Published online Apr 14, 2021. doi: 10.3748/wjg.v27.i14.1369
Peer-review started: January 21, 2021
First decision: February 10, 2021
Revised: February 23, 2021
Accepted: March 17, 2021
Article in press: March 17, 2021
Published online: April 14, 2021
Processing time: 78 Days and 2 Hours
Infection with the hepatitis B virus (HBV) is still a major global health threat as 250 million people worldwide continue to be chronically infected with the virus. While patients may be treated with nucleoside/nucleotide analogues, this only suppresses HBV titre to sub-detection levels without eliminating the persistent HBV covalently closed circular DNA (cccDNA) genome. As a result, HBV infection cannot be cured, and the virus reactivates when conditions are favorable. Interferons (IFNs) are cytokines known to induce powerful antiviral mechanisms that clear viruses from infected cells. They have been shown to induce cccDNA clearance, but their use in the treatment of HBV infection is limited as HBV-targeting immune cells are exhausted and HBV has evolved multiple mechanisms to evade and suppress IFN signalling. Thus, to fully utilize IFN-mediated intracellular mechanisms to effectively eliminate HBV, instead of direct IFN administration, novel strategies to sustain IFN-mediated anti-cccDNA and antiviral mechanisms need to be developed. This review will consolidate what is known about how IFNs act to achieve its intracellular antiviral effects and highlight the critical interferon-stimulated gene targets and effector mechanisms with potent anti-cccDNA functions. These include cccDNA degradation by APOBECs and cccDNA silencing and transcription repression by epigenetic modifications. In addition, the mechanisms that HBV employs to disrupt IFN signalling will be discussed. Drugs that have been developed or are in the pipeline for components of the IFN signalling pathway and HBV targets that detract IFN signalling mechanisms will also be identified and discussed for utility in the treatment of HBV infections. Together, these will provide useful insights into design strategies that specifically target cccDNA for the eradication of HBV.
Core Tip: Hepatitis B virus (HBV) infection remains an incurable disease affecting millions worldwide. Treatment with interferons (IFNs) can eliminate the virus by clearing its persistent genome, covalently closed circular DNA (cccDNA), from infected cells. However, its clinical efficacy is limited as HBV proteins antagonize IFN signalling. Other current therapeutics do not target cccDNA thus cannot eliminate HBV. Therefore, new drugs based on the knowledge of how IFNs cause cccDNA degradation and silencing, as well as insights into how HBV antagonizes IFN-mediated mechanisms are needed. This review summarizes what is known about these processes and highlights drugs and developing therapeutics targeted against them for HBV eradication.
- Citation: Goh ZY, Ren EC, Ko HL. Intracellular interferon signalling pathways as potential regulators of covalently closed circular DNA in the treatment of chronic hepatitis B. World J Gastroenterol 2021; 27(14): 1369-1391
- URL: https://www.wjgnet.com/1007-9327/full/v27/i14/1369.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i14.1369
Chronic hepatitis B (CHB) is a major global health problem affecting about 250 million people worldwide, resulting in 800000 deaths per year[1]. It is the main etiological factor and cause of mortality for liver cirrhosis and hepatocellular carcinoma (HCC). Despite the availability of a prophylactic vaccine and antiviral therapies, the very high numbers of chronic carriers for the causative hepatitis B virus (HBV) indicates that current treatment regimens are inadequate in eliminating HBV. The primary reason for this is due to the stability and persistence of its genomic DNA[2,3], the covalently closed circular DNA (cccDNA), even under long-term therapy. This pivotal viral DNA template is solely responsible for generating all HBV transcripts[4] and viral proteins. Therefore, sub-detection levels of continually present cccDNA in infected cells and tissues act as template reservoirs for HBV to reactivate and persist long after achieving the treatment endpoint of expressing sub-detection levels of HBV antigens in patient sera. Given its importance in maintaining chronicity of HBV infection, experts in the field have come to a consensus that a curative regime can only be achieved through cccDNA elimination or permanently silencing cccDNA[5,6].
Of the 2 treatment options available, oral administration of nucleoside/nucleotide analogues (NAs) that target the HBV polymerase/reverse transcriptase (pol/RT) are better tolerated in patients to achieve reduced HBV titres[7]. However, NAs do not directly target cccDNA and therefore HBV reactivation persists. In contrast, the less tolerated interferon (IFN) treatment has been shown to be directly efficacious in promoting cccDNA clearance or epigenetically silencing cccDNA[8-10]. This is not surprising, as IFNs are a group of cytokines released from host cells as natural defence against external stimuli such as viral pathogens[11]. IFNs bind to their cognate receptors to elicit an intracellular signalling cascade that activates a set of IFN-stimulated genes (ISGs) with antiviral, immunomodulatory and anti-proliferative functions[12]. As such, IFNs are often used to treat viral infections against a range of viruses including HBV[13], hepatitis C virus[14], and West Nile virus[15]. Multiple lines of evidence have shown that IFN treatment effectively inhibits cccDNA function and eliminates cccDNA with great potency in vitro and in selected CHB patients[16,17]. However, the underlying mechanisms of its antiviral functions, especially on cccDNA are poorly understood. This is further compounded by the myriad of adverse effects associated with IFN treatments, such as neuropsychiatric disorders and neutropenia[18,19], greatly limiting its use in HBV therapy. Moreover, HBV has multiple mechanisms that counteract IFN signalling, dampening the antiviral effect of IFN treatment[20,21] to result in poor antiviral response in some patients[22,23]. To overcome and enhance the sub-optimal antiviral response from direct IFN administration, a better understanding of the precise mechanisms of how IFNs target cccDNA and how HBV can overcome these is necessary. By targeting host factors and HBV products that modulate intracellular IFN signalling and its antiviral and anti-cccDNA effectors, curative therapies with fewer undesirable pleiotropic adverse effects can be developed. This review will highlight these mechanisms and targets for which therapeutic agents can be and have been developed.
cccDNA is a stable non-integrated mini chromosome formed in the nucleus of HBV-infected cells[24]. HBV enters cells by engaging the sodium taurocholate cotransporting polypeptide (NTCP) receptor[25] and utilizing epidermal growth factor receptor for internalization[26]. Following cell entry, HBV releases its core nucleocapsid into the cytoplasm while the genomic material carried within, the partially double-stranded relaxed circular DNA (rcDNA) is transported into the nucleus where it is converted into cccDNA through a series of biochemical steps involving multiple host proteins[27-29]. cccDNA contains 4 overlapping open reading frames from which all HBV transcripts are transcribed. These include the 0.7 kb mRNA for the HBx protein, 2.4 kb and 2.1 kb mRNAs that encode three different forms (L, M, and S) of envelope surface proteins (HBs), 3.5 kb pre-Core mRNA that codes for the p22 pre-core protein, a precursor for HBe and another multi-functional 3.5 kb pre-genomic RNA (pgRNA) that is both the template for rcDNA synthesis and also encodes components of the core particle, the core protein (HBc) which forms the viral capsid and the HBV pol/RT[30]. pgRNA and pol/RT are encapsidated by HBc to form the core particle, where reverse transcription takes place within to form the full-length (-) strand of rcDNA, followed by the incomplete synthesis of (+) strand to generate the partially double-stranded rcDNA. This newly synthesized rcDNA can then be recycled into the nucleus to generate more cccDNA, maintaining nuclear cccDNA pool and contributing to cccDNA persistence. Alternatively, core particles can be enveloped for secretion to generate progeny virions[31]. Since cccDNA plays a central role in this replicative cycle, it is clear to see that HBV would cease to replicate or persist when cccDNA is eliminated.
cccDNA exists in low copies and persists long after antiviral treatment, accounting for HBV reactivation after cessation of treatment[32]. cccDNA is found in every phase of the natural course of HBV infection, even in patients who underwent HBs seroconversion to produce protective anti-HBs antibodies after effective antiviral treatment. Seroconversion or the loss of HBs is an important end goal of HBV therapy as it is associated with positive long-term clinical outcomes such as improvement in liver function and reducing the risk of HCC[33]. Surprisingly, there is currently no international standard for HBV cccDNA quantitation, and a universally endorsed HBV cccDNA assay is also lacking. Thus, the kinetics and amount of cccDNA in infected cells are not clearly defined as the cccDNA copy numbers reported merely reflect the assay used in a publication and does not facilitate comparison between studies. Cell culture models suggest that cccDNA persists up to 40 d in infected HepG2 cells, at up to 12.5 copies per infected cell[31]. Studies in human liver biopsies show that the copies of cccDNA per cell varies greatly by >5000-fold, from very low copies of 0.03 copies per cell to very high levels of 173.1 copies per cell, and that this is correlated with HBV reactivation status[3]. Consistent with the correlation between high cccDNA copies and CHB status, HBe-positive patients have a higher level of cccDNA compared to HBe-negative patients. Moreover, very low cccDNA copies of 1.5 copies per infected hepatocyte is sufficient to cause virus persistence[34]. High cccDNA levels has also been shown to be associated with increased risk of liver inflammation[35]. Collectively, high cccDNA levels is associated with greater HBV titres, increased risk of HBV reactivation and higher risk of developing HBV-associated liver diseases, emphasizing the need for cccDNA elimination to achieve a complete cure from HBV infection.
The standard of care for hepatitis B is based on two therapeutic strategies: the use of NAs and IFN-α or its pegylated form (PEG-IFN)[36,37]. To date, six NAs have been approved for treatment, including lamivudine, adefovir, entecavir, telbivudine, tenofovir, and tenofovir alafenamide. As NAs are taken orally, they are easy to administer hence promote patient compliance. NAs suppress viral replication by targeting HBV pol/RT activity to disrupt rcDNA synthesis[38]. By suppressing HBV load, NAs can alleviate HBV-associated liver diseases, regress fibrosis[39] and reduce the risk of developing HCC[40]. However, NAs cannot cure HBV infection as loss of virological markers such as HBs is rarely achieved, and seroconversion rates are negligible (Table 1). As a result, HBV reactivation rate is high, with >50% patients showing flares in virological markers (e.g. HBV DNA) and biochemical markers for liver damage [e.g. alanine aminotransferase (ALT)]. This is often accompanied with irreversible liver decompensation, resulting in death even when re-introduced to lamivudine[41]. Thus, to avoid HBV reactivation, CHB patients are often put on long-term (often >10 years) or even life-long NA therapy.
| Ref. | Treatment | % loss of HBV markers | % seroconvert | % normal ALT | % HBV reactivation | |||||
| n | Schedule | HBs | HBe | rcDNA | cccDNA | α-HBs | α-HBe | |||
| Reijnders et al[131] | 132 | NA: 16-43 mo | 3 | 42 | - | - | - | 35 | - | 56 |
| Song et al[132] | 98 | NA: 6-22 mo | - | 35 | - | - | - | 35 | - | 49 |
| Jeng et al[133] | 691 | NA: 1-8 yr | 6 | n.a. | n.a. | - | 4 | n.a. | - | 79 |
| Liem et al[134] | 45 | NA: ≥ 1 yr | 2 | n.a. | - | - | - | - | - | 71 |
| 22 | NA: ≥ 2 yr | 4 | n.a. | - | - | - | - | - | 18 | |
| Marcellin et al[135] | 181 | NA: 48 wk | 0 | n.a. | 29 | - | 0 | - | 44 | - |
| 177 | PEG-IFN: 48 wk | 4 | n.a. | 43 | - | 3 | - | 59 | - | |
| Lau et al[136] | 272 | NA: 48 wk | 0 | 21 | 22 | - | 0 | 19 | 28 | - |
| 271 | PEG-IFN: 48 wk | 3 | 34 | 32 | - | 3 | 32 | 41 | - | |
| van Zonneveld et al[137] | 165 | IFN: 16 wk | 23 | 33 | 43 | - | - | - | 62 | 13 |
| Niederau et al[138] | 103 | IFN: 4-6 mo | 10 | 51 | 51 | - | - | 51 | 50 | - |
| 53 | Untreated | 0 | 13 | 9 | - | - | - | 9 | - | |
| Liu et al[139] | 38 | IFN: 48 wk | - | - | - | 47 | - | - | Low | - |
| 38 | PEG-IFN: 48 wk | - | - | - | 63 | - | - | High | - | |
However, long-term administration of NAs leads to drug resistance, with nearly 65% of CHB patients becoming resistant when treated with lamivudine for 5 years[42]. This is because NAs target the error-prone HBV pol/RT, generating progeny virions that may carry escape mutations such as rtM204V/I, rtL180M, rtA181T/V, or rtL80V/I in the pol/RT protein sequence, preventing the mutant pol/RT to incorporate NAs into the nascent rcDNA hence escape rcDNA chain termination. As a result, prolonged treatment with NAs allows these “fitter” escape mutants to accumulate, generating drug resistant HBV strains[43,44]. As such, new generations of NAs need to be designed to tackle the problem of HBV mutation. Entecavir and tenofovir are such new generation NAs that are currently favoured over first-generation lamivudine and adefovir due to their higher potency and lower occurrence of drug resistance[45]. Only time will tell if the lower drug resistance rates indeed hold true for these newer NAs. It is more important to note that, NAs do not target cccDNA as the established cccDNA pool does not require HBV pol/RT for maintenance. Therefore, NAs cannot cure HBV infection.
