Published online Jul 14, 2020. doi: 10.3748/wjg.v26.i26.3720
Peer-review started: February 27, 2020
First decision: May 21, 2020
Revised: June 2, 2020
Accepted: June 18, 2020
Article in press: June 18, 2020
Published online: July 14, 2020
Processing time: 138 Days and 2.6 Hours
Hepatocellular carcinoma (HCC) is characterized by high heterogeneity in both intratumoral and interpatient manners. While interpatient heterogeneity is related to personalized therapy, intratumoral heterogeneity (ITH) largely influences the efficacy of therapies in individuals. ITH contributes to tumor growth, metastasis, recurrence, and drug resistance and consequently limits the prognosis of patients with HCC. There is an urgent need to understand the causes, characteristics, and consequences of tumor heterogeneity in HCC for the purposes of guiding clinical practice and improving survival. Here, we summarize the studies and technologies that describe ITH in HCC to gain insight into the origin and evolutionary process of heterogeneity. In parallel, evidence is collected to delineate the dynamic relationship between ITH and the tumor ecosystem. We suggest that conducting comprehensive studies of ITH using single-cell approaches in temporal and spatial dimensions, combined with population-based clinical trials, will help to clarify the clinical implications of ITH, develop novel intervention strategies, and improve patient prognosis.
Core tip: We summarize the comprehensive studies of intratumoral heterogeneity (ITH) of hepatocellular carcinoma, including the different aspects, various dimensions, and clinical significance of ITH.
- Citation: Zhang Q, Lou Y, Bai XL, Liang TB. Intratumoral heterogeneity of hepatocellular carcinoma: From single-cell to population-based studies. World J Gastroenterol 2020; 26(26): 3720-3736
- URL: https://www.wjgnet.com/1007-9327/full/v26/i26/3720.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i26.3720
Liver cancer is one of the leading causes of cancer-related death worldwide, and approximately 90% of these deaths are attributed to hepatocellular carcinoma (HCC)[1]. With several non-surgical therapeutic options (e.g., transcatheter arterial chemoembolization, radiofrequency ablation, stereotactic body radiation therapy, and systemic therapy) available, liver resection and liver transplantation remain the mainstay of cure of HCC. However, less than 30% of patients are eligible for surgery, and a considerable number of them will succumb to rapid tumor recurrence or metastasis[2]. Furthermore, HCC can demonstrate primary or secondary resistance to other interventions, including chemotherapy, targeted therapy, and immune checkpoint blockade. However, despite many cellular and molecular mechanisms revealed, the refractory nature of HCC to treatments is still far from understood. Thus, new perspectives are warranted to decode this intractable issue.
Intratumoral heterogeneity (ITH), first described in the 1830s by German physiologists Johannes Muller and Rudolf Virchow[3], is now found to be ubiquitous in all types of tumors and accounts for nearly all aspects of tumor progression[4]. Currently, people have realized that heterogeneity is not only a distinct morphological profile but also implicates genetics, epigenetics, transcriptomics, proteomics, posttranslational modification (PTM)-omics, metabolomics, and diverse tumor microenvironments (TMEs). Systematic ITH causes a wide functional divergence, providing a platform to perform natural selection in the particular TME and promoting the malignant phenotypes of HCC. Fortunately, the rapid development of next-generation sequencing (NGS) and mass spectrometry techniques has made elegant analysis of ITH a reality in recent years. Several advances in HCC have been achieved to help us gain in-depth insight into the evolutionary mode of tumorigenesis and development, the regulatory pattern of the signaling network, and the interaction between tumor cells and their microenvironment. Nevertheless, how to integrate the discoveries and implications of ITH into clinical practice is rarely discussed.
In this review, we provide an overview of ITH in HCC from microscopic to macrocosmic approaches to explore the clinical implications of ITH. Based on a systemic search of relevant studies from single-cell to population-based designs, we summarize the common patterns of ITH in HCC and propose a practicable way to guide the clinical management of this deadly disease.
Although heterogeneity at the DNA, RNA, and protein levels is frequently investigated, cells are actually the fundamental element of tumor heterogeneity that may impact tumor progression. Heterogeneity itself does not have a malignancy-specific implication since all healthy, transformed, and cancerous cells sustain heterogeneity to some extent. In the liver, hepatocytes are organized from the central vein to the portal node, showing a strong regularity of the gene expression profile[5,6]. For example, the expression of Alb, Glul, Cyp2e1, Cyp2f2, and other hepatocyte markers in different locations can be extremely different. Such cellular heterogeneity presented by highly regulated gene expression and exquisite cell spatial distribution is required for the normal physiological function of the liver. When hepatic cells transform, precancerous nodules show heterogeneous genetic and epigenetic patterns as well[7].
For cancer, pathologists first used microscopes to identify the diversity among tumor cells. Due to the limitation of observational technologies, it took more than a century to describe and explore cellular heterogeneity through pathological sections and immunohistological staining[3], by which ITH cannot be quantitatively evaluated. Thanks to the emergence of single-cell analytic approaches (e.g., single-cell sequencing, single-cell immunoblotting, single-cell mass spectrometry, and single-cell multiomic techniques) in the last decade, it is currently possible to establish a clear panorama of cellular heterogeneity[8-11]. Hou et al[12] developed scTrio-seq, a single-cell triple omics sequencing technique, and provided the first piece of evidence regarding the genetic heterogeneity in HCC at a single-cell level. Two subpopulations with distinct biological functions were observed by unsupervised hierarchical clustering based on genomic copy-number variations (CNVs), although the cells in the same subset were also not identical[12]. Since then, cellular heterogeneity has been repeatedly corroborated in HCC from genomic, transcriptomic, and other dimensions[13-16] (Table 1).
