Published online Sep 21, 2020. doi: 10.3748/wjg.v26.i35.5328
Peer-review started: July 6, 2020
First decision: July 28, 2020
Revised: August 7, 2020
Accepted: August 26, 2020
Article in press: August 26, 2020
Published online: September 21, 2020
Processing time: 73 Days and 1.4 Hours
Our previous study demonstrated that RBBP4 was upregulated in colon cancer and correlated with poor prognosis of colon cancer and hepatic metastasis. However, the potential biological function of RBBP4 in colon cancer is still unknown.
To investigate the biological role and the potential mechanisms of RBBP4 in colon cancer progression.
Real-time polymerase chain reaction and western blot analysis were used to detect the expression of RBBP4 in colon cancer cell lines. The cell proliferation and viability of SW620 and HCT116 cells with RBBP4 knockdown was detected by Cell Counting Kit-8 and 5-ethynyl-2’-deoxyuridine staining. The transwell assay was used to detect the invasion and migration capabilities of colon cancer cells with RBBP4 knockdown. Flow cytometry apoptosis assay was used to detect the apoptosis of colon cancer cells. Western blotting analysis was used to detect the expression of epithelial-mesenchymal transition and apoptosis related markers in colon cancer. The nuclear translocation of β-catenin was examined by Western blotting analysis in colon cancer cells with RBBP4 knockdown. The TOPFlash luciferase assay was used to detect the effect of RBBP4 on Wnt/β-catenin activation. The rescue experiments were performed in colon cancer cells treated with Wnt/β-catenin activator LiCl and RBBP4 knockdown.
We found that RBBP4 was highly expressed in colon cancer cell lines. The 5-ethynyl-2’-deoxyuridine assay showed that knockdown of RBBP4 significantly inhibited cell proliferation. RBBP4 inhibition reduced cell invasion and migration via regulating proteins related to epithelial-mesenchymal transition. Knockdown of RBBP4 significantly inhibited survivin-mediated apoptosis. Mechanistically, the TOPFlash assay showed that RBBP4 knockdown increased activity of the Wnt/β-catenin pathway. Meanwhile, RBBP4 knockdown suppressed nuclear translocation of β-catenin. With Wnt/β-catenin activator, rescue experiments suggested that the role of RBBP4 in colon cancer progression was dependent on Wnt/β-catenin pathway.
RBBP4 promotes colon cancer development via increasing activity of the Wnt/β-catenin pathway. RBBP4 may serve as a novel therapeutic target in colon cancer.
Core Tip: Our previous study demonstrated upregulation of RBBP4 in colon cancer and correlation of poor prognosis with colon cancer and hepatic metastasis. This study explored the potential biological function of RBBP4 in colon cancer. We found that RBBP4 was highly expressed in colon cancer cell lines. Knockdown of RBBP4 significantly inhibited cell proliferation and survivin-mediated apoptosis and suppressed nuclear translocation of β-catenin. RBBP4 inhibition reduced cell invasion and migration via regulating proteins related to epithelial–mesenchymal transition. Mechanistically, RBBP4 knockdown increased activity of the Wnt/β-catenin pathway. RBBP4 may serve as a novel therapeutic target in colon cancer.
- Citation: Li YD, Lv Z, Zhu WF. RBBP4 promotes colon cancer malignant progression via regulating Wnt/β-catenin pathway. World J Gastroenterol 2020; 26(35): 5328-5342
- URL: https://www.wjgnet.com/1007-9327/full/v26/i35/5328.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i35.5328
Colon cancer is one of the most common malignancies in developed countries. There are more than one million new cases of colon cancer worldwide and 608000 deaths every year[1]. Although the treatment of colon cancer has made great progress, including surgery or radiotherapy and chemotherapy, the prognosis of patients with colon cancer has shown no marked progress in recent years[2]. The 5-year relative survival rate of patients with stage IV disease is slightly higher than 10%[3,4]. Although considerable efforts have been made in the past few years to clarify the mechanisms underlying the development and progression of colon cancer, it is still far from completely understood. Hence, it is necessary to explore the further mechanisms involved in the pathogenesis of colon cancer and to develop new therapeutic targets.
RBBP4 is a new, 48-kD tumor-specific protein found in HeLa cell lysates[5,6]. RBBP4 belongs to a highly conserved subfamily of nucleoproteins with four WD repeat sequences. RBBP4 binds to retinoblastoma protein in vivo and in vitro, hence the name[7]. The RBBP4 gene encodes a protein that is part of several chromatin-modified protein complexes, such as nucleosome remodeling and deacetylation complex[8], polycomb repressor complex 2[9], and SIN3-chromatin modulating complexes[10], which influence gene transcription, and regulates cell cycle and proliferation[11]. In recent decades, accumulated research has demonstrated that RBBP4 plays a key role in the pathogenesis of cancers, such as liver[12], breast[13], and gastric[14,15] cancers. In our previous study, we proved that RBBP4 is upregulated in colon cancer and may serve as a novel predictor for poor prognosis of colon cancer and liver metastasis[16]. However, its potential role and mechanisms in colon cancer have not been reported.
Therefore, the present study aimed to explore the potential role of RBBP4 in colon cancer aggravation and the underlying molecular mechanisms. We detected expression of RBBP4 in colon cancer cell lines, then investigated the role of RBBP4 in colon cancer cell proliferation, migration, invasion, and apoptosis and finally explored the molecular mechanisms of RBBP4 in colon cancer malignancy characteristics. This study clarifies the role of RBBP4 in colon cancer development through its effect on the Wnt/b-catenin signaling pathway.
A normal human colon cell line (NCM640) and colon cancer cell lines (SW620, HT29, LoVo, SW480, and HCT-116) were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). These cell lines originated from the American Type Culture Collection (ATCC, Manassas, VA, United States). All the cell lines were cultured in the corresponding medium according to the suggestion of ATCC with 10% fetal bovine serum (FBS). All the cells were maintained in a humidified incubator with 5% CO2 at 37 °C. The Wnt/β-catenin activator LiCl was obtained from Sigma-Aldrich (Munich, Germany).
The plasmids of the human RBBP4 gene, siRNA targeting human RBBP4, and their controls were synthesized by Genechem (Shanghai, China). The cells were transfected with siRNAs, plasmids, or their controls using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, United States). In total, 2 × 105–3 × 105 cells were transfected with 100 pmol siRNA or 2 μg plasmid DNA. Western blotting was used to detect the transfection efficiency 24 h and 48 h after transfection, and real-time polymerase chain reaction was used for verification.
