Published online Feb 7, 2020. doi: 10.3748/wjg.v26.i5.478
Peer-review started: October 9, 2019
First decision: December 5, 2019
Revised: December 20, 2019
Accepted: January 8, 2020
Article in press: January 8, 2020
Published online: February 7, 2020
Processing time: 120 Days and 16.1 Hours
Statistics indicate that the incidence of Crohn’s disease (CD) is rising in many countries. The poor understanding on the pathological mechanism has limited the development of effective therapy against this disease. Previous studies showed that long noncoding RNAs (lncRNAs) could be involved in autoimmune diseases including CD, but the detailed molecular mechanisms remain unclear.
To identify the differentially expressed lncRNAs in the intestinal mucosa associated with CD, and to characterize their pathogenic role(s) and related mechanisms.
The differential expression of lncRNAs was screened by high-throughput RNA sequencing, and the top candidate genes were validated in an expanded cohort by real-time PCR. The regulatory network was predicted by bioinformatic software and competitive endogenous RNA analysis, and was characterized in Caco-2 and HT-29 cell culture using methods of cell transfection, real-time PCR, Western blotting analysis, flow cytometry, and cell migration and invasion assays. Finally, these findings were confirmed in vivo using a CD animal model.
The 3' end of lncRNACNN3-206 and the 3’ UTR of Caspase10 contain high-affinity miR212 binding sites. lncRNACNN3-206 expression was found to be significantly increased in intestinal lesions of CD patients. Activation of the lncRNACNN3-206-miR-212-Caspase10 regulatory network led to increased apoptosis, migration and invasion in intestinal epithelial cells. Knockdown of lncRNACNN3-206 expression alleviated intestinal mucosal inflammation and tissue damage in the CD mouse model.
lncRNACNN3-206 may play a key role in CD pathogenesis. lncRNACNN3-206 could be a therapeutic target for CD treatment.
Core tip: We describe new findings on the overexpression of lncRNACNN3-206 in intestinal lesions of Crohn’s disease (CD) patients by high throughput sequencing. Moreover, forced overexpression of lncRNACNN3-206 led to apoptosis of Caco-2 and HT-29 cells. Since the 3' end of lncRNACNN3-206 and the 3’ UTR of Caspase10 contain high-affinity binding sites of miR-212, whereby lncRNACNN3-206 regulates Caspase10 expression and cell apoptosis/invasion through miR-212 sponging. This mechanism was supported by results from cell lines and the CD mouse model. In conclusion, lncRNACNN3-206 may play a key role in CD pathogenesis, and could be a therapeutic target for CD treatment.
- Citation: Li N, Shi RH. lncRNACNN3-206 activates intestinal epithelial cell apoptosis and invasion by sponging miR-212, an implication for Crohn's disease. World J Gastroenterol 2020; 26(5): 478-498
- URL: https://www.wjgnet.com/1007-9327/full/v26/i5/478.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i5.478
Long non-coding RNAs (lncRNAs) are gene transcripts longer than 200 nucleotides that lack protein-coding function[1]. Studies have shown that lncRNAs can regulate chromatin modification, gene expression, nuclear RNA transport and other cell activities on the epigenetic, gene transcription and post-transcriptional levels[2,3]. A competitive endogenous RNA (ceRNA) mechanism has been proposed, which describes lncRNAs acting as sponges that absorb microRNAs (miRNAs). This in turn indirectly blocks miRNA-mediated downregulation of target genes[4]. Indeed, alterations in the levels of lncRNA, relevant miRNAs, and concomitant changes in downstream target gene expression and functions have been observed in cancer cells and other pathological processes[5].
Crohn's disease (CD) is a chronic, recurrent, inflammatory disorder of the digestive tract, with clinical manifestations of abdominal pain, diarrhea, enterelcosis and intestinal obstruction. CD lesions are often segmentally distributed, potentially affecting any part of the digestive tract. While the terminal ileum is most commonly affected, lesions in the colon and anus are also observed[6]. Statistics show that the incidence rates of CD are climbing in Europe, the United States and Asian countries. In recent decades, CD incidence and prevalence rates in the last decade were 1.21 per 100000 persons/year and 2.29 per 100,000 persons in China, respectively. These rates are higher than those from 1950-2002, which was 0.28 per 100000 persons/years and 1.38 per 100000 persons, respectively[7]. Due to a poor understanding of the etiology and pathogenesis of this disease, current therapy is limited to anti-symptomatic and surgical treatments. The recurrence rate within 3 years after surgery is as high as 80-100%[8,9]. Exploration of the cellular and molecular mechanisms is urgently required to develop new therapeutic modalities against this disease.
While many studies have indicated that lncRNAs might participate in the pathogenesis of autoimmune diseases, very few studies have focused on whether lncRNAs contribute to CD occurrence. Previous studies using high-throughput gene sequencing techniques have shown that the serum of CD patients contain multiple differentially expressed lncRNAs[10,11]. While these differentially expressed lncRNAs could be used as biomarkers for the diagnosis and/or prognosis of CD, their sources remain unclear. In this study, we collected tissue samples of ileal lesions from CD patients, as well as normal intestinal mucosal tissue samples from healthy individuals. The expression patterns of lncRNAs and mRNAs were compared between the lesions and normal ileal tissue samples by microarray hybridization. Bioinformatic analysis was performed to predict the gene regulatory network of lncRNAs-miRNAs-mRNAs. Biological functions of the lncRNAs, relevant miRNAs and target genes, as well as the corresponding regulatory mechanisms, were investigated comprehensively using cell culture and animal models.
The collection and use of clinical specimens were approved by the Institutional Review Board of the Affiliated Zhongda Hospital of Southeast University. All participating patients and normal control subjects signed the written informed consent form. Tissue samples were obtained from inflammatory lesions from the colon and ileum of patients with active CD (n = 40). Normal control tissue samples were obtained from individuals upon physical examination (n = 40) who were undergoing screening colonoscopies. Patients were accrued between May 2017 and May 2018 in the Zhongda Hospital affiliated with Southeast University (Jiangsu, China). The following types of patients were excluded: (1) Patients with various immune diseases; (2) Patients with acute or chronic inflammation; (3) Patients with hematological diseases; and (4) Patients with high fever or who had used anti-inflammatory drugs, steroids or opiates. The basic demographic and clinical characteristics of CD and control patients are shown in Table 1. All tissue samples were snap-frozen in liquid nitrogen and stored at -80 °C until use.