| No | Time | Patients (n) | Cells (n) | Methods | Findings | Ref. |
| Studies on cancer cells | ||||||
| 1 | 2016 | 1 | 25 T | scTrio-seq (DNA, DNA methylation, mRNA) | Two subpopulations were identified based on CNVs, DNA methylome, or mRNA. | [12] |
| 2 | 2018 | 3 | 96 T + 15 N | Single-cell WGS | HCCs can be of monoclonal or polyclonal origins. Models of late dissemination and early seeding have a role in HCC progression. | [13] |
| 3 | 2018 | 1 | 118 CSCs + 860 unsorted | mRNA (SMART-Seq +10X) | Different CSC subsets contain distinct molecular signatures, and are associated with prognosis. | [14] |
| 4 | 2019 | 1 | 139 T | mRNA (C1) | EPCAM+ cells had upregulated expression of multiple oncogenes and sustain CSC property. | [16] |
| Studies on immune cells | ||||||
| 5 | 2017 | 6 | 5063 T cells | mRNA (Smart-Seq2) | 11 T cell subsets were identified based on their molecular and functional properties. | [80] |
| 6 | 2019 | 16 | CD45+ cells (66187 + 11134) | mRNA (10X + Smart-Seq2) | 40 immune cell subsets were identified, as well as their distinct roles in HCC development. | [81] |
| Studies on both | ||||||
| 7 | 2019 | set1: 13 set2: 6 | set1: 5115set2: 4831 | mRNA (10X) | Tumors with higher transcriptomic diversity were associated with higher VEGFA expression, lower cytolytic activities, and worse outcome. | [17] |
| 8 | 2019 | 4 | 19625 | mRNA (microwell-seq) | The extent of heterogeneity in both tumor and immune cells varies among patients. | [72] |
| 9 | 2020 | 2 | 38553 | mRNA (10X) | Cancer cells from the same tumor were divided into different Hoshida subclasses and had different effects on immune infiltration. | [64] |
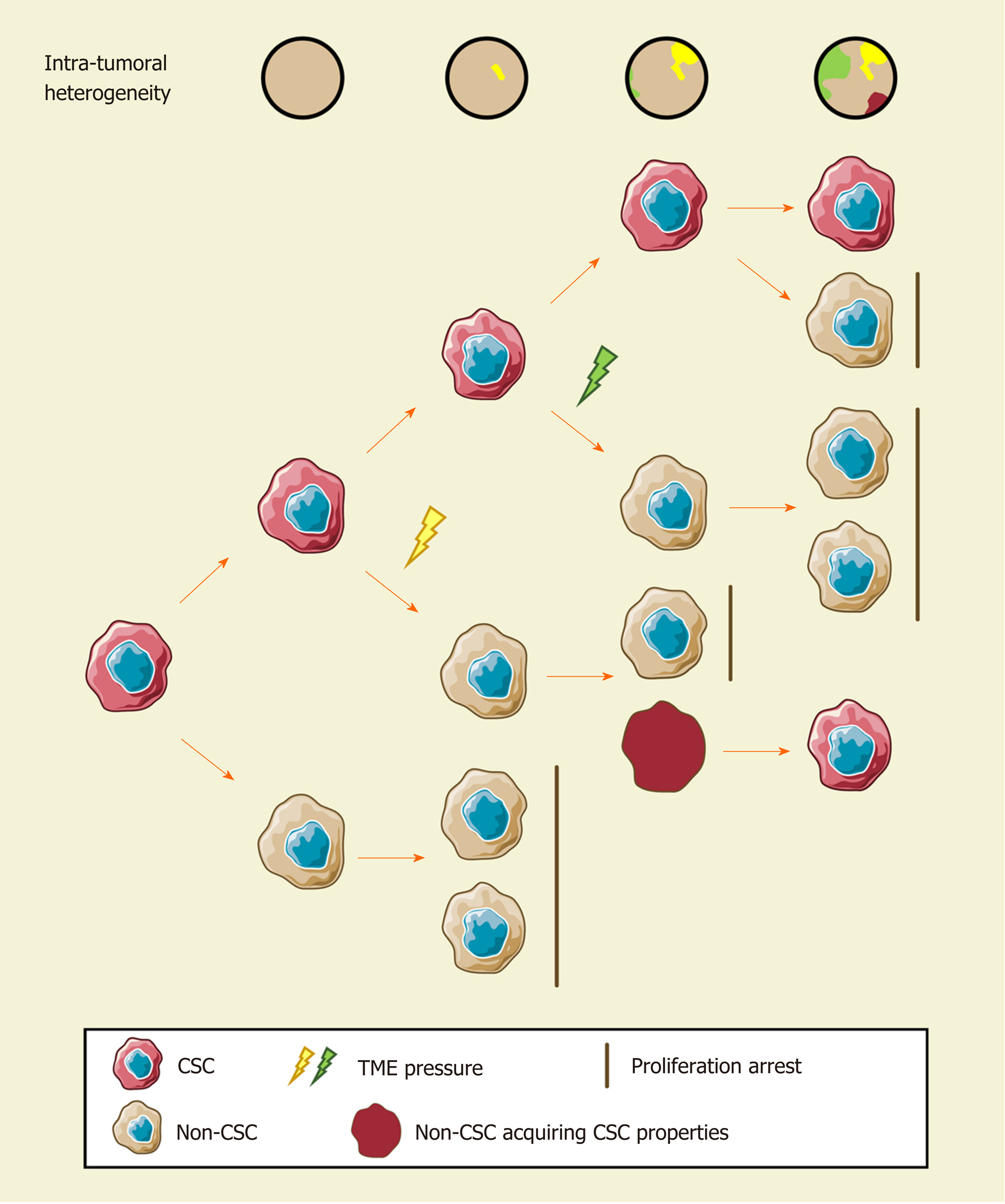
Since cellular heterogeneity varies greatly among individuals[14,17], it is of great value to find the key subpopulations of cancer cells with greater influence on ITH for further studies. Hence, a subset of cancer cells with stemness features, also known as cancer stem cells (CSCs), has attracted much attention[18-21]. Hepatic CSCs can be identified by various cell surface markers. For instance, EpCAM[22-25], CD90 (THY-1)[25-27], CD24[14,25,28], CD133 (Prominin-1)[14,25,28,29], CD13 (ANPEP)[29,30], CD44[16,25,30], CD47[30,31], and others have been identified in HCC in independent studies using single-cell sequencing[14,16]. More importantly, single-cell transcriptomic profiling indicated that stemness gene expression seemed to be consistent with the cancer diversity score based on principal component analysis and was significantly related to clinical outcomes[16,17,26]. These findings coincide with the “cancer stem cell model”, which is favored to explain the origin of ITH recently[32,33] (Figure 1). Predominantly, this model demonstrates that only CSCs possess the capability of self-renewal, differentiation, and maintenance of tumor growth[34-36]. Similar to the normal tissue hierarchy, the hierarchical organization in cancer results from the punctuated proliferation of CSCs and consequently shapes.
Accumulating evidence likely reflects the fact that, even as an extremely tiny subset, CSCs can still be divided into various subpopulations with distinct cell membrane markers, molecular makeups, and functional phenotypes. Recently, elegant studies by Zheng et al[14] and Ma et al[17] provided direct evidence of hepatic CSC heterogeneity and its clinical implications at a single-cell resolution. CSC sorting by CD24, CD133, and/or EpCAM proliferated divergently in both normoxia and hypoxia and showed a distinctive spatial distribution within HCC[14]. These findings demonstrated the uneven distribution of functionally diverse CSC subpopulations across different regions, and the current understanding of CSCs may only reflect the features of specific subgroups[26,37-42]. Moreover, CSC identity is not immutable[14,19]. Under the influence of genetic stochastic mutation and recombination, epigenetic modifications, and field effects in the TME, reversible transformation can occur between CSCs and non-CSCs, which establishes new hierarchical CSC clones to enrich ITH tremendously[32,43,44]. This ability to reconstruct heterogeneity cautions against therapies targeting CSC-related membrane epitopes and increases the difficulty of ITH management.
However, CSCs are not an undruggable subset. Growing evidence suggests that plenty of environmental factors, such as hypoxia, inflammation, and DNA damage, play crucial roles in CSC phenotypes mediating in various cancers[45,46]. As an illustration, hypoxia has received considerable attention in recent years[47]. Similar to oxygen gradients between arteries and veins in liver lobules, abnormal vascular perfusion in HCC induces differential activities of the HIF signaling pathway in an oxygen-dependent manner, and consequently stemness. Hence, it is a promising therapeutic strategy for treatment of CSCs and heterogeneity.
Further identification of CSC subpopulations and the regulatory mechanism of the CSC transformation process is of paramount importance to clarify the roles of CSCs in the development and management of ITH. An advanced understanding of CSCs may create new opportunities for overcoming cellular heterogeneity.