Total RNA from colon cancer tissues and cells was extracted using TRIzol reagent (Invitrogen). Total RNA (0.5 μg) was reversed transcribed to cDNA using PrimeScript™ RT reagent Kit with gDNA Eraser (Takara, Dalian, China). SYBR Green Polymerase Chain Reaction Master mix (Takara) was used to determine the mRNA level of RBBP4 on an ABI 7900Fast Real-time Detection System (Applied Biosystems, Carlsbad, CA, United States) in 20 μL reaction system. All reactions were performed in triplicate. The relative expression of RBBP4 was normalized to the internal reference GAPDH. The 2-DDCT method was used to analyze the data. The primers used in the study were as follows: GAPDH forward primer: 5'-ATG GGG AAG GTG AAG GTC G-3', GAPDH reverse primer: 5'-GGG GTC ATT GAT GGC AAC AAT A-3'; RBBP4 forward primer: 5'-GCT ATG GGC TTT CTT GGA-3', and RBBP4 reverse primer: 5'-CAC AGG CAG ATG GTA TGG-3'.
Cell viability was assessed by Cell Counting Kit-8 (CCK-8) assay (Dojindo Laboratories, Kumamoto, Japan). Cells were seeded in 96-well plates at 3 × 103 cells per well with 200 μL culture medium. With supernatant removed, 10 μL CCK-8 reagent in 100 μL medium was added to each well at 0 h, 24 h, 48 h, 72 h, and 96 h. The plates were incubated in the dark at 37 °C for 2 h, and absorbance at 450 nm was detected with a microplate reader (BioTek, Winooski, VT, United States). The experiments were performed in triplicate.
5-Ethynyl-2’-deoxyuridine (EdU) assay was performed using Click-iT™ EdU Imaging Kit with Alexa Fluor™ 488 Azides (Invitrogen). Briefly, 1 × 105 cells were plated in six-well plates and incubated at room temperature overnight. The cells were incubated with 10 μM EdU for 1 h at 37 °C and fixed in 3.7% paraformaldehyde. After permeabilization with 0.5% Triton X-100 in phosphate buffered saline for 20 min, the cells were reacted with 1 × Click-iT® reaction cocktail for 20 min. The nuclei were labeled with Hoechst 33342 for 30 min and photographed under a fluorescence microscope. All studies were conducted in triplicate.
Total proteins were extracted using the ice cold radioimmunoprecipitation assay buffer with cocktail protease and phosphatase inhibitors (Cell Signaling Technology, Danvers, MA, United States). NE-PER™ Nuclear and Cytoplasmic Extraction Reagent (Thermo Scientific, Waltham, MA, United States) was used to extract the nuclear and cytoplasmic proteins. Proteins were quantified using a BCA protein assay kit (Thermo Scientific). Then equivalent proteins were separated by 10% SDS-PAGE and transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA, United States). The membranes were probed with antibodies against RBBP4 (ab92344), survivin (ab134170), GAPDH (ab181620), β-catenin (ab32572), pro-caspase-3 (ab32150) (Abcam, Cambridge, MA, United States), Cleaved caspase-3 (9661), E-cadherin (14472), N-cadherin (13116), vimentin (5741), and histone H3 (14269) (Cell Signaling Technology) overnight at 4 °C and then incubated with horseradish-peroxidase-conjugated secondary antibodies (Cell Signaling Technology) for 1 h at room temperature. The bands were visualized by ECL kit (Millipore).
The migration and invasion assays were performed by the transwell method. For the invasion assay, the cells were plated on Matrigel-coated upper chambers (24-well inserts; pore size, 8 mm; BD Biosciences, San Jose, CA, United States). For the migration assay, the cells were plated on uncoated upper chambers. In the lower wells, medium was replaced with fresh medium with 5% FBS. The cells were incubated for 24 h in medium containing 1% FBS, trypsinized, and suspended in medium containing 1% FBS at a final concentration of 1 × 106 cells/mL. Then, 200 μL cell suspension was placed in each of the upper wells, and the chamber was incubated at 37 °C for 24 h. Cells were fixed and stained with hematoxylin and eosin. The nonmigrating cells from the upper surface of the filter were wiped with a cotton swab. The cells that migrated to the lower side of the filter were counted and photographed with an optical inverted microscope. Five random fields in each assay were counted and averaged.
For the assessment of apoptosis, an Annexin V–FITC/propidium iodide (PI) apoptosis detection kit (BD Biosciences) was used. Colon cancer cells were collected in six-well plates at 1 × 106 cells/mL. After transfection for 48 h, the cells were trypsinized and washed once with phosphate buffered saline. After centrifugation at 1000 r/min for 5 min, the cells were stained with 5 μL Annexin V–FITC and PI in the dark condition for 30 min, and then were analyzed by flow cytometry (BD Biosciences). At least 10000 events were recorded for each sample. The apoptosis data were analyzed by FlowJo V10 software (Tree Star, San Francisco, CA, United States).
The TOPFlash assay was performed using the T-cell factor Reporter Plasmid Kit (Millipore). The ratio of luciferase activities from a T-cell factor-responsive reporter (pTOPFlash) vs a control luciferase reporter gene construct (pFOPFlash) was determined 48 h after transfection with Lipofectamine 2000 in SW480 and HCT116 cells. Luciferase activities were normalized for transfection efficiency by cotransfection with a β-galactosidase-expressing vector. The cells were transfected with siRBBP4 plasmid or RBBP4 plasmid or control plasmids. The cells were harvested after 24 h and processed for luciferase and β-galactosidase activities, and the data were normalized to β-galactosidase levels.
Statistical analyses were performed using GraphPad Prism version 7.0 (GraphPad Software, La Jolla, CA, United States) software and verified by SPSS version 20.0 (SPSS, Chicago, IL, United States). Each experiment was performed at least in triplicate, and the results were expressed as the mean ± standard deviation. Student’s t test and one-way analysis of variance were conducted to analyze the differences between groups, and a P < 0.05 was considered statistically significant.