| Sex/age in yr | Tissue | Diagnosis | Duration in yr | Treatment |
| F/18 | Colon | CD (active) | 1 | 5-ASA + prednisone |
| M/21 | Ileum | CD (active) | 5 | Infliximab + azathioprine |
| F/35 | Ileum | CD (active) | 10 | Infliximab + prednisone |
| F/37 | Colon | CD (active) | 6 | Prednisone |
| M/50 | Colon | CD (active) | 16 | Infliximab + azathioprine |
| M/23 | Colon | CD (active) | 3 | Prednisone |
| F/28 | Colon | CD (active) | 4 | 5-ASA + prednisone |
| M/19 | Colon | CD (active) | 1 | 5-ASA |
| M/22 | Ileum | CD (active) | 3 | 5-ASA + prednisone |
| M/31 | Colon | CD (active) | 5 | Prednisone |
| F/24 | Colon | CD (active) | 10 | Infliximab + prednisone |
| M/49 | Ileum | CD (active) | 15 | Infliximab + azathioprine |
| M/22 | Ileum | CD (active) | 3 | 5-ASA + prednisone |
| M/24 | Colon | CD (active) | 2 | 5-ASA + prednisone |
| F/15 | Colon | CD (active) | 1 | 5-ASA |
| F/42 | Ileum | CD (active) | 20 | Infliximab + azathioprine |
| M/32 | Ileum | CD (active) | 5 | 5-ASA + azathioprine |
| M/40 | Ileum | CD (active) | 5 | 5-ASA + prednisone |
| F/33 | Colon | CD (active) | 7 | Infliximab + prednisone |
| M/35 | Ileum | CD (active) | 3 | Azathioprine |
| M/36 | Ileum | CD (active) | 5 | Azathioprine |
| F/28 | Ileum | CD (active) | 4 | Azathioprine |
| F/33 | Colon | CD (active) | 5 | 5-ASA + azathioprine |
| F/50 | Ileum | CD (active) | Prednisone | |
| F/45 | Ileum | CD (active) | Azathioprine | |
| M/37 | Colon | CD (active) | 6 | Infliximab + prednisone |
| M/43 | Colon | CD (active) | 16 | Infliximab + prednisone |
| M/22 | Colon | CD (active) | 4 | Infliximab + azathioprine |
| M/18 | Colon | CD (active) | 2 | Infliximab + prednisone |
| M/15 | Colon | CD (active) | 5 | Infliximab + azathioprine |
| F/52 | Colon | CD (active) | 10 | Infliximab + prednisone |
| M/23 | Colon | CD (active) | 4 | 5-ASA + prednisone |
| M/45 | Ileum | CD (active) | 3 | 5-ASA |
| F/23 | Colon | CD (active) | 4 | 5-ASA + prednisone |
| F/33 | Colon | CD (active) | 5 | Prednisone |
| F/39 | Colon | CD (active) | 6 | Infliximab + prednisone |
| F/29 | Colon | CD (active) | 9 | Infliximab + azathioprine |
| F/14 | Ileum | CD (active) | 1 | 5-ASA+ prednisone |
| F/23 | Colon | CD (active) | 6 | Azathioprine |
| F/49 | Colon | CD (active) | 15 | Infliximab + azathioprine |
| F/45 | Ileum | NC | 0 | NA |
| M/37 | Ileum | NC | 0 | NA |
| M/39 | Colon | NC | 0 | NA |
| M/22 | Ileum | NC | 0 | NA |
| M/22 | Colon | NC | 0 | NA |
| M/15 | Ileum | NC | 0 | NA |
| F/49 | Colon | NC | 0 | NA |
| M/23 | Ileum | NC | 0 | NA |
| M/40 | Ileum | NC | 0 | NA |
| F/23 | Colon | NC | 0 | NA |
| F/39 | Colon | NC | 0 | NA |
| F/43 | Colon | NC | 0 | NA |
| M/25 | Colon | NC | 0 | NA |
| M/27 | Ileum | NC | 0 | NA |
| F/15 | Colon | NC | 0 | NA |
| F/23 | Colon | NC | 0 | NA |
| M/32 | Colon | NC | 0 | NA |
| M/10 | Colon | NC | 0 | NA |
| F/33 | Colon | NC | 0 | NA |
| M/35 | Ileum | NC | 0 | NA |
| M/37 | Colon | NC | 0 | NA |
| F/28 | Colon | NC | 0 | NA |
| F/33 | Ileum | NC | 0 | NA |
| F/50 | Ileum | NC | 0 | NA |
| F/45 | Colon | NC | 0 | NA |
| M/37 | Colon | NC | 0 | NA |
| M/42 | Colon | NC | 0 | NA |
| M/22 | Ileum | NC | 0 | NA |
| M/18 | Colon | NC | 0 | NA |
| M/15 | Colon | NC | 0 | NA |
| F/52 | Ileum | NC | 0 | NA |
| M/23 | Ileum | NC | 0 | NA |
| M/42 | Ileum | NC | 0 | NA |
| F/23 | Colon | NC | 0 | NA |
| F/35 | Colon | NC | 0 | NA |
| F/35 | Ileum | NC | 0 | NA |
| M/29 | Colon | NC | 0 | NA |
| F/14 | Ileum | NC | 0 | NA |
| F/23 | Ileum | NC | 0 | NA |
| M/42 | Ileum | NC | 0 | NA |
An RNA extraction kit was purchased from Baitek Biological Company (Shenzhen, China). A reverse transcription kit and SYBR Green for quantitative real-time PCR was purchased from Roche (Basel, Switzerland). Primers specific for lncRNACNN3-206, miR-212 and Caspase10 were purchased from Guangzhou Reebok Biological Company. Matrigel matrix was purchased from BD company (Guangzhou, China). Anti-Caspase10, anti-CD4, anti-IFN-γ, anti-IL-4, anti-IL-17, anti-CD25, anti-FoxP3, and anti-β-actin antibodies were purchased from Abcam Biology Company Branch at Shanghai, China. siRNA was designed and provided by the Jima Company (Shanghai, China). 2,4,6-Trinitrobenzene sulfonic acid (TNBS) was purchased from Beijing Yuwei Technology Company (Beijing, China). Pathogen-free male mice weighing 150-180 g were provided by Anhui Medical University Animal Laboratory Center. The human gene chip G4845A containing 27,985 target genes was purchased from Agilent Company (Santa Clara, United States).
The Trizol one-step method was used to extract total RNA from tissue samples. Cy3-dUTP-labeled and Cy5-dUTP-labeled cRNA of CD and normal intestinal mucosa samples were synthesized by the one-step method. The fluorescence-labeled cRNA was purified and fragmented. After hybridization at 65 °C for 17 h, gene chips were washed and dried at room temperature. Scan Array Express3.0 software was used for data analysis. Hybridization results were required to meet the following criteria: both Cy3 and Cy5 signals reached 200, the Cy5/Cy3 ratio was 0.1-10. A more than 10-fold difference between CD and normal samples, and P ≤ 0.05, was considered positive for differential expression.
Tissue samples were fixed with 4% paraformaldehyde for 1-2 h. The sequences of the lncRNACNN3-206 probe were: CCT TTA GCA TCA GTA AGG GAA AGC ATT. Probe in a concentration of 8 ng/µL was added to tissue sections, and incubation was carried out at 37 °C overnight. After washing, tissue sections were incubated with an anti-CK20 antibody at 4 °C overnight. After incubation with secondary antibody (CY3-conjugated goat anti-rabbit antibody) for 50 min at room temperature, DAPI dye solution was added and incubation continued in the dark for 8 min. The sections were observed under a Nikon positive fluorescence microscope, and photographs were taken and analyzed.
The lncRNACNN3-206 sequence was subcloned into a lentiviral vector. After screening with restriction digestion, the vector was sequenced for verification. After cell transfection and confirmation of the efficiency, si-lncRNACNN3-206 (CCT TTA GCA TCA GTA AGG GAA AGC ATT) was also subcloned into a lentiviral vector. HT-29 and Caco-2 cells were maintained in HG DMEM medium. The culture medium was replaced 24 h after lentiviral infection. The transfection efficiencies of the two cell lines were observed under a fluorescence microscope (Supplementary Figure 1). Experiments were performed in five groups: Blank control group, vector control group, lncRNACNN3-206 overexpression (oe-lncRNACNN3-206) group, si-lncRNACNN3-206 group, and scramble-NC group.