IFN-α and PEG-IFN are immunomodulators that augment cell-mediated immunity, part of which includes intracellular antiviral activities that can be executed without the aid of immune cells[46]. The use of PEG-IFN has superseded standard IFN-α as pegylation improves IFN-α half-life, requiring less frequent dosing[47]. More importantly, PEG-IFN-α is more effective in reducing cccDNA levels and also leads to greater rates of ALT normalization with lower HBV reactivation rates (Table 1). When compared to NAs, treatment with IFN and PEG-IFN leads to higher rates of HBe and HBs seroconversion with greater reduction in HBV markers indicative of lower HBV replication rates. Of note, HBs seroconversion is rarely achieved with the use of NAs. Recent clinical studies[48,49] have also confirmed that switching from NA therapy to IFN therapy sustains more significant HBe and HBs losses for longer periods, demonstrating the potency of IFNs in the suppression of HBV replication. However, the use of IFNs in the clinical setting is limited due to the need for high dosage and unpredictably variable patient response, which depends on the status of immune cells, HBV titre and type of HBV. It is also less tolerated in patients due to pleiotropic off-target effects from IFN signalling.
At the tissue level, immune tolerance from chronic infection significantly reduces clinical efficacy of IFN treatment. The extracellular arm of the IFN-mediated antiviral response depends on the activation of HBV-specific CD4+ helper T-cells and CD8+ cytotoxic T-cells to produce cytokines such as IFN-γ and tumour necrosis factor-alpha (TNF-α) that lead to the elimination of HBV-infected cells[50]. However, constant exposure to HBV leads to T-cell exhaustion and an immunotolerant environment[51]. While IFN-γ also induces intracellular antiviral properties, its continued elevation upregulates programmed death-1 (PD-1) immune checkpoint protein on T-cells and also induces its ligand PD-ligand 1 on hepatocytes, leading to immune tolerance as HBV-specific T-cells fail to act on infected cells[52,53]. IFN-γ also promotes the secretion chemokines from hepatic macrophages that retain CD4+ T-cells in the liver and induce apoptosis of HBV-specific T-cells, further contributing to HBV evasion of immune clearance. Many other mechanisms of how HBV-specific immune cells’ antiviral activities have been augmented in chronic HBV infection have been documented and reviewed[54-56], together showing very clearly that IFN treatment alone cannot induce effective clearance of HBV, allowing HBV to persist. Indeed, strategies aimed at restoring extracellular anti-HBV immunity in CHB patients such as with anti-PD-1 therapeutics[57] and adoptive T-cell therapy[58] are being investigated. Since immune tolerance in CHB patients render immune cells non-responsive to IFN therapy, the efficacy of IFN therapy lies heavily on the intracellular arm of IFN-mediated immunity brought about by induction of intracellular antiviral proteins from IFN signaling.
IFNs induce the production of antiviral proteins that specifically disrupt HBV replication and degrade cccDNA. However, HBV has devised multiple strategies that antagonize IFN signalling. These strategies are so effective that even when coupled with NAs, the inability of NAs to reduce the amount of HBV products allows them to continue disrupting IFN signalling, rendering combination therapy efficacious only in selected patients[59,60] but redundant in others[61]. Multiple factors including type of combination therapy, HBV genotype and level of HBV replication greatly affect IFN treatment efficacy[62]. Some studies suggest that sequential NA and IFN therapy is more effective than simultaneous combination therapy[63], a phenomenon which warrants further confirmatory clinical investigation to enhance clinical response rates. HBV itself also significantly affects patient response to IFN therapy, as treatment outcomes are more efficacious in patients carrying the genotype A virus than the genotype D virus[64], and patients are also thrice more responsive to IFN treatment if they carry the genotype B virus than genotype C virus[65]. As further proof to the extent in which HBV alters patient sensitivity to IFN treatment, 30.4% of patients with low end-of-treatment HBs levels (< 10 IU/mL) achieve HBs clearance in a 5-year follow-up, in stark contrast to < 10% of patients achieving HBs clearance[66] when end-of-treatment HBs levels are ≥ 10 IU/mL. This is further supported by the association of greater PEG-IFN response in patients with HBV DNA levels of < 9 Log10 copies/mL sera[67]. Clearly, the full potential of IFN therapy has yet to be harnessed and directed towards HBV and its cccDNA. To achieve this, a strong understanding of how IFNs target cccDNA specifically and how HBV overcomes such mechanisms is necessary. New therapeutic agents and strategies can then be developed to prevent HBV from interfering with IFN signalling and enhance the anti-HBV and anti-cccDNA activities of IFNs to bring about HBV elimination.
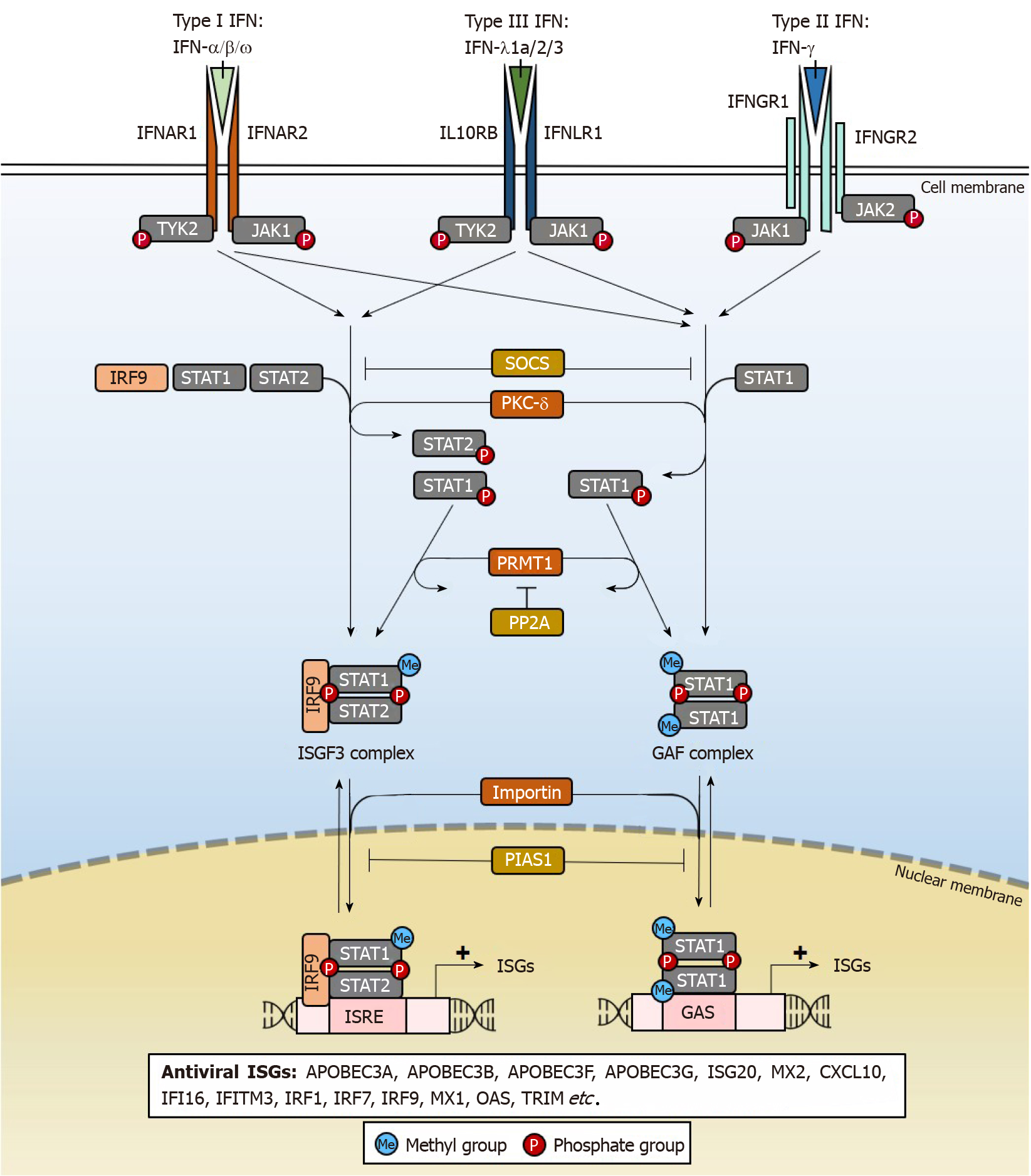
IFNs are key mediators of immunity, comprising a group of cytokines with antiviral properties against a wide range of pathogens. There are 3 types of IFNs (Type I, II, III) (Figure 1) based on the distinct receptors used for signal transduction[68]. Amongst them, only type I IFN-α2a and IFN-α2b are used clinically in the treatment of HBV infections[69,70], although the mechanisms underlying its clinical efficacy remain poorly understood. IFNs from all 3 types have been shown to possess anti-HBV properties (Table 2), but many have poorer patient tolerability than IFN-α. IFNs also differ in anti-HBV potency, but the underlying reasons for such differences are not clear. At high doses, IFNs non-cytolytically purge or silence cccDNA from infected hepatocytes.
| Types of Interferon | Effect on HBV | Remarks | Antiviral ISGs | Ref. | ||||
| cccDNA | rcDNA | HBs | HBe | |||||
| I | IFN-α2a/PEG-IFN-α2a | ↓ | ↓ | ↓ | ↓ | Clinical anti-HBV agent | APOBEC3A1, APOBEC3F1, APOBEC3G1, STAT11, ISG201, TRIM38, MX1, etc. | [140-142] |
| IFN-α2b/PEG-IFN-α2b | ↓ | ↓ | ↓ | ↓ | Clinical anti-HBV agent | IFITM1, IFITM3, TRIM14, RNASEL etc. | [143-145] | |
| IFN-α14 | ↓ | - | ↓ | ↓ | Activates IFN-α and IFN-γ signalling | GBP4, GBP5 | [146] | |
| IFN-β/PEG-IFN-β | ↓ | ↓ | ↓ | ↓ | - | MX1, CXCL10 | [147] | |
| IFN-ω | - | ↓ | ↓ | ↓ | - | IRF1, IRF9, ISG15, OAS | [148-150] | |
| II | IFN-γ | - | - | - | ↓ | - | APOBEC3G1, OAS, IDO | [151-153] |
| III | IFN-λ1a | - | ↓ | ↓ | ↓ | Increased HBe seroconversion | PKR, OAS | [154,155] |
| IFN-λ2 | ↓ | ↓ | - | ↓ | - | APOBEC3A1, APOBEC3B1, APOBEC3G1, MX1, OAS | [156] | |
| IFN-λ3 | - | ↓ | ↓ | ↓ | Increased JAK/STAT signalling | APOBEC3G1, IRF91, IRF7, MX1, OAS, ISG15 etc. | [157-159] | |
The antiviral effects of IFNs are achieved through ISGs[46,68], which are induced through the IFN signalling pathway that activates janus kinase/signal transducers and activators of transcription (JAK/STAT) signalling (Figure 1). After IFNs engage their specific IFN receptors, tyrosine kinase 2 (TYK2) transduces the activating signal to result in phosphorylation of STAT1 and STAT2 transcription factors by protein kinase C-delta (PKC-δ) necessary for nuclear translocation. The heterodimer interacts with IFN regulatory factor 9 (IRF9) to form the ISG factor 3 (ISGF3) transcription factor complex which translocates into the nucleus by importin-dependent mechanisms. ISGF3 can then induce the expression of antiviral ISGs through IFN-stimulated response elements (ISRE) cis regulatory elements with the consensus sequence “AGTTTCNNTTTCN” in ISG gene promoters. Alternatively, phosphorylated STAT1 may homodimerize to form the IFN-gamma-activated factor (GAF) complex that binds to another cis-element, the IFN-gamma activated site with the “TTCN2-4GAA” conserved sequence motif and induce ISG expression in the absence of IRF9[71]. As will be further elaborated, each ISG functions differently to bring about the destruction or silencing of cccDNA and other antiviral effects, accounting for pleiotropic effects of IFN treatment. There are a variety of IFN subtypes, each targeting the activation of different sets of ISGs with antiviral properties that may suppress HBV replication (Table 2). By specifically activating these ISGs relevant for cccDNA degradation or silencing, new drugs can be developed that specifically eliminate HBV. In doing so, adverse effects of IFNs that are not well-tolerated in patients may be eliminated as the activation of ISGs that are irrelevant for suppressing HBV replication are by-passed.
As JAK/STAT signalling is critical for the expression of ISGs, it is tightly regulated by several proteins[72]. Suppressor of cytokine signalling (SOCS) 2 and SOCS3 inhibit the IFN signalling cascade by preventing TYK2 activation and hence STAT1 phosphorylation. STAT1 methylation by protein arginine methyltransferase 1 (PRMT1) prevents its binding to the inhibitor protein inhibitor of activated STAT1 (PIAS1), where PIAS1 functions to sequester STAT1 away from its DNA-binding motifs[73]. The methylation of STAT1 is in turn negatively regulated by protein phosphatase 2A (PP2A), which inhibits PRMT1 activity[74]. These factors can be targeted by HBV to disrupt and dampen the IFN signalling and will be discussed later.
Many of the IFN subtypes with anti-HBV properties induce APOBEC family genes[75], which are cytidine deaminases that perform RNA editing[76]. They have also been well-documented to act directly on cccDNA to result in degradation. IFN-α, which is used in clinical therapy of CHB, has been shown to reduce the expression of cccDNA, HBV RNA, HBs and HBe in primary human hepatocytes (PHH) and HepRG liver cells. The IFN-α mediated loss of cccDNA was found to last up to 15 d[8]. Induced expression of APOBECs can be seen after IFN treatment in liver biopsies of hepatitis B patients, and in HBV-infected chimpanzees[77]. The importance of APOBECs in mediating cccDNA clearance is supported by the fact that the expression of APOBECs correlates with clinical response to IFN.
