Published online Mar 14, 2021. doi: 10.3748/wjg.v27.i10.939
Peer-review started: November 26, 2020
First decision: December 21, 2020
Revised: January 3, 2021
Accepted: February 1, 2021
Article in press: February 1, 2021
Published online: March 14, 2021
Processing time: 105 Days and 3.1 Hours
Hepatocellular carcinoma (HCC) is one of the most prevalent cancers in human populations worldwide. Huanglian decoction is one of the most important Chinese medicine formulas, with the potential to treat cancer.
To investigate the role and mechanism of Huanglian decoction on HCC cells.
To identify differentially expressed genes (DEGs), we downloaded gene expression profile data from The Cancer Genome Atlas Liver Hepatocellular Carcinoma and Gene Expression Omnibus (GSE45436) databases. We obtained phytochemicals of the four herbs of Huanglian decoction from the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform. We also established a regulatory network of DEGs and drug target genes and subsequently analyzed key genes using bioinformatics approaches. Furthermore, we conducted in vitro experiments to explore the effect of Huanglian decoction and to verify the predictions. In particular, the CCNB1 gene was knocked down to verify the primary target of this decoction. Through the identification of the expression levels of key proteins, we determined the primary mechanism of Huanglian decoction in HCC.
Based on the results of the network pharmacological analysis, we revealed 5 bioactive compounds in Huanglian decoction that act on HCC. In addition, a protein-protein interaction network analysis of the target genes of these five compounds as well as expression and prognosis analyses were performed in tumors. CCNB1 was confirmed to be the primary gene that may be highly expressed in tumors and was significantly associated with a worse prognosis. We also noted that CCNB1 may serve as an independent prognostic indicator in HCC. Moreover, in vitro experiments demonstrated that Huanglian decoction significantly inhibited the growth, migration, and invasiveness of HCC cells and induced cell apoptosis and G2/M phase arrest. Further analysis showed that the decoction may inhibit the growth of HCC cells by downregulating the CCNB1 expression level. After Huanglian decoction treatment, the expression levels of Bax, caspase 3, caspase 9, p21 and p53 in HCC cells were increased, while the expression of CDK1 and CCNB1 was significantly decreased. The p53 signaling pathway was also found to play an important role in this process.
Huanglian decoction has a significant inhibitory effect on HCC cells. CCNB1 is a potential therapeutic target in HCC. Further analysis showed that Huanglian decoction can inhibit HCC cell growth by downregulating the expression of CCNB1 to activate the p53 signaling pathway.
Core Tip: The purpose of this study was to investigate the role and mechanism of Huanglian decoction on hepatocellular carcinoma (HCC) cells. The results showed that Huanglian decoction has a significant inhibitory effect on the growth of HCC cells. Huanglian decoction can inhibit HCC cell growth by downregulating the expression of CCNB1 to activate the p53 signaling pathway. CCNB1 is a potential therapeutic target for liver cancer.
- Citation: Li M, Shang H, Wang T, Yang SQ, Li L. Huanglian decoction suppresses the growth of hepatocellular carcinoma cells by reducing CCNB1 expression. World J Gastroenterol 2021; 27(10): 939-958
- URL: https://www.wjgnet.com/1007-9327/full/v27/i10/939.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i10.939
Hepatocellular carcinoma (HCC) is the most frequent primary malignancy of the liver and the third leading cause of cancer mortality worldwide[1-3]. Current treatment options for liver cancer primarily include surgery, local treatment, biological treatment, and various other methods. Recently, molecular targeted therapy has been used to treat some patients with advanced liver cancer, but because targeted drugs are relatively expensive and induce side effects, they have not been fully utilized[4-6]. In the past few years, there has been growing research attention on traditional Chinese medicine (TCM). Previous studies have established that TCM can improve the clinical symptoms of patients, especially for those with advanced liver cancer by prolonging their survival period, and the cost is relatively inexpensive. In particular, TCM formulation contains a variety of chemical components that act on multiple targets and diseases[7-9]. However, due to the complexity and diversity of TCM components, our understanding of the specific molecular mechanisms of Chinese medicine is still largely unknown. This, therefore, limits the development of Chinese medicine. Nevertheless, the recent development of computer technology has allowed us to analyze the components of these medicines and systematically analyze their effects on diseases through network pharmacology, which has greatly promoted our understanding of the molecular network of Chinese medicines and their nature[10-12].
Huanglian decoction is a classical Chinese medicine formula that has the effect of clearing heat and treating abdominal pain[13,14]. This formulation was originally recorded in the classic Chinese medicine book “Treatise on Febrile Diseases” (Chinese name: Shang-Han Lun). In this study, we obtained the formula of Huanglian decoction from the ancient Chinese medicine book (Sheng Ji Zong Lu), which contains one of the original recipes. The formula consists of four Chinese herbal medicines: Coptidis Rhizoma, Zingiberis Rhizoma, Folium Artemisiae Argyi, and Mume Fructus. In addition, this decoction plays a pivotal role in the field of TCM due to its various effects, such as treating colds, white diarrhea, and abdominal pain[15-17]. In some Asian-Pacific regions, this decoction is used as an auxiliary medicine for the treatment of liver cancer. Research has demonstrated that this decoction can exert its effects on the liver[18-20]. However, very few studies have investigated the treatment of HCC with this decoction. Thus, in this respect, we aimed to investigate the therapeutic potential of Huanglian decoction as an HCC treatment.
CCNB1 plays a key role in regulating and forming a complex with CDK1 to promote the transition from the G2 phase of the cell cycle to mitosis[21]. Increasing evidence has shown that CCNB1 is overexpressed in certain human cancers, including colorectal, bladder and stomach cancers[22-24]. In addition, the p53 tumor suppressor pathway also plays a key role in mediating the responses of commonly used cancer treatments, and CCNB1 is significantly correlated with the p53 signaling pathway[25-29]. In this study, we explored the main targets of Huanglian decoction in HCC cells. By reducing CCNB1 expression, we studied the effects of Huanglian decoction on the cell cycle and apoptosis, and the potential functional pathways were also explored. The detailed flow chart of this study is depicted in Figure 1. This study provides novel insights into the mechanism of Huanglian decoction treatment in HCC.
For this study, we downloaded the gene expression profile data of HCC patients from the Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). This dataset includes 347 tumors and 50 non-tumor samples. Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) is a public functional genomics data repository. The GSE45436 dataset contained 93 HCC tissue samples and 41 non-cancerous samples. We retrieved data from publicly available databases, hence it was not applicable for additional ethical approval. Then, we selected PLC/PRF/5 and HepG2 cells for the following experiments. Specifically, the cells were purchased from the Chinese National Infrastructure of Cell Line Resource. The culture media used was Dulbecco's Modified Eagle's medium (Thermo Fisher Scientific, Waltham, MA, United States), supplemented with 10% fetal bovine serum (FBS). Finally, the cells were placed in an incubator (37 °C; 5% CO2) as previously described.
Here, we screened differentially expressed genes (DEGs) between HCC and noncancerous samples using the "limma" R package. The log2FC (fold change) > 1 or < -1 and adjusted P values < 0.05 were considered statistically significant. To identify the DEGs shared by the TCGA and GEO databases, we used TCGA data in the experimental group, while GEO data were applied in the verification group. Afterward, the intersections were obtained and visualized using the website (http://bioinformatics.psb.ugent.be/webtools/Venn/).
For this experiment, we retrieved all phytochemicals of the four components of Huanglian decoction from the TCM System Pharmacology Database and Analysis Platform (TCMSP) (http://tcmspw.com/tcmsp.php). Based on the standards recommended by the TCMSP, we selected an oral bioavailability (OB) index ≥ 30% and drug-likeness (DL) index ≥ 0.18 to determine the pharmacological properties of the compound. Notably, compounds that met all these criteria were considered biologically active ingredients[30-32].
To comprehensively understand the molecular mechanisms of Huanglian decoction, we collectively analyzed DEGs and active ingredients of the drug. Using Cytoscape version 3.7.2, we constructed Huanglian decoction-compound, compound-target genes, and target gene-disease networks[33-35]. In addition, to further understand the relationship between target genes, we constructed a protein-protein interaction (PPI) network according to the information retrieved from the Search Tool for the Retrieval of Interacting Genes (STRING; https://string-db.org/) online database.
