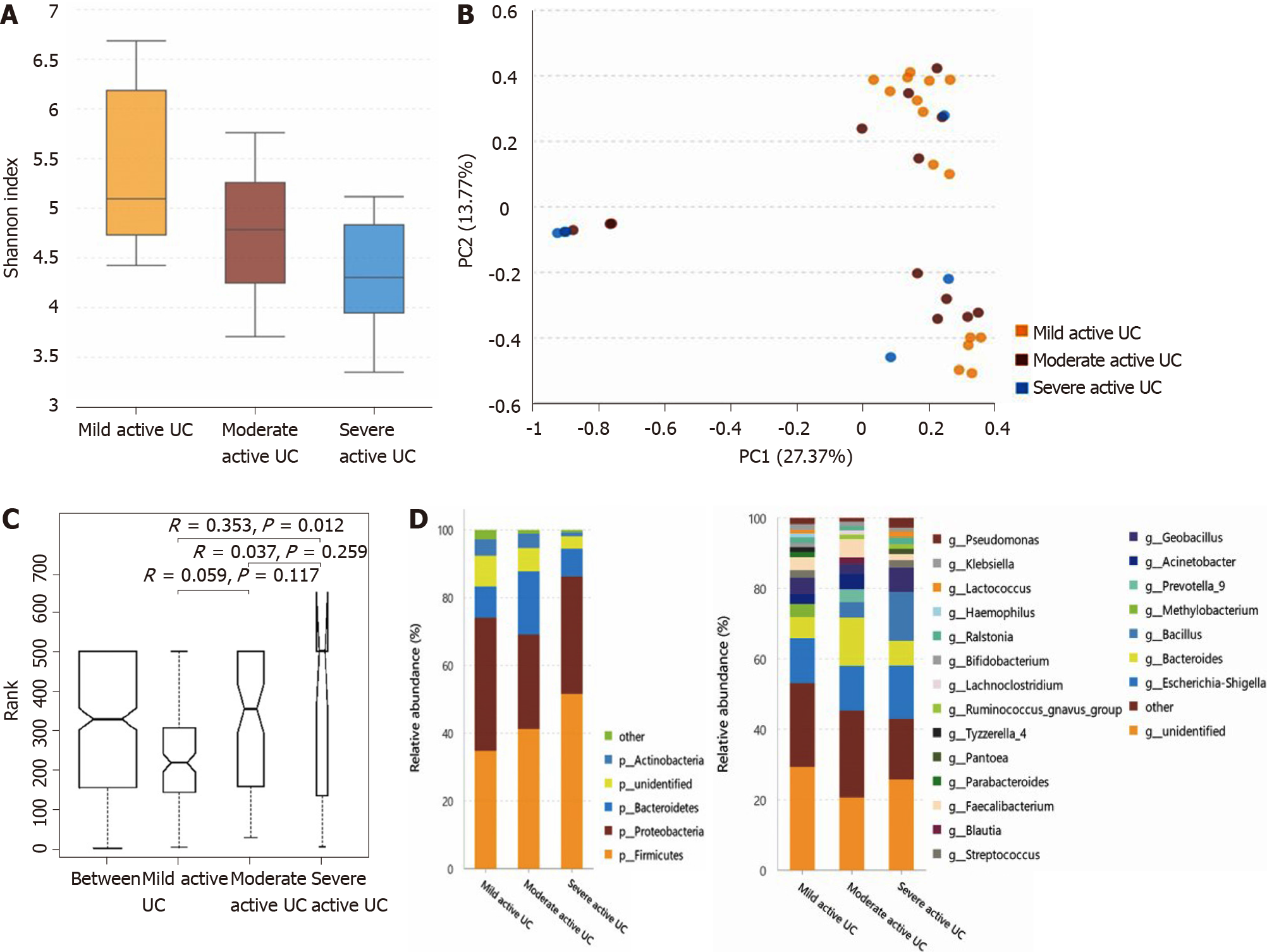
The α-diversity continuously decreases, and some bacteria show a consistently changing pattern with increasing disease activity in patients with active UC: As shown in Figure 3A, the α-diversity of the intestinal microbiota gradually decreased with increasing inflammation degree (the Shannon index of mild, moderate, and severe active UC was 3.74 ± 0.84, 3.08 ± 0.95, and 2.66 ± 1.14, respectively), and the difference between the mild and moderate groups (P = 0.025), and between the mild and severe groups (P = 0.003) was significant, though that between the moderate and severe groups was not obvious (P = 0.235). Significant differences were observed through PCA (Figure 3B) and ANOSIM (Figure 3C). The microbiota compositions of different groups are shown in Figure 3D (the relative abundance and P value are listed in Supplementary Table 7). At the phylum level, the abundance of Proteobacteria showed a significantly increasing trend with deteriorating disease conditions, and that of Bacteroidetes gradually decreased, although only the difference between the mild and severe groups was obvious. At the genus level, some bacterial genera that differed significantly between the non-IBD controls and patients with active UC displayed consistently changing patterns when the disease condition worsened, including a further increase in Escherichia-Shigella in moderate-severe active UC and an obvious reduction of Faecalibacterium, Roseburia, and Bacteroides in severe active UC. The relative abundance of Eubacterium rectale group gradually decreased when the inflammation worsened, although this genus did not differ significantly between the non-IBD controls and patients with UC. Decreased Blautia in moderate active UC and reduced Parasutterella and Lachnospira in severe active UC compared with the mild UC group were also observed.