Besides IFN-α and IFN-γ, other pathways induced by pro-inflammatory cytokines including TNF-α and lymphotoxin-β (LT-β) have also been shown to induce APOBECs and affect the stability of cccDNA. In agreement with its anti-cccDNA functions[78], APOBECs induced through LT-β signalling with LTβR agonist antibody was shown to clear 90% of cccDNA in HBV-infected cultured liver cells with good in vivo tolerability[8]. As the clinical dose of IFN-α used to achieve cccDNA clearance is relatively high, this suggests that inducing expression of APOBECs through LT-β signalling is an alternative means for cccDNA clearance with potentially fewer adverse effects. While its clinical efficacy and safety remain to be studied, upregulation of natural ligands for LTβR in HBV-infected liver tissues have been reported[79]. IFN-γ and TNF-α also exhibit anti-cccDNA effects to degrade cccDNA[80] levels without causing cell death in HBV-infected chimpanzees, PHH and cultured liver cells.
Following IFN receptor or LTβR activation, the upregulated IFN-α induced APOBEC3A (A3A) and LTβR activation-induced APOBEC3B (A3B) are brought to cccDNA by HBc (Figure 2). Similarly, IFN-γ induced the expression of both A3A and A3B while TNF-α induced the expression of A3B only, whereas knockdown of A3A and A3B abrogated the antiviral effects of IFN-γ and TNF-α. HBc amino acids 77-149 are crucial for this interaction, which brings the APOBECs into close proximity with cccDNA for deamination[8]. cccDNA deamination generates apurinic/apyrimidinic (AP) sites that are recognized by endonucleases[81], which ultimately degrade cccDNA. The dependence on AP sites for cccDNA degradation was confirmed by reduced intact DNA amount when DNA extracted from IFN-γ or TNF-α treated cells were digested with recombinant APE1, an AP endonuclease which specifically recognizes and cleaves AP sites[9]. Surprisingly, knockdown of APE1 in IFN-γ and TNF-α treated HBV infected cells did not show a reduction in cccDNA clearance, suggesting the redundancy of endonucleases in clearing cccDNA. While AP sites can be repaired by the host cells’ DNA repair machinery, this is not observed with IFN-mediated AP site formation in cccDNA. This is most likely due to the concurrent downregulation of the base excision repair enzymes such as thymine DNA glycosylase and Nei-like DNA glycosylase after IFN treatment[77]. These studies indicate that A3A and A3B are key proteins responsible for IFN and LTβR mediated cccDNA clearance. APOBECs also have other important roles in clearing HBV cccDNA. APOBEC3F (A3F) and APOBEC3G (A3G) were also found to have anti-HBV properties[82]. A3G for example inhibits pgRNA packaging to reduce virion formation[83]. The mechanism of action for A3F has not been well-characterized.
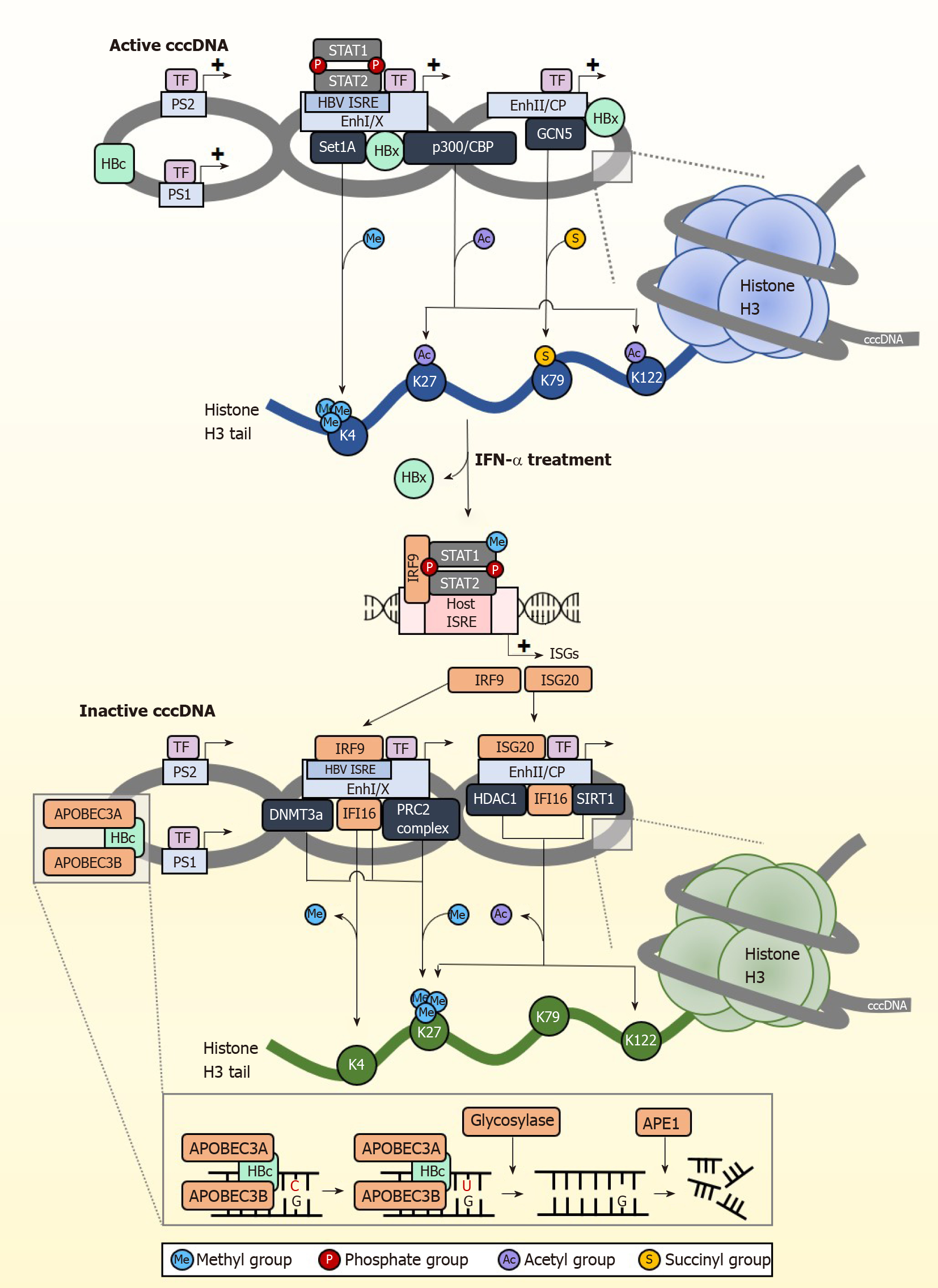
Apart from eliminating cccDNA, permanently silencing cccDNA is another strategy for the development of anti-HBV therapy. Several studies have shown that IFNs control the epigenetic silencing of cccDNA[84,85], spurring growing interest in controlling cccDNA transcriptional activity through epigenetic modifications[86]. HBV cccDNA exists as episomal chromatin wound around cellular histones that undergo post-translational modifications (PTMs)[87], altering cccDNA chromatin compaction hence accessibility to transcription regulators (Figure 2). Some of the histone marks associated with active cccDNA transcription include H3K4me3, H3K27ac, H3K122ac, and repressive PTMs include H3K27me3. Of note, the distribution of these histone marks varies across different types of HBV-producing samples[10]. In the HepG2-NTCP cell model, histone PTM enrichment is mainly confined to the pre-core/core promoter (CP) region whereas in PHH, they are found throughout the genome with the greatest modifications at the X and pre-S1 promoter regions. In contrast, HBV-infected liver tissues have few PTMs at the CP but accumulate them near pre-S2 and the X promoter regions. The reason and basis for this variability is at present unclear.
cccDNA transcription activity is heavily influenced by the state of epigenetic modification by host cellular factors and PTMs. The cccDNA mini chromosome is heavily modified by various activating PTMs under normal circumstances (Figure 2). H3K4 methyltransferase Set domain containing 1A (Set1A) is recruited to cccDNA promoter sites by HBx, depositing the activating PTM H3K4me3 to drive active transcription[88]. By modulating the expression of Set1A, the relative expression of H3K4me3 can be fine-tuned. IFN treatment disrupts cccDNA transcription activity by downregulating these epigenetic PTMs that support transcription. In studies using PHH, IFN-α specifically reduced trimethylation of H3K4 and acetylation of H3K27 and H3K122 on cccDNA chromatin to inhibit transcription of HBV RNA, but had negligible effect on epigenetic modification for the control promoters of ACTB and Nanog in the host genome[10].
IFNs also result in cccDNA transcriptional repression through active recruitment of complexes that confer transcription inhibitory epigenetic modifications. The repressive H3K27me3 PTM is induced by IFN-α through increased binding of polycomb repressive complex 2 (PRC2) to cccDNA[84]. The importance of H3K27me3 in inhibiting cccDNA transcription was also confirmed in a separate study showing that upregulation of DNA methyltransferase 3a hypermethylates HBV cccDNA to repress transcription[89]. IFNs have also been recently shown to inhibit succinylation of cccDNA histones, adding on to the transcriptionally repressed state brought about by hypoacetylation and/or methylation[90]. This involves the succinylation of H3K79 by GCN5 histone succinyltransferase (also known as lysine acetyltransferase 2A), which corresponds to higher HBV replication. They further showed that low levels of succinylated H3K79 from GCN5-specific knockdown resulted in significantly reduced cccDNA levels, and that expression of GCN5 and cccDNA correlate well in HBV-infected individuals. More importantly, IFN-α treatment could overcome the effects of overexpressed GCN5 to reduce the levels of succinylated cccDNA, indicating that IFNs act upstream to control GCN5 function and repress cccDNA transcription, the specific mechanism of which remains to be elucidated. Taken together, IFNs epigenetically silence cccDNA function through the recruitment of epigenetic factor complexes that add repressive PTMs or remove activating PTMs so that cccDNA enters a transcriptionally repressed chromatin structural state. Importantly, IFNs can also do so by inducing ISGs that bring about both the removal of activating PTMs and the addition of suppressive PTMs. For example, IFI16 reduces cccDNA transcription activity[91] by recruiting histone deacetylase 1 (HDAC1) and Sirtuin 1 (SIRT1) to increase repressive H3K27me3 PTMs on cccDNA, and concurrently impairs the recruitment of p300/CBP to prevent the addition of activating PTMs on cccDNA.
Interestingly, cccDNA also carries the ISRE cis-element, and this is critical for establishing IFN-mediated epigenetic changes on cccDNA. The HBV ISRE is located at the enhancer I/X promoter region[92,93], and is recognized by the ISGF3 complex and the ISGs IRF1 and IRF7. In transcriptionally active cccDNA, the HBV ISRE is bound by phosphorylated and unphosphorylated STAT1 and STAT2 transcription factors to active cccDNA (Figure 2). IFN treatment induces redistribution of the STAT proteins from the HBV ISRE towards the IFN signalling pathway, resulting in antiviral effects against HBV cccDNA by the upregulation of cccDNA-targeting ISGs. One of these ISGs is IRF9, which binds directly to the HBV ISRE element to suppress cccDNA transcription[94]. When the HBV ISRE is mutated, loss of IRF9 binding was shown to abrogate IFN-induced suppression of cccDNA transcription. This is clinically significant, as mutations in the HBV ISRE affects CHB patient response to IFN treatment[95] to render IFN treatment less effective. In addition, the HBV ISRE sequence is HBV genotype dependent, thus its sequence-dependent functionality partially accounts for differences in patient responder rates between carriers of HBV genotypes B and C[96]. Another ISG, ISG20, also directly inhibits transcription from cccDNA by direct binding to the enhancer II/CP region[97]. Higher ISG20 expression level also correlates to better response to IFN-α treatment in CHB patients[98] and viral clearance in HBV-infected chimpanzees[99]. With the withdrawal of activating transcription factors from cccDNA and binding of specific transcription repressors induced by IFN treatment, cccDNA transcription is further suppressed by epigenetic modifications of histones such as histone hypoacetylation which occurs through the recruitment of the HDAC1 and SIRT1, and hypermethylated by the PRC2 complex. Thus, in addition to epigenetic silencing of cccDNA, IFNs also suppress cccDNA transcription activity by generating IFN-induced transcription repressors specific to cccDNA.
IFNs are also known to directly inhibit cccDNA synthesis. The antiviral ISG myxovirus resistance protein 2 has been shown to reduce cccDNA formation when overexpressed[100]. Its specific knockdown abrogates the loss of cccDNA induced by IFN-α, providing confirmation for its role in IFN-α induced reduction in cccDNA levels. It has been proposed that this occurs through inhibiting cccDNA synthesis from rcDNA, as well as from downregulated HBV transcripts.
HBV is not without its defences. It generates multiple factors that dampen the IFN signalling pathway. Inter-individual variability in the balance of these against antiviral IFN-mediated mechanisms could explain why CHB patients do not respond equally well to IFN-treatment. By understanding how HBV proteins antagonize IFN action, therapeutic approaches that interrupt the anti-HBV functions of IFNs can be developed to improve the response and efficacy of IFN treatments.
