INTRODUCTION
Emerging coronaviruses (CoVs), such as the severe acute respiratory syndrome coronavirus (SARS-CoV) and the Middle east respiratory syndrome coronavirus (MERS-CoV), caused, and continue to induce infections in humans, resulting in serious threats to Health Care Systems[1]. In December 2019, a group of patients with pneumonia of an unknown cause was reported in Wuhan, China. On January 7, 2020 the new identified coronavirus was initially named 2019 novel coronavirus by the World Health Organization (WHO)[2]. The virus was later renamed severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the disease was known as coronavirus disease-2019 (COVID-19)[3]. On March 11, 2020 the WHO stated that this disease was a pandemic[4]. As of July 15 2020, 13150645 cases have been registered worldwide leading to 574464 deaths[5].
The distribution and expression of angiotensin converting enzyme type 2 (ACE2) in the human body may be a possible infection route of SARS-CoV-2[6]. Through the developed single-cell transcriptomes and RNA sequencing, the ACE2 RNA expression profile was analyzed at the single-cell level[7]. Increased ACE2 levels were found in various organs such as type II alveolar cells of the lung, but most importantly in the epithelial cells of the stomach and bowel[8,9]. These findings suggested that organs with high ACE2 expression, especially the gastrointestinal tract (GI) should be regarded as a possible target for COVID-19[8].
Thus, this review focuses on the analysis of some key elements regarding the SARS-CoV-2 genome and its general characteristics, in addition to the utility of ACE2 receptors in SARS-CoV-2 replication and pathogenesis, especially in the GI tract.
METHODS AND RESULTS
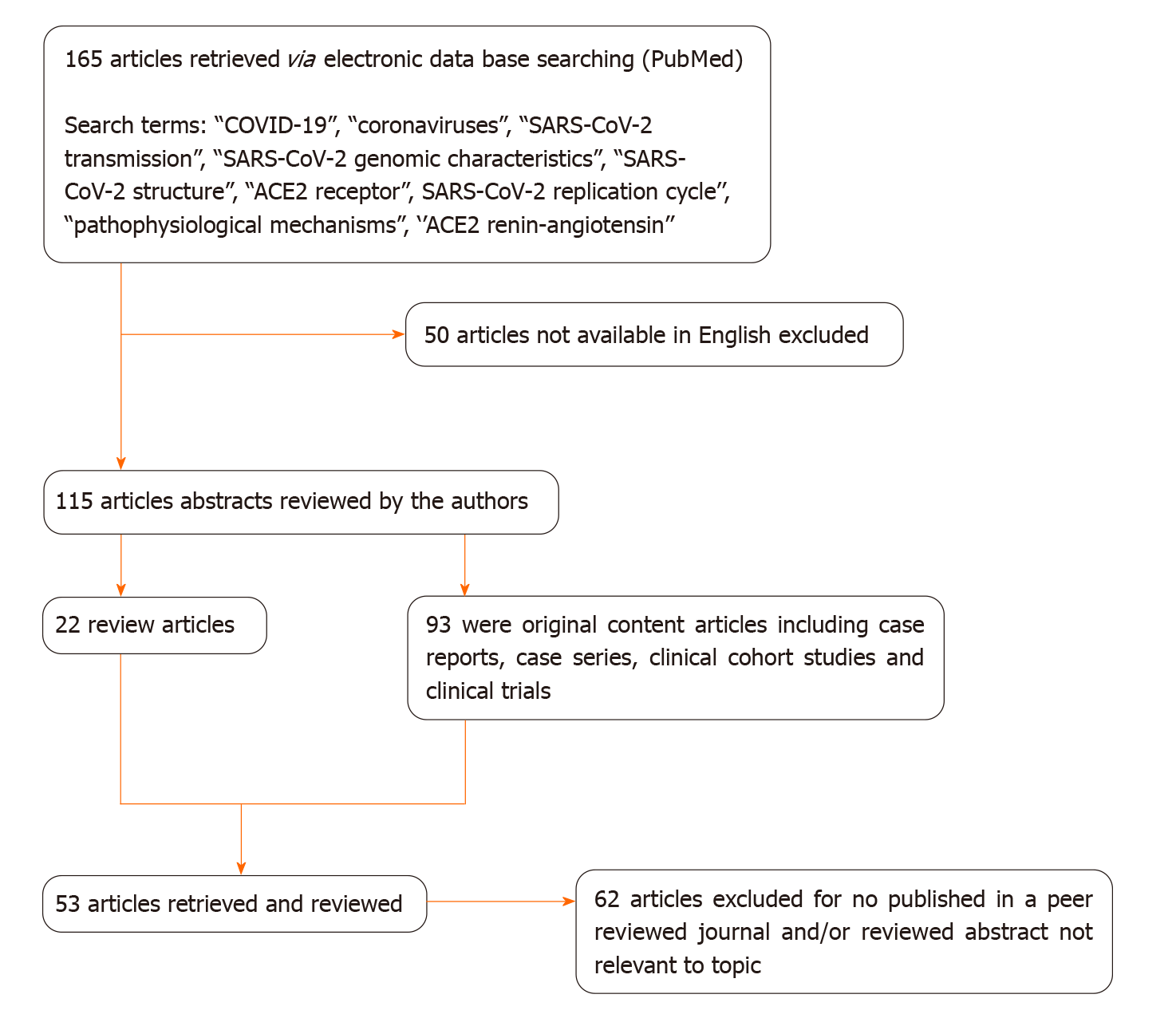