Five ng of total RNA were reverse-transcribed into cDNA. Expression levels of the lncRNACNN3-206, miR-212, and Caspase10 were quantified with an ABI7500 instrument using SYBR PCR master mix reagent, and β-actin was used as an internal reference gene. The primers were: lncRNACNN3-206 forward, CAG ATG GGC ACT AAT AAA GGA GC, and lncRNACNN3-206 backward, TGT AGG AGC AGC ACA GTA TTT GG; miR-212 forward, CTC AAC TGG TGT CGT GGA GTC GG, and miR-212 backward, ACA CTC CAG CTG GGA CCT TGG CT; Caspase10 forward,CTC GCT TCG GCA GCA CA, and Caspase10 backward, AAC GCT TCA CGA ATT TGC GT; β-actin forward, CAG ATG GGC ACT AAT AAA GGA GC, and β-actin backward, TGT AGG AGC AGC ACA GTA TTT GG. The relative mRNA expression levels were calculated using the △△Ct method.
Total cellular proteins were extracted from the five groups of transfected cells, and 30 µg of protein was loaded onto an 8% SDS-PAGE gel. Resolved proteins were electrically transferred onto PDVF membranes. Non-specific binding was blocked with 5% skim milk for 2 h. Primary antibody was added, and incubation was performed overnight at room temperature. Membranes were rinsed with PBS before incubation with the secondary antibody for 2 h at room temperature. The ECL luminescence system was used for imaging development. Quantity One analysis software (Bio-Rad Company, United States) was used for densitometry analysis. The membranes were stripped and β-actin levels were detected, and these results were used as a control for protein loading. The relative expression levels of target proteins were presented as the ratio of target protein versus β-actin protein.
The entire 3’ end of human lncRNACNN3-206 3’ and the untranslated regions (3’-UTR) of Caspase10, which contain predicted miR-212 binding sites (seed sequences: GCC AAG GU for Caspase10 and CCU GGC UGA GAC UGU UAC for lncRNACNN3-206), were amplified by PCR using human genomic DNA as templates. PCR products were inserted into the p-MIR-reporter plasmid (Ambion, Austin, TX, United States). To ensure specificity, the lncRNACNN3-206 3’ end seed sequence was mutated to ACC GAA GCA AGG UUG UCA, and the Caspase10 3’ UTR seed sequence was mutated to CGA UUA CG. For luciferase reporter assays, Caco-2 and HT-29 cells were cultured in 24-well plates, and 1 µg of firefly luciferase reporter plasmid DNA, or 1 µg of psiCHECK-2 expression plasmid DNA (Ambion Company), and 100 pmol of pre-miR-212 or scrambled negative control (NC) RNA were transfected into cells using Lipofectamine 2000. The psiCHECK-2 vector plasmid was used as a transfection control. Twenty-four hours after transfection, the cells were assayed using a luciferase detection kit (Promega, 09 Madison, WI, United States), results from reporter plasmid containing the wild type or mutated miR-212 binding sequences in lncRNACNN3-206 and Caspase10 were compared.
Caco-2 and HT-29 cells were grown to a density of 1 × 105 per well on 6-wells plates, and transfected with plasmid expressing lncRNACNN3-206 (OE-lncRNACNN3-206), interfering RNA (si-lncRNACNN3-206), or scramble RNA as a control. Transfection was performed in triplicates for each group. At 48 h post-transfection, cells were collected and fixed with 200 µL of fixation solution (70% ethanol, 5% fetal bovine serum) for 1 h. To determine cell apoptosis, 400 µL of buffer-suspended cells were incubated with 5 µA of Annexin V-FITC at 2-8 °C in the dark for 15 min. Ten microlitre of propidium iodide (PI) was added for nuclei staining. All cells were analyzed 1 h after staining.
A thin layer of matrigel was overlaid on the upper surface of 6.5 mm Trans-well chambers. Matrigel was allowed to solidify by incubating plates for 4 h at room temperature. Culture medium supplemented with 10% FBS was placed in the lower chamber. Caco-2 or HT-29 cells (5 × 104) were seeded in the upper chamber of the Trans-well apparatus. Cells were fixed with 95% ethanol for 20 min, stained with a 0.5% crystal violet solution for 10 min, and washed with water. Invading cells were counted under an inverted microscope. The experiment was repeated three times.
The CD mouse model was constructed with the use of 2,4,6-trinitrobenzenesulfonic acid (TNBS) following the protocols developed by Kremer et al[12]. Sixty female BALB/C mice were randomly divided into the normal control group (n = 15), TNBS group (n = 15), TNBSc + si-lncRNACNN3-206 group (n = 15), and TNBS + Scramble group (n = 15). The “si-lncRNACNN3-206” lentivirus expressed the si-lncRNACNN3-206 that was successfully used in cell culture for the knockdown of lncRNACNN3-206 expression. The control group was fed routine chow, and the CD model was established in the last three groups using TNBS. The mice were shaved and externally treated with 3.75 mg of TNBS (48% ethanol dissolution). The mice were anesthetized with pentobarbital sodium at the 7th, 14th and 21th d after external TNBS application and fasting for 24 h. The mice were placed in the prone position, and a 20 G trocar was inserted into the colon 4 cm from the anus. Immediately, 100 µL of ethanol solution containing TNBS was slowly injected into the intestinal cavity through a trocar-fixed syringe, and TNBS was administered. The mice were inverted for l min, followed by routine postoperative feeding. Supplementary Figure 2 illustrates the scheme for model establishment. In the third and fourth groups, the mice were injected intraperitoneally[13] with 500 µL of 3.0 × 106 PFU virus expressing si-lncRNACNN3-206 or scramble RNAs, respectively. The first (blank control) and second groups (model control) were injected with an equal volume of saline. After the first treatment, weekly disease activity index (DAI) scoring was performed as previously described[14]. The scoring criteria were as follows: 1 point for bloody stool, 1 point for loose stool, 1 point for erect fur, and 1 point for diarrhea or severe rectal prolapse.
Colon tissue samples approximately 1 cm in length, 3 cm away from the anus, were collected for examination of pathological changes in the CD model. The tissue samples were fixed with 10% neutral formalin, embedded with paraffin, sectioned and stained with hematoxylin and eosin (H&E), and the histopathological changes in the colon were observed under a microscope according to the protocol previously described[15].
SPSS 20.0 statistical software was used for data analysis. Quantitative data are expressed as the mean ± SD. ANOVA was performed to test if there is a significant difference among experimental groups. If the P value is less than or equal to 0.05, a student t-test was performed for one-one comparison between different groups. Paired t-tests were used to analyze data from gene chip screening to identify differentially expressed lncRNAs and mRNAs. P ≤ 0.05 was set as a criterion for statistical significance.