HBV disrupts the IFN signalling cascade at various steps. HBx[21] and HBe[20] downregulate the expression of IFN receptors, IFNAR1 and IL10RB, disrupting the initiation of type I and type III IFN signalling (Figure 3). HBe and HBx achieve this by upregulating SOCS2 and SOCS3 respectively, suppressing the IFN signalling cascade and inhibiting TYK2 activation (Figure 1). As SOCS2 also prevents STAT1 phosphorylation, STAT1 is prevented from entering the nucleus to induce ISG expression. Since SOCS2 and SOCS3 are the most direct upstream inhibitors of the IFN signaling pathway, their increased expression significantly attenuates IFN-dependent transcription of ISGs to compromise the therapeutic efficacy of IFNs. It was found that overexpression of HBe alone leads to 6-fold reduction in phosphorylated STAT1 nuclear translocation, hence downregulating protein kinase R (PKR) and oligoadenylate synthetase (OAS) gene expression by 50%[20]. Similarly, HBx overexpression alone doubled SOCS3 expression and increased PP2A expression by 5-fold, significantly reducing PKR and OAS expression by 3-fold[101]. It is thus clear to see, that the combined effects of expressing HBx and HBe in infected cells, especially in cells with high viral titre and virological markers, will compromise the clinical efficacy of direct IFN therapy. Other HBV proteins act further down the IFN signalling cascade to prevent STAT1 nuclear translocation, further disrupting ISG expression. HBV pol/RT inhibits PKC-δ, preventing STAT1 phosphorylation and competitively binding to importin[102,103]. HBV pol/RT also significantly reduces the induction of IRF9 critical for the generation of ISGs through formation of the ISGF3 complex. p22 also binds importin[104] to prevent STAT1 nuclear translocation. In the nucleus, HBx and HBs upregulate PP2A, which inhibits PRMT1 hence reduces STAT1 methylation, enabling PIAS1 to sequester phosphorylated STAT1 away from its DNA-binding elements in vitro and in liver tissues from HBV infected patients[105]. As HBV proteins specifically target multiple regulatory elements of IFN signalling to block ISG expression, the expression of anti-cccDNA and antiviral ISGs would therefore be significantly reduced in the presence of HBV, rendering the effort to eliminate HBV by direct IFN administration futile. Therefore, to circumvent HBV mechanisms that antagonize IFN signalling, therapies aimed at targeting HBV molecules and components of the IFN-signalling pathway would greatly enhance the efficacy of IFN treatment for the elimination and suppression of cccDNA and HBV.
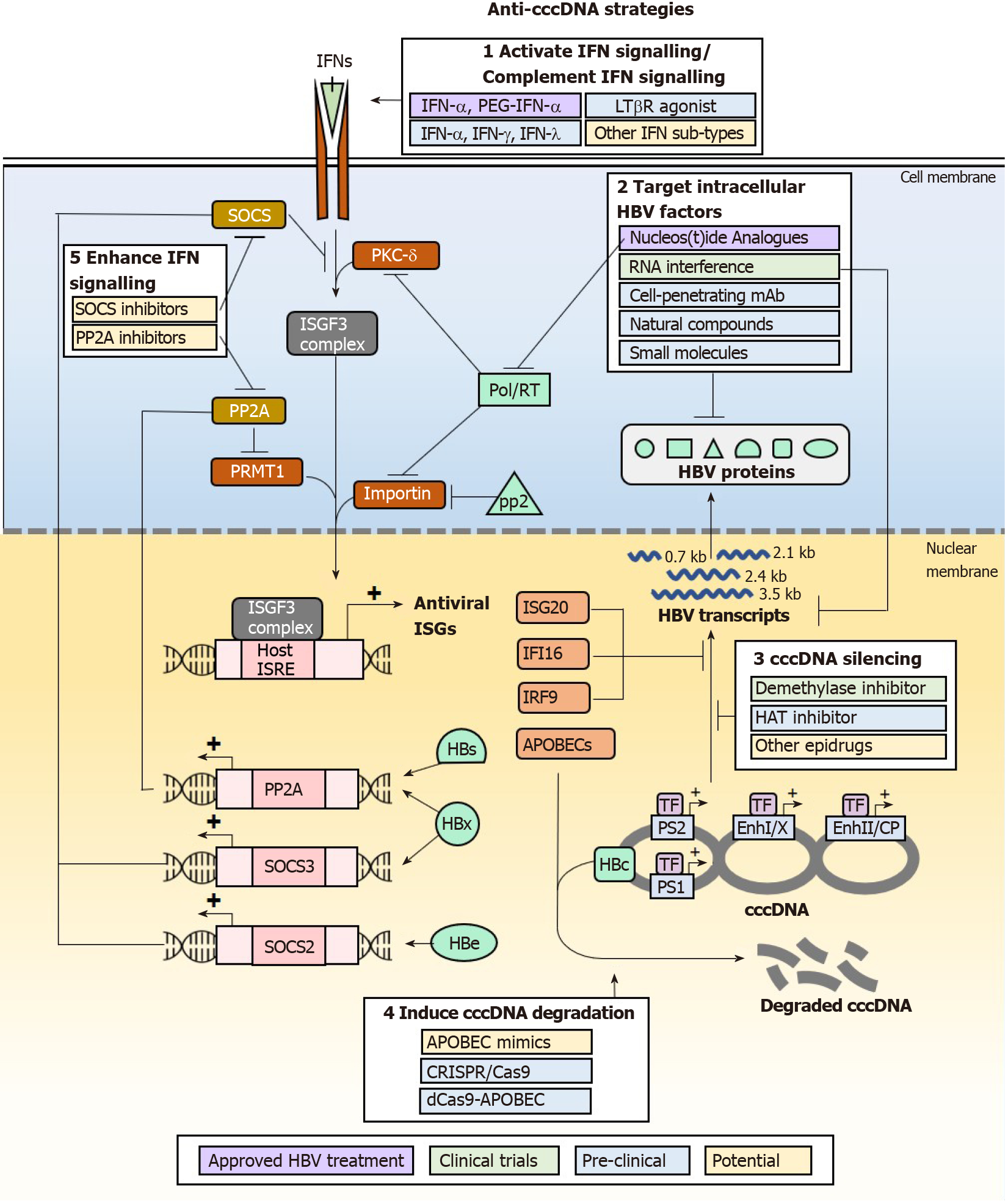
IFNs may eliminate and suppress cccDNA formation for a therapeutic cure of HBV infections, but IFN therapy has not been particularly efficacious due to the adverse effects from pleiotropic off-target effects and the existence of multiple HBV mechanisms that antagonize the IFN signalling pathway and downstream effector functions. As these greatly limit the clinical use of IFNs for treating HBV infections, alternative approaches that specifically enhance the anti-HBV effects of IFNs and suppress the antagonistic effects of HBV proteins can be developed to achieve cccDNA elimination and suppression without adverse effects from IFN signalling. Multiple classes of drugs that are in development or have been developed, that act on such targets may have the beneficial effect by downregulating HBV cccDNA (Figure 3). They act by modulating the functions of IFN signalling pathway components or HBV proteins that antagonize IFN signalling and its anti-cccDNA effector functions. Of note, as HBV disruption of IFN response is multi-factorial, combination therapy may be necessary to achieve cccDNA elimination for a true HBV cure.
Perhaps the most obvious strategy to eliminate cccDNA is to enhance and sustain the expression of APOBECs which actively result in cccDNA degradation. While APOBEC mimics are not yet available, APOBEC expression can be induced through multiple pathways including activation of IFN signalling using other IFN sub-types such as IFN-γ and IFN-λ, and alternative pathways involving TNF-α and LTβR activation. IFN-λ3 was found to be specifically upregulated in patients treated with adefovir or tenofovir, and further shown in cell culture models to be effective in reducing HBs by inducing ISGs[106]. Studies in HepaRG differentiated hepatocytes also show that IFN-β, IFN-λ1 and IFN-λ2 induce longer-lasting APOBEC expression than IFN-α2, and are just as efficient in mediating cccDNA degradation[107]. IFN-γ and TNF-α have also been shown to upregulate APOBEC expression. In particular, the utility of IFN-γ in the treatment of HBV infections can be explored as it is currently used clinically for the treatment of hepatic fibrosis[108]. An LTβR agonist antibody has already been developed and was shown to be safe in mice at lower dosing requirements than clinical IFN-α. This could be further developed and tested in clinical settings for efficacy and safety in the treatment of HBV infections.
The IFN response may also be strengthened by reducing the action of negative regulators of the IFN signalling pathway, primarily by acting on SOCS2 or SOCS3 and PP2A. This directly inhibits the degradation of IFN signaling, allowing ISGF3 and GAF complexes to form hence carry out transcription of ISGs. Thiazolidinediones such as pioglitazone and rosiglitazone are known to reduce the expression of SOCS proteins[109], and consequently would prevent the loss of IFN signalling transduction hence upregulate APOBEC expression in the presence of appropriate IFNs. They are currently used in the treatment of type II diabetes, thus may be readily re-purposed for the treatment of HBV infections to reduce cccDNA levels. The first-in-class small molecule inhibitor of PP2A, LB-100, would theoretically achieve similar effects of inducing APOBEC expression. It was found to be well-tolerated in patients with solid tumours in phase I clinical trials[110].
As cccDNA function is highly dependent on its epigenetic PTM status, “epidrugs” are also being investigated for their efficacy in silencing cccDNA function. HAT inhibitor C646, which is widely used in cancer studies, has been shown to reduce H3K27ac and H3K122ac thus silence cccDNA for reduced HBV transcription[10]. The pro-drug GS-5801 (Gilead), which specifically inhibits KDM5 (lysine demethylase 5) has been shown to suppress cccDNA transcription by removing H3K4me3[111]. Given the importance of other epigenetic modifiers such as GCN5 and p300/CBP for cccDNA transcriptional activation, and SIRT1, HDAC1 for transcriptional repression and silencing, other unexplored epidrugs that inhibit the transcription activation group or activate the transcription repression and silencing group may also be tested for efficacy in directly silencing cccDNA. While epidrugs directly silence cccDNA in vitro, their greatest challenge in utility as an anti-HBV therapeutic depends on their ability to specifically target cccDNA in infected cells while sparing host genome to avoid carcinogenesis and toxicity[112,113]. In addition, due to compromised functionality of HBV-infected livers, many drugs that have safe profiles in the treatment of non-liver diseases are contraindicated in CHB. For example, the Food and Drug Administration-approved DNMT inhibitor, 5-azacytidine commonly used in the treatment of hematologic malignancies is contraindicated in patients with advanced liver cancer to control HBV infection due to hepatotoxicity[114]. Thus, epidrugs need to be carefully evaluated for safety and suitability in the treatment of HBV infections.
To avoid drug-induced toxicity, approaches that directly target HBV products (cccDNA, RNA and proteins) are being developed. One such approach is to directly degrade cccDNA using CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats)/CRISPR-associated protein 9) DNA editing machinery, where guide RNA-directed gene editing of cccDNA specifically results in its erroneous repair by non-homologous end-joining which cleaves and/or mutates cccDNA. This has been shown to effectively reduce cccDNA copies and is followed by concomitant clearance of HBs, HBc and HBe expression in mice with humanized liver[115,116] and multiple human liver cell lines[117,118]. While the cccDNA-targeting system is being developed and improved for clinical use, a major concern for this strategy is the generation of off-target double-strand breaks in the host genome that may facilitate HBV genomic integration to cause liver cancer. A suggested solution for this is to tether impaired Cas9 to APOBECs so that cccDNA may be mutated by base-changes instead of strand-breaks[119], highlighting again the importance of APOBECs in the role of targeted cccDNA degradation.
Another approach that has potentially fewer off-target effects is to specifically destroy HBV proteins that disrupt the IFN signalling pathway. Cell-penetrating monoclonal antibodies (mAb) have been developed for such purpose. For example, the genetically engineered antibody 9D11-Tat, formed by fusing a cell-penetrating peptide on the C-terminus of the heavy chain of a mAb specific to HBx, can target HBx for proteosomal degradation[120]. Alternatively, the transcripts for HBV proteins may be targeted for degradation by small interfering RNA (siRNA) duplexes[121]. siRNA therapeutics are already in clinical trials and currently assessed for efficacy and safety. siRNA candidates ARC-520, JNJ-3989 (Arrowhead Pharmaceuticals), and GSK3389404 (Ionis/GlaxoSmithKline)[122-124] have displayed promising efficacy in clearing viral transcripts to reduce HBs and HBV DNA production. Small molecules may also achieve the same effect by reducing HBV transcripts. Of note, the small molecule RG7834 was found to be even more potent than using the current clinical therapeutic agent entecavir in reducing HBs levels in cell culture model and HBV-infected human liver chimeric uPA-SCID mice[125,126]. With increased interest in natural compounds and their potential antiviral functions, curcumin was shown to reduce the expression of HBe and HBs[127] which antagonizes the IFN signalling cascade. Another natural compound, Dicoumarol, can enhance HBx degradation. It inhibits NQO1 to de-stabilize its interaction with HBx, rendering it susceptible to the action of proteasomes[128]. With such promising new candidate drugs for tackling HBV, combination therapy that couples IFN treatment with these HBV protein-targeting drugs have been proposed to be hopeful for achieving cccDNA elimination without giving HBV the chance to evade the IFN response.