We conducted a literature search in the PubMed database, including most of the published studies in English and Chinese language since the emergence of the pandemic. The following keywords were used for the literature search: “COVID-19”, “coronaviruses”, “SARS-CoV-2 transmission”, “SARS-CoV-2 genomic characteristics”, “SARS-CoV-2 structure”, “ACE2 receptor”, SARS-CoV-2 replication cycle’’, “pathophysiological mechanisms”, and ‘’ACE2 renin-angiotensin’’. One hundred and sixty five publications fulfilled the search requirements. Of these, 115 articles were chosen due to the English language criteria. From these 115 articles, 93 were original articles and 22 were review articles, which included case series, case reports, clinical trials and clinical cohort studies. We reviewed all the abstracts and found 53 full-text articles appropriate for the current study (Figure 1). Two authors (DA and GM) independently reviewed the abstract and full text of the related articles.
Figure 1 Decision tree for literature research strategy.
COVID-19: Coronavirus disease 2019; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2; ACE2: Angiotensin converting enzyme type 2.
ORIGIN AND DIVERSITY OF CORONAVIRUSES
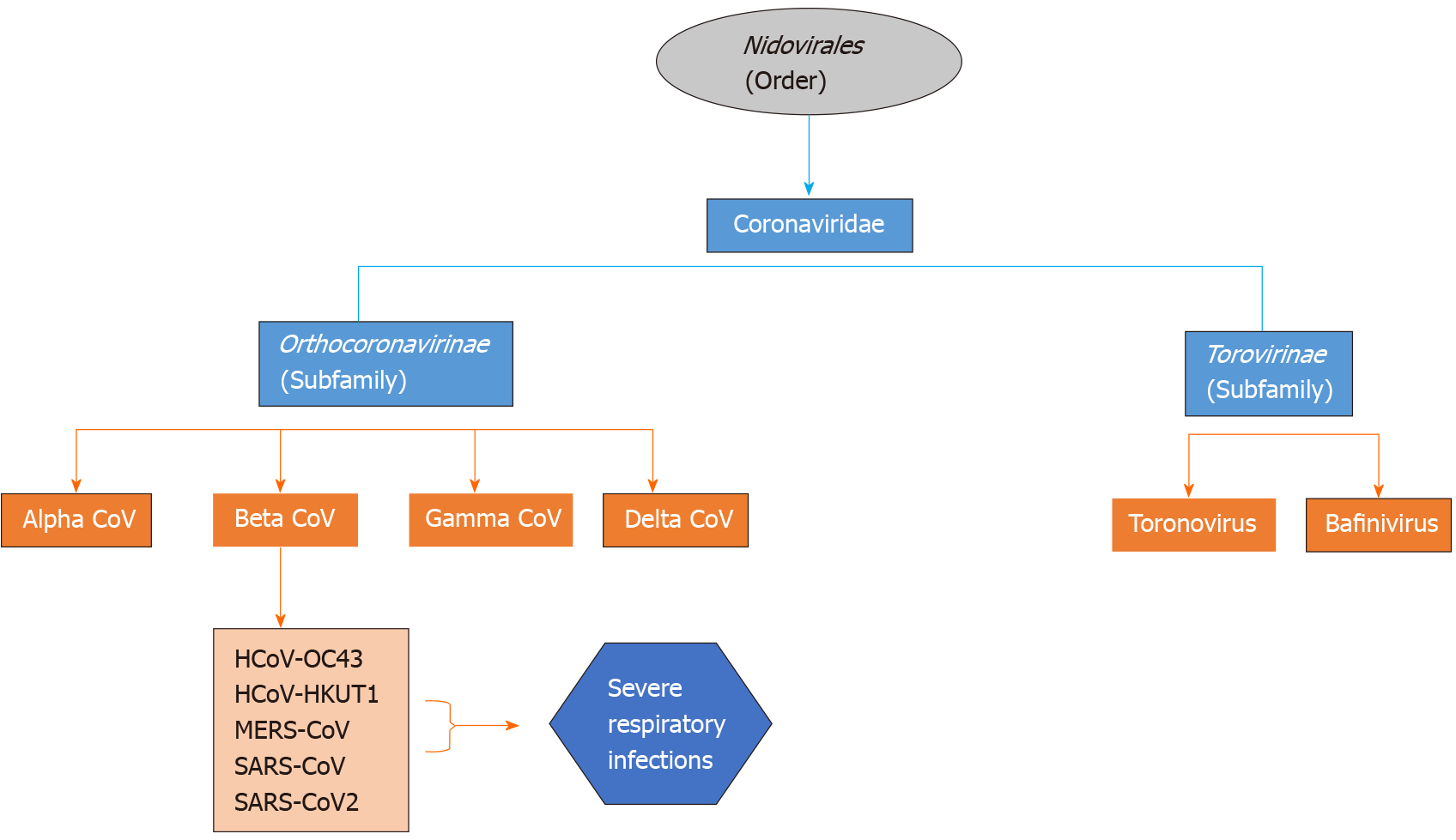
Coronaviruses are enveloped non-segmented positive-sense RNA viruses being members of the family of Coronaviridae, which is the largest class within the order Nidovirales, incorporating two subfamilies: Torovirinae and Orthocoronavirinae (Figure 2). The subfamily Orthocoronavirinae is classified into four genera: alphacoronavirus (alphaCoV), betacoronavirus (betaCoV), gammacoronavirus (gammaCoV) and deltacoronavirus (deltaCoV)[10,11]. CoVs mainly reside in birds and mammals and are frequently encountered in bats, camels and others. Genomic characterization has demonstrated that the most probable gene sources of alphaCoVs and betaCoVs are bats and rodents, and avian species for gammaCoVs and deltaCoVs. Regarding the manifestations of this group of viruses, they are able to cause enteric, neurological, hepatic and respiratory diseases in various animals[1,4].
Figure 2 Classification scheme of the various types of coronaviruses within the family Coronaviridae.