Key genes in the network were extracted, and survival analyses were performed using R software ("survival" packages) to identify genes associated with prognosis, followed by univariate and multivariate Cox analyses of genes associated with prognosis.
To explore the potential molecular mechanisms of key genes, we performed a Gene Set Enrichment Analysis (GSEA) using the GSEA software version 4.0.3. The number of random sample permutations was set at 1000, while the significance threshold was set at P < 0.05[36-39].
The formula of Huanglian decoction used in this study is different from the previously reported formula. This formulation consisted of 50 g of Coptidis Rhizoma, Zingiberis Rhizoma (25 g), Folium Artemisiae Argyi (25 g), and Mume Fructus (10 g). All the above-mentioned herbs were acquired from the Chinese TongRenTang pharmacy. Briefly, all herbs were mixed and soaked for 30 min, then boiled in a casserole (1000 mL of distilled water) for 2 h. Subsequently, the supernatant was centrifuged at 8000 rpm (4293 × g) for 30 min, at which point the whole procedure was repeated twice and the supernatants were mixed together and then evaporated to dryness. Finally, the precipitate was redissolved in distilled water to a concentration of 10 mg/mL and filtered using a 0.22 μm pore-size filter and stored at 4 °C. The active compound of Huanglian Decoction (berberine hydrochloride) was approximately 0.26 mg/mL, which was analyzed by high-performance liquid chromatography (Alliance 2695, Waters, Milford, MA, United States) on an Inertsil ODS-2 C18 analytical column (4.6 mm × 250 mm, 5 μm) with a detection wavelength of 345 nm. It was eluted with acetonitrile-0.05 moL/L phosphoric acid aqueous solution (50:50), the flow rate was 1.0 mL/min, and the column temperature was 30 °C.
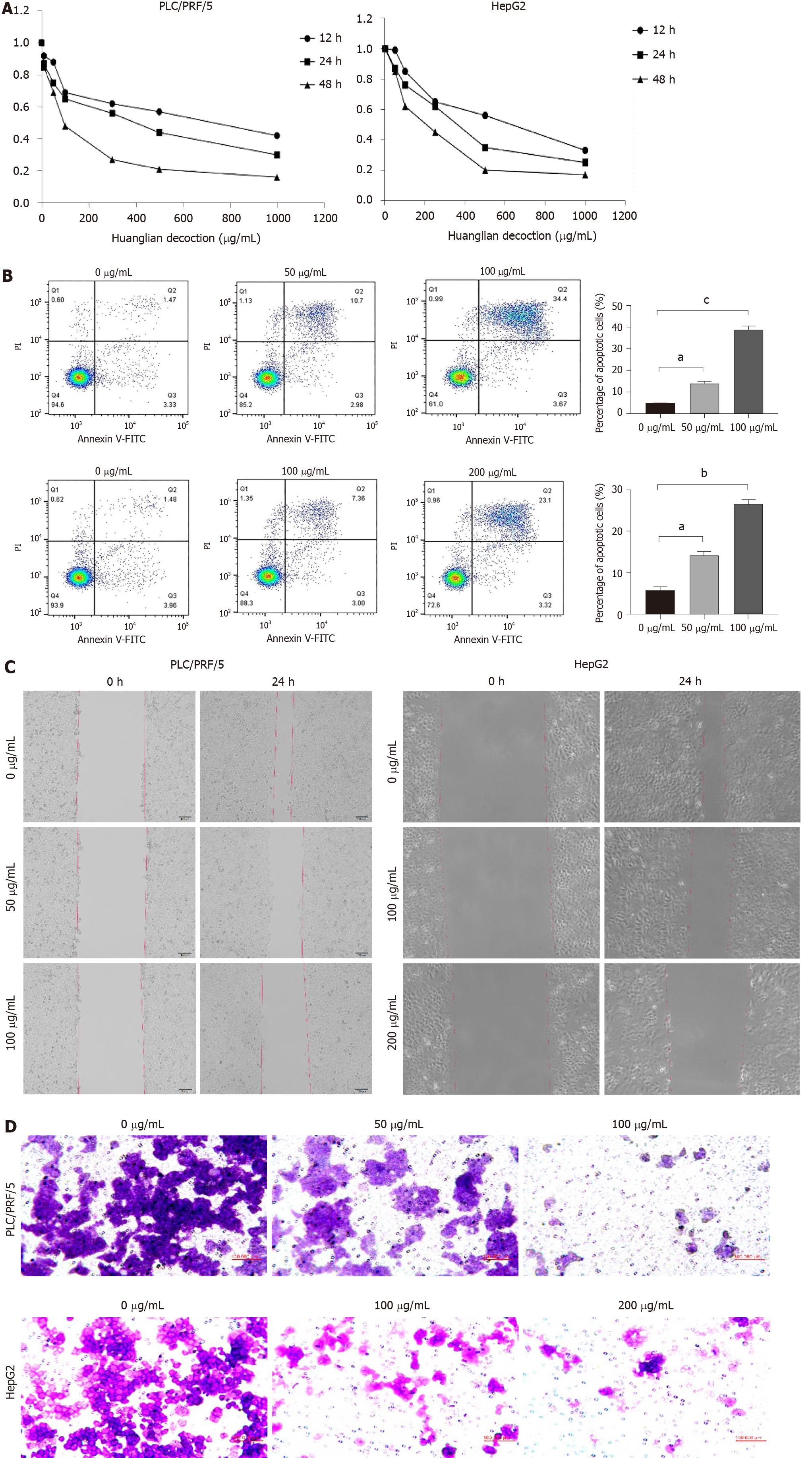
For the wound healing assay, we treated cells with different concentrations of Huanglian decoction (50 and 100 μg/mL for PLC/PRF/5 cells, 100 and 200 μg/mL for HepG2 cells) and then analyzed wound healing[40-42]. On the other hand, for transwell invasion assay, we treated HCC cells (1 × 105 cells/well) under different conditions, then fixed them with 4% paraformaldehyde for 2 h, followed by staining the invaded cells with crystal violet solution for 30 min. Finally, we counted the stained cells under an optical microscope.
SiRNA targeting CCNB1 (CCNB1-siRNA, GAAUUCUGCACUAGUUCAA), was designed and synthesized by Nanjing InvivoGene Biotechnology Co., Ltd. (Nanjing, China). When the cells reached 70%-80% confluence, we performed siRNA transfection using Lipofectamine 2000 (Invitrogen, United States) according to the manufacturer's instructions.
To identify apoptotic cells, HCC cells were treated under different conditions. Using an Annexin V-fluorescein isothiocyanate Apoptosis Detection kit (Thermo Fisher Scientific, Waltham, MA, United States) after 48 h, we performed Annexin V and PI staining. Cell apoptosis was detected using a flow cytometer (arachidonyl-2'-chloroethylamide, NovoCyteTM).
Cells were seeded (4 × 105 cells/well) in 60 mm dishes with 10% FBS and incubated overnight at 37 °C in a 5% CO2 incubator. The HCC cells were then treated under different conditions (48 h). The cells were collected, incubated in ice-cold 70% ethanol, and fixed overnight at 4 °C. The cells were then washed twice with poly (butylene succinate) (PBS) and incubated with 20 mg/mL RNase A and 200 mg/mL propidium iodide in PBS at room temperature for 30 min in the dark. Finally, flow cytometry was used for the analysis.
HCC cells in the logarithmic phase were seeded in a 10 cm dish. When the cells occupied 80% of the dish, they were treated with Huanglian decoction for 24 h in culture medium containing 10% FBS. The harvested cells were then disrupted, and the protein concentration was determined using a dye-binding protein assay kit according to the manufacturer’s instruction manual. After centrifugation at 12000 rpm (9660 × g) for 30 min, the supernatants were collected for protein measurement. After the samples were immersed in water at a temperature of 95 °C for 10 min, the proteins were separated using gel electrophoresis and transferred to a polyvinylidene fluoride membrane. Thereafter, the membranes were incubated overnight at 4 °C with the primary antibody [Rabbit monoclonal (Y106) to Cyclin B1, ab32053, 1/30000 dilution, Abcam, United States] and then further incubated with the secondary antibody (Goat Anti-Rabbit immunoglobulin G H&L (horseradish peroxidase), ab205718, 1/5000 dilution, Abcam, United States). The results were then observed after X-ray exposure, development, and fixation of membranes in a dark environment.