Neither NAs nor IFNs alone can achieve effective HBV elimination in majority of HBV carriers. However, their mechanisms of action complement one another to potentially achieve viral elimination. NAs are effective in suppressing viral titre in most patients, allowing IFNs to effectively mount a cellular immune response in a less immunotolerant environment when HBV titre is decreased, and concurrently allow IFN-mediated intracellular antiviral mechanisms to effectively act against HBV and its cccDNA with less antagonistic effects from decreased HBV titre. As such, combination therapy is increasingly explored, with many experimenting the types of combinations that can be administered (Table 3). Combinations explored include NA monotherapy followed by IFN monotherapy or vice versa, periods of monotherapy followed by periods of IFN and NA co-administration, or co-therapy followed by monotherapy. Interestingly, the efficacies of combination therapy differ greatly. When NA monotherapy is switched to IFN monotherapy, higher rates of HBe and HBs seroconversion are observed together with lower relapse rates. Simultaneous administration of NAs and IFNs for more than 24 wk, followed by sustained NA treatment gives very high HBe seroconversion rate of 50%, accompanied by the loss of HBs expression in 16% patients. This is a remarkable feat considering the loss of HBs is usually less than 5% with NA or IFN monotherapy (Table 1). In contrast, multiple reports show that simultaneous administration of NAs and IFN yield conflicting results, with many studies showing little benefit from adding IFNs into the regime of NA treatment[129,130]. Further large-scale clinical studies are needed to ascertain the differences in these findings.
| Ref. | Treatment | % loss of HBV markers | % seroconvert | % normal ALT | % HBV reactivation | |||||
| Type | Schedule | HBs | HBe | rcDNA | cccDNA | α-HBs | α-HBe | |||
| Hagiwara et al[160] | Sim | Combi: 48 wk | 4 | 60 | 62 | 85 | - | 60 | 77 | 38 |
| Zhang et al[63] | Sim | Combi: 48 wk | 9 | - | 58 | - | - | 30 | 73 | - |
| Mono | IFN: 48 wk | 6 | - | 31 | - | - | 25 | 56 | - | |
| Add | IFN: 12 wk; +Combi: 36 wk; +NA: 12 wk | 16 | - | 72 | - | - | 50 | 78 | - | |
| Add | IFN: 24 wk; +Combi: 24 wk; +NA: 24 wk | 9 | - | 69 | - | - | 31 | 78 | - | |
| Huang et al[48] | Swi | NA: ≥ 2 yr +IFN: 60 wk | 33 | 91 | n.a. | - | 26 | 65 | n.a. | - |
| Mono | NA: ≥ 2 yr | 0 | 38 | n.a. | - | 0 | 22 | n.a. | - | |
| Zhou et al[49] | Swi | NA: ≥ 4 yr +IFN: 48 wk | 36 | n.a. | n.a. | - | 27 | n.a. | - | 25 |
| Mono | NA: ≥ 5 yr | 4 | n.a. | n.a. | - | 0 | n.a. | - | 58 | |
It is widely accepted that elimination or silencing of cccDNA is necessary to cure HBV infections so that no active template remains for HBV to reactivate. It is possible for IFNs to achieve this through induction of ISGs, primarily APOBECs that specifically target cccDNA for degradation. IFN signalling also results in the recruitment of epigenetic modifiers that render cccDNA in a transcriptionally repressed and silent state. However, HBV prevents these by inhibiting IFN signalling through upregulating negative regulators of IFN signal transduction such as SOCS2, and sequestration of factors that transduce IFN signalling. Thus, to achieve an anti-HBV response that targets cccDNA specifically, drugs that enhance and sustain the anti-cccDNA effects of IFN signalling and drugs that target HBV products that disrupt these IFN-mediated mechanisms need to be developed. Fortunately, many of the anti-cccDNA mechanisms are not unique to HBV infection. Drugs known to modulate these IFN-mediated pathways can be re-purposed and tested for efficacy in suppressing HBV replication and cccDNA levels. These include LTβR agonists and other cytokines that serve as alternative pathways to strengthen IFN-mediated APOBEC expression, inhibitors in the treatment of type II diabetes that act on SOCS proteins to prevent dampening of IFN signalling transduction, and epidrugs that render cccDNA transcriptionally silent. These drugs may be used together with current HBV therapies, new anti-HBV drugs in the pipelines such as cell-penetrating antibodies, cccDNA-targeting CRISPR/Cas9 systems and even natural compounds that directly target HBV proteins that disrupt IFN signalling for degradation. Hopefully, the success of these novel strategies would materialize for the eradication of HBV.
We thank Zhuo Ziyi for her assistance and comments on the figures.
| 1. | World Health Organization. Hepatitis B, 2020 [cited Feb 23, 2021]. Available from: https://www.who.int/news-room/fact-sheets/detail/hepatitis-b. |
| 2. | Abdelhamed AM, Kelley CM, Miller TG, Furman PA, Isom HC. Rebound of hepatitis B virus replication in HepG2 cells after cessation of antiviral treatment. J Virol. 2002;76:8148-8160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 3. | Chen Y, Sze J, He ML. HBV cccDNA in patients' sera as an indicator for HBV reactivation and an early signal of liver damage. World J Gastroenterol. 2004;10:82-85. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 37] [Cited by in RCA: 40] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 4. | Tuttleman JS, Pourcel C, Summers J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell. 1986;47:451-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 502] [Cited by in RCA: 535] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 5. | Alter H, Block T, Brown N, Brownstein A, Brosgart C, Chang KM, Chen PJ, Chisari FV, Cohen C, El-Serag H, Feld J, Gish R, Glenn J, Greten T, Guo H, Guo JT, Hoshida Y, Hu J, Kowdley KV, Li W, Liang J, Locarnini S, Lok AS, Mason W, McMahon B, Mehta A, Perrillo R, Revill P, Rice CM, Rinaudo J, Schinazi R, Seeger C, Shetty K, Tavis J, Zoulim F. A research agenda for curing chronic hepatitis B virus infection. Hepatology. 2018;67:1127-1131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 77] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 6. | Lucifora J, Protzer U. Attacking hepatitis B virus cccDNA--The holy grail to hepatitis B cure. J Hepatol. 2016;64:S41-S48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 150] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 7. | Dienstag JL. Benefits and risks of nucleoside analog therapy for hepatitis B. Hepatology. 2009;49:S112-S121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 89] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 8. | Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, Remouchamps C, Chou WM, Thasler WE, Hüser N, Durantel D, Liang TJ, Münk C, Heim MH, Browning JL, Dejardin E, Dandri M, Schindler M, Heikenwalder M, Protzer U. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 2014;343:1221-1228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 640] [Cited by in RCA: 764] [Article Influence: 63.7] [Reference Citation Analysis (0)] |
| 9. | Xia Y, Stadler D, Lucifora J, Reisinger F, Webb D, Hösel M, Michler T, Wisskirchen K, Cheng X, Zhang K, Chou WM, Wettengel JM, Malo A, Bohne F, Hoffmann D, Eyer F, Thimme R, Falk CS, Thasler WE, Heikenwalder M, Protzer U. Interferon-γ and Tumor Necrosis Factor-α Produced by T Cells Reduce the HBV Persistence Form, cccDNA, Without Cytolysis. Gastroenterology. 2016;150:194-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 276] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 10. | Tropberger P, Mercier A, Robinson M, Zhong W, Ganem DE, Holdorf M. Mapping of histone modifications in episomal HBV cccDNA uncovers an unusual chromatin organization amenable to epigenetic manipulation. Proc Natl Acad Sci USA. 2015;112:E5715-E5724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 194] [Article Influence: 17.6] [Reference Citation Analysis (1)] |
| 11. | Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778-809, table of contents. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2029] [Cited by in RCA: 2026] [Article Influence: 81.0] [Reference Citation Analysis (0)] |
| 12. | Maher SG, Sheikh F, Scarzello AJ, Romero-Weaver AL, Baker DP, Donnelly RP, Gamero AM. IFNalpha and IFNlambda differ in their antiproliferative effects and duration of JAK/STAT signaling activity. Cancer Biol Ther. 2008;7:1109-1115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 133] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 13. | Müller R, Baumgarten R, Markus R, Schulz M, Wittenberg H, Hintsche-Kilger B, Fengler JD, Von Wussow P, Meisel H, Klein H. Treatment of chronic hepatitis B with interferon alfa-2b. J Hepatol. 1990;11 Suppl 1:S137-S140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 14. | Palumbo E. Pegylated interferon and ribavirin treatment for hepatitis C virus infection. Ther Adv Chronic Dis. 2011;2:39-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 55] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 15. | Lewis M, Amsden JR. Successful treatment of West Nile virus infection after approximately 3 weeks into the disease course. Pharmacotherapy. 2007;27:455-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 16. | Chuaypen N, Sriprapun M, Praianantathavorn K, Payungporn S, Wisedopas N, Poovorawan Y, Tangkijvanich P. Kinetics of serum HBsAg and intrahepatic cccDNA during pegylated interferon therapy in patients with HBeAg-positive and HBeAg-negative chronic hepatitis B. J Med Virol. 2017;89:130-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 17. | Mu D, Yuan FC, Chen Y, Jiang XY, Yan L, Jiang LY, Gong JP, Zhang DZ, Ren H, Liao Y. Baseline value of intrahepatic HBV DNA over cccDNA predicts patient's response to interferon therapy. Sci Rep. 2017;7:5937. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 18. | Sleijfer S, Bannink M, Van Gool AR, Kruit WH, Stoter G. Side effects of interferon-alpha therapy. Pharm World Sci. 2005;27:423-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 211] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 19. | Hindi NN, Saleh MI. Patient characteristics associated with peglyated interferon alfa-2a induced neutropenia in chronic hepatitis C patients. Clin Exp Pharmacol Physiol. 2018;45:636-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 20. | Yu Y, Wan P, Cao Y, Zhang W, Chen J, Tan L, Wang Y, Sun Z, Zhang Q, Wan Y, Zhu Y, Liu F, Wu K, Liu Y, Wu J. Hepatitis B Virus e Antigen Activates the Suppressor of Cytokine Signaling 2 to Repress Interferon Action. Sci Rep. 2017;7:1729. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 21. | Cho IR, Oh M, Koh SS, Malilas W, Srisuttee R, Jhun BH, Pellegrini S, Fuchs SY, Chung YH. Hepatitis B virus X protein inhibits extracellular IFN-α-mediated signal transduction by downregulation of type I IFN receptor. Int J Mol Med. 2012;29:581-586. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 22. | Brook MG, McDonald JA, Karayiannis P, Caruso L, Forster G, Harris JR, Thomas HC. Randomised controlled trial of interferon alfa 2A (rbe) (Roferon-A) for the treatment of chronic hepatitis B virus (HBV) infection: factors that influence response. Gut. 1989;30:1116-1122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 76] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 23. | Heijtink RA, Janssen HL, Hop WC, Osterhaus AD, Schalm SW. Interferon-alpha therapy for chronic hepatitis B: early response related to pre-treatment changes in viral replication. J Med Virol. 2001;63:217-219. [PubMed] |
| 24. | Bock CT, Schwinn S, Locarnini S, Fyfe J, Manns MP, Trautwein C, Zentgraf H. Structural organization of the hepatitis B virus minichromosome. J Mol Biol. 2001;307:183-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 288] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 25. | Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. 2012;1:e00049. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1280] [Cited by in RCA: 1649] [Article Influence: 117.8] [Reference Citation Analysis (1)] |
| 26. | Iwamoto M, Saso W, Sugiyama R, Ishii K, Ohki M, Nagamori S, Suzuki R, Aizaki H, Ryo A, Yun JH, Park SY, Ohtani N, Muramatsu M, Iwami S, Tanaka Y, Sureau C, Wakita T, Watashi K. Epidermal growth factor receptor is a host-entry cofactor triggering hepatitis B virus internalization. Proc Natl Acad Sci USA. 2019;116:8487-8492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 191] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 27. | Cui X, McAllister R, Boregowda R, Sohn JA, Cortes Ledesma F, Caldecott KW, Seeger C, Hu J. Does Tyrosyl DNA Phosphodiesterase-2 Play a Role in Hepatitis B Virus Genome Repair? PLoS One. 2015;10:e0128401. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 69] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 28. | Qi Y, Gao Z, Xu G, Peng B, Liu C, Yan H, Yao Q, Sun G, Liu Y, Tang D, Song Z, He W, Sun Y, Guo JT, Li W. DNA Polymerase κ Is a Key Cellular Factor for the Formation of Covalently Closed Circular DNA of Hepatitis B Virus. PLoS Pathog. 2016;12:e1005893. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 168] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 29. | Long Q, Yan R, Hu J, Cai D, Mitra B, Kim ES, Marchetti A, Zhang H, Wang S, Liu Y, Huang A, Guo H. The role of host DNA ligases in hepadnavirus covalently closed circular DNA formation. PLoS Pathog. 2017;13:e1006784. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 100] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 30. | Seeger C, Mason WS. Molecular biology of hepatitis B virus infection. Virology. 2015;479-480:672-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 569] [Cited by in RCA: 655] [Article Influence: 59.5] [Reference Citation Analysis (1)] |