SARS-CoV: Severe acute respiratory syndrome coronavirus; MERS-CoV: Middle East respiratory syndrome coronavirus; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2; HCoV: Human coronavirus; CoV: Coronavirus.
It is estimated that approximately 2% of the population are asymptomatic carriers of a coronavirus, which in turn is responsible for 5% to 10% of respiratory infections[1]. The first recognized CoVs were Infectious Bronchitis Viruses causing respiratory disorders in chickens and the human CoVs (HCoVs), HCoV-OC43 and HCoV-229E, are responsible for the common cold. Since the emergence of HCoV-OC43 and HCoV-229E, seven other HCoVs have been recognized including HCoV-229E, HCoV-OC43, HCoV-NL63 and HCoV-HKU1[4,10]. The other two known betaCoVs, the SARS-CoV and the MERS-CoV are implicated in human respiratory infections with a case fatality rate of 10% and 35%, respectively[12,13].
Genetic sequence analysis has shown that the genome sequence of SARS-CoV-2 has a 79.0% similarity to SARS-CoV and 51.8% to MERS-CoV. This was the primary reason that the newly identified coronavirus was renamed SARS-CoV-2. In addition, it was shown that SARS-CoV-2 is 96% identical to the bat coronavirus (RaTG13) genome. Thus, bats have been considered the natural hosts of SARS-CoV-2. Nevertheless, the potential amplifying mammalian host, intermediate between human and host remains unidentified[14,15].
TRANSMISSION OF SARS-COV-2
The emergence of multiple cases of unidentified respiratory tract infection, which appeared initially in China, was assumed to be caused by direct exposure to a seafood market. Zoonotic transmission was considered to be the primary mechanism. Several studies have indicated that bats could be a potential reservoir of SARS-CoV-2. Nevertheless, it remains unclear whether SARS-CoV-2 emerged from the seafood market. Therefore, it was concluded that human-to-human interactions were also a potential source of transmission of the virus[10,16].