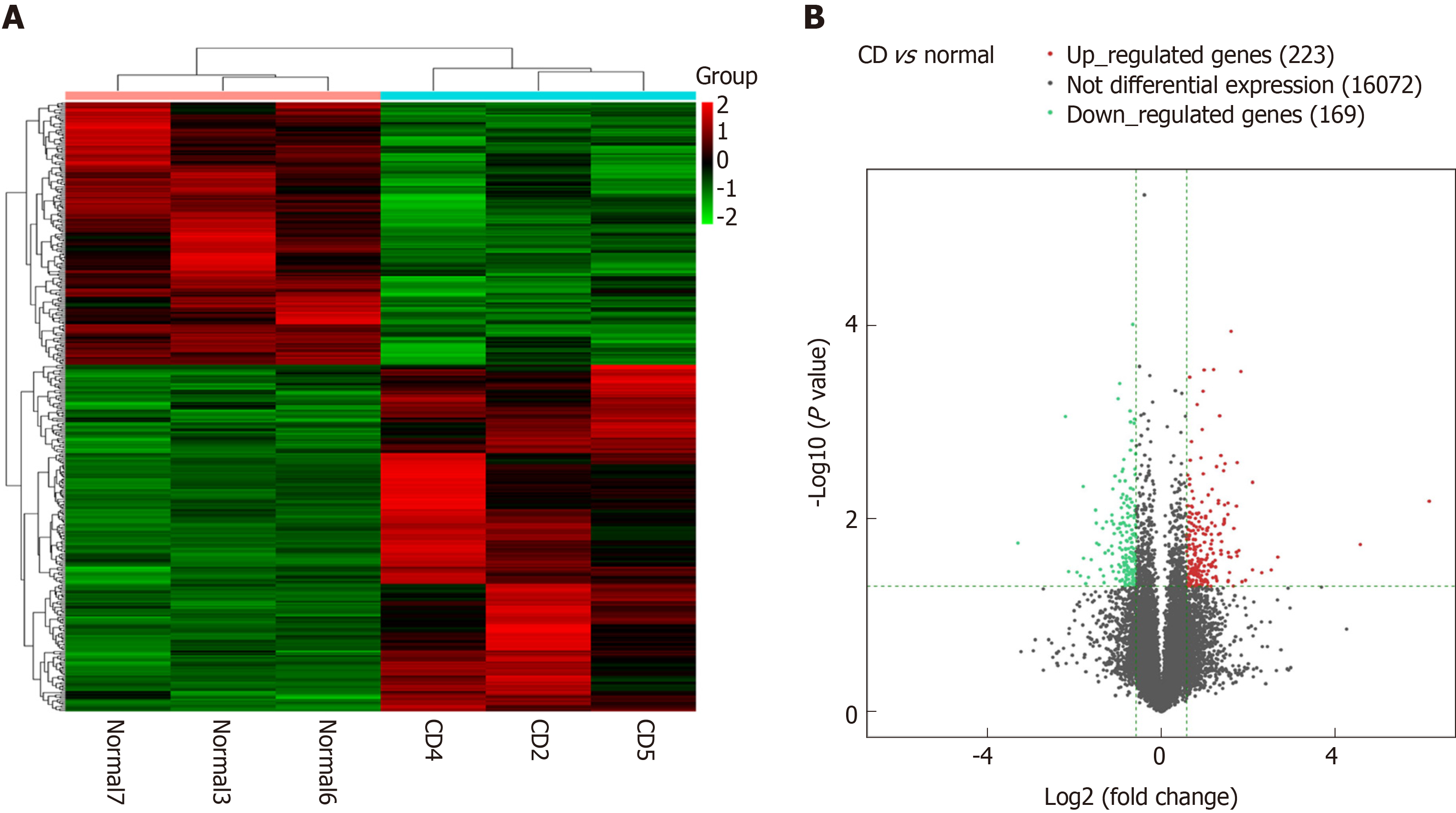
Using microarray hybridization technique, the transcriptome was compared in three pairs of intestinal mucosa samples of CD patients and normal subjects. Among the 51,388 genes screened, 400 lncRNAs were found to be differentially expressed (more than 10-fold changes, P ≤ 0.05), with 243 significantly upregulated and 157 downregulated, in CD lesion tissues (Figure 1). The five most upregulated and five most downregulated lncRNAs were validated by real-time PCR in an expanded clinical cohort of 40 CD diseased intestinal mucosa and 40 normal intestinal mucosa samples. The results showed that while the pattern of changes was similar to those found in microarray analysis, four out of the ten lncRNAs were confirmed to have significant differential expression in CD lesions (Supplementary Figure 3). Among these lncRNAs, lncRNACNN3-206 and lncRNA-ZNF292 were identified to be the most upregulated in CD lesions. The overexpression of these two lncRNAs in the expanded cohort is shown in Supplementary Figure 4.
lncRNACNN3-206, which showed the greatest difference, was chosen for further investigation. lncRNACNN3-206 was documented by NCBI and Ensemble as a non-coding gene (Supplementary Figure 5A). Information from the Ensemble database showed that the lncRNACNN3-206 transcript contained 793 base pairs. The full-length sequence of the lncRNA was obtained by using the 5' race and 3' race technique. Our PCR experiments using flanking primers showed that a band of approximately 800 bp, representing lncRNACNN3-206, was obtained from multiple CD lesions (Supplementary Figure 5B).
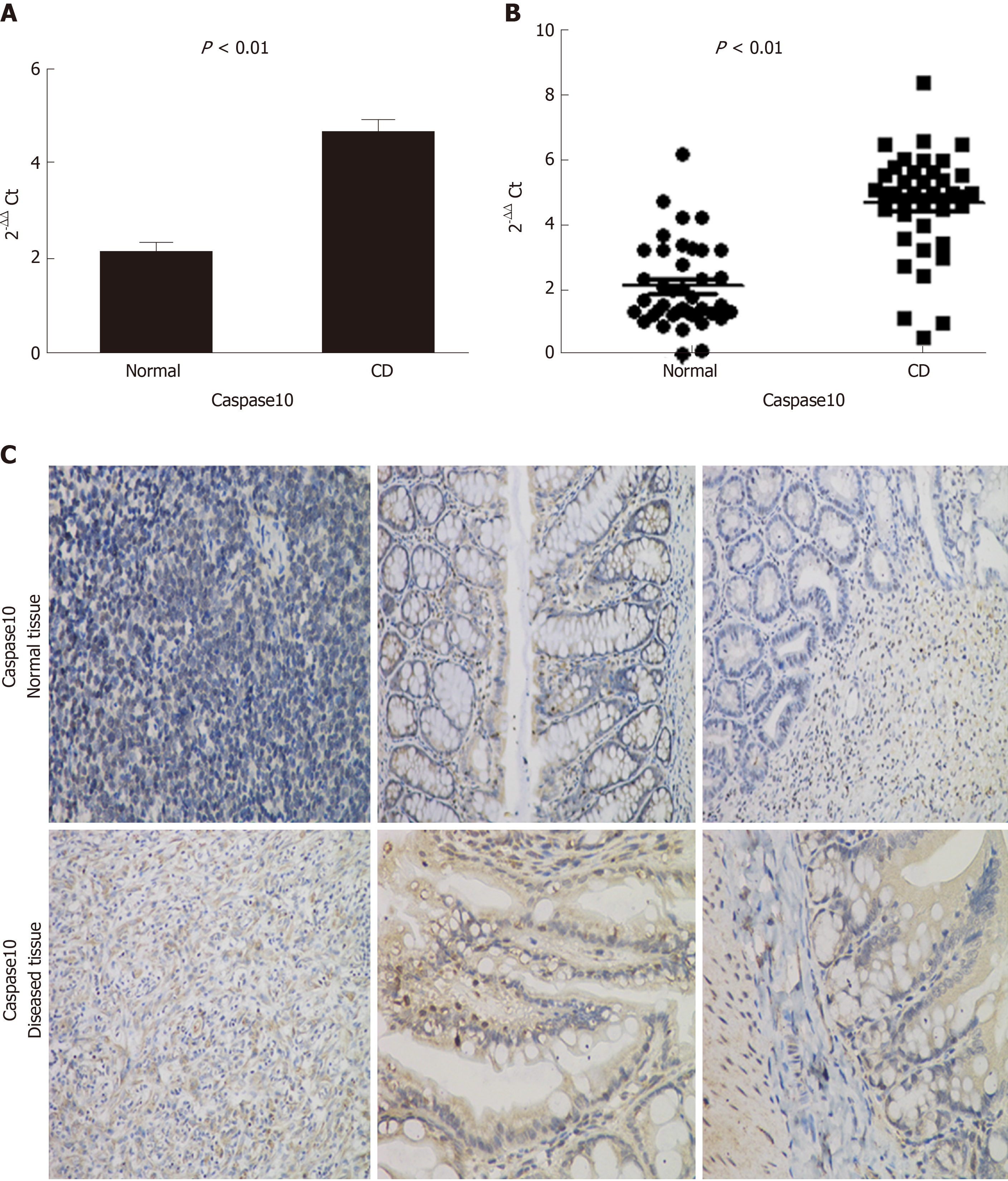
The mRNAs (Supplementary Figure 6) targeted by lncRNACNN3-206 were predicted by CNC analysis, and the differentially expressed genes involved in inflammatory activation and apoptosis signaling pathways were identified by searching the GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. Cross-checking the two lists and prediction of genes containing lncRNACNN3-206 binding sites (below) led to the selection of Caspase10, FOXP3, TGF-β, MHC-II, IL-22 and ATG5 as candidate downstream genes for further investigation. Real-time PCR (Figure 2A) and immunohistochemical experiments (Figure 2B) were used to determine the expression levels of these genes in the diseased and normal intestinal mucosa, and the overexpression of Caspase10 was found to be the most significant. Fluorescence in situ hybridization (FISH) experiments confirmed that lncRNACNN3-206 was highly expressed in the intestinal mucosa associated with CD (Figure 3).
The FISH assay was performed with 40 intestinal mucosa tissues from CD lesions and 40 normal intestinal mucosa tissues. An lncRNACNN3-206-specific fluorescent probe (5'-FAM-CCT TTA GCA TCA GTA AGG GAA GCA TT-FAM, green) was used to determine the expression of lncRNACNN3-206 in the CD patient intestinal mucosa samples. While lncRNACNN3-206 was found to be highly expressed in the intestinal mucosa of CD lesions, low expression of the lncRNACNN3-206 could be detected in epithelial cells of normal intestinal mucosa (Figure 3). Further observation under microscopy showed that the signals were mainly distributed in the cytoplasm of intestinal epithelial cells. TargetScan, miRBase, and PicTar databases were used to predict that miR-212 had multiple high-affinity binding sites in the lncRNACNN3-206 and Caspase10 gene sequences (Figure 4A).