| 31. | Ko C, Chakraborty A, Chou WM, Hasreiter J, Wettengel JM, Stadler D, Bester R, Asen T, Zhang K, Wisskirchen K, McKeating JA, Ryu WS, Protzer U. Hepatitis B virus genome recycling and de novo secondary infection events maintain stable cccDNA levels. J Hepatol. 2018;69:1231-1241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 183] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 32. | Werle-Lapostolle B, Bowden S, Locarnini S, Wursthorn K, Petersen J, Lau G, Trepo C, Marcellin P, Goodman Z, Delaney WE 4th, Xiong S, Brosgart CL, Chen SS, Gibbs CS, Zoulim F. Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology. 2004;126:1750-1758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 691] [Cited by in RCA: 744] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 33. | Arase Y, Ikeda K, Suzuki F, Suzuki Y, Saitoh S, Kobayashi M, Akuta N, Someya T, Hosaka T, Sezaki H, Kobayashi M, Kumada H. Long-term outcome after hepatitis B surface antigen seroclearance in patients with chronic hepatitis B. Am J Med. 2006;119:71.e9-71.16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 113] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 34. | Laras A, Koskinas J, Dimou E, Kostamena A, Hadziyannis SJ. Intrahepatic levels and replicative activity of covalently closed circular hepatitis B virus DNA in chronically infected patients. Hepatology. 2006;44:694-702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 161] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 35. | Liang LB, Zhu X, Yan LB, Du LY, Liu C, Liao J, Tang H. Quantitative intrahepatic HBV cccDNA correlates with histological liver inflammation in chronic hepatitis B virus infection. Int J Infect Dis. 2016;52:77-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 36. | Cornberg M, Lok AS, Terrault NA, Zoulim F; 2019 EASL-AASLD HBV Treatment Endpoints Conference Faculty. Guidance for design and endpoints of clinical trials in chronic hepatitis B - Report from the 2019 EASL-AASLD HBV Treatment Endpoints Conference‡. J Hepatol. 2020;72:539-557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 258] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 37. | Ghany MG. Current treatment guidelines of chronic hepatitis B: The role of nucleos(t)ide analogues and peginterferon. Best Pract Res Clin Gastroenterol. 2017;31:299-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 61] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 38. | Zoulim F, Durantel D. Antiviral therapies and prospects for a cure of chronic hepatitis B. Cold Spring Harb Perspect Med. 2015;5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 126] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 39. | Rockey DC. Liver Fibrosis Reversion After Suppression of Hepatitis B Virus. Clin Liver Dis. 2016;20:667-679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (1)] |
| 40. | Hosaka T, Suzuki F, Kobayashi M, Seko Y, Kawamura Y, Sezaki H, Akuta N, Suzuki Y, Saitoh S, Arase Y, Ikeda K, Kobayashi M, Kumada H. Long-term entecavir treatment reduces hepatocellular carcinoma incidence in patients with hepatitis B virus infection. Hepatology. 2013;58:98-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 519] [Cited by in RCA: 554] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 41. | Lim SG, Wai CT, Rajnakova A, Kajiji T, Guan R. Fatal hepatitis B reactivation following discontinuation of nucleoside analogues for chronic hepatitis B. Gut. 2002;51:597-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 95] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 42. | Lok AS, Lai CL, Leung N, Yao GB, Cui ZY, Schiff ER, Dienstag JL, Heathcote EJ, Little NR, Griffiths DA, Gardner SD, Castiglia M. Long-term safety of lamivudine treatment in patients with chronic hepatitis B. Gastroenterology. 2003;125:1714-1722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 584] [Cited by in RCA: 592] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 43. | Lei J, Wang Y, Wang LL, Zhang SJ, Chen W, Bai ZG, Xu LY. Profile of hepatitis B virus resistance mutations against nucleoside/nucleotide analogue treatment in Chinese patients with chronic hepatitis B. Virol J. 2013;10:313. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 44. | Fung J, Lai CL, Seto WK, Yuen MF. Nucleoside/nucleotide analogues in the treatment of chronic hepatitis B. J Antimicrob Chemother. 2011;66:2715-2725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 146] [Article Influence: 9.7] [Reference Citation Analysis (2)] |
| 45. | Kwon H, Lok AS. Hepatitis B therapy. Nat Rev Gastroenterol Hepatol. 2011;8:275-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 213] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 46. | Tan G, Song H, Xu F, Cheng G. When Hepatitis B Virus Meets Interferons. Front Microbiol. 2018;9:1611. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 75] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 47. | Craxi A, Cooksley WG. Pegylated interferons for chronic hepatitis B. Antiviral Res. 2003;60:87-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 39] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 48. | Huang J, Zhang K, Chen W, Liao J, Luo X, Chen R. Switching to PegIFNα-2b leads to HBsAg loss in patients with low HBsAg levels and HBV DNA suppressed by NAs. Sci Rep. 2017;7:13383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 49. | Zhou Y, Yan R, Ru GQ, Yu LL, Yao J, Wang H. Pegylated-interferon consolidation treatment versus nucleos(t)ide analogue consolidation treatment in non-cirrhotic hepatitis B patients with hepatitis B e antigen seroconversion: an open-label pilot trial. Hepatol Int. 2019;13:422-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 50. | Bertoletti A, Ferrari C. Innate and adaptive immune responses in chronic hepatitis B virus infections: towards restoration of immune control of viral infection. Gut. 2012;61:1754-1764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 338] [Cited by in RCA: 363] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 51. | Li X, Liu X, Tian L, Chen Y. Cytokine-Mediated Immunopathogenesis of Hepatitis B Virus Infections. Clin Rev Allergy Immunol. 2016;50:41-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 74] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 52. | Zeng Z, Li L, Chen Y, Wei H, Sun R, Tian Z. Interferon-γ facilitates hepatic antiviral T cell retention for the maintenance of liver-induced systemic tolerance. J Exp Med. 2016;213:1079-1093. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 53. | Boni C, Fisicaro P, Valdatta C, Amadei B, Di Vincenzo P, Giuberti T, Laccabue D, Zerbini A, Cavalli A, Missale G, Bertoletti A, Ferrari C. Characterization of hepatitis B virus (HBV)-specific T-cell dysfunction in chronic HBV infection. J Virol. 2007;81:4215-4225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 649] [Cited by in RCA: 752] [Article Influence: 39.6] [Reference Citation Analysis (1)] |
| 54. | Fisicaro P, Barili V, Rossi M, Montali I, Vecchi A, Acerbi G, Laccabue D, Zecca A, Penna A, Missale G, Ferrari C, Boni C. Pathogenetic Mechanisms of T Cell Dysfunction in Chronic HBV Infection and Related Therapeutic Approaches. Front Immunol. 2020;11:849. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 125] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 55. | Ye B, Liu X, Li X, Kong H, Tian L, Chen Y. T-cell exhaustion in chronic hepatitis B infection: current knowledge and clinical significance. Cell Death Dis. 2015;6:e1694. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 208] [Cited by in RCA: 299] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 56. | Maini MK, Burton AR. Restoring, releasing or replacing adaptive immunity in chronic hepatitis B. Nat Rev Gastroenterol Hepatol. 2019;16:662-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 116] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 57. | Gane E, Verdon DJ, Brooks AE, Gaggar A, Nguyen AH, Subramanian GM, Schwabe C, Dunbar PR. Anti-PD-1 blockade with nivolumab with and without therapeutic vaccination for virally suppressed chronic hepatitis B: A pilot study. J Hepatol. 2019;71:900-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 268] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 58. | Kah J, Koh S, Volz T, Ceccarello E, Allweiss L, Lütgehetmann M, Bertoletti A, Dandri M. Lymphocytes transiently expressing virus-specific T cell receptors reduce hepatitis B virus infection. J Clin Invest. 2017;127:3177-3188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 96] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 59. | Zheng C, Yan H, Zeng J, Cai S, Wu X. Comparison of pegylated interferon monotherapy and de novo pegylated interferon plus tenofovir combination therapy in patients with chronic hepatitis B. Infect Drug Resist. 2019;12:845-854. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 60. | Marcellin P, Ahn SH, Ma X, Caruntu FA, Tak WY, Elkashab M, Chuang WL, Lim SG, Tabak F, Mehta R, Petersen J, Foster GR, Lou L, Martins EB, Dinh P, Lin L, Corsa A, Charuworn P, Subramanian GM, Reiser H, Reesink HW, Fung S, Strasser SI, Trinh H, Buti M, Gaeta GB, Hui AJ, Papatheodoridis G, Flisiak R, Chan HL; Study 149 Investigators. Combination of Tenofovir Disoproxil Fumarate and Peginterferon α-2a Increases Loss of Hepatitis B Surface Antigen in Patients With Chronic Hepatitis B. Gastroenterology. 2016;150:134-144.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 283] [Cited by in RCA: 286] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 61. | Wu D, Ning Q. Toward a Cure for Hepatitis B Virus Infection: Combination Therapy Involving Viral Suppression and Immune Modulation and Long-term Outcome. J Infect Dis. 2017;216:S771-S777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 62. | Revill PA, Tu T, Netter HJ, Yuen LKW, Locarnini SA, Littlejohn M. The evolution and clinical impact of hepatitis B virus genome diversity. Nat Rev Gastroenterol Hepatol. 2020;17:618-634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 122] [Article Influence: 20.3] [Reference Citation Analysis (2)] |
| 63. | Zhang K, Cao H, Liang J, Shu X, Sun H, Li G, Xu Q. CONSORT: Effects of adding adefovirdipivoxil to peginterferon alfa-2a at different time points on HBeAg-positivepatients: A prospective, randomized study. Medicine (Baltimore). 2016;95:e4471. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 64. | Erhardt A, Blondin D, Hauck K, Sagir A, Kohnle T, Heintges T, Häussinger D. Response to interferon alfa is hepatitis B virus genotype dependent: genotype A is more sensitive to interferon than genotype D. Gut. 2005;54:1009-1013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 192] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 65. | Kao JH, Wu NH, Chen PJ, Lai MY, Chen DS. Hepatitis B genotypes and the response to interferon therapy. J Hepatol. 2000;33:998-1002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 347] [Cited by in RCA: 350] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 66. | Wu S, Luo W, Wu Y, Chen H, Peng J. HBsAg quantification predicts off-treatment response to interferon in chronic hepatitis B patients: a retrospective study of 250 cases. BMC Gastroenterol. 2020;20:121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 67. | Buster EH, Hansen BE, Lau GK, Piratvisuth T, Zeuzem S, Steyerberg EW, Janssen HL. Factors that predict response of patients with hepatitis B e antigen-positive chronic hepatitis B to peginterferon-alfa. Gastroenterology. 2009;137:2002-2009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 301] [Cited by in RCA: 330] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 68. | Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2205] [Cited by in RCA: 2733] [Article Influence: 130.1] [Reference Citation Analysis (0)] |
| 69. | Tseng TC, Kao JH, Chen DS. Peginterferon α in the treatment of chronic hepatitis B. Expert Opin Biol Ther. 2014;14:995-1006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 70. | Woo ASJ, Kwok R, Ahmed T. Alpha-interferon treatment in hepatitis B. Ann Transl Med. 2017;5:159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 67] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 71. | Au-Yeung N, Mandhana R, Horvath CM. Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. JAKSTAT. 2013;2:e23931. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 197] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 72. | Rawlings JS, Rosler KM, Harrison DA. The JAK/STAT signaling pathway. J Cell Sci. 2004;117:1281-1283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1190] [Cited by in RCA: 1366] [Article Influence: 62.1] [Reference Citation Analysis (0)] |