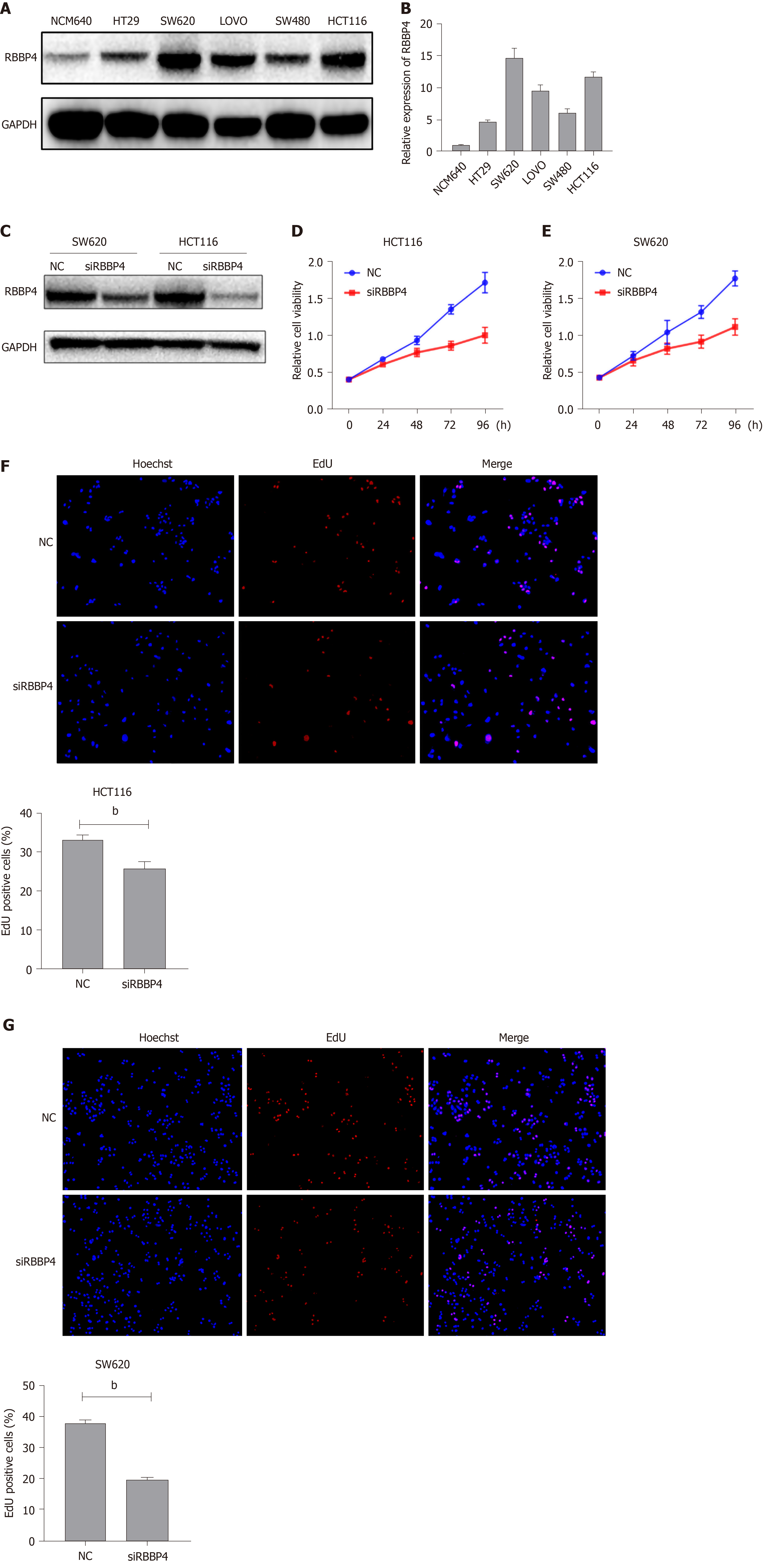
In our previous study, we detected the expression pattern of RBBP4 in colon cancer tissues and proved that RBBP4 was upregulated. In the present study, we examined the protein and mRNA levels of RBBP4 in five human colon cancer cell lines and a normal human colon cell line. mRNA and protein levels of RBBP4 were significantly higher in colon cancer cell lines compared with the normal human colon cell line. We selected SW480 and HCT116 cells with high RBBP4 levels for subsequent experiments (Figure 1A and 1B).
To investigate the biological function of RBBP4 in colon cancer cells, we knocked down RBBP4 via siRNA-mediated gene silencing. The knockdown efficiency was determined by Western blotting (Figure 1C). We then examined the role of RBBP4 in colon cancer cell viability using the CCK-8 assay. Cell viability was decreased in both HCT116 and SW620 cells after RBBP4 knockdown (Figure 1D and 1E). EdU proliferation assay showed that RBBP4 knockdown significantly reduced proliferation of colon cancer cells (Figure 1F and 1G). These results showed that RBBP4 played an essential role in the growth of colon cancer cells.
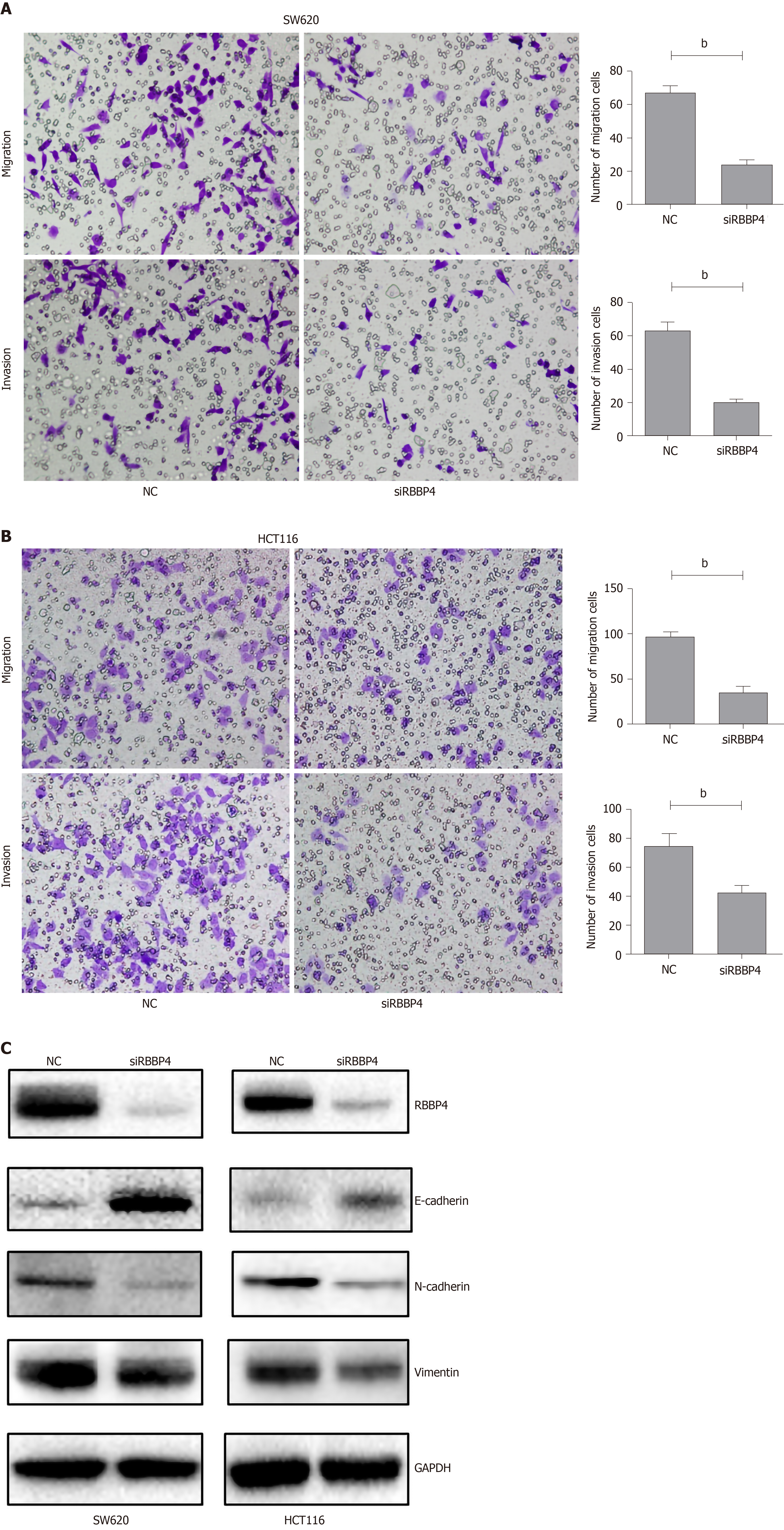
We examined the effect of RBBP4 knockdown on colon cancer cell migration and invasion in vitro using the transwell assay. The number of migrated and invasive cells in RBBP4 knockdown HCT116 and SW620 colon cancer cells was less than that in the control group (Figure 2A and 2B). The epithelial-mesenchymal transition (EMT) pathway has been proved to play a key role in tumor migration and invasion[17]. To investigate the molecular mechanisms of RBBP4 in regulating colon cancer cell migration and invasion, we performed Western blotting to detect expression of EMT-related proteins including N-cadherin, E-cadherin, and vimentin. RBBP4 knockdown markedly decreased mesenchymal proteins, but upregulated epidermal protein expression (Figure 2C). These results indicated that RBBP4 regulated colon cancer cell migration and invasion via the EMT pathway.