Based on three independent experiments, data are expressed as mean ± SEM. All statistical analyses were implemented using R software version 3.5.0 (R Foundation for Statistical Computing, Vienna, Austria) and GraphPad Prism version 8.0.1 (GraphPad Software Inc., United States). If not specified above, a level of P < 0.05 was considered statistically significant.
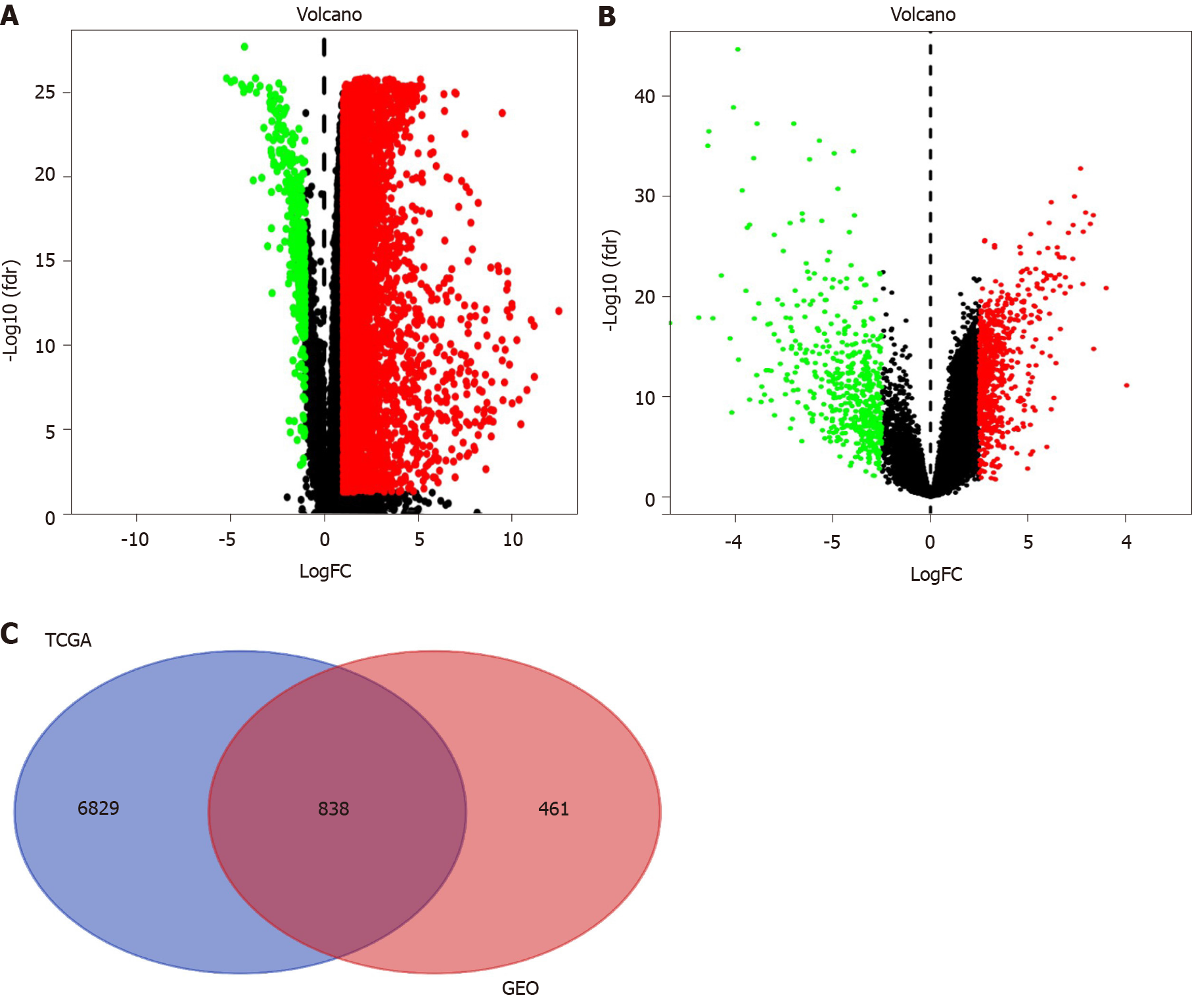
We conducted this study as illustrated in the flow chart (Figure 1). To identify the common DEGs in the TCGA and GEO datasets, we performed differential gene analysis on the two datasets. We identified a total of 7667 DEGs at the mRNA level in tumor tissues (n = 374) compared with normal tissues (n = 50) based on the analysis of the TCGA dataset (Figure 2A). On the other hand, we identified a total of 1299 DEGs at the mRNA level in tumor tissues (n = 93) compared with normal tissues (n = 41) according to the GEO dataset analysis (Figure 2B). In summary, these results show an overlap of 838 DEGs (Figure 2C), between the TCGA and GEO datasets.
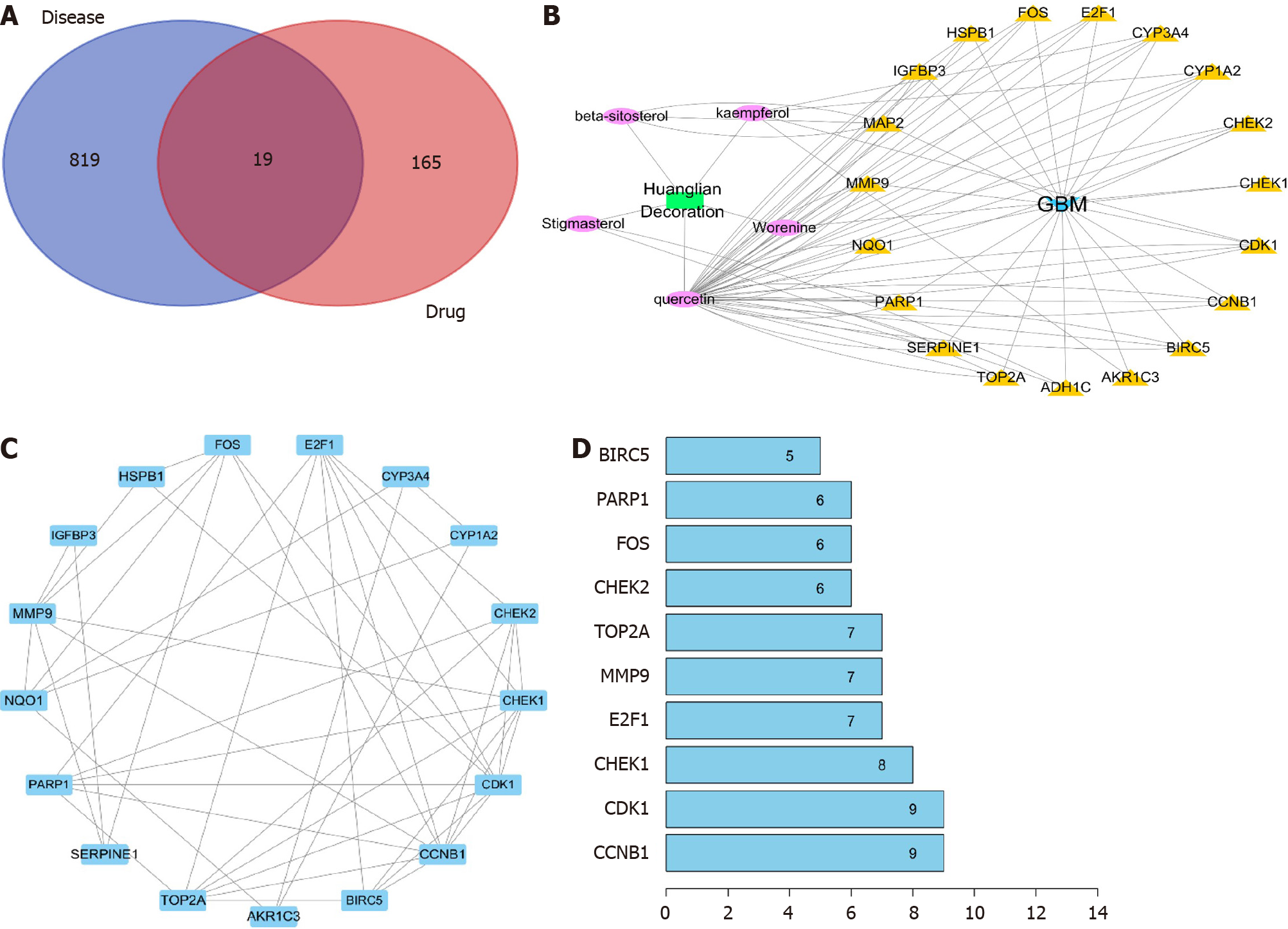
The four main active ingredients in Huanglian decoction are Coptidis Rhizoma, Zingiberis Rhizoma, Folium Artemisiae Argyi, and Mume Fructus. We retrieved the main chemical constituents of the four components from the TCMSP analytical platform and selected OB ≥ 30% and DL index ≥ 0.18 to determine the druggability of the compounds. Overall, we identified 765 bioactive compounds of the four main herbs, which were reduced to 24 after deleting duplicates (Supplementary Table 1).