For functional studies, two luciferase reporter plasmids containing the 3' end of the lncRNACNN3-206 or the 3' UTR of Caspase10 were constructed. Co-transfection with miR-212 and a reporter vector containing the 3’ end of lncRNACNN3-206 or 3' UTR of Caspase10 led to a significant inhibition in luciferase activity. This inhibition was not observed when the 3’ end of lncRNACNN3-206 was mutated, or the miR-212 was replaced with mimic, non-specific, RNA sequences, demonstrating a specific inhibition of lncRNACNN3-206 3' end activity by miR-212. Similar experiments were designed and performed for the 3' UTR of Caspase10, and the same pattern of changes in reporter activity were observed. These results strongly suggested that miR-212 may bind to the 3' end of lncRNACNN3-206 and 3’ UTR of Caspase10 (Figure 4B). Thus, a ternary mechanism based on functional interactions among lncRNACNN3-206, miR-212, and Caspase10 was proposed (Figure 4C). The effective level of miR-212 would be decreased through binding to lncRNACNN3-206, and the decreased miR-212 level might lead to a de-repression of Caspase10 expression by miR-212.
Caco-2 and HT-29 cell lines were transfected with lentiviral vectors expressing lncRNACNN3-206 (oe-lncRNACNN3-206) or an siRNA that inhibits lncRNACNN3-206 levels (si-lncRNACNN3-206). The un-transfected cells (Blank), cells transfected with vector (Vector), and vector producing a nonspecific, scramble RNA (Scramble-NC) were included as controls. After transfection, the expression of three RNAs of lncRNACNN3-206, miR-212, and Caspase10 were determined in the five groups by real-time PCR using primers as described under Materials and Methods. As shown in Supplementary Figure 7, we found that overexpression of lncRNACNN3-206 led to decreased levels of miR-212 in cells, but increased levels of Caspase10 mRNA. After lncRNACNN3-206 expression was knocked down, miR-212 levels were increased, while Caspase10 mRNA levels were decreased. These observations provided further support for the proposed mechanism that lncRNACNN3-206 overexpression could absorb miR-212, leading to the degradation of miR-212, resulting in a decrease in intracellular free miR-212 levels, thereby alleviating the inhibition of Caspase10 by miR-212, and eventually significantly increasing mRNA levels of Caspase10 (Supplementary Figure 7). In the two cell lines, Caspase10 protein levels in the five groups were determined by Western blotting. The results showed that Caspase10 protein levels were significantly increased when lncRNACNN3-206 was overexpressed. In contrast, when lncRNACNN3-206 expression was knocked down, levels of Caspase10 were decreased (Supplementary Figure 8). These findings were consistent with the mRNA levels, and provide support for the proposed regulatory pathway as illustrated in Figure 4C.
Manipulation of lncRNACNN3-206 levels leads to changes in cell apoptosis: To determine the cellular function of lncRNACNN3-206, flow cytometry was performed following lncRNACNN3-206 overexpression. As shown in Figure 5A, the apoptosis rates of Caco-2 and HT-29 cells increased significantly 48 h after transfection, with the lentivirus expressing lncRNACNN3-206 in comparison with the blank, vector, and scramble control groups. Accompanying cell apoptosis and increased necrosis were observed. On the contrary, inhibition of lncRNACNN3-206 by transfection with the lentivirus expressing si-lncRNACNN3-206 led to decreased apoptosis in both cell lines. These results indicated that lncRNACNN3-206, through its regulation of miR212, may indirectly modulate Caspase10 levels by promoting cell apoptosis.
lncRNACNN3-206 regulates the invasive growth of intestinal epithelial cells: Previous studies suggested that cellular levels of Caspase10 may correlate with cell invasion[14]. We determined the levels of cell invasion following manipulation of lncRNACNN3-206 expression in Caco-2 and HT-29 cell culture. As shown in Figure 5B, compared to the three control groups, overexpression of lncRNACNN3-206 led to increased cell invasion. Reduction of lncRNACNN3-206 levels by transfecting with si-lncRNACNN3-206 led to decreased cell invasion. It is noteworthy that penetration of intestinal epithelial cells into the serosal layer is considered a pathological feature of CD, and may be related to the formation of scar stenosis and intestinal fistulas. From this point of view, upregulation of lncRNACNN3-206 may contribute to the pathogenesis of CD by its effects on cell migration and invasion.