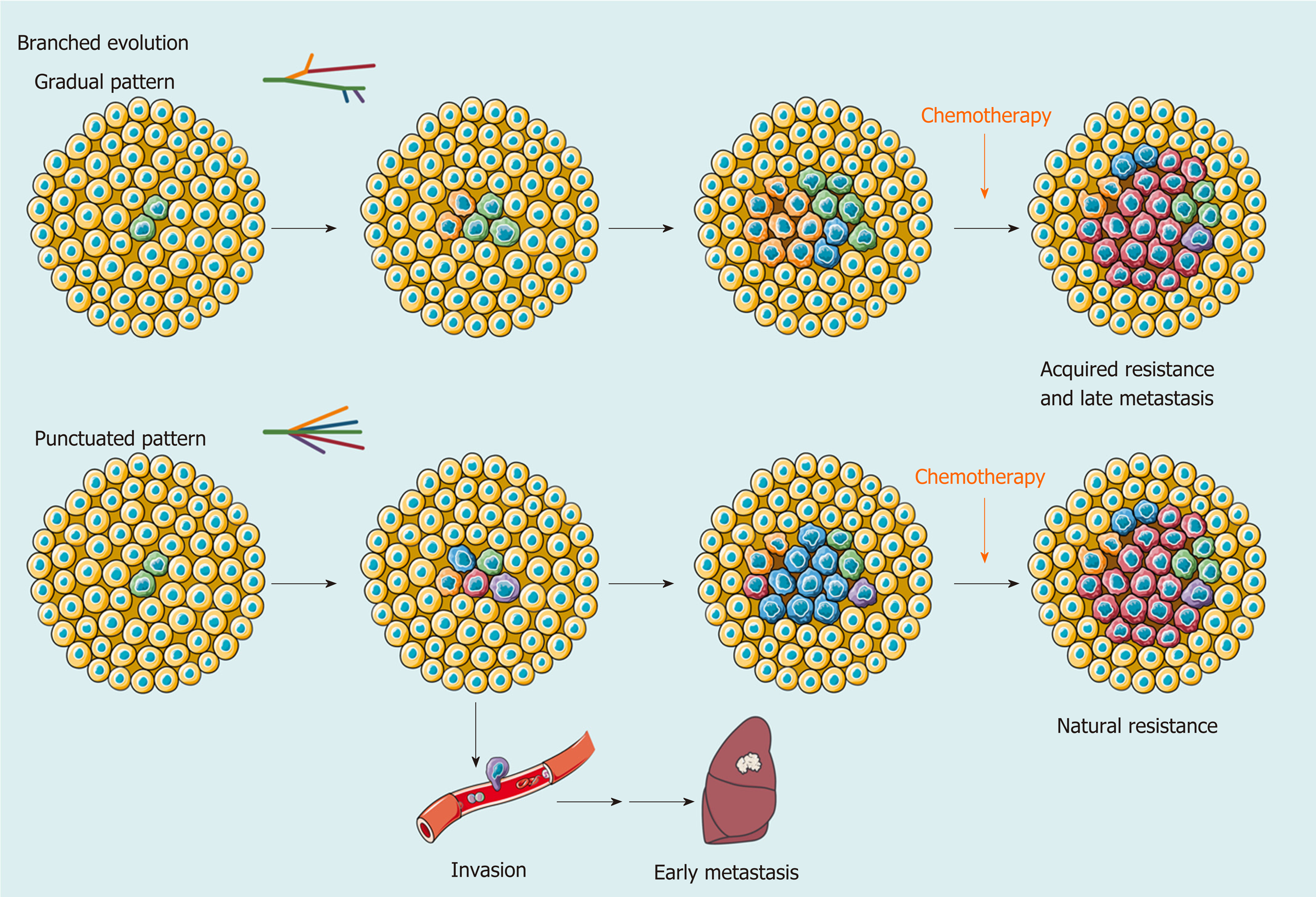
In addition to single-cell technologies, multiregional sampling, the method to sample biopsies from multiple regions within a single tumor, is the most common investigational strategy to determine the extent of ITH[48-50]. This strategy is based on the “clonal evolution model”, which was first proposed by Nowell[51] in 1976, and became the most accepted hypothesis to depict the process of ITH in addition to the “CSC model”. In this theory, carcinogenesis is considered to be a stochastic process. Numerous random genetic alterations created by genomic instability and continuous stimulation of carcinogens results in the emergence of subclones. The subclones with a survival advantage in a spatiotemporally specific microenvironment will establish dominance, while the non-adaptive subclones will be eliminated. Upon the selective pressure of the TME, the complex architecture of ITH is preserved and dynamically reorganized[52,53] (Figure 2).
Using a multiregional sampling strategy, spatially neighboring clones that are supposed to be genetically more similar can be pooled and compared with bulk samples from other regions. In early studies, ITH in morphology and genomics was poorly described[54-56]. Subsequently, bulk whole-genome sequencing (WGS)[57-59], whole-exome sequencing (WES)[59-62], targeted sequencing[63], single-cell sequencing[13,64], and DNA methylation profiling[61,65] were introduced into heterogeneity studies to quantify genetic alterations and provide a more comprehensive blueprint of hepatic ITH. Public, shared, and private genetic changes were defined to construct an evolutionary tree of tumor subclones[66]. As expected, the variable extent of ITH and the branch evolutionary model are well recapitulated[13,57-59], and subclonal populations that are spatially closer tend to be genetically more similar in both planar and solid models[59,65], which suggests the existence of a common ancestral clone and a continuous clonal expansion pattern with the accumulation of mutations. In the meantime, the time when potential driver mutations formed can be estimated. Similar to the mutation profile of HCC, the most common alterations, such as key driver mutations (e.g., TP53, CTNNB1, and TERT) and CNVs (e.g., amplification in 1q and deletions in 4q and 16q), are mostly identified in the trunks[57-59,61,63], revealing that they may represent an early event in HCC progression. However, an uneven distribution of driver mutations in TP53 and CTNNB1 across different regions was also observed in a minority of patients[58,62,63,65,67,68]. These findings indicated that this heterogeneous disease can be either monoclonal or polyclonal in origin. Additional mutations in AXIN1, RB1, KIT1, FAT4, and other HCC-related genes involved in clonal evolution could dramatically change marker expression (e.g., CK7, β-catenin, and GS) and functional phenotypes[60-62,65,67,68]. It is the functional diversity that promotes tumor development and, more importantly, worsens clinical outcome[60]. Unlike genetic alterations, ITH of genomic methylation appears prior to tumor initiation and forms clonal expansions in nonmalignant tissues[7,63]. In regard to cancer, regulation of DNA-methylated heterogeneity acts in a conservative manner, largely driven by the tumor itself and significantly related to tumor progression[13].
Clearly, clonal diversity changes dynamically with tumor evolution, but ITH is not necessarily correlated with positive changes. In support of this hypothesis, the findings of several studies have suggested that various genetic alterations in the single key gene across regions (e.g., loss-of-function and stop-gain mutations in CTNNB1) embody convergent evolution of HCC under microenvironmental stress[57,65]. However, one size does not fit all in regard to clonal evolution. A study collected 286 samples on a plane of a single HCC and drew the most elaborate clonal map of HCC to date[69]. More than 100 million mutations are defined, illustrating astonishing genetic heterogeneity and a non-Darwinian mode during HCC development. In addition, another work using single-cell genome sequencing likely demonstrated the punctuated pattern[13], which is characterized by the early occurrence of numerous genetic alterations[70].