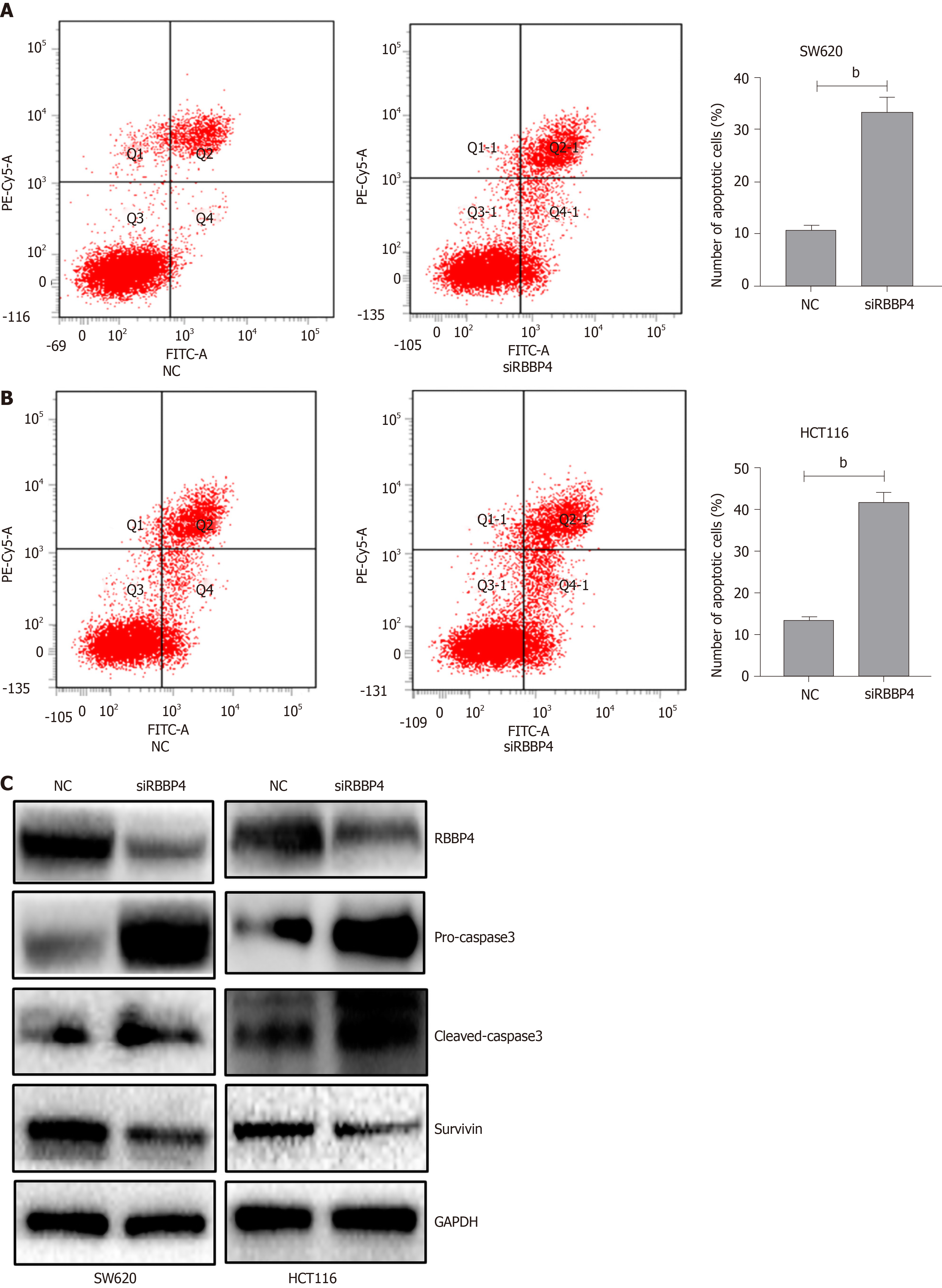
To investigate the mechanism underlying RBBP4-knockdown-induced antiproliferation, we detected the apoptotic rate of colon cancer cells by flow cytometry. Apoptosis rate in HCT116 and SW620 cells with RBBP4 knockdown increased by 42.0% ± 2.2% and 33.3% ± 2.91%, respectively, compared with the control cells (12.95% ± 1.40% and 10.83% ± 0.93%, P < 0.01, Figure 3A and 3B). To investigate the molecular mechanisms of RBBP4 in regulating colon cancer cell apoptosis, we detected apoptosis-related proteins, and showed that survivin, an antiapoptotic protein, was downregulated after RBBP4 knockdown, thus leading to the increase of pro-caspase-3 and cleaved caspase-3.