SARS-CoV-2 human-to-human transmission takes place primarily among individuals closely related (i.e., family and friends), after encountering COVID-19 patients or carriers. According to the medical records of case series in Wuhan city and data from the local CDCs, it was reported that among non-residents of Wuhan, 72.3% exhibited a preceding contact with Wuhan inhabitants, and 31.3% had recently travelled to this city[17]. Moreover, it was reported that the incubation time of the virus might generally be within 3 to 7 d and up to 2 wk as the greatest time period from infection to symptomatology was 12.5 d[16,18]. As with other respiratory pathogens such as rhinovirus and influenza, it is mainly assumed that transmission occurs via respiratory droplets due to coughing and sneezing[1]. Fomites are also regarded as the main source of infectious particles, but there is some doubt regarding this. Fomites include both porous and nonporous surfaces or objects, which can be contaminated with pathogenic microorganisms that might be used as vehicles in the transmission process[19]. Additionally, in a symptomatic male, SARS-CoV-2 RNA was discovered in a stool specimen while the serum specimen was negative[18]. Chinese researchers were able to isolate SARS-CoV-2 from a swab sample of confirmed patient’s feces, demonstrating the potential for fecal-oral transmission[18]. Finally, it seems that possible COVID-19 sources are also asymptomatic people[20,21].
GENOMIC AND STRUCTURE CHARACTERISTICS OF SARS-COV-2
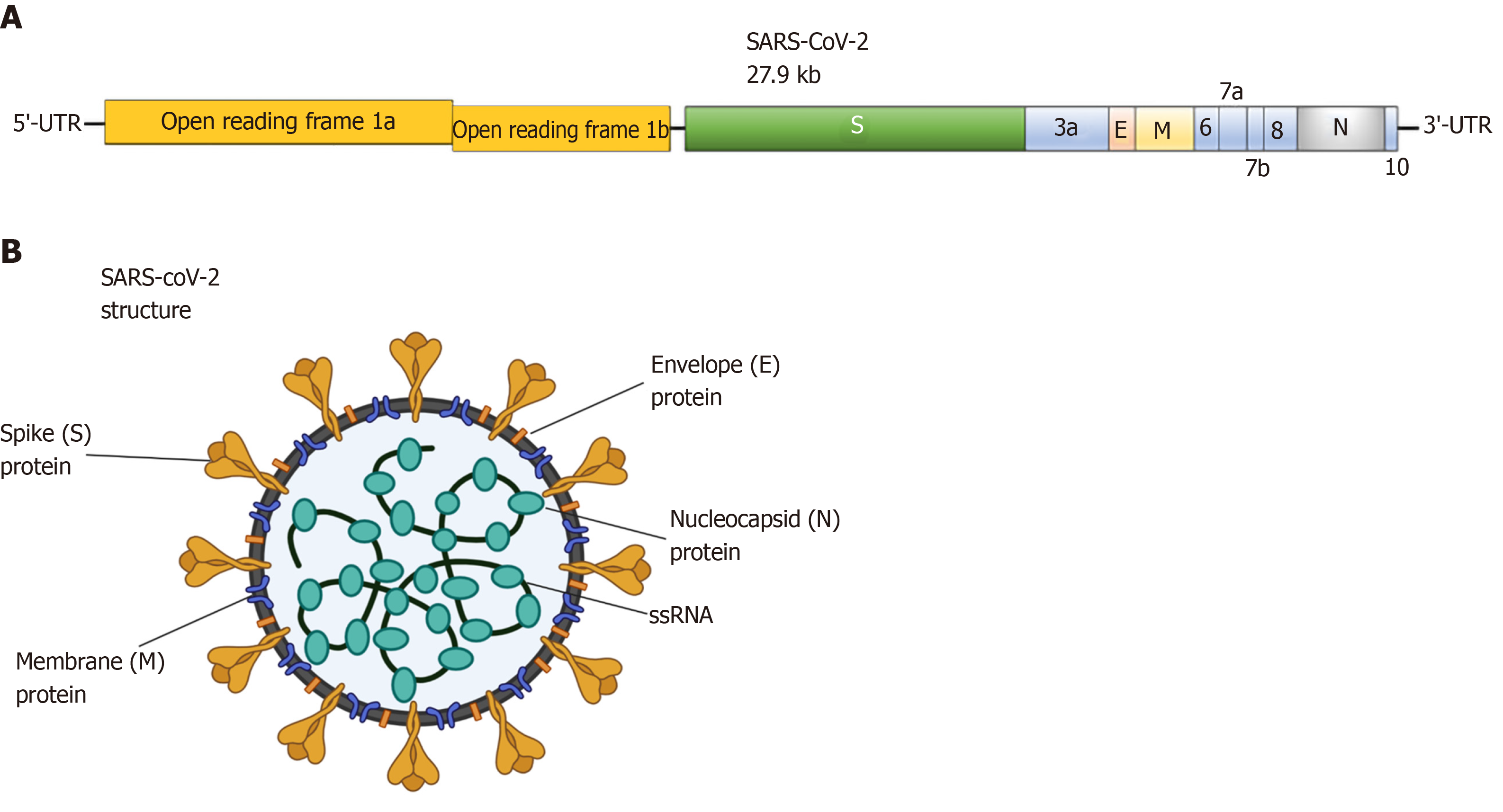
Coronaviruses are spherical and have spike protein projections on their surface resulting in a crown-like morphology, leading to their nomenclature[22,23]. SARS-CoV-2 has a round form with a diameter of approximately 60-140 nm[1]. In general, CoVs are viruses enveloped by a lipid bilayer. Their structure is composed of spike (S) glycoprotein, protein matrix (M), nucleocapsid (N) protein and small envelope (E) protein (Figure 3). The M, S and E proteins are enclosed in the viral envelope, while N protein binds with the viral RNA forming the nucleocapsid[13,15]. The M protein is responsible for the virus outline and with the assistance of E protein leads to the formation of the mature viral envelope, orchestrating in parallel, the assembly of the virus[24]. The S protein is responsible for the formation of homotrimeric spikes of SARS-CoV-2 surface inducing the entry into host cells[25,26].