To further understand the target genes of Huanglian decoction, we constructed a regulatory network. First, the DEGs of disease and the target gene of Huanglian decoction were subjected to intersection analysis, and 19 common genes were found (Figure 3A and Supplementary Table 2). Then, we constructed the Huanglian decoction-compound, compound-target genes, and target gene-disease networks. As shown in Figure 3B, we uncovered 5 biologically active substances (Quercetin, Stigmasterol, beta-sitosterol, Worenine, and kaempferol) and 19 target genes (HSPB1, CCNB1, CYP1A2, BIRC5, NQO1, CDK1, FOS, CHEK1, TOP2A, E2F1, CHEK2, MAP2, SERPINE1, ADH1C, AKR1C3, IGFBP3, CYP3A4, MMP9, and PARP1). Finally, we constructed a protein interaction network for these 19 genes (Figure 3C) and screened the top 10 genes according to the number of nodes (Figure 3D).
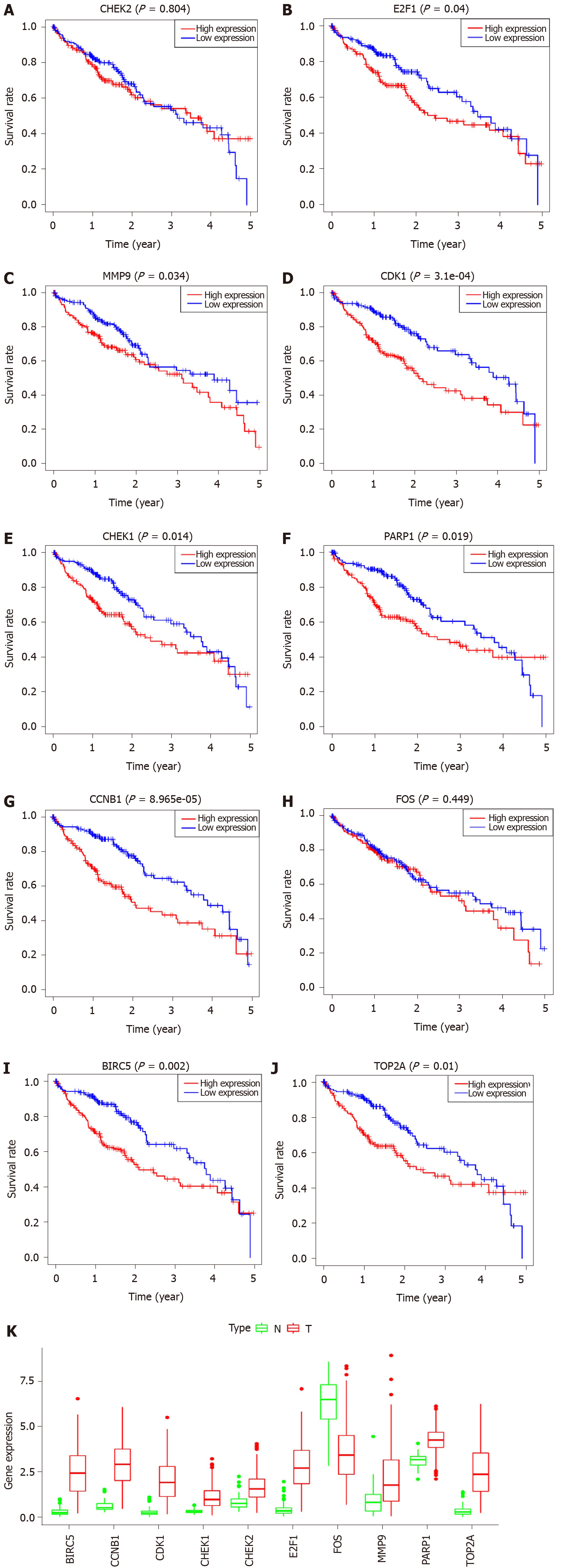
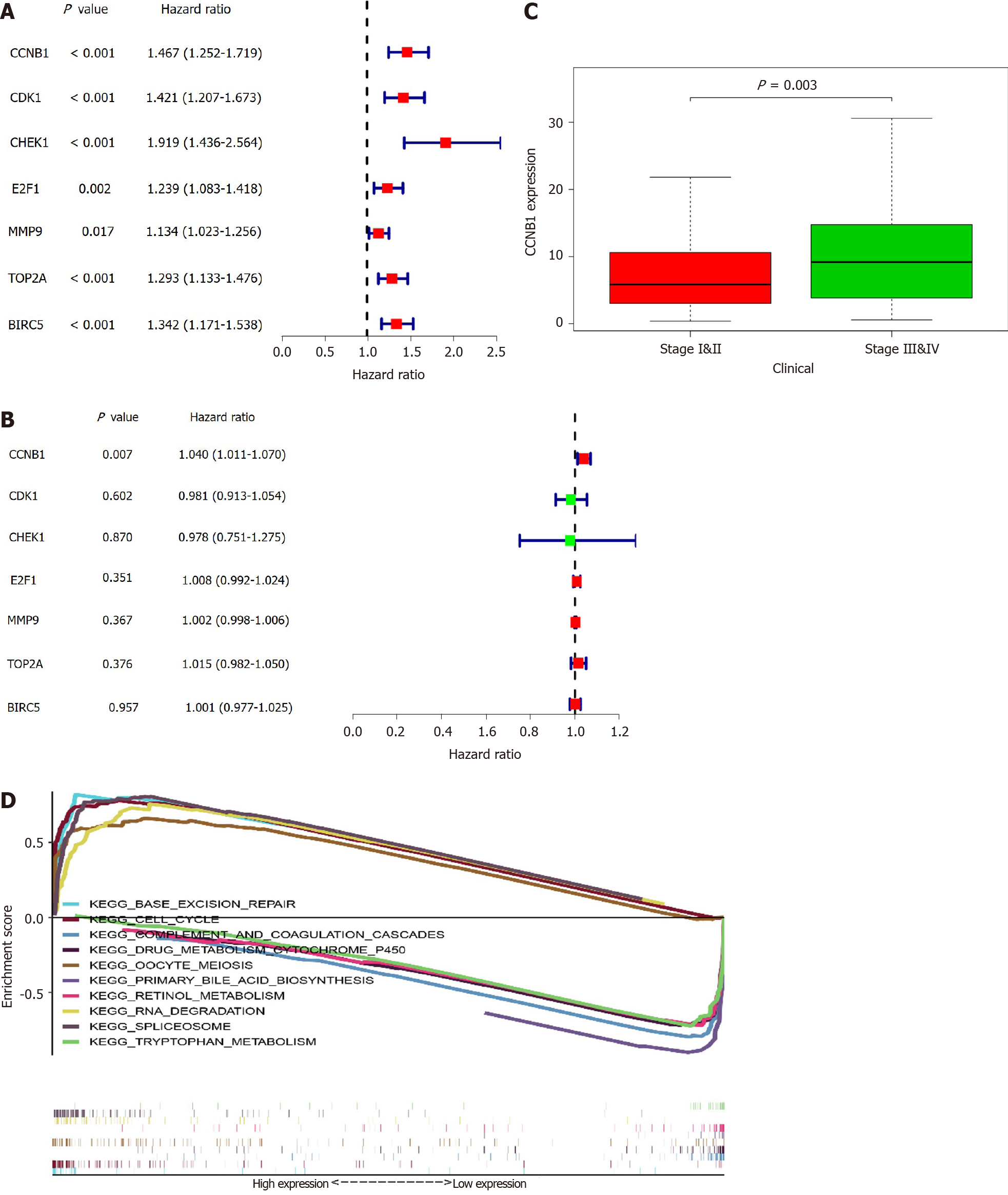
In all, 10 genes were identified as hub genes. The survival analysis of the hub genes was evaluated using a Kaplan-Meier curve (Figure 4). Overall, we identified 7 genes that were significantly associated with prognosis [E2F1 (P = 0.005), MMP9 (P = 0.014), CDK1 (P = 1.36e-04), CHEK1 (P = 7.838e-04), CCNB1 (P = 0.002), BIRC5 (P = 4.06e-05), and TOP2A (P = 0.001)]. We then analyzed the expression of these genes in tumors and normal tissues and found that all except FOS were highly expressed in tumors.