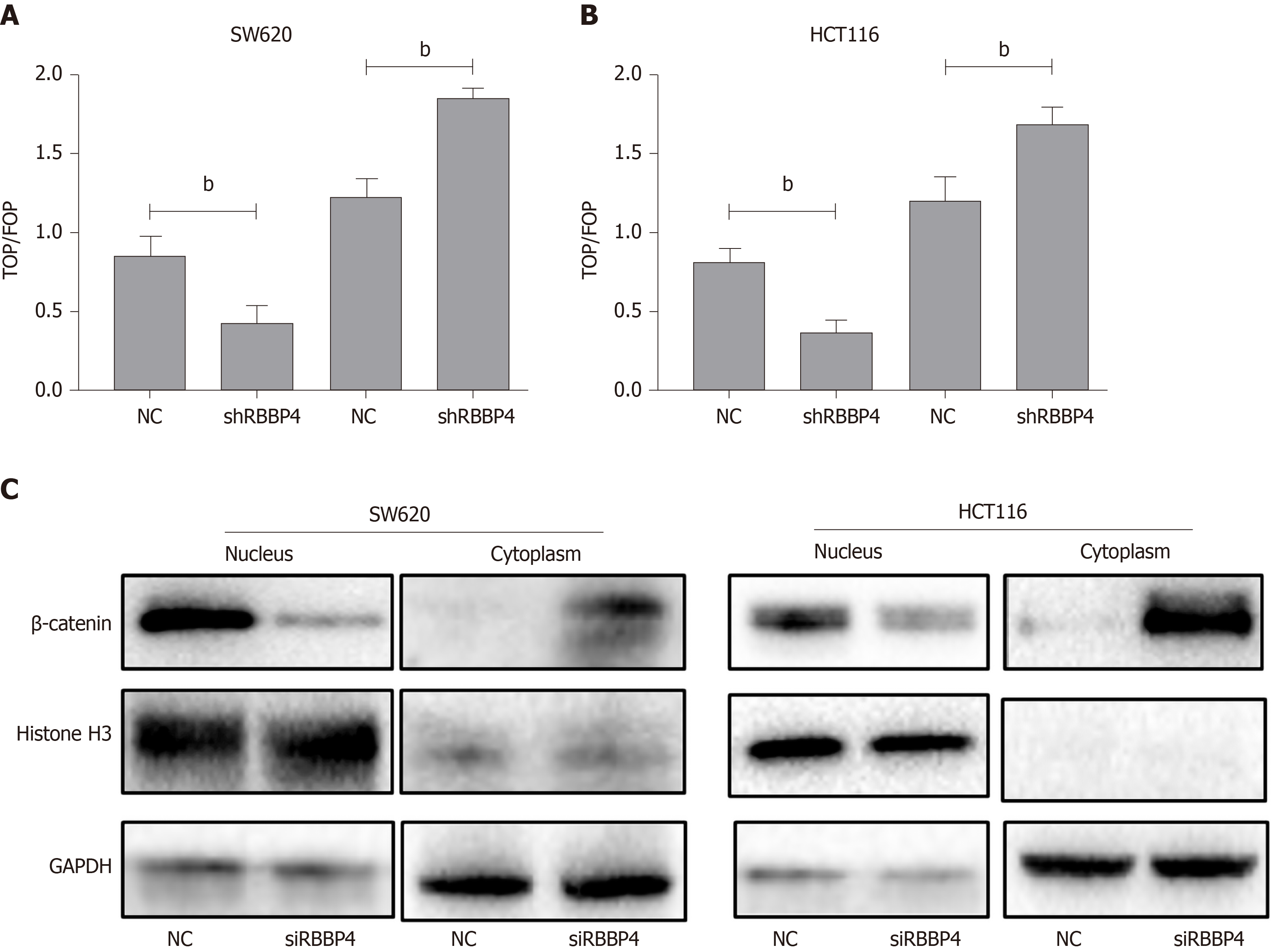
The Wnt/β-catenin pathway is one of the important signaling pathways inducing EMT, and survivin is a known downstream target of the pathway[17-19]. We hypothesized that the biological role of RBBP4 was executed through the Wnt/β-catenin pathway. To clarify this hypothesis, the TOP/FOP flash luciferase reporter assays were used. Compared with the control cells, overexpression of RBBP4 led to an increase of TOP flash luciferase reporter activity in HCT116 and SW620 cells (Figure 4A and 4B). However, RBBP4 knockdown inhibited the activity of the TOP flash luciferase reporter (Figure 4A and 4B). As reported previously, β-catenin nuclear translocation is an essential event for Wnt/β-catenin pathway activation. To elucidate further the underlying mechanism, we examined the influence of RBBP4 on β-catenin nuclear translocation in colon cancer cells. The level of β-catenin in the nucleus was decreased, while that in cytoplasm was increased by RBBP4 knockdown (Figure 4C). All these data indicated that activity of the Wnt/β-catenin pathway was regulated by RBBP4 via regulation of the nuclear translocation of the β-catenin protein in colon cancer cells.