Changes in the morphology and DAI scores of the TNBS model: The construction of a CD mouse model using TNBS was described in Materials and Methods. The morphological changes and general status of mice in each group were carefully monitored. The TNBS and TNBS + scramble groups exhibited weight loss, diarrhea, feces containing blood and uneven hair distribution (5 wk after TNBS administration). The TNBS + si-lncRNACNN3-206 mice had mild diarrhea and erect hair on the 1st and 2nd d. Beginning on the 3rd wk, their diarrhea disappeared, and their fur was smooth. After mice were injected with TNBS, DAI scores were recorded once every week. As shown in Figure 6A, DAI scores continually increased in all three groups until the 4th wk. After that point, while the anti-scramble groups displayed higher DAI scores than the control group (TNBS), the TNBS + si-lncRNACNN3-206 group had lower DAI scores. These observations indicated that si-lncRNACNN3-206 could rescue some of the CD symptoms in TNBS mice, and improve the overall body status of these mice.
Effects of lncRNACNN3-206 knockdown on CD lesions: As shown in Figure 6B, mice were anesthetized, and the colon and ileocecal section were dissected. The colonic morphology of each group was observed at the end of the experiment. The colons from mice in the TNBS and TNBS + anti-scramble groups were significantly shortened, and hyperemia was observed in the ileocecal and segmental colon. No obvious contracture was observed in the colons from mice in the TNBS + si-lncRNACNN3-206 group. The ileocecal colons in this group had mild segmenting congestion, which was similar to that of the control group.
To examine pathological changes, the CD-like lesions in the TNBS model were dissected, fixed, and sectioned for H&E staining. As shown in Figure 7A, in the TNBS + anti-scramble and TNBS groups, the colonic lesion tissue had a moderate to severe infiltration of inflammatory cells, mainly mononuclear macrophages and neutrophils, with most of them found in the submucosa and muscular layer with severe local crypt damage and ulcers. Hyperplasia was seen in the areas with contracture. However, the TNBS + si-lncRNACNN3-206 group showed significantly less severe colonic inflammation than the control groups. The above comparison indicated that si-lncRNACNN3-206 significantly reduced the pathological damage of the colon elicited by TNBS.
Expression and localization of lncRNACNN3-206 in the colon wall of the TNBS and TNBS + si-lncRNACNN3-206 groups were determined by FISH assay. In the TNBS group, lncRNACNN3-206 expression was mainly detected in the submucosa and muscular layer, while in the TNBS + si-lncRNACNN3-206 group, lncRNACNN3-206 expression was observed in the mucosal layer at a significantly reduced level (Figure 7B). This result confirmed the constitutional expression of lncRNACNN3-206 in colon tissue, and, more importantly, the efficacy of si-lncRNACNN3-206 for knocking down lncRNACNN3-206 levels.
Four mice were randomly selected from four groups. Caspase10 expression in the cytoplasm of cells in colon tissues from the four mice was determined by Western blotting (Figure 7C). The expression level of Caspase10 in the lncRNACNN3-206 interference group was significantly lower than that in the TNBS and scramble groups. The expression levels of β-actin from different groups were comparable, indicating similar protein loading. Densitometry analysis and calculation of the ratio between Caspase10 and β-actin levels showed that while administration of TNBS led to increased Caspase10 expression, and interference by si-lncRNACNN3-206 led to a significant reduction of Caspase10. This result was consistent with that of an in vitro study using cell culture. Since Caspase10 is important for cell apoptosis and cell invasion, its change following interference of lncRNACNN3-206 may partially explain the alleviation of CD-like symptoms by si-lncRNACNN3-206.
CD4+ T lymphocytes represent the main effector cells during the inflammation of intestinal mucosal[16]. T helper lymphocytes 1 (Th1) and Th2, which differentiate from CD4+ T cells, are antagonistic to each other. The Thl lymphocyte plays a dominant role in CD patients and mediates strong cellular immunity[17]. Additionally, some scholars found that Th17 promoted intestinal inflammation and induced the occurrence of autoimmune diseases, while Treg inhibited intestinal mucosal inflammation[18]. To determine changes in CD4+ T lymphocytes, one mouse was randomly taken from each group, and the four CD4+ T lymphocyte subtypes (Th1, Th2, Th17 and Treg) in the spleen were counted by flow cytometry. As shown in Figure 8, it was found that the Th1/Th2 ratio in the TNBS group was greater than 1, while the Th1/Th2 ratio in the TNBS + si-lncRNACNN3-206 group was lower than 1. In addition, the Th17/Treg ratio in the TNBS group was above 1, but the Th17/Treg ratio in the TNBS + si-lncRNACNN3-206 group was below 1. Thus, changes in the CD4+ T lymphocyte ratio of the TNBS model recapitulated those observed in CD patients. Moreover, upregulation of lncRNACNN3-206 may be involved in CD-associated inflammation, and knockdown of lncRNACNN3-206 could partially alleviate inflammation in the CD mouse model.
It is estimated that more than 4 million cases of inflammatory bowel disease (IBD) will be diagnosed each year worldwide[19]. Since the disease is common among teenagers and young adults, it could affect the physical and mental development of the human population[20]. The high treatment cost of this lifelong disease causes a heavy economic burden to patients, their families and society as a whole. A large proportion of IBD patients are CD cases. Unfortunately, there is currently no curative treatment for IBD. In the past 20 years, multiple hypotheses about CD pathogenesis have been proposed, which emphasize specific role(s) for immune dysfunction[21], gene mutations, epigenetic changes[22], persistent infection by intestinal bacteria[23], or food antigen-based allergies[24]. In addition, some scholars have proposed that mental stress could promote CD onset, but with only circumstantial evidence[25]. The underlying connections among these factors remain unclear.
LncRNAs are broadly expressed in all eukaryotic cells. Their regulation, function and disease-causing mechanisms have become hot research topics in recent years. Previous studies have implicated lncRNAs in the pathogenesis of CD. Chen et al[10] examined lncRNA levels in the peripheral blood of CD patients using microarray analysis, and found 1,211 upregulated and 777 downregulated lncRNAs. In the current study, we determined the expression of lncRNAs in pathological intestinal mucosal tissue samples from CD patients and normal intestinal mucosal tissue samples from non-CD patients. A total of 400 differentially expressed lncRNAs were detected in the intestinal mucosal tissue samples from CD patients, with 243 upregulated and 157 downregulated. The smaller number of lncRNAs identified in our study compared to Chen’s study may reflect the high complexity of blood components, to which many different cells/tissues may contribute. An important function of lncRNAs is their regulation of downstream genes. Since we used inflammatory lesion tissues to isolate RNA and perform gene chip hybridization, it is reasonable to assume that many of the lncRNA alterations we observed may be directly or indirectly related to altered mRNA and protein expression patterns during inflammation in the intestinal mucosa of CD patients.