| 73. | Mowen KA, Tang J, Zhu W, Schurter BT, Shuai K, Herschman HR, David M. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell. 2001;104:731-741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 369] [Cited by in RCA: 380] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 74. | Duong FH, Christen V, Berke JM, Penna SH, Moradpour D, Heim MH. Upregulation of protein phosphatase 2Ac by hepatitis C virus modulates NS3 helicase activity through inhibition of protein arginine methyltransferase 1. J Virol. 2005;79:15342-15350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 77] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 75. | Bonvin M, Achermann F, Greeve I, Stroka D, Keogh A, Inderbitzin D, Candinas D, Sommer P, Wain-Hobson S, Vartanian JP, Greeve J. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology. 2006;43:1364-1374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 213] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 76. | Blanc V, Davidson NO. APOBEC-1-mediated RNA editing. Wiley Interdiscip Rev Syst Biol Med. 2010;2:594-602. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 97] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 77. | Li Y, Xia Y, Han M, Chen G, Zhang D, Thasler WE, Protzer U, Ning Q. IFN-α-mediated Base Excision Repair Pathway Correlates with Antiviral Response Against Hepatitis B Virus Infection. Sci Rep. 2017;7:12715. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 78. | Banks TA, Rickert S, Benedict CA, Ma L, Ko M, Meier J, Ha W, Schneider K, Granger SW, Turovskaya O, Elewaut D, Otero D, French AR, Henry SC, Hamilton JD, Scheu S, Pfeffer K, Ware CF. A lymphotoxin-IFN-beta axis essential for lymphocyte survival revealed during cytomegalovirus infection. J Immunol. 2005;174:7217-7225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 72] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 79. | Haybaeck J, Zeller N, Wolf MJ, Weber A, Wagner U, Kurrer MO, Bremer J, Iezzi G, Graf R, Clavien PA, Thimme R, Blum H, Nedospasov SA, Zatloukal K, Ramzan M, Ciesek S, Pietschmann T, Marche PN, Karin M, Kopf M, Browning JL, Aguzzi A, Heikenwalder M. A lymphotoxin-driven pathway to hepatocellular carcinoma. Cancer Cell. 2009;16:295-308. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 323] [Cited by in RCA: 327] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 80. | Wieland SF, Spangenberg HC, Thimme R, Purcell RH, Chisari FV. Expansion and contraction of the hepatitis B virus transcriptional template in infected chimpanzees. Proc Natl Acad Sci U S A. 2004;101:2129-2134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 176] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 81. | Marenstein DR, Wilson DM, Teebor GW. Human AP endonuclease (APE1) demonstrates endonucleolytic activity against AP sites in single-stranded DNA. DNA Repair (Amst). 2004;3:527-533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 70] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 82. | Rösler C, Köck J, Kann M, Malim MH, Blum HE, Baumert TF, von Weizsäcker F. APOBEC-mediated interference with hepadnavirus production. Hepatology. 2005;42:301-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 124] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 83. | Turelli P, Mangeat B, Jost S, Vianin S, Trono D. Inhibition of hepatitis B virus replication by APOBEC3G. Science. 2004;303:1829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 347] [Cited by in RCA: 375] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 84. | Belloni L, Allweiss L, Guerrieri F, Pediconi N, Volz T, Pollicino T, Petersen J, Raimondo G, Dandri M, Levrero M. IFN-α inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J Clin Invest. 2012;122:529-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 422] [Cited by in RCA: 495] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 85. | Liu F, Campagna M, Qi Y, Zhao X, Guo F, Xu C, Li S, Li W, Block TM, Chang J, Guo JT. Alpha-interferon suppresses hepadnavirus transcription by altering epigenetic modification of cccDNA minichromosomes. PLoS Pathog. 2013;9:e1003613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 133] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 86. | Hong X, Kim ES, Guo H. Epigenetic regulation of hepatitis B virus covalently closed circular DNA: Implications for epigenetic therapy against chronic hepatitis B. Hepatology. 2017;66:2066-2077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 163] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 87. | Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrito G, Raimondo G, Levrero M. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology. 2006;130:823-837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 391] [Cited by in RCA: 381] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 88. | Alarcon V, Hernández S, Rubio L, Alvarez F, Flores Y, Varas-Godoy M, De Ferrari GV, Kann M, Villanueva RA, Loyola A. The enzymes LSD1 and Set1A cooperate with the viral protein HBx to establish an active hepatitis B viral chromatin state. Sci Rep. 2016;6:25901. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 89. | Vivekanandan P, Daniel HD, Kannangai R, Martinez-Murillo F, Torbenson M. Hepatitis B virus replication induces methylation of both host and viral DNA. J Virol. 2010;84:4321-4329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 113] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 90. | Yuan Y, Yuan H, Yang G, Yun H, Zhao M, Liu Z, Zhao L, Geng Y, Liu L, Wang J, Zhang H, Wang Y, Zhang XD. IFN-α confers epigenetic regulation of HBV cccDNA minichromosome by modulating GCN5-mediated succinylation of histone H3K79 to clear HBV cccDNA. Clin Epigenetics. 2020;12:135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 91. | Yang Y, Zhao X, Wang Z, Shu W, Li L, Li Y, Guo Z, Gao B, Xiong S. Nuclear Sensor Interferon-Inducible Protein 16 Inhibits the Function of Hepatitis B Virus Covalently Closed Circular DNA by Integrating Innate Immune Activation and Epigenetic Suppression. Hepatology. 2020;71:1154-1169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 92. | Alcantara FF, Tang H, McLachlan A. Functional characterization of the interferon regulatory element in the enhancer 1 region of the hepatitis B virus genome. Nucleic Acids Res. 2002;30:2068-2075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 93. | Tur-Kaspa R, Teicher L, Laub O, Itin A, Dagan D, Bloom BR, Shafritz DA. Alpha interferon suppresses hepatitis B virus enhancer activity and reduces viral gene transcription. J Virol. 1990;64:1821-1824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 47] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 94. | Nakao K, Nakata K, Yamashita M, Tamada Y, Hamasaki K, Ishikawa H, Kato Y, Eguchi K, Ishii N. p48 (ISGF-3gamma) is involved in interferon-alpha-induced suppression of hepatitis B virus enhancer-1 activity. J Biol Chem. 1999;274:28075-28078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 95. | Lu JJ, Chen EQ, Yang JH, Zhou TY, Liu L, Tang H. A mutation in the interferon regulatory element of HBV may influence the response of interferon treatment in chronic hepatitis B patients. Virol J. 2012;9:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 96. | Guo Y, Lu H, Xu L, Idris NFB, Li Y, Hu J, Huang A, Tu Z. The response of hepatitis B virus genotype to interferon is associated with a mutation in the interferon-stimulated response element. Medicine (Baltimore). 2019;98:e18442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 97. | Park YK, Lee SY, Lee AR, Kim KC, Kim K, Kim KH, Choi BS. Antiviral activity of interferon-stimulated gene 20, as a putative repressor binding to hepatitis B virus enhancer II and core promoter. J Gastroenterol Hepatol. 2020;35:1426-1436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 98. | Xiao C, Qin B, Chen L, Liu H, Zhu Y, Lu X. Preactivation of the interferon signalling in liver is correlated with nonresponse to interferon alpha therapy in patients chronically infected with hepatitis B virus. J Viral Hepat. 2012;19:e1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 99. | Wieland S, Thimme R, Purcell RH, Chisari FV. Genomic analysis of the host response to hepatitis B virus infection. Proc Natl Acad Sci USA. 2004;101:6669-6674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 520] [Cited by in RCA: 565] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 100. | Wang YX, Niklasch M, Liu T, Wang Y, Shi B, Yuan W, Baumert TF, Yuan Z, Tong S, Nassal M, Wen YM. Interferon-inducible MX2 is a host restriction factor of hepatitis B virus replication. J Hepatol. 2020;72:865-876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 101. | Tsunematsu S, Suda G, Yamasaki K, Kimura M, Izumi T, Umemura M, Ito J, Sato F, Nakai M, Sho T, Morikawa K, Ogawa K, Tanaka Y, Watashi K, Wakita T, Sakamoto N. Hepatitis B virus X protein impairs α-interferon signaling via up-regulation of suppressor of cytokine signaling 3 and protein phosphatase 2A. J Med Virol. 2017;89:267-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 102. | Chen J, Wu M, Zhang X, Zhang W, Zhang Z, Chen L, He J, Zheng Y, Chen C, Wang F, Hu Y, Zhou X, Wang C, Xu Y, Lu M, Yuan Z. Hepatitis B virus polymerase impairs interferon-α-induced STA T activation through inhibition of importin-α5 and protein kinase C-δ. Hepatology. 2013;57:470-482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 108] [Article Influence: 8.3] [Reference Citation Analysis (2)] |
| 103. | Wu M, Xu Y, Lin S, Zhang X, Xiang L, Yuan Z. Hepatitis B virus polymerase inhibits the interferon-inducible MyD88 promoter by blocking nuclear translocation of Stat1. J Gen Virol. 2007;88:3260-3269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 76] [Article Influence: 4.2] [Reference Citation Analysis (1)] |
| 104. | Mitra B, Wang J, Kim ES, Mao R, Dong M, Liu Y, Zhang J, Guo H. Hepatitis B Virus Precore Protein p22 Inhibits Alpha Interferon Signaling by Blocking STAT Nuclear Translocation. J Virol. 2019;93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 105. | Christen V, Duong F, Bernsmeier C, Sun D, Nassal M, Heim MH. Inhibition of alpha interferon signaling by hepatitis B virus. J Virol. 2007;81:159-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 92] [Article Influence: 4.8] [Reference Citation Analysis (2)] |
| 106. | Murata K, Asano M, Matsumoto A, Sugiyama M, Nishida N, Tanaka E, Inoue T, Sakamoto M, Enomoto N, Shirasaki T, Honda M, Kaneko S, Gatanaga H, Oka S, Kawamura YI, Dohi T, Shuno Y, Yano H, Mizokami M. Induction of IFN-λ3 as an additional effect of nucleotide, not nucleoside, analogues: a new potential target for HBV infection. Gut. 2018;67:362-371. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 155] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 107. | Bockmann JH, Stadler D, Xia Y, Ko C, Wettengel JM, Schulze Zur Wiesch J, Dandri M, Protzer U. Comparative Analysis of the Antiviral Effects Mediated by Type I and III Interferons in Hepatitis B Virus-Infected Hepatocytes. J Infect Dis. 2019;220:567-577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 108. | Weng HL, Wang BE, Jia JD, Wu WF, Xian JZ, Mertens PR, Cai WM, Dooley S. Effect of interferon-gamma on hepatic fibrosis in chronic hepatitis B virus infection: a randomized controlled study. Clin Gastroenterol Hepatol. 2005;3:819-828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 70] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 109. | Feng X, Tang H, Leng J, Jiang Q. Suppressors of cytokine signaling (SOCS) and type 2 diabetes. Mol Biol Rep. 2014;41:2265-2274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 110. | Chung V, Mansfield AS, Braiteh F, Richards D, Durivage H, Ungerleider RS, Johnson F, Kovach JS. Safety, Tolerability, and Preliminary Activity of LB-100, an Inhibitor of Protein Phosphatase 2A, in Patients with Relapsed Solid Tumors: An Open-Label, Dose Escalation, First-in-Human, Phase I Trial. Clin Cancer Res. 2017;23:3277-3284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 95] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 111. | Gilmore S, Tam D, Dick R, Appleby T, Birkus G, Willkom M, Delaney WE, Notte GT, Feierbach B. Antiviral activity of GS-5801, a liver-targeted prodrug of a lysine demethylase 5 inhibitor, in a hepatitis B virus primary human hepatocyte infection model. J Hepatol. 2017;66:S690-S691. [DOI] [Full Text] |
| 112. | Morel D, Jeffery D, Aspeslagh S, Almouzni G, Postel-Vinay S. Combining epigenetic drugs with other therapies for solid tumours - past lessons and future promise. Nat Rev Clin Oncol. 2020;17:91-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 322] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 113. | Patnaik S, Anupriya. Drugs Targeting Epigenetic Modifications and Plausible Therapeutic Strategies Against Colorectal Cancer. Front Pharmacol. 2019;10:588. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 77] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 114. | Vogler WR, Miller DS, Keller JW. 5-Azacytidine (NSC 102816): a new drug for the treatment of myeloblastic leukemia. Blood. 1976;48:331-337. [PubMed] |
| 115. | Stone D, Long KR, Loprieno MA, De Silva Feelixge HS, Kenkel EJ, Liley RM, Rapp S, Roychoudhury P, Nguyen T, Stensland L, Colón-Thillet R, Klouser LM, Weber ND, Le C, Wagoner J, Goecker EA, Li AZ, Eichholz K, Corey L, Tyrrell DL, Greninger AL, Huang ML, Polyak SJ, Aubert M, Sagartz JE, Jerome KR. CRISPR-Cas9 gene editing of hepatitis B virus in chronically infected humanized mice. Mol Ther Methods Clin Dev. 2021;20:258-275. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 116. | Ramanan V, Shlomai A, Cox DB, Schwartz RE, Michailidis E, Bhatta A, Scott DA, Zhang F, Rice CM, Bhatia SN. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci Rep. 2015;5:10833. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 228] [Cited by in RCA: 237] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 117. | Seeger C, Sohn JA. Targeting Hepatitis B Virus With CRISPR/Cas9. Mol Ther Nucleic Acids. 2014;3:e216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 220] [Cited by in RCA: 233] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 118. | Kennedy EM, Bassit LC, Mueller H, Kornepati AVR, Bogerd HP, Nie T, Chatterjee P, Javanbakht H, Schinazi RF, Cullen BR. Suppression of hepatitis B virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA-guided DNA endonuclease. Virology. 2015;476:196-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 188] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 119. | Yang YC, Chen YH, Kao JH, Ching C, Liu IJ, Wang CC, Tsai CH, Wu FY, Liu CJ, Chen PJ, Chen DS, Yang HC. Permanent Inactivation of HBV Genomes by CRISPR/Cas9-Mediated Non-cleavage Base Editing. Mol Ther Nucleic Acids. 2020;20:480-490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 75] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 120. | Zhang JF, Xiong HL, Cao JL, Wang SJ, Guo XR, Lin BY, Zhang Y, Zhao JH, Wang YB, Zhang TY, Yuan Q, Zhang J, Xia NS. A cell-penetrating whole molecule antibody targeting intracellular HBx suppresses hepatitis B virus via TRIM21-dependent pathway. Theranostics. 2018;8:549-562. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 64] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 121. | Hu B, Zhong L, Weng Y, Peng L, Huang Y, Zhao Y, Liang XJ. Therapeutic siRNA: state of the art. Signal Transduct Target Ther. 2020;5:101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 535] [Cited by in RCA: 1044] [Article Influence: 174.0] [Reference Citation Analysis (0)] |