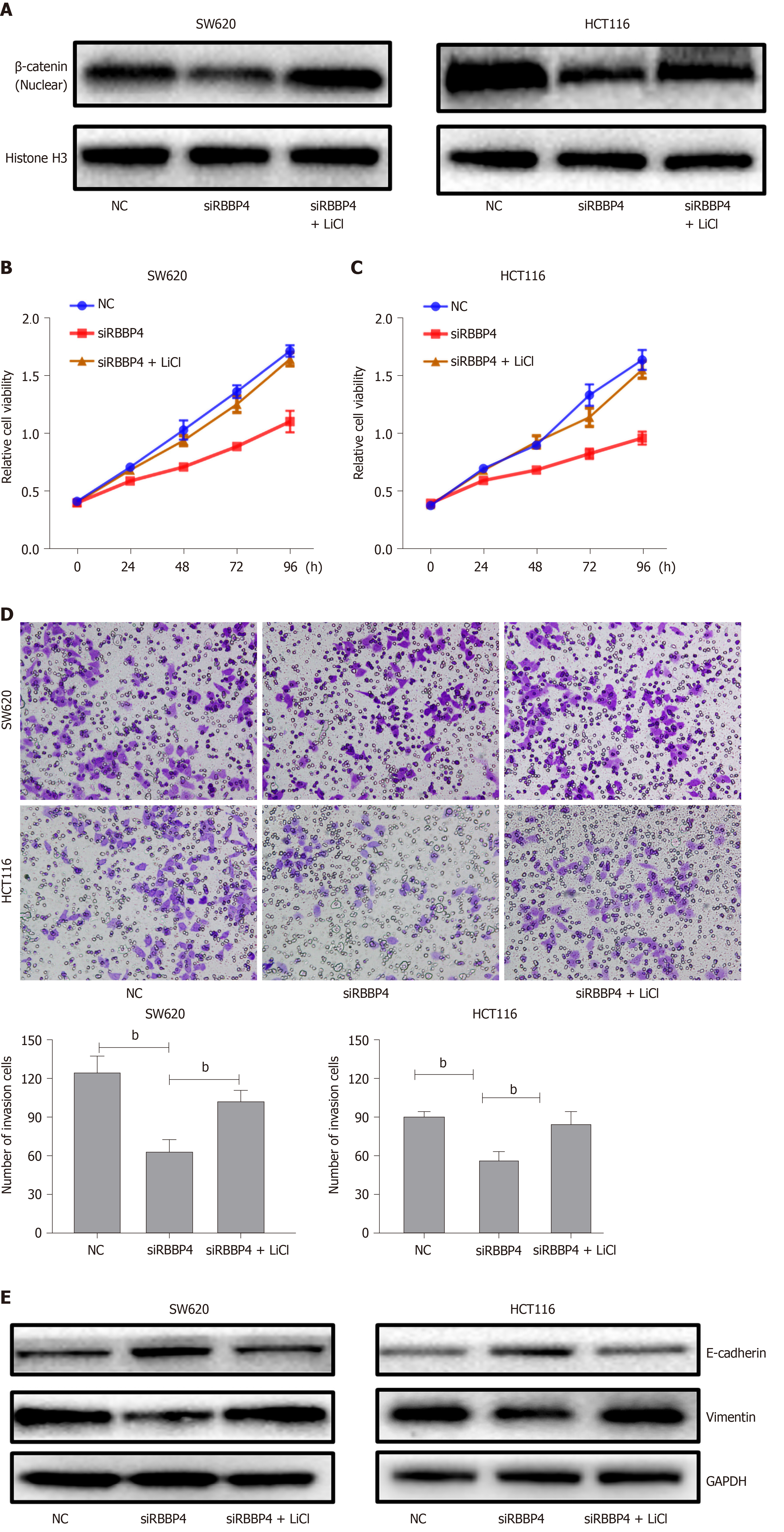
To examine whether the function of RBBP4 in colon cancer was mediated by the Wnt/β-catenin pathway, we used the Wnt/β-catenin pathway activator LiCl in a rescue experiment. The role of RBBP4 knockdown on the inhibition of β-catenin nuclear translocation was partly reversed by LiCl (Figure 5A). CCK-8 assays showed that the viability of HCT116 and SW620 cells treated with RBBP4 siRNA was significantly enhanced when they were cotreated with LiCl compared with untreated cells (P < 0.05, Figure 5B and 5C). The transwell assay showed that the inhibition of invasion by RBBP4 knockdown was partly reversed by LiCl (P < 0.05, Figure 5C and 5D). The expression of EMT-related proteins was also partly reversed by LiCl compared with the shRBBP4 group (Figure 5E). All these results suggested that the role of RBBP4 in colon cancer progression is mediated by the Wnt/β-catenin pathway.
RBBP4, also known as RbAp48, is named for its ability to bind to retinoblastoma proteins in vivo and in vitro[20]. Previous studies showed that the RBBP4 protein is a component of a variety of complexes involved in chromatin assembly, remodeling, and nucleosome modification, such as SIN3[21], polycomb repressor complex 2[22,23], histone acetyltransferase 1[24], and chromatin assembly factor 1[25] and plays a different role in each complex. It has been demonstrated that RBBP4 expression is upregulated and correlated with the malignant phenotypes in many types of human tumors, such as lung cancer[26], liver cancer[27], thyroid carcinoma[28], and acute lymphoblastic leukemia[29]. However, little is known about RBBP4 in colon cancer.
In our previous study, we demonstrated that RBBP4 was upregulated in colon cancer tissues, and elevated RBBP4 level was correlated with poor prognosis and liver metastasis. Nevertheless, the detailed molecular biological function of RBBP4 and the potential mechanisms in colon cancer are unclear. In the present study, our evidence indicated that RBBP4 knockdown decreased the proliferation, apoptosis, and aggressiveness of colon cancer cells, suggesting its oncogenic functions in colon cancer progression. RBBP4 promoted the nuclear accumulation of β-catenin, thus activating the Wnt/β-catenin signaling pathway. The role of RBBP4 in colon cancer progression was partially dependent on Wnt/β-catenin signaling pathway.
Abnormalities in the Wnt signaling pathway are associated with a variety of tumor types, including colon cancer[30,31]. The Wnt pathway is classified into canonical and noncanonical pathways; the former of which is β-catenin dependent[32]. For the canonical pathway, in the absence of Wnt ligands, free cytoplasmic β-catenin binds to cytoplasmic complexes containing Adenomatous Polyposis Coli, axin, casein kinase 1a, and glycogen synthase kinase 3b, which promotes the phosphorylation of β-catenin leading to β-catenin ubiquitination and subsequent proteasomal degradation. The interaction between Wnt and Frizzled leads to the activation of the Disheveled family proteins. The activated Disheveled proteins cause the inhibition of glycogen synthase kinase 3b, resulting in the accumulation of free cytoplasmic β-catenin, which is then transported to the nucleus. In the nucleus, β-catenin binds to various transcription factors, such as T-cell factor and lymphoid enhancer factor 1, to activate Wnt target genes[33-35]. However, the relationship between RBBP4 and the Wnt pathway is poorly understood. In our study, for the first time we proved that RBBP4 could enhance the nuclear translocation of β-catenin and activate the Wnt/β-catenin pathway in colon cancer.
Studies over the past decade have shown that cells that harbor functionally impaired mutations of Wnt signaling cascades, such as Adenomatous Polyposis Coli, β-catenin, and axin, are thought to be prevalent in colon cancer[36]. The mutations lead to abnormal transcriptional induction of Wnt/β-catenin downstream genes[37]. Many target genes of Wnt/β-catenin have been identified, such as survivin[38]. Survivin was recently identified as an inhibitor of apoptosis that directly inhibits caspase-3 and caspase-7 activity[39]. The role of survivin in colorectal tumorigenesis has been shown. We found that RBBP4 knockdown inhibited the level of survivin, thus inducing apoptosis of colon cancer cells. This result proved that RBBP4 regulates the Wnt/β-catenin pathway.
Liver metastasis is an important characteristic of colon cancer, and EMT plays a central role[40]. A previous study showed that activation of Wnt/β-catenin signaling results in expression of target genes that lead to the dedifferentiated phenotype and EMT of colon cancer cells[41]. The nuclear translocation of β-catenin was reported to induce Slug and inhibit E-cadherin transcription in colon cancer[42]. However, the relationship between RBBP4 and EMT has not been clarified. In our study, we found that RBBP4 knockdown markedly decreased mesenchymal proteins but upregulated expression of epidermal proteins, indicating inhibition of the EMT pathway. This process may be mediated by the Wnt/β-catenin pathway.