Figure 3 Severe acute respiratory syndrome coronavirus 2 schematic structure.
A: Schematic representation of the genomic organization of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2); B: Schematic diagram of the structural features of SARS-CoV-2 and its primary structural proteins. The above diagrams were created with Biorender.com. Kb: kilobase pair; ORF: Open reading frame; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2; S: Spike protein; E: Envelope protein; M: Membrane protein; N: Nucleocapsid protein.
The genomic size of CoVs ranges from 26 to 32 kilobases (kb)[27]. More specifically, according to genomic analysis of a new RNA virus strain initially named Wuhan-hu-1 coronavirus (WHCV) it was found that one strain of SARS-CoV-2 is 29.9 kb in size[28]. However, MERS-CoV and SARS-CoV have positive RNA genomes of 30.1 kb and 27.9 kb in size. Viral RNA codes for structural and nonstructural proteins. The structural proteins are formed within the 3’ end of the RNA genome. On the contrary, the 5΄ two-thirds of the genome codes for nonstructural proteins (nsps), essential for replication[11,27].
The aforementioned genomic content is initially utilized as a template for the synthesis of polyprotein 1a/b (pp1a/ppqab), encoding for nsps for the formation of the replication-transcription complex organized in double-membrane vesicles resulting in the production of subgenomic RNAs (sgRNAs). These subgenomic messenger RNAs (mRNAs) share the same leader sequence consisting of 70-90 nucleotides at their 5’ ends and the same 3’ ends. Transcription termination and the resulting acquisition of a leader RNA appears at transcription sequences, found between the open reading frames (ORFs). These particular minus-strand sgRNAs act as templates for the formation of subgenomic mRNAs[15,29].
The genome and the subgenomes of a typical CoV include at least six ORFs. Two-thirds of the viral RNA, primarily found in the first ORFs (ORF1a/b) encodes 16 nsps, except gammaCoV which lacks nsp1. Two polypeptides, pp1a and pp1b, are formed from the 1- frameshift between ORF1a and ORF1b. The two polypeptides are further processed by the virally encoded main protease or chymotrypsin-like protease. The remaining ORFs are responsible for encoding structural and accessory proteins. The remaining genome encodes the four critical structural proteins, S, M, E and N, as well as a number of additional proteins interfering with the host innate immune response (Figure 3)[10,15,29].