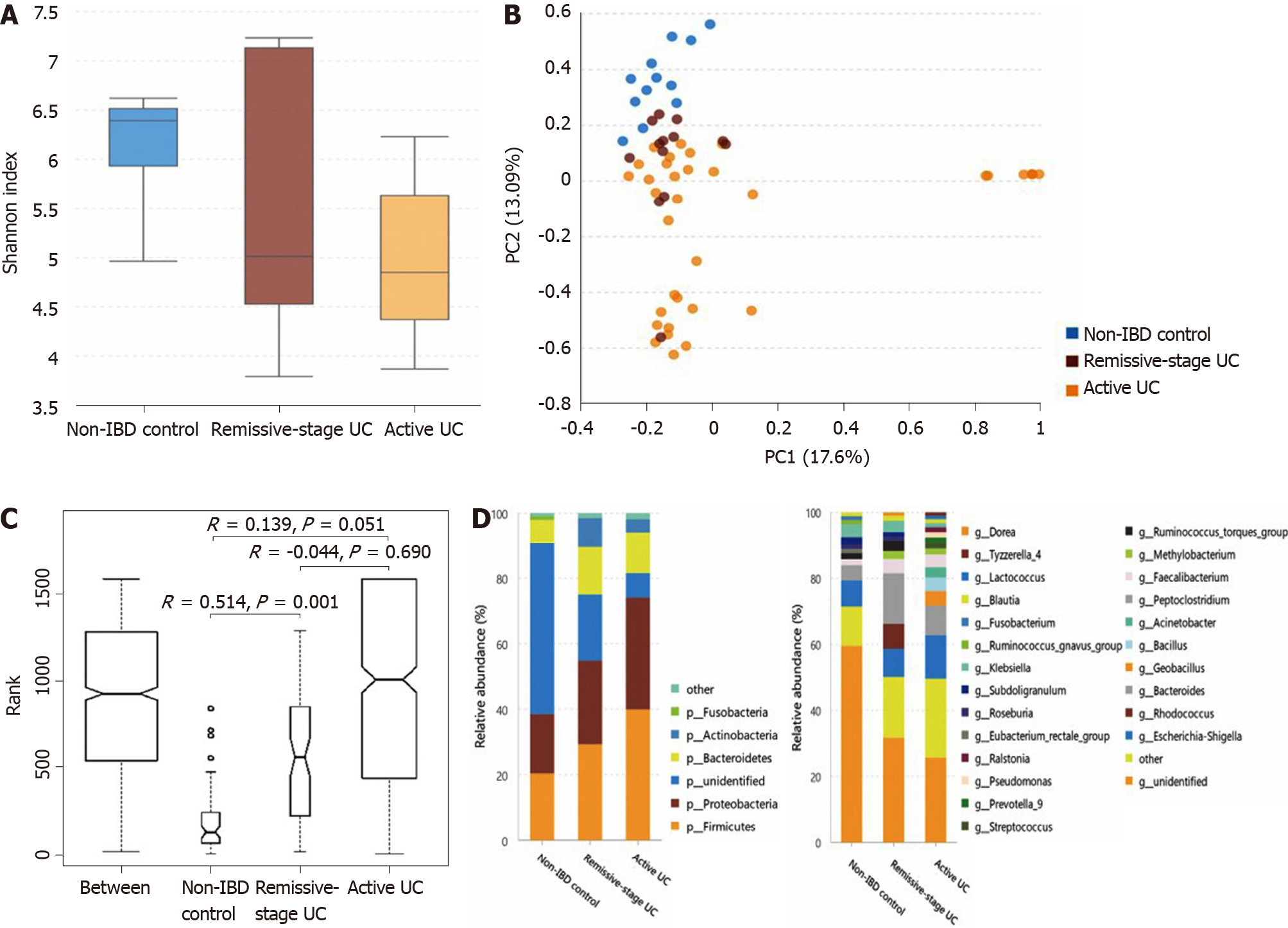
The α-diversity of the mucosal microbiota decreases in patients with active UC, and the microbiota composition differs among groups: As shown in Figure 4A, the α-diversity of the mucosal microbiota in the non-IBD controls (Shannon index, 6.12 ± 0.70) was similar to that in patients with UC in remission (Shannon index, 5.63 ± 1.53; P = 0.682) but was significantly higher than that in patients with active UC (4.99 ± 1.02; P = 0.001). The α-diversity of patients with UC in remission and with active UC showed little difference (P = 0.453). Direct observation of the PCA rectangular coordinates (Figure 4B) indicated that the flora structure of the non-IBD controls was different from that of either UC group. ANOSIM indicated that the difference between the controls and patients with UC in remission was significant (Figure 4C). As shown in Figure 4D (the relative abundance and P value are listed in Supplementary Table 6), at the phylum level, compared with the non-IBD controls, the relative abundances of Firmicutes and Proteobacteria significantly increased in patients with active UC, and that of Actinobacteria was significantly higher in patients with both active UC and UC in remission. The relative abundance of unidentified bacterial phyla obviously decreased in patients with UC in remission and further decreased in patients with active UC. At the genus level, the relative abundances of Geobacillus, Lactococcus, Pseudomonas, Methylobacterium, Acinetobacter, Streptococcus, Bacillus, and Ralstonia increased in the active UC group compared with the non-IBD controls, while those of Ruminococcus torques and the unidentified bacterial genera were reduced. When comparing the non-IBD controls and patients with UC in remission, the relative abundances of Eubacterium rectale and the unidentified genera were lower in the latter, while those of Methylobacterium, Rhodococcus, Peptoclostridium, and Faecalibacterium were higher. The difference between patients with active UC and UC in remission was also obvious, and
The α-diversity of mucosal bacteria further decreases in severe active UC, and the microbiota composition is slightly different among subgroups that vary in inflammation degree: The α-diversity of mucosal bacteria in patients with severe active UC (Shannon index, 3.67 ± 1.25) was significantly lower than that in patients with mild active UC (Shannon index, 5.40 ± 1.01; P = 0.017). However, little difference was observed between the mild and moderate subgroups (Shannon index, 4.80 ± 0.90; P = 0.100) or between the moderate and severe subgroups (P = 0.251) (Figure 5A). Only the flora structure of the mild and severe