In conclusion, the results presented in this study demonstrated that RBBP4 plays an important role in the malignant progression of colon cancer. This is probably induced via inhibiting Wnt/β-catenin pathway activity and relocating β-catenin from the nucleus to the plasma membrane. Further investigation of the functional mechanism of RBBP4 as a tumor oncogene may provide a potential therapeutic strategy for intervention of colon cancer progression.
Our previous study demonstrated that RBBP4 is upregulated in colon cancer and correlated with poor prognosis of colon cancer and hepatic metastasis. However, the potential biological function of RBBP4 in colon cancer is still unknown.
To explore the potential mechanisms underlying colon cancer development and discover biomarkers for the treatment of colon cancer.
To investigate the underlying mechanisms of RBBP4 in colon cancer malignant development.
Real-time polymerase chain reaction and western blot analysis were used to detect the expression of RBBP4 in colon cancer cell lines. The cell proliferation and viability of SW620 and HCT116 cells with RBBP4 knockdown was detected by Cell Counting Kit-8 and 5-ethynyl-2’-deoxyuridine staining. The transwell assay was used to detect the invasion and migration capabilities of colon cancer cells with RBBP4 knockdown. Flow cytometry apoptosis assay was used to detect the apoptosis of colon cancer cells with RBBP4 knockdown. Western blot analysis was used to detect the expression of epithelial-mesenchymal transition and apoptosis related markers in colon cancer with RBBP4 knockdown. The nuclear translocation of β-catenin was examined by western blot analysis in colon cancer cells with RBBP4 knockdown. The TOPFlash luciferase assay was used to detect effect of RBBP4 on Wnt/β-catenin activation. The rescue experiments were performed in colon cancer cells treated with Wnt/β-catenin activator LiCl and RBBP4 knockdown.
We found that RBBP4 was highly expressed in colon cancer cell lines. The 5-ethynyl-2’-deoxyuridine assay showed that knockdown of RBBP4 significantly inhibited cell proliferation. RBBP4 inhibition reduced cell invasion and migration via regulating proteins related to epithelial-mesenchymal transition. Knockdown of RBBP4 significantly inhibited surviving-mediated apoptosis. Mechanistically, the TOPFlash assay showed that RBBP4 knockdown increased activity of the Wnt/β-catenin pathway. RBBP4 knockdown suppressed nuclear translocation of β-catenin. With a Wnt/β-catenin activator, rescue experiments suggested that the role of RBBP4 in colon cancer progression was dependent on the Wnt/β-catenin pathway.
This study demonstrated that RBBP4 promoted colon cancer development via increasing activity of the Wnt/β-catenin pathway. RBBP4 may serve as a novel therapeutic target in colon cancer.
In the future, additional research will be carried out to further explore the important role of RBBP4 and whether RBBP4 knockdown can be employed to enhance the sensitivity of chemotherapy of colon cancer and to develop novel anticancer treatments.
This study was supported by NHC Key Laboratory of Combined Multi-Organ Transplantation. The authors thank the technicians Rong Su and Hui Chen for guiding the experiment.
| 1. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11573] [Cited by in RCA: 13298] [Article Influence: 1662.3] [Reference Citation Analysis (4)] |
| 2. | Ferlay J, Colombet M, Soerjomataram I, Dyba T, Randi G, Bettio M, Gavin A, Visser O, Bray F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer. 2018;103:356-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1625] [Cited by in RCA: 1730] [Article Influence: 216.3] [Reference Citation Analysis (0)] |
| 3. | Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL, Siegel RL. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69:363-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2417] [Cited by in RCA: 3121] [Article Influence: 445.9] [Reference Citation Analysis (0)] |
| 4. | Moghimi-Dehkordi B, Safaee A. An overview of colorectal cancer survival rates and prognosis in Asia. World J Gastrointest Oncol. 2012;4:71-75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 102] [Cited by in RCA: 131] [Article Influence: 9.4] [Reference Citation Analysis (9)] |
| 5. | Tyler JK, Bulger M, Kamakaka RT, Kobayashi R, Kadonaga JT. The p55 subunit of Drosophila chromatin assembly factor 1 is homologous to a histone deacetylase-associated protein. Mol Cell Biol. 1996;16:6149-6159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 108] [Article Influence: 3.6] [Reference Citation Analysis (2)] |
| 6. | Qian YW, Wang YC, Hollingsworth RE, Jones D, Ling N, Lee EY. A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature. 1993;364:648-652. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 220] [Article Influence: 6.7] [Reference Citation Analysis (2)] |
| 7. | Qian YW, Lee EY. Dual retinoblastoma-binding proteins with properties related to a negative regulator of ras in yeast. J Biol Chem. 1995;270:25507-25513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 112] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 8. | Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924-1935. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 814] [Cited by in RCA: 863] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 9. | Kouznetsova VL, Tchekanov A, Li X, Yan X, Tsigelny IF. Polycomb repressive 2 complex-Molecular mechanisms of function. Protein Sci. 2019;28:1387-1399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 65] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 10. | Vermaak D, Wade PA, Jones PL, Shi YB, Wolffe AP. Functional analysis of the SIN3-histone deacetylase RPD3-RbAp48-histone H4 connection in the Xenopus oocyte. Mol Cell Biol. 1999;19:5847-5860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 58] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 11. | Wolffe AP, Urnov FD, Guschin D. Co-repressor complexes and remodelling chromatin for repression. Biochem Soc Trans. 2000;28:379-386. [PubMed] |
| 12. | Li L, Tang J, Zhang B, Yang W, LiuGao M, Wang R, Tan Y, Fan J, Chang Y, Fu J, Jiang F, Chen C, Yang Y, Gu J, Wu D, Guo L, Cao D, Li H, Cao G, Wu M, Zhang MQ, Chen L, Wang H. Epigenetic modification of MiR-429 promotes liver tumour-initiating cell properties by targeting Rb binding protein 4. Gut. 2015;64:156-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 111] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 13. | Guo Q, Cheng K, Wang X, Li X, Yu Y, Hua Y, Yang Z. Expression of HDAC1 and RBBP4 correlate with clinicopathologic characteristics and prognosis in breast cancer. Int J Clin Exp Pathol. 2020;13:563-572. [PubMed] |