To further examine the role of key genes in prognosis, we performed univariate and multivariate Cox analyses of these genes. From the results of the univariate Cox analysis, we noted that CCNB1 [hazard ratio (HR) = 1.467, 95%CI: 1.252-1.719; P < 0.001], CDK1 (HR = 1.421, 95%CI: 1.207-1.673; P < 0.001), CHEK1 (HR = 1.919, 95%CI: 1.436-2.564; P < 0.001), E2F1 (HR = 1.134, 95%CI: 1.083-1.418; P = 0.002), MMP9 (HR = 1.134, 95%CI: 1.023-1.256; P = 0.017), TOP2A (HR = 1.293, 95%CI: 1.133-1.476; P < 0.001), and BIRC5 (HR = 1.342, 95%CI: 1.171-1.538; P < 0.001) were all associated with poorer outcomes (Figure 5A). From the multivariate Cox analysis, we found that only CCNB1 (HR = 1.040, 95%CI: 1.011-1.070; P = 0.007) was associated with a worse prognosis (Figure 5B). These findings revealed that only CCNB1 can serve as an independent prognostic gene. Subsequently, we noted that CCNB1 was associated with tumor development (Figure 5C) and with multiple biological pathways (Table 1 and Figure 5D). Finally, we performed GO (Supplementary Figure 1) and KEGG (Supplementary Figure 2) enrichment analyses on CCNB1 using R software.
| Name | ES | NES | NOM P value | FDR q-value |
| KEGG_CELL_CYCLE | 0.78 | 2.24 | 0.000 | 0.000 |
| KEGG_BASE_EXCISION_REPAIR | 0.82 | 2.23 | 0.000 | 0.000 |
| KEGG_RNA_DEGRADATION | 0.76 | 2.17 | 0.000 | 0.000 |
| KEGG_SPLICEOSOME | 0.81 | 2.16 | 0.000 | 0.000 |
| KEGG_OOCYTE_MEIOSIS | 0.66 | 2.14 | 0.000 | 0.000 |
| KEGG_COMPLEMENT_AND_COAGULATION_CASCADES | -0.80 | -2.27 | 0.000 | 0.000 |
| KEGG_DRUG_METABOLISM_CYTOCHROME_P450 | -0.72 | -2.17 | 0.000 | 0.000 |
| KEGG_RETINOL_METABOLISM | -0.73 | -2.16 | 0.000 | 0.000 |
| KEGG_PRIMARY_BILE_ACID_BIOSYNTHESIS | -0.92 | -2.09 | 0.000 | 0.000 |
| KEGG_TRYPTOPHAN_METABOLISM | 0.73 | -2.09 | 0.000 | 0.000 |
Here, we performed cell viability assays on liver cancer cells, the outcomes of which are depicted in Figure 6A. These findings demonstrate that Huanglian decoction exhibited time- and dose-dependent effects on HCC cell viability. The IC50 value of Huanglian decoction after 48 h of treatment was approximately 100 μg/mL for PLC/PRF/5 and 200 μg/mL for HepG2 cells. Moreover, the apoptosis analysis indicated that this decoction can induce apoptosis in both PLC/PRF/5 and HepG2 cells (Figure 6B). Interestingly, further analysis showed that this decoction can reduce the migration and invasion activities of PLC/PRF/5 and HepG2 cells (Figure 6C and D).
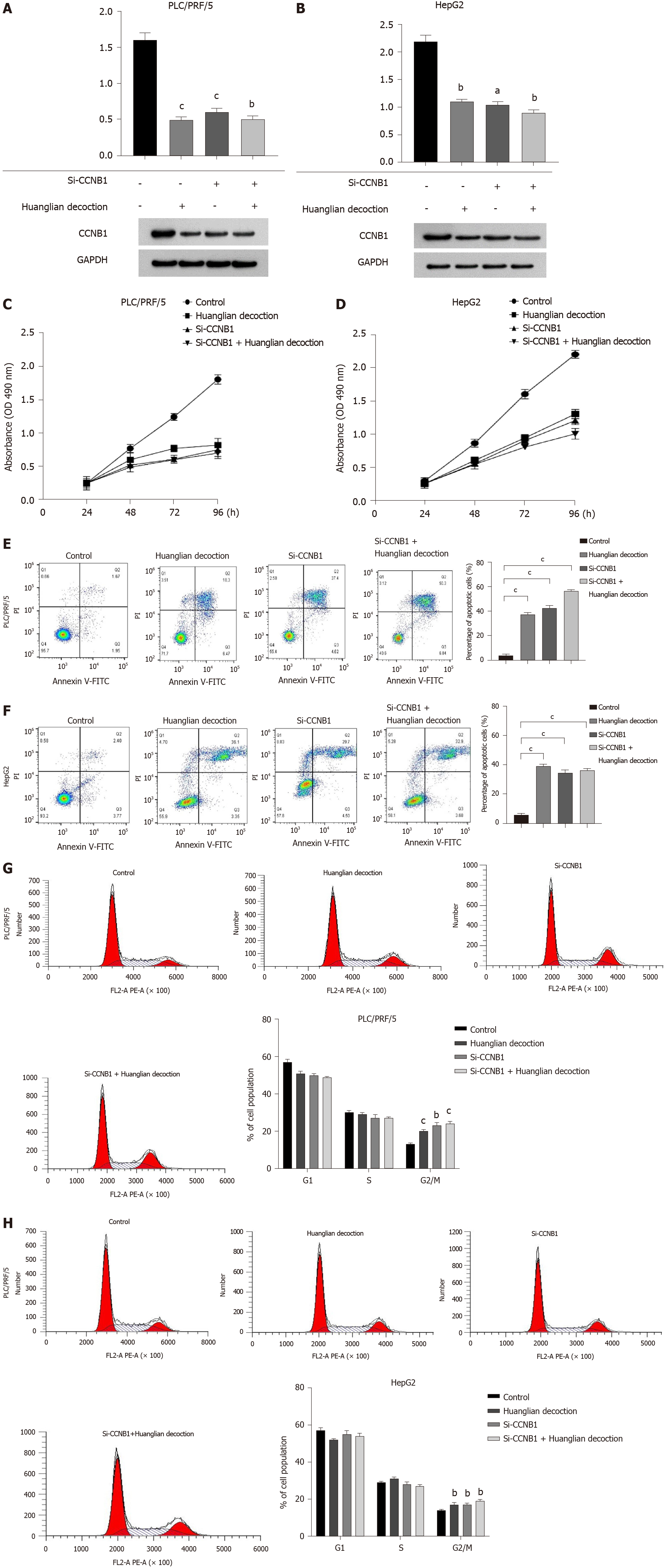
To explore the potential mechanism of Huanglian decoction on HCC cells, we first identified the protein level. Our results highlighted that this decoction can reduce the CCNB1 protein expression levels in both PLC/PRF/5 and HepG2 cells. Furthermore, we noted that after adding this decoction (100 μg/mL for PLC/PRF/5 cells and 200 μg/mL for HepG2 cells) to the si-CCNB1 group, the protein level did not change significantly (Figure 7A and B). Notably, the results of cell proliferation showed that the growth rate of cells was significantly reduced after transfection. Another important finding was that when this decoction was added to the transfected cells, the growth rate of the cells did not change significantly (Figure 7C and D). Remarkably, the findings of cell apoptosis analysis revealed that inhibiting the expression of CCNB1 can promote apoptosis of both PLC/PRF/5 and HepG2 cells, whereas adding Huanglian decoction to the cells after transfection did not significantly promote apoptosis (Figure 7E and F). The cell cycle analysis revealed that Huanglian decoction treatment caused a significant accumulation of cells in G2/M phase in both cell lines (Figure 7G and H), which may be regulated by CCNB1.