groups showed obvious differences according to PCA and ANOSIM (Figure 5B and C). As shown in Figure 5D (the relative abundance and P value are listed in Supplementary Table 7), at the phylum level, the relative abundance of Actinobacteria in patients with severe active UC was significantly lower than that in patients with mild/moderate UC, and fewer unidentified bacterial phyla were observed in patients with severe UC than in patients with mild UC. At the genus level, the relative abundance of Methylobacterium in patients with mild UC was significantly higher than that in patients with moderate/severe UC, and Blautia significantly decreased in patients with severe UC compared with the other two subgroups.
Patients with active UC with different disease extents show slight differences in bacterial α-diversity and composition: Based on the extent of colitis, patients with active UC were divided into two groups: Partial colitis (E1 and E2) and pancolitis groups (E3). The α-diversity of fecal bacteria in these groups was similar, while that of mucosal bacteria was higher in the pancolitis group. Regarding the bacterial composition in stool samples, only the relative abundance of the phylum Fusobacteria and its genus Fusobacterium was significantly higher in the stool of pancolitis patients. Regarding the mucosa sample, the relative abundance of unidentified phyla and the genus Methylobacterium in the “partial colitis” group was significantly lower than that in the pancolitis group (Supplementary Figures 1 and 2, and Supplementary Table 8).
Mucosa samples from inflamed and noninflamed sites have similar intestinal microbiota structures: Nineteen pairs of mucosa samples from inflamed and noninflamed sites of one subject were collected. There was no significant difference between the microbiota of mucosa samples from the inflamed and noninflamed sites as evaluated by the α-diversity, PCA, ANOSIM, or microbiota composition at the phylum and genus levels (Supplementary Figure 3).
The α-diversity and composition of stool fungal microbiota slightly differ among groups: There was little difference between the fecal fungal microbiota of patients with UC and the non-IBD controls or among the active UC groups with different degrees of inflammation as evaluated by α-diversity, PCA, or ANOSIM. Regarding the fungal composition, only a few fungal genera differed significantly, including a higher relative abundance of Mycosphaerella, Malassezia, and/or Suillus in patients with active UC, more Xeromyces and Yarrowia in patients with severe active UC, and more Rhizopus in patients with moderate active UC (Supplementary Figures 4 and 5, and Supplementary Table 9).