In this study, lncRNACNN3-206 was found to be the most significantly upregulated lncRNA in CD lesions. The expression level of lncRNACNN3-206 in diseased tissue samples was more than 100-times higher than that in normal tissue samples. Moreover, the co-expressed gene Caspase10 was identified using GO pathway analyses and Pearson's correlation analysis, and the intermediate regulator miR-212 was found based on the ceRNA mechanism and gene prediction websites TargetScan and miRBase. Our studies revealed for the first time the lncRNACNN3-206-miR-212-Capase10 regulatory network and its clinical relevance in cell culture, a CD mouse model and patient samples. The significance of this ternary mechanism was supported by three lines of evidence. First, forced overexpression/knockdown of lncRNACNN3-206 in two intestinal epithelial cell lines showed concomitant changes in the expression levels of miR-212 and Caspase10. Second, experiments in cell lines indicated that changes in lncRNACNN3-206 caused alterations in cell function, including cell apoptosis and invasion. Third, in the CD mouse model, knockdown of lncRNACNN3-206 could partially alleviate the CD-related changes, including histological changes in the mouse colon, corresponding changes in Caspase10 expression, infiltration of inflammatory cells, and alterations in Th1/Th2 and Th17/Treg ratios.
This study revealed that lncRNACNN3-206 regulates epithelial cell apoptosis. lncRNACNN3-206 appeared to downregulate miR-212, affecting its binding to the Caspase10 3’ UTR, resulting in inhibition of Caspase10 expression. When a high level of Caspase10 was released into the cytoplasm, Caspase3 and Caspase7 could be activated, leading to increased cell apoptosis and invasion. Murthy et al[26] found that lncRNA ATG could activate Caspase10 and Caspase3, initiate the apoptosis cascade in intestinal epithelial cells, and induce intestinal mucosal destruction as well as promote a continuous inflammatory response. Intriguingly, activation of Caspase-3 could also lead to the degradation of lncRNA ATG. Such a negative feedback mechanism may be required for the homeostasis of intestinal mucosal cells and the maintenance of normal intestinal function. It is unclear if there is also negative feedback in the lncRNACNN3-206-miR-212-Capase10 regulatory network. In addition, our attempts to determine the physical binding of miR-212 to the 3’ end of lncRNACNN3-206 and the 3’ UTR of Caspase10 have failed after repeated efforts. The lack of evidence of physical interactions among lncRNACNN3-206, miR-212 and Capase10 mRNA constitutes a major limitation of the current study.
This study also found that overexpression of lncRNACNN3-206 in intestinal epithelial cells led to increased cell apoptosis and invasion. These changes in cell function may play important roles in CD pathogenesis. Pathological intestinal epithelial cells invading the submucosa and muscularis may cause an infiltration of lymphocytes and mononuclear macrophages, and a release of inflammatory cytokines, ultimately resulting in inflammation of the entire intestinal wall. Hoshi et al[15] found that Caspase10 expression in invasive gastric cancer cells was significantly higher than that in normal tumor-adjacent cells. This phenomenon indicated that Caspase10 may promote the invasion of gastric cancer cells[26]. While our results on Caspase10 are consistent with previous findings, whether lncRNACNN3-206 could promote the invasion of intestinal epithelial cells through regulation of migration-related genes, such as TGF-β, requires further studies to clarify.
It was reported that increased numbers and activation of Th17 cells play an important role in CD inflammation[27]. lncRNACNN3-206 interference in intestinal epithelial cells in mice decreased the proportion of Th17 cells in the CD4+ T cell population in the mouse spleen, indicating that lncRNACNN3-206 was involved in the systemic inflammatory response in the CD mouse model. Tregs are able to inhibit an inflammatory reaction in the intestinal mucosa, and Th17 cells and Tregs are mutually transforming[28]. Studies have shown that in the presence of IL-6 and/or IL-23, TGF-β promotes the differentiation of naive T lymphocytes into Th17 cells, which secrete the pro-inflammatory cytokine IL-17, and promote the development of autoimmunity and inflammation[29]. Future studies should focus on whether lncRNACNN3-206 could promote the secretion of IL-6 and/or IL-23 by intestinal epithelial cells. An answer to this question will help us better understand the initiation of CD pathogenesis by lncRNACNN3-206 upregulation.
In conclusion, our studies identified lncRNACNN3-206 as a highly upregulated non-coding RNA in the intestinal lesion tissues of CD patients. Manipulation of lncRNACNN3-206 in cell culture led to significant changes in Caspase10 expression, cell apoptosis and invasion activity. Experiments in a CD mouse model demonstrated that knockdown of lncRNACNN3-206 could alleviate some CD manifestations, most likely through inhibition of intestinal epithelial cell apoptosis and inflammatory responses. These findings do not only facilitate our understanding of the pathologic mechanism, but also have strong clinical implications. The upregulated lncRNACNN3-206 gene could be a new therapeutic target for the prevention and treatment of CD patients.
Crohn's disease (CD) is a chronic, recurrent, inflammatory disorder of the digestive tract, with clinical manifestations of abdominal pain, diarrhea, enterelcosis and intestinal obstruction. Statistics showed that CD incidence rates are climbing in Europe and the United States, as well as in Asian countries.
Due to a poor understanding of the etiology and pathogenesis of this disease, current therapy is limited to anti-symptomatic and surgical treatments. The recurrence rate within 3 years after surgery is as high as 80-100%. Exploration of the cellular and molecular mechanisms is urgently required to develop new therapeutic modalities against this disease. While many studies have indicated that long noncoding RNAs (lncRNAs) might participate in the pathogenesis of autoimmune diseases, very few studies focused on whether lncRNAs may contribute to CD occurrence. Previous studies using high-throughput gene sequencing techniques have shown that the serum of CD patients contained multiple differentially expressed lncRNAs. While these differentially expressed lncRNAs could be used as biomarkers for the diagnosis and/or prognosis of CD, their sources remain unclear.
Our goal is to identify the differentially expressed lncRNAs in the intestinal mucosa associated with CD, and to characterize their pathogenic role(s) and related mechanisms.
In this study, we collected tissue samples of the ileal lesions from CD patients and normal intestinal mucosal tissue samples isolated during physical examinations of healthy patients. The expression patterns of lncRNAs and mRNAs were compared between the lesions and normal ileal tissue samples by microarray hybridization. Bioinformatic analysis was performed to predict the gene regulatory network of lncRNAs-miRNAs-mRNAs. The biological functions of the lncRNAs, relevant miRNAs and target genes, as well as the related regulatory mechanisms, were comprehensively investigated using cell culture and animal models.
Our studies identified lncRNACNN3-206 as a highly upregulated non-coding RNA in the intestinal lesions of CD patients. Manipulation of lncRNACNN3-206 in cell culture led to significant changes in Caspase10 expression, cell apoptosis and invasion activity. Experiments using a CD mouse model demonstrated that lncRNACNN3-206 knockdown could alleviate some CD manifestations, most likely through inhibition of intestinal epithelial cell apoptosis and inflammatory responses.
Overexpression of lncRNACNN3-206 in intestinal epithelial cells led to increased cell apoptosis and invasion. These changes in cell function may play important roles in CD pathogenesis. These findings do not only facilitate our understanding of the pathological mechanisms but also have strong clinical implications. The upregulated lncRNACNN3-206 gene could be a new therapeutic target for the prevention and treatment of CD.