Another focus in ITH studies based on multiregional sampling is tracking the origins of different nodules to clarify the significance of ITH in invasion and metastasis. Comparing the primary lesions with intrahepatic metastases, tumor thrombi, and satellite nodules found that migration is likely to occur either at the early or late stage of HCC development, while satellite nodules appear at the late stage[57,59,65]. Multicentral lesions, regarded as a unique subtype of HCC, share few common driver mutations, which implies a parallel evolutionary pattern[57,62,63]. These findings provide a theoretical basis for accurate differentiation between intrahepatic metastases and multicentric lesions in clinical practice[58,63].
Generally, a multiregional sampling strategy improves the ability to explore the symbiotic relationship between clonal heterogeneity and tumor evolution and enriches the value of ITH investigations in precision medicine. However, it can be impractical to obtain multiregional samples from an advanced tumor in clinical practice due to the high risk of bleeding after biopsy. A clinically friendly approach for ITH investigation is urgently needed.
Although studies of genetic and epigenetic heterogeneity reflect the life history of tumors, functional diversity is the factor affecting clinical outcome directly. The concept of “functional heterogeneity” was proposed in 2018 to integrate information on genomics, nongenomics, stemness, and TME heterogeneity[71]. Here, we adopt this concept to summarize the functional and microenvironmental characteristics of ITH and discuss its significance in tumor progression and treatment.
Transcriptomic[58,62,72] and proteomic profilings[72,73] take the lead in describing functional heterogeneity. Unsurprisingly, ITH at the RNA and protein levels is apparent. However, the gene expression profile does not seem to strictly follow the law of genetic alterations[58,62,72,73] and has poor relevance to tumor geometry[73]. Due to the distinct gene expression profiles across regions, neither the origin of metastatic clones[58] nor the potential targets for clinical intervention[72,73] can be distinguished; however, the pivotal role of TME in functional heterogeneity is still reflected[62,72,73].
Owing to the breakthrough of immunotherapy, local immunity in HCC has attracted growing attention. Multiplex immunohistological staining was first used to decipher tumor infiltrating immune cells (TICs) in solid tumors[74]. Apparently, ITH of the microenvironment is neatly illustrated by the density, distribution, and composition of TICs and is likely correlated with tumor differentiation[74], which reveals the coordination of the tumoral functional phenotype and immune infiltration. To deeply explore the interaction between TICs and ITH, various algorithms have been developed to quantify the composition of immune infiltration using sequencing data[75-78] and expand the effectiveness of immune function assessment enormously[62,79]. Coincidentally, the latest two studies integrated DNA-seq, RNA-seq, T-cell receptor (TCR)-seq, neoantigen analysis, and other methods to uncover the role of functional heterogeneity in immune evasion[64,79]. Cancer cells across different regions present different immunogenicity and escape immune surveillance in distinct ways, such as a decrease in immune cell infiltration, loss of heterozygosity of human leukocyte antigen, low TCR clonality, and immunoediting[64]. This finding suggested that the coevolution of tumor and local immunity is quite ubiquitous in HCC and could partially explain the low response rate of immune checkpoint blockade monotherapy in HCC.
Similar to previous studies on clonal evolution[58,63], ITH in intrahepatic metastases and multicentral lesions was compared at an immunological resolution to evaluate the potential effect of immunotherapy[79]. Uneven composition of immune cell infiltration was observed, such as abundant TAM infiltration in intrahepatic metastases and rich CD8+ T-cell infiltration with a higher level of inhibitory immune checkpoint expression in multicentral lesions, which suggested that patients with multicentric HCC could be more likely to benefit from immune checkpoint blockade[79].
In addition, single-cell sequencing is also applied to TME research for a better understanding of immunological heterogeneity. Systematic works completed by Zemin Zhang’s group provided a dynamic atlas of the immune landscape of HCC[80,81]. When integrating tumoral single-cell transcriptome data, the HCC cells of different Hoshida subclasses in a single tumor induced regional variance in the antitumoral immune response[64]. More importantly, the extent of ITH was responsible for hypoxia adaptation, VEGF expression, and drug resistance[17], which may be considered in the choice of transarterial (chemo)embolization, sorafenib, lenvatinib, or other antivascular therapies.
Our group also made some efforts to provide a comprehensive landscape of ITH of HCC in multiple dimensions[72]. The research integrating genomic, transcriptomic, proteomic, metabolomic, and immunomic (mass cytometry) data together demonstrated a heterogeneous interactive network of HCC cells, metabolites, and TICs and proposed a novel clinically friendly immunophenotypic classification to improve the clinical management of patients with HCC. We found that tumor cell heterogeneity and microenvironmental heterogeneity, which included diverse TICs, were closely related to each other and showed coevolution. Thus, the evaluation of the functional heterogeneity of HCC requires comprehensive investigation and currently lacks accurate and practical biomarkers.
The significance of heterogeneity has been demonstrated in accumulating studies, but large-scale population-based studies are still needed to verify the effectiveness of ITH investigation in clinical practice (Table 2). A well-known prospective cohort study on non–small cell lung cancer (NSCLC) included 100 patients with NSCLC and validated the feasibility of predicting clinical outcome and recurrence with ITH[82]. ITH may also play a prognostic role in HCC since genome instability is a common feature of cancer[59]. A clinical trial aiming to investigate the clinical trajectories of HCC during its progression (NCT03267641) is critical to understand the dynamic change and clinical relevance of ITH. However, it is still ongoing with limited information to disclose. Current limited evidence reveals that heterogeneity evaluation of recurrent and primary tumors may serve as a predictor of clinical outcome[65,83].