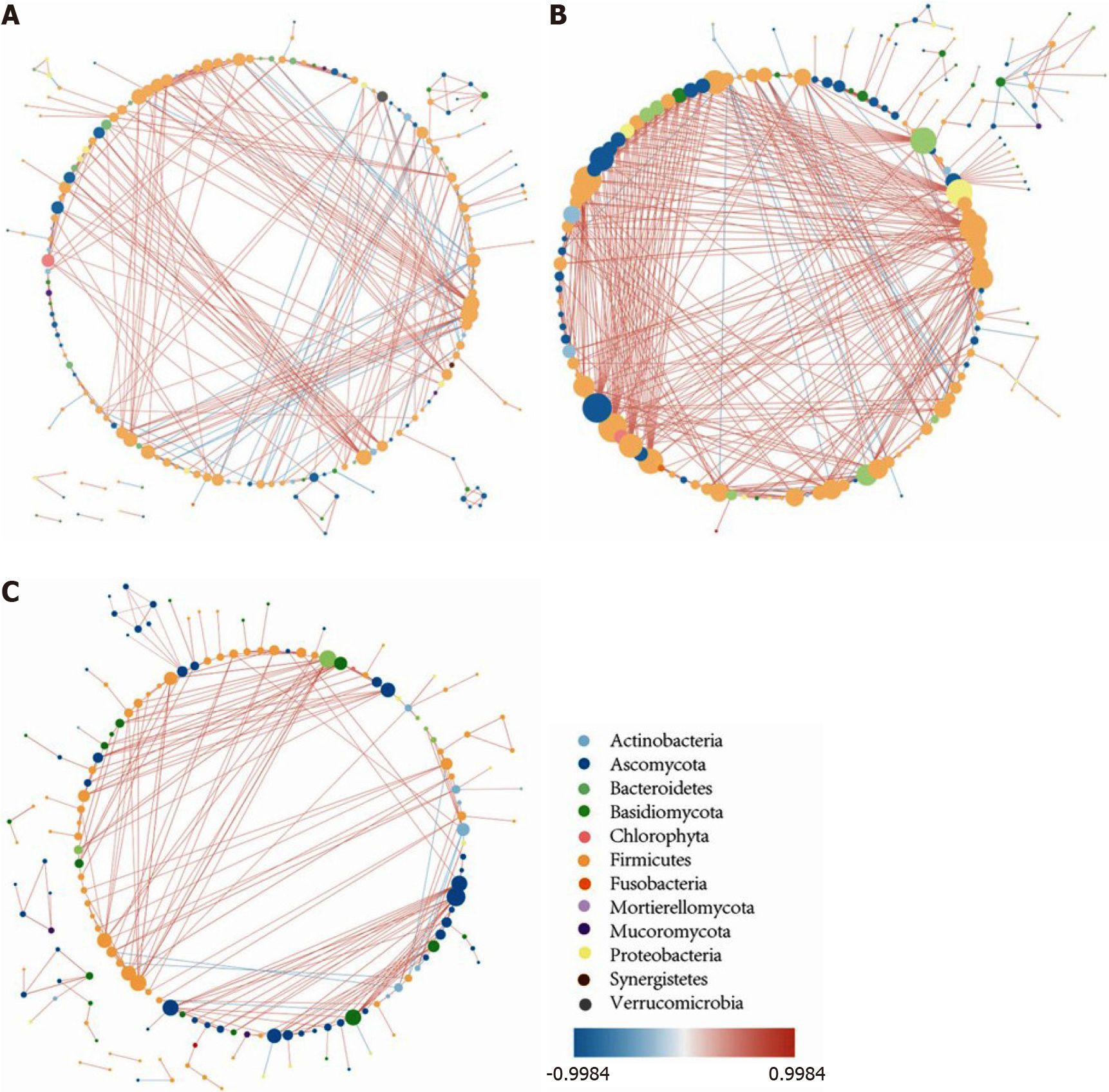
The intestinal intermicrobial correlation changes in patients with UC compared with non-IBD controls: The intestinal intermicrobial correlations at the genus level in the non-IBD controls and the UC in remission and active UC groups are shown in a correlation network (Figure 6). In all of the groups, most of the bacteria-bacteria correlations existed in Firmicutes, the majority of the bacteria-fungi correlations occurred in Firmicutes and Ascomycota, and the Ascomycota-Ascomycota/Basidiomycota correlations were the major fungi-fungi correlations. Compared with the controls, the number of significant correlations increased in patients with UC in remission but decreased in patients with active UC. The most obvious change occurred in the number of bacteria-fungi correlations, while the number of fungi-fungi correlations was insignificant among the three groups.
To further investigate the change in bacteria-fungi correlations, we focused on the bacterium-fungus pairs in which at least one of the two genera had a relative abundance higher than 0.01, and the correlations with a P value smaller than 0.05 are shown in the heat map (Supplementary Figure 6). In the non-IBD controls, bacterial genera from nine bacterial phyla were correlated with fungal genera from four fungal phyla, while for patients with UC in remission, the number of involved bacterial phyla decreased to five, and for patients with active UC, the number of phyla was further reduced to three. In addition, the correlation intensity of the related bacterium-fungus pairs was distributed evenly among the controls, while the correlation intensity of some microbe pairs in patients with UC became prominent, indicating that the bacteria-fungi interaction became single and circumscribed in patients with UC.
In this study, the intestinal bacteria flora of patients with UC was analyzed, and three important results were acquired. First, the fecal bacteria and mucosal bacteria of patients with UC were different, and the former was characterized by a lower α-diversity, lower relative abundances of Proteobacteria and unidentified phyla, and higher proportions of Bacteroidetes and Firmicutes. Significant differences were also shown in 16 bacterial genera. This result agrees with a previous study showing that the sample origin (biopsy vs stool) is the single most influential factor on the microbial community structure in IBD patients[10]. Another study also reported fecal-specific bacterial species[11]. Bowel preparation impacted the mucosal microbiota α-diversity (mainly presented as decreased diversity) and the abundance of certain bacteria[25], which may partially contribute to the difference between biopsy and fecal samples. The vastly distinct environment for the colonization of microbes may be the principal factor.