Based on primary data regarding the WHCV virus strain previously mentioned, it was shown that WHCV shares a phylogenetic and genomic resemblance to SARS-CoV, especially in the S-glycoprotein gene, emphasizing the ability of direct person spread[28]. In line with this, most proteins of SARS-CoV-2 are almost identical to SARS-CoVs with slight differences. Another study demonstrated that the mutation in both nsp2 and nsp3 are essential in the differentiation mechanism of the virus[30,31]. Therefore, further investigations are anticipated regarding the difference in transmission and host tropism between SARS-CoV and SARS-CoV-2. Finally, according to another Chinese research study analyzing the SARS-CoV-2 genotypes of patients from different areas, an increased mutation rate of the virus in many individuals was observed[32].
ACE2 ROLE IN SARS-COV-2 REPLICATION AND PATHOGENESIS
Various cellular receptors have been described as receptors of CoVs, such as ACE2[13,33]. ACE2 is a zinc-dependent carboxypeptidase and is expressed in vascular endothelial, cardiovascular and renal tissue, testes and epithelia of the small intestine[34]. The ACE2 genes codes for an 805 amino acid type I transmembrane glycoprotein and spans 18 exons. Furthermore, ACE2 includes a short intracellular cytoplasmic tail and a longer extracellular area exhibiting carboxyl-monopeptidase activity. The carboxyl-terminal domain of this particular receptor is approximately 48% homologous with collectrin, a protein expressed in the kidney[35].
ACE2 is a recognized cellular receptor for SARS-CoV regulating both human-to-human and cross-species spread[10]. Different studies have confirmed that SARS-CoV-2 exhibits similar and even, greater affinity to ACE2 receptors than SARS-CoV S protein[36,37]. Since the interaction of ACE2 receptor and S glycoprotein from SARS-CoV-2 is essential for virus entry, virus-receptor binding activity is undergoing exhaustive investigation via various methods. More specifically, the methodical identification of β-CoV receptors demonstrated that ACE2 positive human cells not expressing human aminopeptidase N or dipeptidyl peptidase-4 improved viral entry[10].
The virion S-glycoprotein binds to the human ACE2 receptor. After membrane infusion, the viral genome is liberated into the cytoplasm inducing translation of the two polyproteins, followed by the transcription of sgRNAs and lastly, its replication[13]. Newly formed envelope glycoproteins are directed in the Golgi apparatus or endoplasmic reticulum membranes leading to the formation of nucleocapsid proteins, envelope glycoproteins and genomic RNA, which in turn are accumulated and form viral unit buds. Finally, virion-containing vesicles release the virus[10,13].
DEFICIENCY OF ACE2
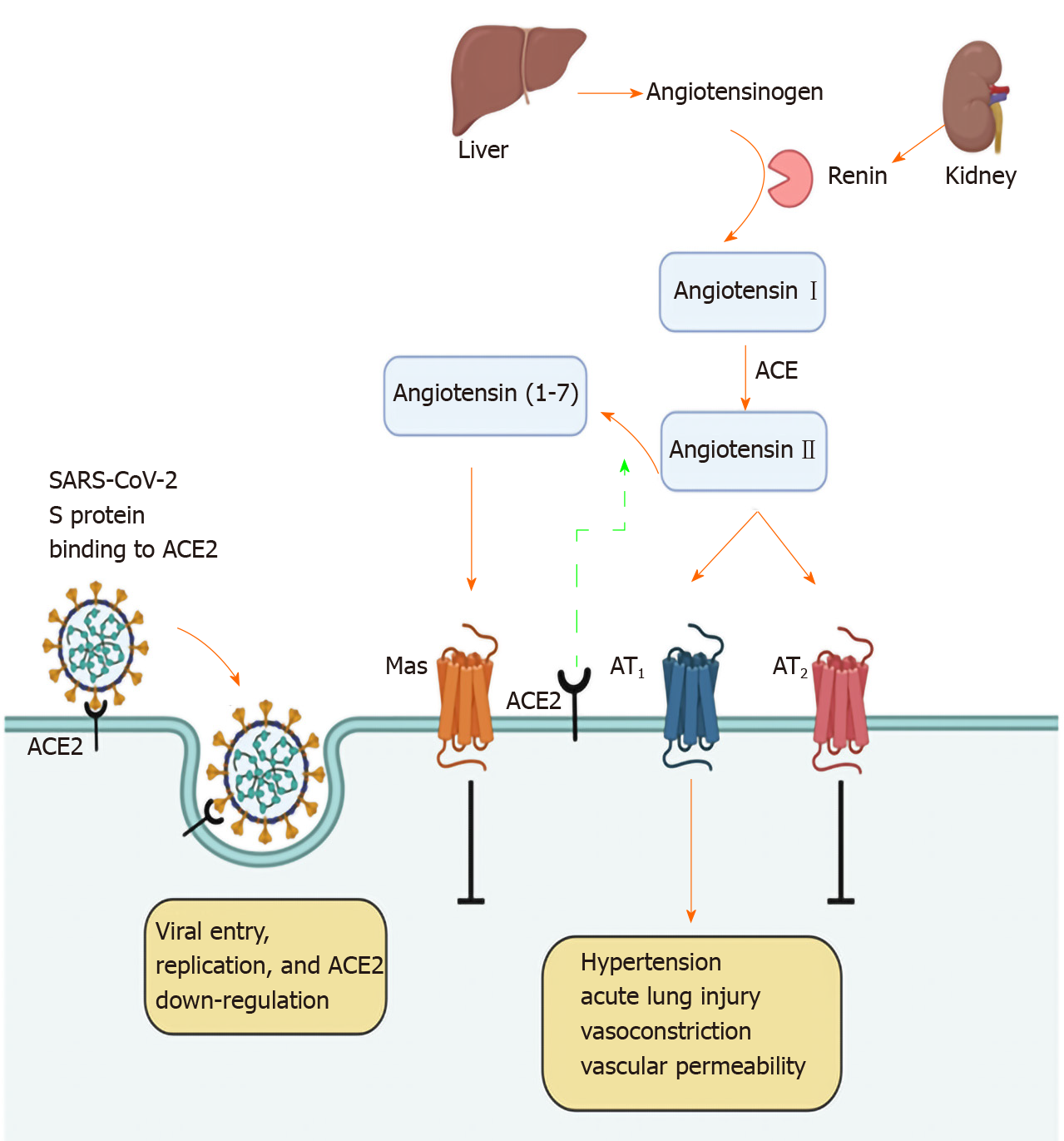
ACE2 deficiency has been implicated in several pathologies such as acute lung injury or cardiovascular and renal disease due to the activation of the renin-angiotensin system (RAS)[38]. The RAS is an important regulatory cascade operating both at a systemic or circulatory level and at a local tissue level.