| No | Time | Patients (n) | Samples(n) | Methods | Findings | Ref. |
| Studies on cancer cells | ||||||
| 1 | 2001 | 11 | 29 T + 11 N | IHC and TP53/CTNNB1 sequencing | Heterogeneity existed in small HCC, accompanied by increased proliferative activity. | [68] |
| 2 | 2015 | 23 | 120 T | IHC and TP53/CTNNB1 sequencing | Intratumor heterogeneity may contribute to treatment failure and drug resistance in many cases of HCC. | [67] |
| 3 | 2015 | 1 | 286 T | WES/Genotyping | 20 unique cell clones were defined by WES. The size distribution of the clones revealed a non-Darwinian evolution model. | [69] |
| 4 | 2016 | 1 | 8 HCC + 3 ICC +N | WES | IM showed similarity to a primary nodule and indicated that it could be an early event in HCC. | [60] |
| 5 | 2016 | 10 | 43 T + 10 N | WGS/WES | Ubiquitous mutations ranged from 8% to over90%. Satellite nodules occurred late in HCC. | [57] |
| 6 | 2017 | 11 | 52 T + 6 N + 11 B | WES + DNA methylation | 29% of putative driver mutations were present in the branches. DNA methylation heterogeneity was largely driven by the cancer self. | [61] |
| 7 | 2017 | 9 | 51 T + 9 N | WGS/WES | Tumor physically closer tend to be genetically more similar. HCC arose from ancestral clones and genetic lineages diverged as tumor grew. | [59] |
| 8 | 2017 | 23 | 49 T | WGS + RNA-seq | Genetic diagnosis is good for an effective choice of therapeutic strategy and IM/MC determination. | [58] |
| 9 | 2017 | 59 | 31 N + 120 T | TDS | Trunk events in early stages (TERT, TP53, and CTNNB1 mutations) were ubiquitous across different regions. | [63] |
| 10 | 2017 | 5 | 32 T + 5 N + cfDNA | WES + TDS | Single region TDS identified 70% of the total mutations, while the cfDNA covered 47.2% of total. | [116] |
| 11 | 2017 | 10 | 55 T | WES | 53.8% of oncogenic alterations varied among subclones. Targetable alterations were identified in subregions from 4 HCCs. | [121] |
| 12 | 2018 | 5 | 15 T + 5 N | Proteomics | Diagnostic outcome may drastically differ if different sectors of tumor are analyzed. | [73] |
| 13 | 2019 | 6 | 34 T + 5 N | WES + RNA-seq | Largest tumor contained higher proportion of ancestral clones. RNA expression pattern was associated with E-S grade | [62] |
| 14 | 2019 | 5 | 36 | WES + RNA-seq + proteomics + metabolomics + CyTOF | Comprehensive intratumoral heterogeneity exists in all dimensions, and the novel immunoclassification of HCC facilitates prognostic prediction and may guide therapy. | [72] |
| 15 | 2019 | 113 | 356 (T + N) | WGS/WES/TDS + DNA methylation | Intratumoral heterogeneity revealed interactions between genomic and epigenomic features associated with tumor progression and recurrence. | [65] |
| 16 | 2019 | 88 | 230 T | IHC and TERT promoter sequencing | Distinct marker expression in different nodules. Limited heterogeneity in metastasis compared to primary sites. | [83] |
| Studies on immune cells | ||||||
| 17 | 2018 | 124 | 919 T | Multiplex IHC | Varying degrees of intratumor heterogeneity of the immune microenvironment were observed. | [74] |
| 18 | 2019 | 13 | 79 T | IHC + RNA-seq | A single-region sample might be reliable for the evaluation of tumor immune infiltration in approximately 60%-70% of patients with HCC. | [117] |
| 19 | 2019 | 15 | 47 T | WES + RNA-seq + TCR-seq + IHC + immunopeptidomes | Genetic structure, neoepitope landscape, T cell profile and immunoediting status collectively shape tumor evolution. | [79] |
| 20 | 2020 | 14 | 51 T + 20 N | WES + RNA-seq + TCR-seq + SNP array + immunofluorescence | The different components of the tumor ecosystem interact during cancer evolution, and promote heterogeneity in liver cancer. | [64] |
To clarify the clinical implication of heterogeneity, researchers have developed different methods to quantify ITH with single bulk sequencing data from public databases to predict survival and recurrence. With single-region bulk DNA data, the heterogeneity score (or diversity score) calculated by mutant allele tumor heterogeneity and clonality approaches significantly distinguished patients with a higher ITH signature and poor prognosis[61,64]. It is remarkable that tumor clonality outperforms mutational burden in predicting prognosis in the TCGA-HCC dataset[64], indicating the crucial value of ITH in HCC. Other methods, such as the mITH method for DNA methylation data[61], the ITH gene signature algorithm for RNA data[17,64], and ITH-related models of CSCs and immune evasion[79], shared similar results that a higher level of ITH was associated with limited survival, which is also observed in many other types of cancer[84-86]. Moreover, evidence across a wide range of cancer types illustrated that ITH fostered resistance to various treatments[82,87-89]. In HCC, sorafenib-targeted genetic alterations have been constantly validated as a subclonal change[62], which may explain the low objective response rate to sorafenib in the clinic[90,91]. Furthermore, the response to immunotherapy is also manifested by functional heterogeneity. Tumors with higher ITH tend to secrete more VEGFA to promote immune suppression and limit the clinical efficacy of immune checkpoint blockade[17]. Similar phenomena were also observed in other tumors[92,93].
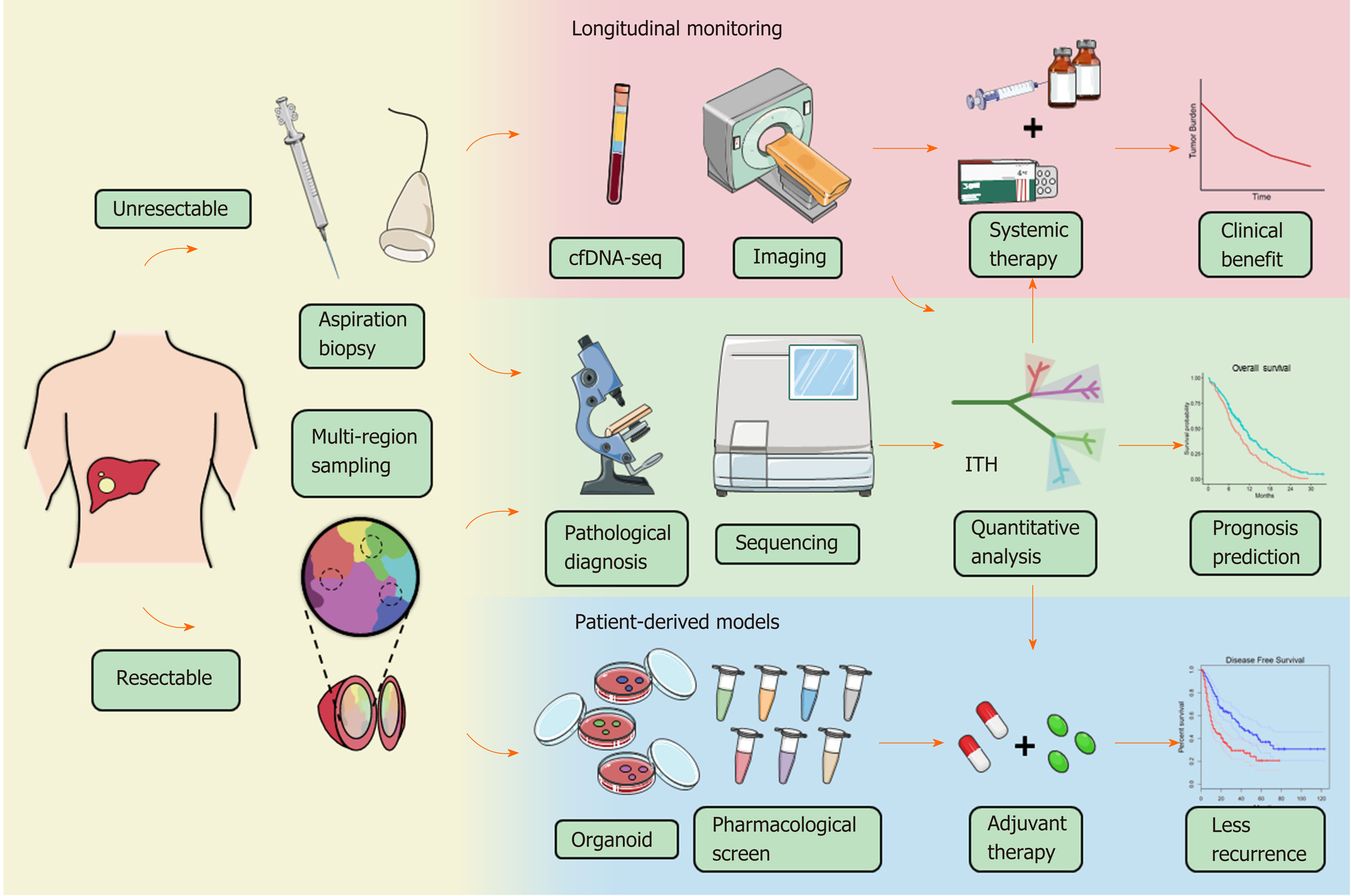
In the future, more stable, practicable, and cost-effective techniques should be introduced to large-scale population-based studies to broaden the scope of the clinical application of ITH. Based on current evidence, liquid biopsy and advanced imaging are promising candidates. Cell-free DNA (cfDNA) is one of the most notable minimally invasive examinations for cancer in recent years and is used to evaluate tumor load, tumor mutational burden, tumor recurrence, drug resistance, and patient survival[94-96]. There is evidence suggesting that cfDNA can reflect ITH in HCC and offer some insight into tumor evolution[97,98]. Using cfDNA sequencing, practical genetic heterogeneity assessment in advanced HCCs with a large size or multiple lesions becomes possible, although many technical and clinical confirmations are warranted before it can be accepted by physicians. cfDNA sequencing also creates the possibility for dynamic observation of ITH either combined with or even substitute sequential sampling[99]. By tracking the dynamic evolution of HCC, people can make more precise interventions for this heterogeneous disease and may obtain better efficacy. In addition, multiparametric magnetic resonance imaging and computed tomography have been proven effective in reflecting the histologic and genomic features and prognosis of HCC[100,101]. In lung cancer, breast cancer, and esophageal cancer, a quantitative ultrasound system and positron emission tomogra-phy/computed tomography are qualified to image ITH[102-105]. Combined with liver-specific contrast agents and radiomic analysis, the digital imaging system may bring ITH assessment into a new chapter[106-109]. However, more clinical trials are expected to test the potential role of these approaches in benefit-gain for HCC patients.