Second, obvious distinctions were observed between the non-IBD controls and patients with UC. Notably, the intestinal bacteria in patients with UC in remission and active UC may be regarded as two dissimilar populations owing to their significantly different α-diversity and flora structure. The former was more similar to that of the non-IBD controls, as reflected by a smaller decrease in α-diversity and smaller changes in the bacterial composition. Moreover, the bacterial composition of the active and relieved UC groups even showed opposite changing patterns in some circumstances; for example, Proteobacteria and its genus Escherichia-Shigella increased in the feces of active UC but decreased in UC in remission, while Firmicutes and some of its members changed oppositely. Previous studies have confirmed a depletion of α-diversity in patients with UC, but the changes in bacterial composition were inconsistent. Some studies have reported a depletion of fecal Firmicutes and its members in patients with UC compared with non-IBD controls or co-habitation healthy partners[10,15,17,26,27], while others have reported opposite alterations[7]. The analysis of fecal Bacteroids and their members in patients with UC compared with non-IBD controls also reached different results[7,11,15,17,27]. The changes in mucosal bacteria in patients with UC are even more complicated and inconsistent than those in fecal ones[6,12,13,16]. Such confusing results may partially be attributed to the different sample sizes, ages, and races of subjects[14]; sampling biogeography; and the luminal pH values[10]. Another possible reason may be that many studies took the active and the quiescent patients with UC as a whole and analyzed them without distinction. This opinion was supported by our study as well as other studies that found different changing patterns of certain bacteria in active and quiescent UC[9,28]. Patients with UC even displayed different bacterial compositions before and after treatment, with the latter being more similar to non-IBD controls[8].
Third, the intestinal bacteria displayed a gradually decreased α-diversity and consistently changing pattern of certain bacteria when the inflammation state of active UC increased, which was represented by a gradual increase in Proteobacteria and Escherichia-Shigella and decrease in Bacteroidetes, Faecalibacterium, Roseburia, Bacteroides, and Eubacterium rectale group in stool, as well as less Methylobacterium and Blautia in the mucosa. Previous studies also found that the patients with active UC with intestinal bacteria dominated by some kinds of bacteria (the members of Proteobacteria) exhibited more severe disease conditions than those enriched in other genera (Blautia, Ruminococcus, and other members belonging to Firmicutes)[17,18,29].
Despite the obvious alteration in intestinal bacteria along with the changed disease activity of UC, the bacterial composition only showed a slight or no difference when the disease type or biopsy site differed. These results agreed with most of the previous studies[11,16,19,20] except one finding that showed a significant difference between inflamed and noninflamed sites[6].
There were some commonalities among the changes of intestinal bacteria in this study. The bacteria with “protective effects” decreased while those with “deleterious effects” increased in active UC, especially in patients with moderate-severe inflammation, while in UC in remission, some of these bacteria changed in the opposite way, indicating a “correction effect” of some “protective” bacteria in recovering patients. The most representative change was the reduction of a series of genera affiliated with Lachnospiraceae and Ruminococcaceae in active UC. These are important bacteria for producing butyric acid, a substance beneficial to IBD in terms of anti-inflammation and intestinal barrier defense[30]. In addition, a decrease in the genus Alistipes in active UC was reported to produce bacterial sulfonolipids with potential anti-inflammatory effects[31]. Most of the genera that increased in the active UC and/or moderate-severe UC groups were opportunistic pathogens that may be harmful to gastrointestinal health. For example, Escherichia-Shigella[32] and Prevotella[33] promote inflammatory cytokine expression. In addition, Fusobacterium, which was more abundant in the feces of pancolitis patients, was enriched in patients with gut inflammation and was proportionally associated with colorectal cancer[34]. However, it is difficult to determine whether the change in microbiota is a cause or a consequence of UC since the bacteria depleted in patients were sometimes strictly anaerobic and failed to survive in the broken intestinal environment[30].
This study also detected fecal fungi and the intermicrobiota correlations. The diversity of fecal fungi was similar among groups, and the fungal composition only showed subtle differences at the genus level, which was in accordance with a previous study[17] but contrary to another[5]. The number of intermicrobiota correlations increased for UC in remission and decreased in active UC, with the most obvious change in the bacteria-fungi correlation, and the distribution of the correlations became uneven in patients with UC. The meaning of these results is still unclear, and more studies are needed to explore the complex changes in the intestinal fungi of IBD patients.