Within the RAS, regulation is carried out via protease cascades generating various bioactive peptides (Figure 4). The angiotensinogen originating from the liver and cleaved by renin is converted to angiotensin I (Ang I)[38]. The latter is subsequently, converted to angiotensin II (Ang II) by ACE, which is expressed in endothelial cells of vessels and in organs such as the heart, kidney, brain or the lung[39]. Ang II displays a major bioactive peptide within the RAS, which mediates its effects via the interaction with two G protein-coupled receptors, angiotensin II receptor type 1 and 2 (AT1 and AT2), activating downstream signaling[38,40].
Figure 4 Interaction between severe acute respiratory syndrome coronavirus 2 and the renin-angiotensin system.
The above diagram was created with Biorender.com. SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2; ACE2: Angiotensin converting enzyme type 2; ACE: Angiotensin converting enzyme; AT: Angiotensin II receptor type.
Regarding the respiratory tract, elevated levels of Ang II stimulate the production of pulmonary fibrosis and hypertension. Local tissue-based RAS exacerbates lung injury, and worsens acute respiratory distress syndrome[41]. ACE inhibitors, along with AT1 antagonists have been associated with reduced pulmonary hypertension and reduced SARS-Co-V spike protein-induced lung injury.
ACE2 AND THE GASTROINTESTINAL LUMEN
ACE2 has also been found to be associated with the function of the gut, since it is expressed in intestinal epithelial cells. ACE2 function is required for the maintenance of antimicrobial peptide expression, ecology of the gut microbiome and amino acid homeostasis[42,43]. ACE2, as mentioned before, is a chimeric protein, which is homologous with collectrin. The discontinuance of collectrin function in mice is linked with near-complete downregulation of renal neutral amino acid transporters such as B0AT1; therefore, affecting renal amino acid re-absorption[42]. However, studies have shown that ACE2 can bind and stabilize B0AT1 on the surface of intestinal cells[38]. Moreover, both of these proteins are responsible for regulation of the recruitment of neutral amino acids in the bowel where the expression of collectrin is not found[38]. Furthermore, mutations in the slc6a encoding B0AT1 result in Hartnup’s disease[42]. The primary characteristic of this disease is the neutral aminoaciduria resembling the findings from collectrin-deficient mice. Most patients are commonly asymptomatic, even though certain conditions such as infections or malnutrition lead to pellagra-like symptoms[38]. Interestingly, dietary tryptophan is, mainly taken up via the B0AT1/ACE2 transport route on the cell surface located in the small gut. Tryptophan must be acquired from food, as is needed for the in vivo formation of nicotinamide preventing its deficiency (namely pellagra), by promoting the activation of mammalian target of rapamycin (mTOR), either directly via the tryptophan-nicotinamide pathway or through nutrient sensing, inducing the secretion of antimicrobial peptides such as a-defensins[42,44].