Here, we take ITH into clinical consideration and discuss how to embody the implication of functional heterogeneity in HCC patient management. At present, only a few traditional indicators are well accepted and widely adopted by clinicians in HCC. Tumoral macrovascular invasion (MVI)[110-112] and TNM classification[113,114] systems, for example, are normally used for disease staging, therapy decision making, and prediction of prognosis. However, the demand for precision medicine forces clinicians to biopsy every tumor to obtain a molecular makeup for individual treatment. How to ensure the representativeness of biopsy samples under the influence of heterogeneity becomes an important and unavoidable problem. A study on renal cancer proposed that at least three distinct regions of the tumor should be sampled to detect five key genes with an accuracy of 90% or above[115]. In HCC, researchers carried out WES, targeted deep sequencing, and cfDNA sequencing to identify the effects of sampling strategy on genetic mutation detection[116]. The results indicated that targeted sequencing with single sampling could identify approximately 70% of the total genetic mutations, while cfDNA had merely 47.2%. Another study revealed that 60%-70% of patients with HCC obtained reliable results of immune cell infiltration with a single sample[117]. Altogether, these results suggested that ITH did affect clinical examination at least to some extent, and multiregional sampling might diminish or even eliminate such influence. Considering the clinical risk of the sampling strategy, it is of great value to find more effective methods for patients with advanced HCC.
ITH may also act as a novel measure combined with other indicators to establish a more comprehensive and effective system for several clinical scenarios. ITH quantification can be performed in silico, as mentioned above, using high-throughput sequencing data and radiomic data. Acquiring data from cfDNA or imaging can not only provide information on the entire tumor but is also minimally invasive or noninvasive, cost-effective, and temporally dynamic, which brings infinite possibility to clinical practice. In support of this strategy, the findings of a previous study highlighted the positive correlation between VEGFA expression and ITH, which leads to severe immune exhaustion in more heterogeneous tumors. This phenomenon indeed provides an explanation for the combination therapy of immune checkpoint inhibitors and anti-VEGF molecules to improve therapeutic efficacies (NCT03434379, NCT03006926, NCT03418922)[118,119], suggesting that ITH investigation may screen out HCC patients who benefit from anti-VEGF therapies.
The use of ITH to propose effective intervention strategies is the most attractive aim of the translational study of ITH. Pan-cancer therapeutic strategies targeting CSCs or clonal evolution have been fully described in previous reviews. The former includes inhibition of CSC signaling pathways, CSC ablation using antibody-drug conjugates, and epigenetic therapies[43]; the latter includes clonal events targeting therapies, attenuating or exploiting genome instability, and exploiting evolutionary constraints[66]. To provide a model for drug sensitivity assessment of heterogeneous clones, Nuciforo et al[120] established an organoid system with needle biopsies and succeeded in preserving the heterogeneity of original cancers. In another study, WES of 55 samples from multiple regions of ten HCC patients indicated that 40% of patients could be targeted by existing pharmacologic agents, and the significance was verified in vitro[121]. Our group developed a potential intervention strategy based on a novel immunoclassification according to ITH; however, the validity still needs to be verified in clinical trials[72]. Indeed, there are few treatments against ITH currently published regarding HCC and other types of liver cancer, and more research is thus urgently needed to explore the feasible strategy for HCC treatment and determine the roles of ITH in HCC management (Figure 3).
In general, ITH is a crucial issue that cannot be avoided in HCC progression and management. Although great efforts have been made to obtain a more comprehensive understanding of heterogeneity, the field of hepatic ITH-targeting therapeutics is still in its infancy. Heterogeneous clones and relevant ecosystems are spatially distinct and dynamically change over time, which poses many challenges to clinicians and scientists. Therefore, particular attention should be paid to several aspects in further research. First, integrative analysis of multi-omics data should be performed to establish a systematic relationship between genetic evolution and immune evasion in HCC. Second, longitudinal monitoring of ITH alterations should be conducted for a high-resolution trajectory of clonal evolution. Finally, prospective population-based studies on HCC should be performed to accelerate the translation of ITH assessment into clinical practice. With an improved understanding of ITH, the evolutionary trajectory of HCC is likely to be predictable in the near future. Through the entire process of tracking tumor development, people may rewrite the clinical management of HCC and ultimately find solutions to completely change the outcome of HCC patients.