| 122. | Lopatin U. Drugs in the Pipeline for HBV. Clin Liver Dis. 2019;23:535-555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 123. | Yuen MF, Schiefke I, Yoon JH, Ahn SH, Heo J, Kim JH, Lik Yuen Chan H, Yoon KT, Klinker H, Manns M, Petersen J, Schluep T, Hamilton J, Given BD, Ferrari C, Lai CL, Locarnini SA, Gish RG. RNA Interference Therapy With ARC-520 Results in Prolonged Hepatitis B Surface Antigen Response in Patients With Chronic Hepatitis B Infection. Hepatology. 2020;72:19-31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 109] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 124. | Han K, Cremer J, Elston R, Oliver S, Baptiste-Brown S, Chen S, Gardiner D, Davies M, Saunders J, Hamatake R, Losos J, Leivers M, Hood S, van der Berg F, Paff M, Ritter JM, Theodore D. A Randomized, Double-Blind, Placebo-Controlled, First-Time-in-Human Study to Assess the Safety, Tolerability, and Pharmacokinetics of Single and Multiple Ascending Doses of GSK3389404 in Healthy Subjects. Clin Pharmacol Drug Dev. 2019;8:790-801. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 125. | Han X, Zhou C, Jiang M, Wang Y, Wang J, Cheng Z, Wang M, Liu Y, Liang C, Wang J, Wang Z, Weikert R, Lv W, Xie J, Yu X, Zhou X, Luangsay S, Shen HC, Mayweg AV, Javanbakht H, Yang S. Discovery of RG7834: The First-in-Class Selective and Orally Available Small Molecule Hepatitis B Virus Expression Inhibitor with Novel Mechanism of Action. J Med Chem. 2018;61:10619-10634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 126. | Mueller H, Wildum S, Luangsay S, Walther J, Lopez A, Tropberger P, Ottaviani G, Lu W, Parrott NJ, Zhang JD, Schmucki R, Racek T, Hoflack JC, Kueng E, Point F, Zhou X, Steiner G, Lütgehetmann M, Rapp G, Volz T, Dandri M, Yang S, Young JAT, Javanbakht H. A novel orally available small molecule that inhibits hepatitis B virus expression. J Hepatol. 2018;68:412-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 114] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 127. | Wei ZQ, Zhang YH, Ke CZ, Chen HX, Ren P, He YL, Hu P, Ma DQ, Luo J, Meng ZJ. Curcumin inhibits hepatitis B virus infection by down-regulating cccDNA-bound histone acetylation. World J Gastroenterol. 2017;23:6252-6260. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 68] [Cited by in RCA: 68] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 128. | Cheng ST, Hu JL, Ren JH, Yu HB, Zhong S, Wai Wong VK, Kwan Law BY, Chen WX, Xu HM, Zhang ZZ, Cai XF, Hu Y, Zhang WL, Long QX, Ren F, Zhou HZ, Huang AL, Chen J. Dicoumarol, an NQO1 inhibitor, blocks cccDNA transcription by promoting degradation of HBx. J Hepatol. 2021;74:522-534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 97] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 129. | Hsu CW, Su WW, Lee CM, Peng CY, Chuang WL, Kao JH, Chu HC, Huang YH, Chien RN, Liaw YF. Phase IV randomized clinical study: Peginterferon alfa-2a with adefovir or entecavir pre-therapy for HBeAg-positive chronic hepatitis B. J Formos Med Assoc. 2018;117:588-597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 130. | Janssen HL, van Zonneveld M, Senturk H, Zeuzem S, Akarca US, Cakaloglu Y, Simon C, So TM, Gerken G, de Man RA, Niesters HG, Zondervan P, Hansen B, Schalm SW; HBV 99-01 Study Group; Rotterdam Foundation for Liver Research. Pegylated interferon alfa-2b alone or in combination with lamivudine for HBeAg-positive chronic hepatitis B: a randomised trial. Lancet. 2005;365:123-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 918] [Cited by in RCA: 910] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 131. | Reijnders JG, Perquin MJ, Zhang N, Hansen BE, Janssen HL. Nucleos(t)ide analogues only induce temporary hepatitis B e antigen seroconversion in most patients with chronic hepatitis B. Gastroenterology. 2010;139:491-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 152] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 132. | Song BC, Suh DJ, Lee HC, Chung YH, Lee YS. Hepatitis B e antigen seroconversion after lamivudine therapy is not durable in patients with chronic hepatitis B in Korea. Hepatology. 2000;32:803-806. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 257] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 133. | Jeng WJ, Chen YC, Chien RN, Sheen IS, Liaw YF. Incidence and predictors of hepatitis B surface antigen seroclearance after cessation of nucleos(t)ide analogue therapy in hepatitis B e antigen-negative chronic hepatitis B. Hepatology. 2018;68:425-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 265] [Article Influence: 33.1] [Reference Citation Analysis (0)] |
| 134. | Liem KS, Fung S, Wong DK, Yim C, Noureldin S, Chen J, Feld JJ, Hansen BE, Janssen HLA. Limited sustained response after stopping nucleos(t)ide analogues in patients with chronic hepatitis B: results from a randomised controlled trial (Toronto STOP study). Gut. 2019;68:2206-2213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 119] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 135. | Marcellin P, Lau GK, Bonino F, Farci P, Hadziyannis S, Jin R, Lu ZM, Piratvisuth T, Germanidis G, Yurdaydin C, Diago M, Gurel S, Lai MY, Button P, Pluck N; Peginterferon Alfa-2a HBeAg-Negative Chronic Hepatitis B Study Group. Peginterferon alfa-2a alone, lamivudine alone, and the two in combination in patients with HBeAg-negative chronic hepatitis B. N Engl J Med. 2004;351:1206-1217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 874] [Cited by in RCA: 862] [Article Influence: 39.2] [Reference Citation Analysis (0)] |
| 136. | Lau GK, Piratvisuth T, Luo KX, Marcellin P, Thongsawat S, Cooksley G, Gane E, Fried MW, Chow WC, Paik SW, Chang WY, Berg T, Flisiak R, McCloud P, Pluck N; Peginterferon Alfa-2a HBeAg-Positive Chronic Hepatitis B Study Group. Peginterferon Alfa-2a, lamivudine, and the combination for HBeAg-positive chronic hepatitis B. N Engl J Med. 2005;352:2682-2695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1188] [Cited by in RCA: 1182] [Article Influence: 56.3] [Reference Citation Analysis (1)] |
| 137. | van Zonneveld M, Honkoop P, Hansen BE, Niesters HG, Darwish Murad S, de Man RA, Schalm SW, Janssen HL. Long-term follow-up of alpha-interferon treatment of patients with chronic hepatitis B. Hepatology. 2004;39:804-810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 253] [Article Influence: 11.5] [Reference Citation Analysis (2)] |
| 138. | Niederau C, Heintges T, Lange S, Goldmann G, Niederau CM, Mohr L, Häussinger D. Long-term follow-up of HBeAg-positive patients treated with interferon alfa for chronic hepatitis B. N Engl J Med. 1996;334:1422-1427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 609] [Cited by in RCA: 574] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 139. | Liu S, Zheng H, Huang Y, Li B, Dong Z. The effect of peginterferon alpha-2a vs. interferon alpha-2a on intrahepatic covalently closed circular DNA in HBeAg-positive chronic hepatitis B patients. Clin Res Hepatol Gastroenterol. 2016;40:304-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 140. | Sun J, Hou JL, Xie Q, Li XH, Zhang JM, Wang YM, Wang H, Lai JY, Chen SJ, Jia JD, Sheng JF, Chan HL, Wang JF, Li MK, Jiang M, Popescu M, Sung JJ. Randomised clinical trial: efficacy of peginterferon alfa-2a in HBeAg positive chronic hepatitis B patients with lamivudine resistance. Aliment Pharmacol Ther. 2011;34:424-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 141. | Tan G, Yi Z, Song H, Xu F, Li F, Aliyari R, Zhang H, Du P, Ding Y, Niu J, Wang X, Su L, Qin FX, Cheng G. Type-I-IFN-Stimulated Gene TRIM5γ Inhibits HBV Replication by Promoting HBx Degradation. Cell Rep. 2019;29:3551-3563.e3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 61] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 142. | Liu Y, Nie H, Mao R, Mitra B, Cai D, Yan R, Guo JT, Block TM, Mechti N, Guo H. Interferon-inducible ribonuclease ISG20 inhibits hepatitis B virus replication through directly binding to the epsilon stem-loop structure of viral RNA. PLoS Pathog. 2017;13:e1006296. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 114] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 143. | Hoofnagle JH, Peters M, Mullen KD, Jones DB, Rustgi V, Di Bisceglie A, Hallahan C, Park Y, Meschievitz C, Jones EA. Randomized, controlled trial of recombinant human alpha-interferon in patients with chronic hepatitis B. Gastroenterology. 1988;95:1318-1325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 323] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 144. | Cheng J, Zhao Q, Zhou Y, Tang L, Sheraz M, Chang J, Guo JT. Interferon Alpha Induces Multiple Cellular Proteins That Coordinately Suppress Hepadnaviral Covalently Closed Circular DNA Transcription. J Virol. 2020;94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 145. | Dill MT, Makowska Z, Trincucci G, Gruber AJ, Vogt JE, Filipowicz M, Calabrese D, Krol I, Lau DT, Terracciano L, van Nimwegen E, Roth V, Heim MH. Pegylated IFN-α regulates hepatic gene expression through transient Jak/STAT activation. J Clin Invest. 2014;124:1568-1581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 146. | Chen J, Li Y, Lai F, Wang Y, Sutter K, Dittmer U, Ye J, Zai W, Liu M, Shen F, Wu M, Hu K, Li B, Lu M, Zhang X, Zhang J, Li J, Chen Q, Yuan Z. Functional Comparison of Interferon-α Subtypes Reveals Potent Hepatitis B Virus Suppression by a Concerted Action of Interferon-α and Interferon-γ Signaling. Hepatology. 2021;73:486-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 69] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 147. | Tsuge M, Uchida T, Hiraga N, Kan H, Makokha GN, Abe-Chayama H, Miki D, Imamura M, Ochi H, Hayes CN, Shimozono R, Iwamura T, Narumi H, Suzuki T, Kainoh M, Taniguchi T, Chayama K. Development of a Novel Site-Specific Pegylated Interferon Beta for Antiviral Therapy of Chronic Hepatitis B Virus. Antimicrob Agents Chemother. 2017;61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 148. | Hagelstein J, Kist A, Stremmel W, Galle PR. Antiviral potential of interferon-omega on hepatitis B virus replication in human hepatoma cells. Arzneimittelforschung. 1998;48:343-347. [PubMed] |
| 149. | Hajnická V, Fuchsberger N, Liptáková H, Stancek D, Kontsek P. Interferon-omega suppresses hepatitis B surface antigen production in human hepatoma cell line. Acta Virol. 1996;40:221-222. [PubMed] |
| 150. | Künzi MS, Pitha PM. Role of interferon-stimulated gene ISG-15 in the interferon-omega-mediated inhibition of human immunodeficiency virus replication. J Interferon Cytokine Res. 1996;16:919-927. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 50] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 151. | Kakumu S, Ishikawa T, Mizokami M, Orido E, Yoshioka K, Wakita T, Yamamoto M. Treatment with human gamma interferon of chronic hepatitis B: comparative study with alpha interferon. J Med Virol. 1991;35:32-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 22] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 152. | Mao R, Zhang J, Jiang D, Cai D, Levy JM, Cuconati A, Block TM, Guo JT, Guo H. Indoleamine 2,3-dioxygenase mediates the antiviral effect of gamma interferon against hepatitis B virus in human hepatocyte-derived cells. J Virol. 2011;85:1048-1057. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 113] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 153. | Lau JY, Bain VG, Naoumov NV, Smith HM, Alexander GJ, Williams R. Effect of interferon-gamma on hepatitis B viral antigen expression in primary hepatocyte culture. Hepatology. 1991;14:975-979. [PubMed] |
| 154. | Chan HLY, Ahn SH, Chang TT, Peng CY, Wong D, Coffin CS, Lim SG, Chen PJ, Janssen HLA, Marcellin P, Serfaty L, Zeuzem S, Cohen D, Critelli L, Xu D, Wind-Rotolo M, Cooney E; LIRA-B Study Team. Peginterferon lambda for the treatment of HBeAg-positive chronic hepatitis B: A randomized phase 2b study (LIRA-B). J Hepatol. 2016;64:1011-1019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 86] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 155. | Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol. 2006;80:4501-4509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 460] [Cited by in RCA: 505] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 156. | Bockmann JH, Xia Y, Stadler D, Protzer U. Type III interferons induce cccDNA degradation similar to type I interferons in HBV-infected HepaRG cells. Z Gastroenterol. 2015;53:a5-54. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 157. | Guo X, Chen D, Cai Q, Huang Z, Xu W, Peng L, Chen P. Minicircle DNA vector expressing interferon-lambda-3 inhibits hepatitis B virus replication and expression in hepatocyte-derived cell line. BMC Mol Cell Biol. 2020;21:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 158. | Pagliaccetti NE, Chu EN, Bolen CR, Kleinstein SH, Robek MD. Lambda and alpha interferons inhibit hepatitis B virus replication through a common molecular mechanism but with different in vivo activities. Virology. 2010;401:197-206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 67] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 159. | Makjaroen J, Somparn P, Hodge K, Poomipak W, Hirankarn N, Pisitkun T. Comprehensive Proteomics Identification of IFN-λ3-regulated Antiviral Proteins in HBV-transfected Cells. Mol Cell Proteomics. 2018;17:2197-2215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 160. | Hagiwara S, Nishida N, Watanabe T, Ida H, Sakurai T, Ueshima K, Takita M, Komeda Y, Nishijima N, Osaki Y, Kudo M. Sustained antiviral effects and clearance of hepatitis surface antigen after combination therapy with entecavir and pegylated interferon in chronic hepatitis B. Antivir Ther. 2018;23:513-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Manuscript source: Invited manuscript
Specialty type: Infectious diseases
Country/Territory of origin: Singapore
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Bowden S, Gu J, Wongkajornsilp A, Zhu WF S-Editor: Zhang H L-Editor: A P-Editor: Liu JH