There were some limitations in this study. First, the sample size was insufficient, especially for the mucosa samples. Second, non-IBD controls were asymptomatic patients with colonic polyps rather than healthy people owing to ethical issues. Since the patients with colonic polyps generally had a higher onset age than patients with UC, the average ages of the controls and patients with UC were unmatched. It has been reported that age influences gut microbiota in adults, which is mainly manifested as decreases in the families of Firmicutes and Bifidobacteria, as well as an increase in Proteobacteria members, indicating a more proinflammatory gut microbiota phenotype in elderly people[35]. Therefore, the difference between non-IBD controls and patients with UC may be more obvious than that presented in this study. Third, many studies have implied that the gut fungal composition was significantly affected by lifestyle[36,37], but due to practical reasons, our study did not collect the information of the lifestyle in this population, which can be a confounding factor. Finally, the sequencing method in our study can only reach the genus level, and further research using the metagenomic sequencing is required to acquire more details at the species level.
Patients with active UC have a significantly different intestinal microbiota than non-IBD controls, while the difference between patients with UC in remission and non-IBD control is smaller. Disease degree of patients with active UC significantly affects the gut microbiota, and some bacteria show consistently changing patterns when the disease worsens. Some “beneficial” bacteria decrease while some “detrimental” bacteria increase in patients with active UC, especially in patients with moderate-severe inflammation. A difference was also observed in the intestinal bacteria flora collected from paired stool and mucosa samples of patients with UC. The fecal fungal microbiota does not show evident changes compared with the controls. However, significantly different intermicrobial correlations and an unevenly distributed bacteria-fungi interaction were observed.
The etiology of ulcerative colitis (UC) is still unclear, and dysbacteriosis may be a crucial environmental factor. Numerous studies have focused on the variations in intestinal microbiota diversity and composition of patients with UC.
Existing studies concerning UC and intestinal microbiota have arrived at various conclusions. A further understanding of the constitution and changes of the intestinal microbiota in patients with UC is necessary to elucidate the possible pathogenesis and investigate potential treatments.
To analyze the relationship between different clinical features and the intestinal microbiota, including bacteria and fungi, in Chinese patients with UC.
Gene amplicon sequencing of 16S rRNA gene and fungal internal transcribed spacer gene was used to detect the intestinal microbiota composition. Alpha diversity, principal component analysis, similarities analysis, and Metastats analysis were employed to evaluate differences among groups.
A total of 89 patients with UC and 33 controls were enrolled. A significant difference existed between the fecal and mucosal bacteria, and the α-diversity of intestinal bacteria and the relative abundance of some families decreased with the increasing severity of bowel inflammation. More intermicrobial correlations in UC in remission than in active patients were observed, and the bacteria-fungi correlations became single and uneven in patients with UC.
Patients with active UC have a significantly different intestinal microbiota in terms of various sample types and disease activities. Significantly different intermicrobial correlations and an unevenly distributed bacteria-fungi interaction were observed.
There is still a need to collect subjects to expand the sample size and further include data from other parts of China. And metagenomic sequencing method may be needed for more details at the species level in future research.
We thank Dr. Tao-Tao Han for review of the statistical methods of this study.
| 1. | Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2894] [Cited by in RCA: 3414] [Article Influence: 179.7] [Reference Citation Analysis (12)] |
| 2. | Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW, Kaplan GG. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 2012; 142: 46-54. quiz e30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3789] [Cited by in RCA: 3601] [Article Influence: 257.2] [Reference Citation Analysis (6)] |
| 3. | Ng SC, Tang W, Ching JY, Wong M, Chow CM, Hui AJ, Wong TC, Leung VK, Tsang SW, Yu HH, Li MF, Ng KK, Kamm MA, Studd C, Bell S, Leong R, de Silva HJ, Kasturiratne A, Mufeena MNF, Ling KL, Ooi CJ, Tan PS, Ong D, Goh KL, Hilmi I, Pisespongsa P, Manatsathit S, Rerknimitr R, Aniwan S, Wang YF, Ouyang Q, Zeng Z, Zhu Z, Chen MH, Hu PJ, Wu K, Wang X, Simadibrata M, Abdullah M, Wu JC, Sung JJY, Chan FKL; Asia–Pacific Crohn's and Colitis Epidemiologic Study (ACCESS) Study Group. Incidence and phenotype of inflammatory bowel disease based on results from the Asia-pacific Crohn's and colitis epidemiology study. Gastroenterology. 2013;145:158-165.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 515] [Cited by in RCA: 615] [Article Influence: 47.3] [Reference Citation Analysis (3)] |