| 1. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12667] [Cited by in RCA: 15534] [Article Influence: 2589.0] [Reference Citation Analysis (6)] |
| 2. | Belghiti J, Kianmanesh R. Surgical treatment of hepatocellular carcinoma. HPB (Oxford). 2005;7:42-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 121] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 3. | Zellmer VR, Zhang S. Evolving concepts of tumor heterogeneity. Cell Biosci. 2014;4:69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 60] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 4. | Greaves M. Evolutionary determinants of cancer. Cancer Discov. 2015;5:806-820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 316] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 5. | Treyer A, Müsch A. Hepatocyte polarity. Compr Physiol. 2013;3:243-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 202] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 6. | Halpern KB, Shenhav R, Matcovitch-Natan O, Toth B, Lemze D, Golan M, Massasa EE, Baydatch S, Landen S, Moor AE, Brandis A, Giladi A, Avihail AS, David E, Amit I, Itzkovitz S. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature. 2017;542:352-356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 568] [Cited by in RCA: 799] [Article Influence: 88.8] [Reference Citation Analysis (0)] |
| 7. | Hlady RA, Zhou D, Puszyk W, Roberts LR, Liu C, Robertson KD. Initiation of aberrant DNA methylation patterns and heterogeneity in precancerous lesions of human hepatocellular cancer. Epigenetics. 2017;12:215-225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 8. | Navin NE. The first five years of single-cell cancer genomics and beyond. Genome Res. 2015;25:1499-1507. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 263] [Cited by in RCA: 269] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 9. | Packer J, Trapnell C. Single-Cell Multi-omics: An Engine for New Quantitative Models of Gene Regulation. Trends Genet. 2018;34:653-665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 68] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 10. | Liu L, Liu C, Quintero A, Wu L, Yuan Y, Wang M, Cheng M, Leng L, Xu L, Dong G, Li R, Liu Y, Wei X, Xu J, Chen X, Lu H, Chen D, Wang Q, Zhou Q, Lin X, Li G, Liu S, Wang Q, Wang H, Fink JL, Gao Z, Liu X, Hou Y, Zhu S, Yang H, Ye Y, Lin G, Chen F, Herrmann C, Eils R, Shang Z, Xu X. Deconvolution of single-cell multi-omics layers reveals regulatory heterogeneity. Nat Commun. 2019;10:470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 163] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 11. | Kang CC, Lin JM, Xu Z, Kumar S, Herr AE. Single-cell Western blotting after whole-cell imaging to assess cancer chemotherapeutic response. Anal Chem. 2014;86:10429-10436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 79] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 12. | Hou Y, Guo H, Cao C, Li X, Hu B, Zhu P, Wu X, Wen L, Tang F, Huang Y, Peng J. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016;26:304-319. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 370] [Cited by in RCA: 459] [Article Influence: 45.9] [Reference Citation Analysis (0)] |
| 13. | Duan M, Hao J, Cui S, Worthley DL, Zhang S, Wang Z, Shi J, Liu L, Wang X, Ke A, Cao Y, Xi R, Zhang X, Zhou J, Fan J, Li C, Gao Q. Diverse modes of clonal evolution in HBV-related hepatocellular carcinoma revealed by single-cell genome sequencing. Cell Res. 2018;28:359-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 100] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 14. | Zheng H, Pomyen Y, Hernandez MO, Li C, Livak F, Tang W, Dang H, Greten TF, Davis JL, Zhao Y, Mehta M, Levin Y, Shetty J, Tran B, Budhu A, Wang XW. Single-cell analysis reveals cancer stem cell heterogeneity in hepatocellular carcinoma. Hepatology. 2018;68:127-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 274] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 15. | Yan P, Zhou B, Ma Y, Wang A, Hu X, Luo Y, Yuan Y, Wei Y, Pang P, Mao J. Tracking the important role of JUNB in hepatocellular carcinoma by single-cell sequencing analysis. Oncol Lett. 2020;19:1478-1486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 16. | Ho DW, Tsui YM, Sze KM, Chan LK, Cheung TT, Lee E, Sham PC, Tsui SK, Lee TK, Ng IO. Single-cell transcriptomics reveals the landscape of intra-tumoral heterogeneity and stemness-related subpopulations in liver cancer. Cancer Lett. 2019;459:176-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 124] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 17. | Ma L, Hernandez MO, Zhao Y, Mehta M, Tran B, Kelly M, Rae Z, Hernandez JM, Davis JL, Martin SP, Kleiner DE, Hewitt SM, Ylaya K, Wood BJ, Greten TF, Wang XW. Tumor Cell Biodiversity Drives Microenvironmental Reprogramming in Liver Cancer. Cancer Cell. 2019;36:418-430.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 390] [Cited by in RCA: 617] [Article Influence: 88.1] [Reference Citation Analysis (13)] |
| 18. | Brown HK, Tellez-Gabriel M, Heymann D. Cancer stem cells in osteosarcoma. Cancer Lett. 2017;386:189-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 235] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 19. | Dirkse A, Golebiewska A, Buder T, Nazarov PV, Muller A, Poovathingal S, Brons NHC, Leite S, Sauvageot N, Sarkisjan D, Seyfrid M, Fritah S, Stieber D, Michelucci A, Hertel F, Herold-Mende C, Azuaje F, Skupin A, Bjerkvig R, Deutsch A, Voss-Böhme A, Niclou SP. Stem cell-associated heterogeneity in Glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat Commun. 2019;10:1787. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 251] [Cited by in RCA: 396] [Article Influence: 56.6] [Reference Citation Analysis (0)] |
| 20. | MacDonagh L, Gray SG, Breen E, Cuffe S, Finn SP, O'Byrne KJ, Barr MP. Lung cancer stem cells: The root of resistance. Cancer Lett. 2016;372:147-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 126] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 21. | Zhang M, Lee AV, Rosen JM. The Cellular Origin and Evolution of Breast Cancer. Cold Spring Harb Perspect Med. 2017;7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 22. | Ji J, Yamashita T, Budhu A, Forgues M, Jia HL, Li C, Deng C, Wauthier E, Reid LM, Ye QH, Qin LX, Yang W, Wang HY, Tang ZY, Croce CM, Wang XW. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50:472-480. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 423] [Cited by in RCA: 439] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 23. | Zhao X, Parpart S, Takai A, Roessler S, Budhu A, Yu Z, Blank M, Zhang YE, Jia HL, Ye QH, Qin LX, Tang ZY, Thorgeirsson SS, Wang XW. Integrative genomics identifies YY1AP1 as an oncogenic driver in EpCAM(+) AFP(+) hepatocellular carcinoma. Oncogene. 2015;34:5095-5104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 24. | Yamashita T, Budhu A, Forgues M, Wang XW. Activation of hepatic stem cell marker EpCAM by Wnt-beta-catenin signaling in hepatocellular carcinoma. Cancer Res. 2007;67:10831-10839. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 322] [Cited by in RCA: 368] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 25. | Gu Y, Wei X, Sun Y, Gao H, Zheng X, Wong LL, Jin L, Liu N, Hernandez B, Peplowska K, Zhao X, Zhan QM, Feng XH, Tang ZY, Ji J. miR-192-5p Silencing by Genetic Aberrations Is a Key Event in Hepatocellular Carcinomas with Cancer Stem Cell Features. Cancer Res. 2019;79:941-953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 70] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 26. | Yamashita T, Honda M, Nakamoto Y, Baba M, Nio K, Hara Y, Zeng SS, Hayashi T, Kondo M, Takatori H, Yamashita T, Mizukoshi E, Ikeda H, Zen Y, Takamura H, Wang XW, Kaneko S. Discrete nature of EpCAM+ and CD90+ cancer stem cells in human hepatocellular carcinoma. Hepatology. 2013;57:1484-1497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 231] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 27. | Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, Chu PW, Lam CT, Poon RT, Fan ST. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 883] [Cited by in RCA: 937] [Article Influence: 52.1] [Reference Citation Analysis (0)] |