| 14. | Cui F, Zan X, Li Y, Sun W, Yang Y, Ping L. Grifola frondosa Glycoprotein GFG-3a Arrests S phase, Alters Proteome, and Induces Apoptosis in Human Gastric Cancer Cells. Nutr Cancer. 2016;68:267-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 15. | Ding L, Zhao Y, Dang S, Wang Y, Li X, Yu X, Li Z, Wei J, Liu M, Li G. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol Cancer. 2019;18:45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 196] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 16. | Li YD, Lv Z, Xie HY, Zheng SS. Retinoblastoma binding protein 4 up-regulation is correlated with hepatic metastasis and poor prognosis in colon cancer patients. Hepatobiliary Pancreat Dis Int. 2019;18:446-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 17. | Findlay VJ, Wang C, Watson DK, Camp ER. Epithelial-to-mesenchymal transition and the cancer stem cell phenotype: insights from cancer biology with therapeutic implications for colorectal cancer. Cancer Gene Ther. 2014;21:181-187. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 94] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 18. | Siddharth S, Das S, Nayak A, Kundu CN. SURVIVIN as a marker for quiescent-breast cancer stem cells-An intermediate, adherent, pre-requisite phase of breast cancer metastasis. Clin Exp Metastasis. 2016;33:661-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 19. | Bastos LG, de Marcondes PG, de-Freitas-Junior JC, Leve F, Mencalha AL, de Souza WF, de Araujo WM, Tanaka MN, Abdelhay ES, Morgado-Díaz JA. Progeny from irradiated colorectal cancer cells acquire an EMT-like phenotype and activate Wnt/β-catenin pathway. J Cell Biochem. 2014;115:2175-2187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 49] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 20. | Nicolas E, Morales V, Magnaghi-Jaulin L, Harel-Bellan A, Richard-Foy H, Trouche D. RbAp48 belongs to the histone deacetylase complex that associates with the retinoblastoma protein. J Biol Chem. 2000;275:9797-9804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 103] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 21. | Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 444] [Cited by in RCA: 478] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 22. | Qi L, Sun B, Liu Z, Cheng R, Li Y, Zhao X. Wnt3a expression is associated with epithelial-mesenchymal transition and promotes colon cancer progression. J Exp Clin Cancer Res. 2014;33:107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 103] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 23. | Schmitges FW, Prusty AB, Faty M, Stützer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, Bunker RD, Wirth U, Bouwmeester T, Bauer A, Ly-Hartig N, Zhao K, Chan H, Gu J, Gut H, Fischle W, Müller J, Thomä NH. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell. 2011;42:330-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 525] [Cited by in RCA: 576] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 24. | Shang WH, Hori T, Westhorpe FG, Godek KM, Toyoda A, Misu S, Monma N, Ikeo K, Carroll CW, Takami Y, Fujiyama A, Kimura H, Straight AF, Fukagawa T. Acetylation of histone H4 lysine 5 and 12 is required for CENP-A deposition into centromeres. Nat Commun. 2016;7:13465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 62] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 25. | Ng C, Aichinger M, Nguyen T, Au C, Najar T, Wu L, Mesa KR, Liao W, Quivy JP, Hubert B, Almouzni G, Zuber J, Littman DR. The histone chaperone CAF-1 cooperates with the DNA methyltransferases to maintain Cd4 silencing in cytotoxic T cells. Genes Dev. 2019;33:669-683. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 26. | Fukuoka J, Fujii T, Shih JH, Dracheva T, Meerzaman D, Player A, Hong K, Settnek S, Gupta A, Buetow K, Hewitt S, Travis WD, Jen J. Chromatin remodeling factors and BRM/BRG1 expression as prognostic indicators in non-small cell lung cancer. Clin Cancer Res. 2004;10:4314-4324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 170] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 27. | Song H, Xia SL, Liao C, Li YL, Wang YF, Li TP, Zhao MJ. Genes encoding Pir51, Beclin 1, RbAp48 and aldolase b are up or down-regulated in human primary hepatocellular carcinoma. World J Gastroenterol. 2004;10:509-513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 54] [Cited by in RCA: 60] [Article Influence: 2.7] [Reference Citation Analysis (1)] |
| 28. | Pacifico F, Paolillo M, Chiappetta G, Crescenzi E, Arena S, Scaloni A, Monaco M, Vascotto C, Tell G, Formisano S, Leonardi A. RbAp48 is a target of nuclear factor-kappaB activity in thyroid cancer. J Clin Endocrinol Metab. 2007;92:1458-1466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 29. | Casas S, Ollila J, Aventín A, Vihinen M, Sierra J, Knuutila S. Changes in apoptosis-related pathways in acute myelocytic leukemia. Cancer Genet Cytogenet. 2003;146:89-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 30. | Carreira-Barbosa F, Nunes SC. Wnt Signaling: Paths for Cancer Progression. Adv Exp Med Biol. 2020;1219:189-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 31. | Bienz M. The subcellular destinations of APC proteins. Nat Rev Mol Cell Biol. 2002;3:328-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 190] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 32. | Li H, Pamukcu R, Thompson WJ. beta-Catenin signaling: therapeutic strategies in oncology. Cancer Biol Ther. 2002;1:621-625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 56] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 33. | Bullions LC, Levine AJ. The role of beta-catenin in cell adhesion, signal transduction, and cancer. Curr Opin Oncol. 1998;10:81-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 137] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 34. | Willert K, Nusse R. Beta-catenin: a key mediator of Wnt signaling. Curr Opin Genet Dev. 1998;8:95-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 566] [Cited by in RCA: 565] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 35. | Zhong Z, Virshup DM. Wnt Signaling and Drug Resistance in Cancer. Mol Pharmacol. 2020;97:72-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 176] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 36. | Schneikert J, Behrens J. The canonical Wnt signalling pathway and its APC partner in colon cancer development. Gut. 2007;56:417-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 219] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 37. | Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4088] [Cited by in RCA: 4525] [Article Influence: 226.3] [Reference Citation Analysis (1)] |
| 38. | Sakoguchi-Okada N, Takahashi-Yanaga F, Fukada K, Shiraishi F, Taba Y, Miwa Y, Morimoto S, Iida M, Sasaguri T. Celecoxib inhibits the expression of survivin via the suppression of promoter activity in human colon cancer cells. Biochem Pharmacol. 2007;73:1318-1329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 67] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 39. | Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917-921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2342] [Cited by in RCA: 2398] [Article Influence: 82.7] [Reference Citation Analysis (1)] |
| 40. | August DA, Ottow RT, Sugarbaker PH. Clinical perspective of human colorectal cancer metastasis. Cancer Metastasis Rev. 1984;3:303-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 255] [Article Influence: 6.1] [Reference Citation Analysis (17)] |
| 41. | Cheng R, Sun B, Liu Z, Zhao X, Qi L, Li Y, Gu Q. Wnt5a suppresses colon cancer by inhibiting cell proliferation and epithelial-mesenchymal transition. J Cell Physiol. 2014;229:1908-1917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 71] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 42. | Zhu QC, Gao RY, Wu W, Qin HL. Epithelial-mesenchymal transition and its role in the pathogenesis of colorectal cancer. Asian Pac J Cancer Prev. 2013;14:2689-2698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Yu H S-Editor: Yan JP L-Editor: Filipodia P-Editor: Ma YJ