| 4. | Hörmannsperger G, Schaubeck M, Haller D. Intestinal Microbiota in Animal Models of Inflammatory Diseases. ILAR J. 2015;56:179-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 5. | Sokol H, Leducq V, Aschard H, Pham HP, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, Cosnes J, Seksik P, Langella P, Skurnik D, Richard ML, Beaugerie L. Fungal microbiota dysbiosis in IBD. Gut. 2017;66:1039-1048. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 658] [Cited by in RCA: 958] [Article Influence: 106.4] [Reference Citation Analysis (0)] |
| 6. | Sepehri S, Kotlowski R, Bernstein CN, Krause DO. Microbial diversity of inflamed and noninflamed gut biopsy tissues in inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:675-683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 181] [Article Influence: 9.5] [Reference Citation Analysis (2)] |
| 7. | Imhann F, Vich Vila A, Bonder MJ, Fu J, Gevers D, Visschedijk MC, Spekhorst LM, Alberts R, Franke L, van Dullemen HM, Ter Steege RWF, Huttenhower C, Dijkstra G, Xavier RJ, Festen EAM, Wijmenga C, Zhernakova A, Weersma RK. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut. 2018;67:108-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 420] [Cited by in RCA: 588] [Article Influence: 73.5] [Reference Citation Analysis (1)] |
| 8. | Walujkar SA, Kumbhare SV, Marathe NP, Patangia DV, Lawate PS, Bharadwaj RS, Shouche YS. Molecular profiling of mucosal tissue associated microbiota in patients manifesting acute exacerbations and remission stage of ulcerative colitis. World J Microbiol Biotechnol. 2018;34:76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 9. | Takeshita K, Mizuno S, Mikami Y, Sujino T, Saigusa K, Matsuoka K, Naganuma M, Sato T, Takada T, Tsuji H, Kushiro A, Nomoto K, Kanai T. A Single Species of Clostridium Subcluster XIVa Decreased in Ulcerative Colitis Patients. Inflamm Bowel Dis. 2016;22:2802-2810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 126] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 10. | Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1756] [Cited by in RCA: 2122] [Article Influence: 151.6] [Reference Citation Analysis (0)] |
| 11. | Takaishi H, Matsuki T, Nakazawa A, Takada T, Kado S, Asahara T, Kamada N, Sakuraba A, Yajima T, Higuchi H, Inoue N, Ogata H, Iwao Y, Nomoto K, Tanaka R, Hibi T. Imbalance in intestinal microflora constitution could be involved in the pathogenesis of inflammatory bowel disease. Int J Med Microbiol. 2008;298:463-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 252] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 12. | Walker AW, Sanderson JD, Churcher C, Parkes GC, Hudspith BN, Rayment N, Brostoff J, Parkhill J, Dougan G, Petrovska L. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011;11:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 470] [Cited by in RCA: 547] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 13. | Shah R, Cope JL, Nagy-Szakal D, Dowd S, Versalovic J, Hollister EB, Kellermayer R. Composition and function of the pediatric colonic mucosal microbiome in untreated patients with ulcerative colitis. Gut Microbes. 2016;7:384-396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 91] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 14. | Rehman A, Rausch P, Wang J, Skieceviciene J, Kiudelis G, Bhagalia K, Amarapurkar D, Kupcinskas L, Schreiber S, Rosenstiel P, Baines JF, Ott S. Geographical patterns of the standing and active human gut microbiome in health and IBD. Gut. 2016;65:238-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 137] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 15. | Maukonen J, Kolho KL, Paasela M, Honkanen J, Klemetti P, Vaarala O, Saarela M. Altered Fecal Microbiota in Paediatric Inflammatory Bowel Disease. J Crohns Colitis. 2015;9:1088-1095. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 88] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 16. | Forbes JD, Van Domselaar G, Bernstein CN. Microbiome Survey of the Inflamed and Noninflamed Gut at Different Compartments Within the Gastrointestinal Tract of Inflammatory Bowel Disease Patients. Inflamm Bowel Dis. 2016;22:817-825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 109] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 17. | Mar JS, LaMere BJ, Lin DL, Levan S, Nazareth M, Mahadevan U, Lynch SV. Disease Severity and Immune Activity Relate to Distinct Interkingdom Gut Microbiome States in Ethnically Distinct Ulcerative Colitis Patients. mBio. 2016;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 79] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 18. | Magnusson MK, Strid H, Isaksson S, Simrén M, Öhman L. The Mucosal Antibacterial Response Profile and Fecal Microbiota Composition Are Linked to the Disease Course in Patients with Newly Diagnosed Ulcerative Colitis. Inflamm Bowel Dis. 2017;23:956-966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 19. | Zhang M, Liu B, Zhang Y, Wei H, Lei Y, Zhao L. Structural shifts of mucosa-associated lactobacilli and Clostridium leptum subgroup in patients with ulcerative colitis. J Clin Microbiol. 2007;45:496-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 64] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 20. | Fyderek K, Strus M, Kowalska-Duplaga K, Gosiewski T, Wedrychowicz A, Jedynak-Wasowicz U, Sładek M, Pieczarkowski S, Adamski P, Kochan P, Heczko PB. Mucosal bacterial microflora and mucus layer thickness in adolescents with inflammatory bowel disease. World J Gastroenterol. 2009;15:5287-5294. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 107] [Cited by in RCA: 112] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 21. | Schroeder KW, Tremaine WJ, Ilstrup DM. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study. N Engl J Med. 1987;317:1625-1629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1958] [Cited by in RCA: 2326] [Article Influence: 59.6] [Reference Citation Analysis (0)] |