The top 10 lncRNAs with differential expression were screened out by high-throughput sequencing, but only six lncRNAs with true differential expression were identified by PCR validation in expanded sample tissues. All of the subjects had to therefore be definitively confirmed using a large number of clinical tissue samples. Moreover, experimental research insight is given through the signaling pathways predicted by bioinformatics websites, as well as the knowledge of signaling pathways predicted by GO/Pathway analysis, which requires the detection of specific biological gene functions through cell experiments. The predicted function is sometimes experimentally shown to be absent in the cell. In this study, due to limited conditions, the targeted binding of lncRNACNN3-206 and miR212 was demonstrated by luciferase reporter assays. Nevertheless, the better method is RIP technology, which can directly observe the adsorption of miR212 by lncRNA as a sponge.
| 1. | Li X, Wu Z, Fu X, Han W. lncRNAs: insights into their function and mechanics in underlying disorders. Mutat Res Rev Mutat Res. 2014;762:1-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 189] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 2. | Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:6391-6400. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 463] [Cited by in RCA: 532] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 3. | Kovacic JC. Unlocking the Many Secrets of Noncoding RNA. J Am Coll Cardiol. 2015;65:2538-2541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 4. | Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4127] [Cited by in RCA: 5737] [Article Influence: 382.5] [Reference Citation Analysis (0)] |
| 5. | Gezer U, Özgür E, Cetinkaya M, Isin M, Dalay N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol Int. 2014;38:1076-1079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 226] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 6. | Strober W, Fuss I, Mannon P. The fundamental basis of inflammatory bowel disease. J Clin Invest. 2007;117:514-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 992] [Cited by in RCA: 1021] [Article Influence: 53.7] [Reference Citation Analysis (0)] |
| 7. | Zheng JJ, Shi XH, Zhu XS, Huangfu Z, Guo ZR. [A comparative study of incidence and prevalence of Crohn's disease in mainland China in different periods]. Zhonghua Nei Ke Za Zhi. 2011;50:597-600. [PubMed] |
| 8. | Bernstein CN. Treatment of IBD: where we are and where we are going. Am J Gastroenterol. 2015;110:114-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 175] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 9. | Yeo J, Woo HS, Lee SM, Kim YJ, Kwon KA, Park DK, Kim JH, Kim KO, Chung JW. Drug-induced eosinophilic pneumonia in a patient with Crohn's disease: diagnosis and treatment using fraction of exhaled nitric oxide. Intest Res. 2017;15:529-534. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 10. | Chen D, Liu J, Zhao HY, Chen YP, Xiang Z, Jin X. Plasma long noncoding RNA expression profile identified by microarray in patients with Crohn's disease. World J Gastroenterol. 2016;22:4716-4731. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 30] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 11. | Qiao YQ, Huang ML, Xu AT, Zhao D, Ran ZH, Shen J. LncRNA DQ786243 affects Treg related CREB and Foxp3 expression in Crohn's disease. J Biomed Sci. 2013;20:87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 79] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 12. | Kremer B, Mariman R, van Erk M, Lagerweij T, Nagelkerken L. Temporal colonic gene expression profiling in the recurrent colitis model identifies early and chronic inflammatory processes. PLoS One. 2012;7:e50388. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 13. | Macdonald TT. Viral vectors expressing immunoregulatory cytokines to treat inflammatory bowel disease. Gut. 1998;42:460-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 14. | Spencer DM, Veldman GM, Banerjee S, Willis J, Levine AD. Distinct inflammatory mechanisms mediate early versus late colitis in mice. Gastroenterology. 2002;122:94-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 158] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 15. | Hoshi T, Sasano H, Kato K, Yabuki N, Ohara S, Konno R, Asaki S, Toyota T, Tateno H, Nagura H. Immunohistochemistry of Caspase3/CPP32 in human stomach and its correlation with cell proliferation and apoptosis. Anticancer Res. 1998;18:4347-4353. [PubMed] |
| 16. | Dieleman LA, Palmen MJ, Akol H, Bloemena E, Peña AS, Meuwissen SG, Van Rees EP. Chronic experimental colitis induced by dextran sulphate sodium (DSS) is characterized by Th1 and Th2 cytokines. Clin Exp Immunol. 1998;114:385-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 747] [Cited by in RCA: 963] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 17. | Bamias G, Sugawara K, Pagnini C, Cominelli F. The Th1 immune pathway as a therapeutic target in Crohn's disease. Curr Opin Investig Drugs. 2003;4:1279-1286. [PubMed] |
| 18. | Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5130] [Cited by in RCA: 5560] [Article Influence: 278.0] [Reference Citation Analysis (0)] |
| 19. | Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307-317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2019] [Cited by in RCA: 1982] [Article Influence: 132.1] [Reference Citation Analysis (2)] |
| 20. | Zheng JJ, Shi XH, Zhu XS, Huangfu Z, Guo ZR. [A comparative study of incidence and prevalence of Crohn's disease in mainland China in different periods]. Zhonghua Nei Ke Za Zhi. 2011;50:597-600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 21. | Adams AT, Kennedy NA, Hansen R, Ventham NT, OʼLeary KR, Drummond HE, Noble CL, El-Omar E, Russell RK, Wilson DC, Nimmo ER, Hold GL, Satsangi J. Two-stage genome-wide methylation profiling in childhood-onset Crohn's Disease implicates epigenetic alterations at the VMP1/MIR21 and HLA loci. Inflamm Bowel Dis. 2014;20:1784-1793. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 73] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 22. | Smith P, Siddharth J, Pearson R, Holway N, Shaxted M, Butler M, Clark N, Jamontt J, Watson RP, Sanmugalingam D, Parkinson SJ. Host genetics and environmental factors regulate ecological succession of the mouse colon tissue-associated microbiota. PLoS One. 2012;7:e30273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 23. | Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. 2017;14:573-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1021] [Cited by in RCA: 1298] [Article Influence: 144.2] [Reference Citation Analysis (0)] |
| 24. | Papadakis KA, Targan SR. Current theories on the causes of inflammatory bowel disease. Gastroenterol Clin North Am. 1999;28:283-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 68] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 25. | Gray WN, Boyle SL, Graef DM, Janicke DM, Jolley CD, Denson LA, Baldassano RN, Hommel KA. Health-related quality of life in youth with Crohn disease: role of disease activity and parenting stress. J Pediatr Gastroenterol Nutr. 2015;60:749-753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 26. | Murthy A, Li Y, Peng I, Reichelt M, Katakam AK, Noubade R, Roose-Girma M, DeVoss J, Diehl L, Graham RR, van Lookeren Campagne M. A Crohn's disease variant in Atg16l1 enhances its degradation by caspase 3. Nature. 2014;506:456-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 312] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 27. | Huang W, Thomas B, Flynn RA, Gavzy SJ, Wu L, Kim SV, Hall JA, Miraldi ER, Ng CP, Rigo F, Meadows S, Montoya NR, Herrera NG, Domingos AI, Rastinejad F, Myers RM, Fuller-Pace FV, Bonneau R, Chang HY, Acuto O, Littman DR. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature. 2015;528:517-522. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 137] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 28. | Kitani A, Xu L. Regulatory T cells and the induction of IL-17. Mucosal Immunol. 2008;1 Suppl 1:S43-S46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 60] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 29. | Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M, Cheroutre H. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317:256-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1505] [Cited by in RCA: 1622] [Article Influence: 85.4] [Reference Citation Analysis (7)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ierardi E S-Editor: Gong ZM L-Editor: Filipodia E-Editor: Ma YJ