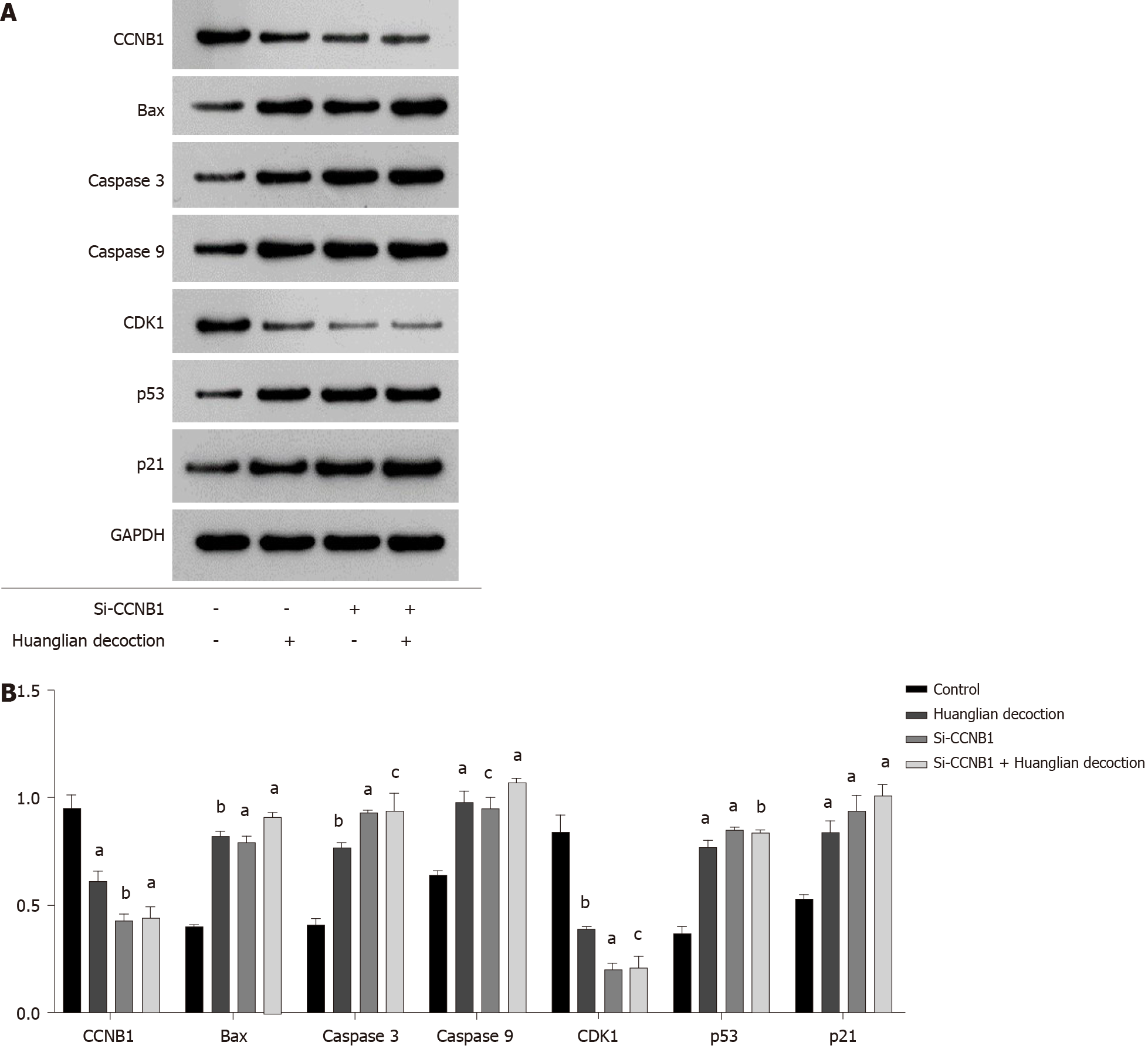
To further explore the effect of Huanglian decoction on HCC cells, we analyzed the expression levels of several key proteins following treatment of PLC/PRF/5 cells with Huanglian decoction for 48 h (Figure 8A). From the results shown in Figure 8B, we found that the expression levels of apoptosis-related proteins (Bax, caspase 3 and caspase 9) in the Huanglian decoction treatment group and the si-CCNB1 group were upregulated.
Western blotting was performed to examine the expression levels of key regulators responsible for cell cycle regulation (G2/M), including CCNB1, CDK1, and p21. The complex CyclinB1/CDK1 formation is the essential player in G2/M transition, and the decrease of CyclinB1/CDK1 formation will induce G2/M arrest. As shown in Figure 8B, the expression levels of CDK1 and CCNB1 in the Huanglian decoction treatment group and the si-CCNB1 group were both decreased, while the expression level of p21 was significantly increased.
It is well known that the p53 pathway plays an important role in the process of cancer treatment. Jin et al[43] noted that CCNB1 silencing significantly promoted activation of the p53 pathway in HCC cells[43]. We have shown that Huanglian decoction works mainly by inhibiting CCNB1, and thus, we speculate that Huanglian decoction may affect the p53 pathway. We first verified the expression of CDK1, p53, and p21 in the p53 pathway and found that in the Huanglian decoction treatment group and the si-CCNB1 group, the expression of CDK1 was significantly reduced, while the expression of p53 and p21 was greatly increased (Figure 8B).
In this study, we found that Huanglian decoction exhibits an effective therapeutic effect on HCC, primarily by inhibiting the expression level of CCNB1. Based on the analysis of TCGA and GEO databases, we identified DEGs in HCC. Subsequently, we also uncovered the main bioactive compounds associated with HCC in Huanglian decoction and key target genes using network pharmacological analysis. Simultaneously, we further screened key genes through PPI network and survival analyses and ultimately, selected 7 genes (E2F1, MMP9, CDK1, CHEK1, CCNB1, BIRC5, and TOP2A) that met the conditions. We then performed univariate and multivariate Cox analyses on these 7 genes. The outcomes indicated that CCNB1 can be used as an independent prognostic indicator. Another important finding is that CCNB1 is involved in the cell cycle, base excision repair, RNA degradation, spliceosome, oocyte meiosis, complement and coagulation cascades, drug metabolism-cytochrome P450, retinol metabolism, primary bile acid biosynthesis, and the tryptophan metabolism signaling pathway of HCC (Figure 5D). Previous studies established that the normal process of cell division occurs through the cell cycle. However, dysregulation of the cell cycle would result in sustained unscheduled cell growth, proliferation, and eventually a hallmark of cancer. Base excision repair is a key genomic maintenance pathway that possesses a tumor-suppressing effect. Therefore, CCNB1 may regulate the development of HCC through these pathways. We also established an in vitro experimental model. Our results show that Huanglian decoction can inhibit the growth of HCC cells in vitro. Further analysis elucidated that this decoction exhibited no obvious inhibitory ability on HCC cells (si-CCNB1). Thus, we speculated that this decoction might inhibit the growth of HCC cells by suppressing the expression of CCNB1.
Induction of apoptosis in cancer cells is a key therapeutic strategy for cancer treatment. Following Huanglian decoction treatment, we found that apoptosis of HCC cells was significantly increased and that the expression of apoptosis-related proteins was increased. The results of the si-CCNB1 group and the si-CCNB1 + Huanglian decoction group showed that Huanglian decoction mainly affected cell apoptosis through CCNB1. The cell cycle is a complex and complicated process. We found that Huanglian decoction can induce G2/M phase arrest. Cyclin B1 plays an important role in G2/M transition and during M phase[43,44]. The cyclin-dependent kinase inhibitor p21 protein plays an important role in G2 phase arrest[45] and has been shown to contribute to cell cycle arrest through transcriptional repression of cell cycle regulatory genes. Therefore, Huanglian decoction may regulate G2/M cell cycle arrest through p21. Our study showed that CCNB1 silencing suppressed the expression of CCNB1 and CDK1 but increased the expression of Bax, caspase 3, caspase 9, p53 and p21. Bax has been reported to be directly activated by p53 in the absence of other proteins to permeabilize mitochondria and initiate the apoptotic program[46-48]. Moreover, p53-induced apoptosis involves triggering the caspase-9 initiator and its downstream caspase-3 executioner. Previous studies have shown that CCNB1 silencing can inhibit cell proliferation and promote cell senescence by activating the p53 signaling pathway in pancreatic cancer[21]. We found that the same phenomenon exists in HCC. Therefore, we speculate that Huanglian decoction can prevent HCC cell progression by inhibiting CCNB1 expression and activating the p53 signaling pathway.