| 22. | Satsangi J, Silverberg MS, Vermeire S, Colombel JF. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut. 2006;55:749-753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1970] [Cited by in RCA: 2445] [Article Influence: 122.3] [Reference Citation Analysis (2)] |
| 23. | Munyaka PM, Eissa N, Bernstein CN, Khafipour E, Ghia JE. Antepartum Antibiotic Treatment Increases Offspring Susceptibility to Experimental Colitis: A Role of the Gut Microbiota. PLoS One. 2015;10:e0142536. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 119] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 24. | White TJ BT, Lee SB. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications. Salt Lake City: Academic Press. 1990: 315–322. |
| 25. | Shobar RM, Velineni S, Keshavarzian A, Swanson G, DeMeo MT, Melson JE, Losurdo J, Engen PA, Sun Y, Koenig L, Mutlu EA. The Effects of Bowel Preparation on Microbiota-Related Metrics Differ in Health and in Inflammatory Bowel Disease and for the Mucosal and Luminal Microbiota Compartments. Clin Transl Gastroenterol. 2016;7:e143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 86] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 26. | Bajer L, Kverka M, Kostovcik M, Macinga P, Dvorak J, Stehlikova Z, Brezina J, Wohl P, Spicak J, Drastich P. Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J Gastroenterol. 2017;23:4548-4558. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 215] [Cited by in RCA: 279] [Article Influence: 31.0] [Reference Citation Analysis (4)] |
| 27. | Chen GL, Zhang Y, Wang WY, Ji XL, Meng F, Xu PS, Yang NM, Ye FQ, Bo XC. Partners of patients with ulcerative colitis exhibit a biologically relevant dysbiosis in fecal microbial metacommunities. World J Gastroenterol. 2017;23:4624-4631. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 20] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 28. | Duranti S, Gaiani F, Mancabelli L, Milani C, Grandi A, Bolchi A, Santoni A, Lugli GA, Ferrario C, Mangifesta M, Viappiani A, Bertoni S, Vivo V, Serafini F, Barbaro MR, Fugazza A, Barbara G, Gioiosa L, Palanza P, Cantoni AM, de'Angelis GL, Barocelli E, de'Angelis N, van Sinderen D, Ventura M, Turroni F. Elucidating the gut microbiome of ulcerative colitis: bifidobacteria as novel microbial biomarkers. FEMS Microbiol Ecol. 2016;92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 122] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 29. | Walujkar SA, Dhotre DP, Marathe NP, Lawate PS, Bharadwaj RS, Shouche YS. Characterization of bacterial community shift in human Ulcerative Colitis patients revealed by Illumina based 16S rRNA gene amplicon sequencing. Gut Pathog. 2014;6:22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 85] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 30. | Plöger S, Stumpff F, Penner GB, Schulzke JD, Gäbel G, Martens H, Shen Z, Günzel D, Aschenbach JR. Microbial butyrate and its role for barrier function in the gastrointestinal tract. Ann N Y Acad Sci. 2012;1258:52-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 333] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 31. | Walker A, Pfitzner B, Harir M, Schaubeck M, Calasan J, Heinzmann SS, Turaev D, Rattei T, Endesfelder D, Castell WZ, Haller D, Schmid M, Hartmann A, Schmitt-Kopplin P. Sulfonolipids as novel metabolite markers of Alistipes and Odoribacter affected by high-fat diets. Sci Rep. 2017;7:11047. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 96] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 32. | Vega-Magaña N, Delgado-Rizo V, García-Benavides L, Del Toro-Arreola S, Segura-Ortega J, Morales ASMZ, Zepeda-Nuño JS, Escarra-Senmarti M, Gutiérrez-Franco J, Haramati J, Bueno-Topete MR. Bacterial Translocation Is Linked to Increased Intestinal IFN-γ, IL-4, IL-17, and mucin-2 in Cholestatic Rats. Ann Hepatol. 2018;17:318-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 33. | Larsen JM. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology. 2017;151:363-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 472] [Cited by in RCA: 995] [Article Influence: 110.6] [Reference Citation Analysis (0)] |
| 34. | Bashir A, Miskeen AY, Hazari YM, Asrafuzzaman S, Fazili KM. Fusobacterium nucleatum, inflammation, and immunity: the fire within human gut. Tumour Biol. 2016;37:2805-2810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 101] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 35. | Tuddenham S, Sears CL. The intestinal microbiome and health. Curr Opin Infect Dis. 2015;28:464-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 129] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 36. | Wu J, Zhang C, Xu S, Xiang C, Wang R, Yang D, Lu B, Shi L, Tong R, Teng Y, Dong W, Zhang J. Fecal Microbiome Alteration May Be a Potential Marker for Gastric Cancer. Dis Markers. 2020;2020:3461315. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 37. | Devkota S. The gut microbiome during acute lifestyle transition. Nat Med. 2020;26:1013-1015. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/Licenses/by-nc/4.0/
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Zhang J S-Editor: Fan JR L-Editor: Wang TQ P-Editor: Liu JH