In an ongoing investigation of single cell-RNA sequencing information from control subjects and subjects with colitis or inflammatory bowel disease (IBD), it was indicated that ACE2 presence in colon cells was emphatically linked with genes inducing immunity, as well as viral infection, but was adversely connected with functions such as viral transcription, phagocytosis etc[45].
Furthermore, in another recent study, the potential pathogen transmission route of the SARS-CoV-2 infection in the GI tract was investigated. Colon tissue from healthy adults and colorectal cancer (CRC) patients was analyzed in terms of RNA expression of ACE2. The data showed that ACE2 is highly expressed in colonic cells, showing a gradual increase from healthy control individuals to CRC patients. Thus, it was suggested that SARS-CoV-2 may be more detrimental to patients with colorectal malignancy than individuals without CRC[46].
Finally, there has been important clinical evidence highlighting an ACE2 imbalance as a major player in poor outcomes (higher mortality rate and disease severity) in COVID-19 patients with pre-existing age-related comorbidities (renal, metabolic and cardiovascular disorders) addressing a possible role of gut microbiota dysbiosis in the disease prognosis[47].
HEPATOBILIARY SYSTEM AND RAS AXIS ACTIVATION
Research has provided proof that RAS also plays an essential role in the pathophysiological mechanisms of chronic hepatobiliary disorders. It was shown in animal experiments that a noticeable up-regulation of intrahepatic RAS components exists, and the inhibition of RAS proved to prevent liver fibrogenetic changes[48,49]. RAS may be considered a dual function system in which the proliferative and antiproliferative functions are, mainly orchestrated by the ACE-ACE2 balance. High levels of Ang II due to up-regulation of ACE, result in the catabolism of Ang 1-7 inducing vasoconstriction and thus, liver injury and pulmonary fibrosis[50-52].
Different studies have shown that an opposite ratio in rat injured liver models and cirrhotic human livers is connected with up-regulation of ACE2 (gene and protein levels) and conversion of Ang II to Ang 1-7, resulting in elevated Ang 1-7 levels in the circulation facilitating vasodilation, and thus, antifibrotic activity in hepatic disorders[39,50,51]. Thus, the antagonism between Ang 1-7/Mas receptor and Ang II may represent an overall counter-regulatory action in the RAS system (Figure 4)[52]. Finally, but importantly, experimental studies have shown that ACE2 might also be upregulated due to oxygen reduction in the tissue. There is accumulating evidence that liver fibrosis is linked with advanced damage of the hepatocellular oxygenation mechanism with the most probable cause encompassing vasoconstriction, intrahepatic shunting and sinusoidal capillarization. Based on this hypothesis, it has been shown that in HepG2 cells, ACE2 mRNA progressively rose during increasing periods of hypoxia[50].
Finally, it has been shown that ACE2 up-regulation might also contribute to the vasodilation of cirrhosis via the degradation of Ang II. This observation might help understand why vasodilation continues to persist in individuals with progressive hepatic disorder, in spite of the stimulation of systemic RAS and why vasoconstrictor responses to Ang II are impaired[50,53].
CONCLUSION
Several studies have indicated the importance of the digestive system as an infection pathway of SARS-CoV-2. ACE2 receptors, highly expressed in the GI tract, have a significant contribution in the pathogenesis and replication of COVID-19, and consequently, in the GI system, as shown in this minireview. Further research is needed to clarify ACE cell function and its activity regarding SARS-CoV-2, as well as its utility concerning the pathophysiology of gastrointestinal and hepatobiliary diseases.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Greece
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Huerta-Franco MR, Lalmuanawma S S-Editor: Zhang H L-Editor: Webster JR P-Editor: Ma YJ