Multiple reports have emphasized that Chinese medicine formula plays an essential role in the treatment of tumors. For instance, Ze-Qi-Tang (ZQT) can inhibit the growth of non-small cell lung cancer cells[49], whereas Jianpi Jiedu decoction can inhibit the tumorigenesis, metastasis, and angiogenesis of colorectal cancer[50]. In addition, Huanglian Jiedu decoction can induce apoptosis and cell cycle arrest and inhibit the migratory and invasive properties of HCC cells[19,51]. These results further support the idea that TCM formulas are potential complementary and alternative treatments for cancer. Nonetheless, our literature search found that most of the TCM formulas related to Huanglian include Huanglian Jiedu, Huanglian-Wendan, and Huanglian-Renshen decoctions and that studies on Huanglian decoction are scarce. Interestingly, a recent article reported that Huanglian decoction exerts a certain therapeutic effect on diabetes, but the formula source of this decoction is somewhat different from the formula used in this study, and thus, it may not have reference value. However, it is important to note that exploration of the differences among various formulas and their potential medicinal value may be the next step. This is a critical area that should be addressed in future research.
Admittedly, our study had some limitations. First, we did not investigate the mechanism of CCNB1 in HCC. Moreover, it would be better if in vivo experiments were performed to validate our findings.
In summary, this study demonstrates that CCNB1 is an independent prognostic indicator in HCC and is significantly associated with a worse prognosis. We confirmed that Huanglian decoction suppressed HCC cell growth in vitro by inhibiting the expression level of the CCNB1 protein. We also explored the mechanism of Huanglian decoction in HCC cells. The results showed that Huanglian decoction can inhibit HCC cell growth by downregulating the expression of CCNB1 to activate the p53 signaling pathway. CCNB1 is a potential therapeutic target for liver cancer. Therefore, this information may help in the development of novel HCC treatments.
Hepatocellular carcinoma (HCC) is the most common primary malignancy of the liver and is the third leading cause of cancer-related mortality worldwide. Huanglian decoction is one of the most important Chinese medicine formulas, with the potential to treat cancer.
In recent years, the application of traditional Chinese medicine in the field of cancer treatment has increased. However, research on Huanglian decoction is still insufficient. We believe that Huanglian decoction may have the potential to treat HCC.
Our research aimed to clarify the effect of Huanglian decoction on HCC cells and to analyze its primary targets and potential functional pathways.
In our study, we analyzed the regulatory network of Huanglian decoction and HCC through bioinformatics and network pharmacology and identified key genes. In vitro experiments were performed to verify the effect of Huanglian decoction on HCC cells, and the CCNB1 gene was knocked down to confirm that it is the main target of Huanglian decoction. Determination of the expression levels of key proteins verified the primary mechanism of Huanglian decoction.
Huanglian decoction can significantly inhibit the growth, migration and invasiveness of HCC cells and can also induce apoptosis and G2/M phase arrest. The results of the network pharmacological analysis showed that the main target of Huanglian decoction in HCC is CCNB1. We verified that Huanglian decoction had an inhibitory effect on liver cancer cells mainly via CCNB1 downregulation. Following Huanglian decoction treatment, the expression levels of Bax, caspase 3, caspase 9, p21 and p53 in HCC cells were increased, while the expression of CDK1 and CCNB1 was significantly decreased. Finally, the p53 signaling pathway plays an important role in this process.
Huanglian decoction has a significant inhibitory effect on HCC cells. Further analysis showed that Huanglian decoction can inhibit HCC cell growth by downregulating CCNB1 expression to activate the p53 signaling pathway.
Huanglian decoction has the potential to treat HCC, and CCNB1 is a key therapeutic target in HCC.
We would like to thank everyone who contributed to this article. We thank Dr. Qin Sun from the Statistics Center of Sichuan Provincial People's Hospital for his assistance in statistical analysis. We thank Dr. Le-Yi Dai from Beijing Foreign Studies University for his assistance in language modification.
| 1. | El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-2576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3846] [Cited by in RCA: 4301] [Article Influence: 226.4] [Reference Citation Analysis (2)] |
| 2. | Armengol C, Sarrias MR, Sala M. Hepatocellular carcinoma: Present and future. Med Clin (Barc). 2018;150:390-397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 3. | Cabrera R, Nelson DR. Review article: the management of hepatocellular carcinoma. Aliment Pharmacol Ther. 2010;31:461-476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 161] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 4. | Qi F, Zhao L, Zhou A, Zhang B, Li A, Wang Z, Han J. The advantages of using traditional Chinese medicine as an adjunctive therapy in the whole course of cancer treatment instead of only terminal stage of cancer. Biosci Trends. 2015;9:16-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 340] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 5. | Ying J, Zhang M, Qiu X, Lu Y. The potential of herb medicines in the treatment of esophageal cancer. Biomed Pharmacother. 2018;103:381-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 6. | Yan Z, Lai Z, Lin J. Anticancer Properties of Traditional Chinese Medicine. Comb Chem High Throughput Screen. 2017;20:423-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 111] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 7. | Cheng CS, Chen J, Tan HY, Wang N, Chen Z, Feng Y. Scutellaria baicalensis and Cancer Treatment: Recent Progress and Perspectives in Biomedical and Clinical Studies. Am J Chin Med. 2018;46:25-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 95] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 8. | Ming-Hua C, Bao-Hua Z, Lei Y. Mechanisms of Anorexia Cancer Cachexia Syndrome and Potential Benefits of Traditional Medicine and Natural Herbs. Curr Pharm Biotechnol. 2016;17:1147-1152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 9. | Wang SF, Wu MY, Cai CZ, Li M, Lu JH. Autophagy modulators from traditional Chinese medicine: Mechanisms and therapeutic potentials for cancer and neurodegenerative diseases. J Ethnopharmacol. 2016;194:861-876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 10. | Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4:682-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2254] [Cited by in RCA: 2948] [Article Influence: 163.8] [Reference Citation Analysis (0)] |
| 11. | Ye H, Wei J, Tang K, Feuers R, Hong H. Drug Repositioning Through Network Pharmacology. Curr Top Med Chem. 2016;16:3646-3656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 64] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 12. | Boezio B, Audouze K, Ducrot P, Taboureau O. Network-based Approaches in Pharmacology. Mol Inform. 2017;36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 238] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 13. | Pan L, Li Z, Wang Y, Zhang B, Liu G, Liu J. Network pharmacology and metabolomics study on the intervention of traditional Chinese medicine Huanglian Decoction in rats with type 2 diabetes mellitus. J Ethnopharmacol. 2020;258:112842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 108] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 14. | Zhang MQ, Wilkinson B. Drug discovery beyond the 'rule-of-five'. Curr Opin Biotechnol. 2007;18:478-488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 253] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 15. | Matter H, Baringhaus KH, Naumann T, Klabunde T, Pirard B. Computational approaches towards the rational design of drug-like compound libraries. Comb Chem High Throughput Screen. 2001;4:453-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 43] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 16. | Tao W, Xu X, Wang X, Li B, Wang Y, Li Y, Yang L. Network pharmacology-based prediction of the active ingredients and potential targets of Chinese herbal Radix Curcumae formula for application to cardiovascular disease. J Ethnopharmacol. 2013;145:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 368] [Cited by in RCA: 479] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 17. | Xie W, Meng X, Zhai Y, Zhou P, Ye T, Wang Z, Sun G, Sun X. Panax Notoginseng Saponins: A Review of Its Mechanisms of Antidepressant or Anxiolytic Effects and Network Analysis on Phytochemistry and Pharmacology. Molecules. 2018;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 100] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 18. | Ohta Y, Sasaki E, Nishida K, Hayashi T, Nagata M, Ishiguro I. Preventive effect of oren-gedoku-to (huanglian-jie-du-tang) extract on progression of carbon tetrachloride-induced acute liver injury in rats. Am J Chin Med. 1997;25:57-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 19. | Huang J, Guo W, Cheung F, Tan HY, Wang N, Feng Y. Integrating Network Pharmacology and Experimental Models to Investigate the Efficacy of Coptidis and Scutellaria Containing Huanglian Jiedu Decoction on Hepatocellular Carcinoma. Am J Chin Med. 2020;48:161-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 58] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 20. | Jia KK, Ding H, Yu HW, Dong TJ, Pan Y, Kong LD. Huanglian-Wendan Decoction Inhibits NF-κB/NLRP3 Inflammasome Activation in Liver and Brain of Rats Exposed to Chronic Unpredictable Mild Stress. Mediators Inflamm. 2018;2018:3093516. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 21. | Zhang H, Zhang X, Li X, Meng WB, Bai ZT, Rui SZ, Wang ZF, Zhou WC, Jin XD. Effect of CCNB1 silencing on cell cycle, senescence, and apoptosis through the p53 signaling pathway in pancreatic cancer. J Cell Physiol. 2018;234:619-631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 140] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 22. | Fang Y, Yu H, Liang X, Xu J, Cai X. Chk1-induced CCNB1 overexpression promotes cell proliferation and tumor growth in human colorectal cancer. Cancer Biol Ther. 2014;15:1268-1279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 129] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 23. | Liu A, Zeng S, Lu X, Xiong Q, Xue Y, Tong L, Xu W, Sun Y, Zhang Z, Xu C. Overexpression of G2 and S phase-expressed-1 contributes to cell proliferation, migration, and invasion via regulating p53/FoxM1/CCNB1 pathway and predicts poor prognosis in bladder cancer. Int J Biol Macromol. 2019;123:322-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 24. | Chen EB, Qin X, Peng K, Li Q, Tang C, Wei YC, Yu S, Gan L, Liu TS. HnRNPR-CCNB1/CENPF axis contributes to gastric cancer proliferation and metastasis. Aging (Albany NY). 2019;11:7473-7491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 90] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 25. | Kanapathipillai M. Treating p53 Mutant Aggregation-Associated Cancer. Cancers (Basel). 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 159] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 26. | Vieler M, Sanyal S. p53 Isoforms and Their Implications in Cancer. Cancers (Basel). 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 95] [Article Influence: 11.9] [Reference Citation Analysis (9)] |
| 27. | Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20:1803-1815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1121] [Cited by in RCA: 1221] [Article Influence: 48.8] [Reference Citation Analysis (0)] |
| 28. | Zhang J, Huang K, O'Neill KL, Pang X, Luo X. Bax/Bak activation in the absence of Bid, Bim, Puma, and p53. Cell Death Dis. 2016;7:e2266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 60] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 29. | Li Q, Zhang L, Jiang J, Zhang Y, Wang X, Zhang Q, Wang Y, Liu C, Li F. CDK1 and CCNB1 as potential diagnostic markers of rhabdomyosarcoma: validation following bioinformatics analysis. BMC Med Genomics. 2019;12:198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 30. | Kibble M, Saarinen N, Tang J, Wennerberg K, Mäkelä S, Aittokallio T. Network pharmacology applications to map the unexplored target space and therapeutic potential of natural products. Nat Prod Rep. 2015;32:1249-1266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 334] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 31. | Danhof M. Systems pharmacology - Towards the modeling of network interactions. Eur J Pharm Sci. 2016;94:4-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 87] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 32. | Bortolomeazzi M, Keddar MR, Ciccarelli FD, Benedetti L. Identification of non-cancer cells from cancer transcriptomic data. Biochim Biophys Acta Gene Regul Mech. 2020;1863:194445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 33. | Wu X, Hasan MA, Chen JY. Pathway and network analysis in proteomics. J Theor Biol. 2014;362:44-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 84] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 34. | Fan Z, Liu H, Xue Y, Lin J, Fu Y, Xia Z, Pan D, Zhang J, Qiao K, Zhang Z, Liao Y. Reversing cold tumors to hot: An immunoadjuvant-functionalized metal-organic framework for multimodal imaging-guided synergistic photo-immunotherapy. Bioact Mater. 2021;6:312-325. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 111] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 35. | Rampersad SN. Multiple applications of Alamar Blue as an indicator of metabolic function and cellular health in cell viability bioassays. Sensors (Basel). 2012;12:12347-12360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 668] [Cited by in RCA: 717] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 36. | Stoddart MJ. Cell viability assays: introduction. Methods Mol Biol. 2011;740:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 150] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 37. | Sakata S, Enoki Y, Tomita S, Kohzuki H. In vitro erythropoietin assay based on erythroid colony formation in fetal mouse liver cell culture. Br J Haematol. 1985;61:293-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 38. | Heyworth CM, Dexter TM, Nicholls SE, Whetton AD. Protein kinase C activators can interact synergistically with granulocyte colony-stimulating factor or interleukin-6 to stimulate colony formation from enriched granulocyte-macrophage colony-forming cells. Blood. 1993;81:894-900. [PubMed] |
| 39. | Eliason JF, Odartchenko N. Colony formation by primitive hemopoietic progenitor cells in serum-free medium. Proc Natl Acad Sci USA. 1985;82:775-779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 28] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 40. | Planz V, Wang J, Windbergs M. Establishment of a cell-based wound healing assay for bio-relevant testing of wound therapeutics. J Pharmacol Toxicol Methods. 2018;89:19-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 41. | Grada A, Otero-Vinas M, Prieto-Castrillo F, Obagi Z, Falanga V. Research Techniques Made Simple: Analysis of Collective Cell Migration Using the Wound Healing Assay. J Invest Dermatol. 2017;137:e11-e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 516] [Article Influence: 57.3] [Reference Citation Analysis (0)] |
| 42. | Luan S, Hao R, Wei Y, Chen D, Fan B, Dong F, Guo W, Wang J, Chen J. A microfabricated 96-well wound-healing assay. Cytometry A. 2017;91:1192-1199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 43. | Jin J, Xu H, Li W, Xu X, Liu H, Wei F. LINC00346 Acts as a Competing Endogenous RNA Regulating Development of Hepatocellular Carcinoma via Modulating CDK1/CCNB1 Axis. Front Bioeng Biotechnol. 2020;8:54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 44. | Xie B, Wang S, Jiang N, Li JJ. Cyclin B1/CDK1-regulated mitochondrial bioenergetics in cell cycle progression and tumor resistance. Cancer Lett. 2019;443:56-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 182] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 45. | Cazzalini O, Scovassi AI, Savio M, Stivala LA, Prosperi E. Multiple roles of the cell cycle inhibitor p21(CDKN1A) in the DNA damage response. Mutat Res. 2010;704:12-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 304] [Cited by in RCA: 358] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 46. | Mirakhor Samani S, Ezazi Bojnordi T, Zarghampour M, Merat S, Fouladi DF. Expression of p53, Bcl-2 and Bax in endometrial carcinoma, endometrial hyperplasia and normal endometrium: a histopathological study. J Obstet Gynaecol. 2018;38:999-1004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 47. | Kim EM, Jung CH, Kim J, Hwang SG, Park JK, Um HD. The p53/p21 Complex Regulates Cancer Cell Invasion and Apoptosis by Targeting Bcl-2 Family Proteins. Cancer Res. 2017;77:3092-3100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 161] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 48. | Yao TH, Pataer P, Regmi KP, Gu XW, Li QY, Du JT, Ge SM, Tu JB. Propranolol induces hemangioma endothelial cell apoptosis via a p53BAX mediated pathway. Mol Med Rep. 2018;18:684-694. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 49. | Xu Z, Zhang F, Zhu Y, Liu F, Chen X, Wei L, Zhang N, Zhou Q, Zhong H, Yao C, Zhu X, Gong C, Zhu S, Zou C. Traditional Chinese medicine Ze-Qi-Tang formula inhibit growth of non-small-cell lung cancer cells through the p53 pathway. J Ethnopharmacol. 2019;234:180-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 50. | Peng W, Zhang S, Zhang Z, Xu P, Mao D, Huang S, Chen B, Zhang C, Zhang S. Jianpi Jiedu decoction, a traditional Chinese medicine formula, inhibits tumorigenesis, metastasis, and angiogenesis through the mTOR/HIF-1α/VEGF pathway. J Ethnopharmacol. 2018;224:140-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 51. | Wang N, Feng Y, Tan HY, Cheung F, Hong M, Lao L, Nagamatsu T. Inhibition of eukaryotic elongation factor-2 confers to tumor suppression by a herbal formulation Huanglian-Jiedu decoction in human hepatocellular carcinoma. J Ethnopharmacol. 2015;164:309-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/Licenses/by-nc/4.0/
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: New Zealand
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Su SB, Wang L, Xia Q S-Editor: Fan JR L-Editor: Webster JR P-Editor: Liu JH