| 28. | Wang R, Li Y, Tsung A, Huang H, Du Q, Yang M, Deng M, Xiong S, Wang X, Zhang L, Geller DA, Cheng B, Billiar TR. iNOS promotes CD24+CD133+ liver cancer stem cell phenotype through a TACE/ADAM17-dependent Notch signaling pathway. Proc Natl Acad Sci USA. 2018;115:E10127-E10136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 130] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 29. | Wu J, Zhu P, Lu T, Du Y, Wang Y, He L, Ye B, Liu B, Yang L, Wang J, Gu Y, Lan J, Hao Y, He L, Fan Z. The long non-coding RNA LncHDAC2 drives the self-renewal of liver cancer stem cells via activation of Hedgehog signaling. J Hepatol. 2019;70:918-929. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 30. | Rodríguez MM, Fiore E, Bayo J, Atorrasagasti C, García M, Onorato A, Domínguez L, Malvicini M, Mazzolini G. 4Mu Decreases CD47 Expression on Hepatic Cancer Stem Cells and Primes a Potent Antitumor T Cell Response Induced by Interleukin-12. Mol Ther. 2018;26:2738-2750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 55] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 31. | Lee TK, Cheung VC, Lu P, Lau EY, Ma S, Tang KH, Tong M, Lo J, Ng IO. Blockade of CD47-mediated cathepsin S/protease-activated receptor 2 signaling provides a therapeutic target for hepatocellular carcinoma. Hepatology. 2014;60:179-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 182] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 32. | Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1349] [Cited by in RCA: 1469] [Article Influence: 97.9] [Reference Citation Analysis (0)] |
| 33. | Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1549] [Cited by in RCA: 1875] [Article Influence: 144.2] [Reference Citation Analysis (0)] |
| 34. | Merlos-Suárez A, Barriga FM, Jung P, Iglesias M, Céspedes MV, Rossell D, Sevillano M, Hernando-Momblona X, da Silva-Diz V, Muñoz P, Clevers H, Sancho E, Mangues R, Batlle E. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 666] [Cited by in RCA: 755] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 35. | Dalerba P, Kalisky T, Sahoo D, Rajendran PS, Rothenberg ME, Leyrat AA, Sim S, Okamoto J, Johnston DM, Qian D, Zabala M, Bueno J, Neff NF, Wang J, Shelton AA, Visser B, Hisamori S, Shimono Y, van de Wetering M, Clevers H, Clarke MF, Quake SR. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol. 2011;29:1120-1127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 569] [Cited by in RCA: 552] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 36. | Zomer A, Ellenbroek SI, Ritsma L, Beerling E, Vrisekoop N, Van Rheenen J. Intravital imaging of cancer stem cell plasticity in mammary tumors. Stem Cells. 2013;31:602-606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 121] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 37. | Cairo S, Wang Y, de Reyniès A, Duroure K, Dahan J, Redon MJ, Fabre M, McClelland M, Wang XW, Croce CM, Buendia MA. Stem cell-like micro-RNA signature driven by Myc in aggressive liver cancer. Proc Natl Acad Sci USA. 2010;107:20471-20476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 152] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 38. | Wang T, Qin ZY, Wen LZ, Guo Y, Liu Q, Lei ZJ, Pan W, Liu KJ, Wang XW, Lai SJ, Sun WJ, Wei YL, Liu L, Guo L, Chen YQ, Wang J, Xiao HL, Bian XW, Chen DF, Wang B. Epigenetic restriction of Hippo signaling by MORC2 underlies stemness of hepatocellular carcinoma cells. Cell Death Differ. 2018;25:2086-2100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 39. | Li L, Liu Y, Guo Y, Liu B, Zhao Y, Li P, Song F, Zheng H, Yu J, Song T, Niu R, Li Q, Wang XW, Zhang W, Chen K. Regulatory MiR-148a-ACVR1/BMP circuit defines a cancer stem cell-like aggressive subtype of hepatocellular carcinoma. Hepatology. 2015;61:574-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 79] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 40. | Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, Jia H, Ye Q, Qin LX, Wauthier E, Reid LM, Minato H, Honda M, Kaneko S, Tang ZY, Wang XW. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012-1024. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 936] [Cited by in RCA: 972] [Article Influence: 57.2] [Reference Citation Analysis (0)] |
| 41. | Sia D, Villanueva A, Friedman SL, Llovet JM. Liver Cancer Cell of Origin, Molecular Class, and Effects on Patient Prognosis. Gastroenterology. 2017;152:745-761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 619] [Cited by in RCA: 871] [Article Influence: 96.8] [Reference Citation Analysis (2)] |
| 42. | Shimada S, Mogushi K, Akiyama Y, Furuyama T, Watanabe S, Ogura T, Ogawa K, Ono H, Mitsunori Y, Ban D, Kudo A, Arii S, Tanabe M, Wands JR, Tanaka S. Comprehensive molecular and immunological characterization of hepatocellular carcinoma. EBioMedicine. 2019;40:457-470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 191] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 43. | Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23:1124-1134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1234] [Cited by in RCA: 2020] [Article Influence: 224.4] [Reference Citation Analysis (0)] |
| 44. | Matak A, Lahiri P, Ford E, Pabst D, Kashofer K, Stellas D, Thanos D, Zatloukal K. Stochastic phenotype switching leads to intratumor heterogeneity in human liver cancer. Hepatology. 2018;68:933-948. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 45. | Saygin C, Matei D, Majeti R, Reizes O, Lathia JD. Targeting Cancer Stemness in the Clinic: From Hype to Hope. Cell Stem Cell. 2019;24:25-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 385] [Article Influence: 48.1] [Reference Citation Analysis (0)] |
| 46. | Yuan H, Yan M, Zhang G, Liu W, Deng C, Liao G, Xu L, Luo T, Yan H, Long Z, Shi A, Zhao T, Xiao Y, Li X. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019;47:D900-D908. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 191] [Cited by in RCA: 696] [Article Influence: 116.0] [Reference Citation Analysis (0)] |
| 47. | Qian J, Rankin EB. Hypoxia-Induced Phenotypes that Mediate Tumor Heterogeneity. Adv Exp Med Biol. 2019;1136:43-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 48. | Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos CR, Martinez P, Phillimore B, Begum S, Rabinowitz A, Spencer-Dene B, Gulati S, Bates PA, Stamp G, Pickering L, Gore M, Nicol DL, Hazell S, Futreal PA, Stewart A, Swanton C. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46:225-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1008] [Cited by in RCA: 1001] [Article Influence: 83.4] [Reference Citation Analysis (0)] |
| 49. | Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883-892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6102] [Cited by in RCA: 6090] [Article Influence: 435.0] [Reference Citation Analysis (0)] |
| 50. | Yates LR, Gerstung M, Knappskog S, Desmedt C, Gundem G, Van Loo P, Aas T, Alexandrov LB, Larsimont D, Davies H, Li Y, Ju YS, Ramakrishna M, Haugland HK, Lilleng PK, Nik-Zainal S, McLaren S, Butler A, Martin S, Glodzik D, Menzies A, Raine K, Hinton J, Jones D, Mudie LJ, Jiang B, Vincent D, Greene-Colozzi A, Adnet PY, Fatima A, Maetens M, Ignatiadis M, Stratton MR, Sotiriou C, Richardson AL, Lønning PE, Wedge DC, Campbell PJ. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med. 2015;21:751-759. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 653] [Cited by in RCA: 646] [Article Influence: 58.7] [Reference Citation Analysis (0)] |
| 51. | Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4428] [Cited by in RCA: 4235] [Article Influence: 84.7] [Reference Citation Analysis (1)] |
| 52. | Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6:924-935. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1190] [Cited by in RCA: 1105] [Article Influence: 55.3] [Reference Citation Analysis (0)] |
| 53. | Craig AJ, von Felden J, Garcia-Lezana T, Sarcognato S, Villanueva A. Tumour evolution in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2020;17:139-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 597] [Article Influence: 99.5] [Reference Citation Analysis (1)] |
| 54. | Okada S, Ishii H, Nose H, Okusaka T, Kyogoku A, Yoshimori M, Sakamoto M, Hirohashi S. Intratumoral DNA heterogeneity of small hepatocellular carcinoma. Cancer. 1995;75:444-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 55. | Saeki R, Nagai H, Kaneko S, Unoura M, Yamanaka N, Okamoto E, Kobayashi K, Matsubara K. Intratumoral genomic heterogeneity in human hepatocellular carcinoma detected by restriction landmark genomic scanning. J Hepatol. 2000;33:99-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 56. | Sirivatanauksorn Y, Sirivatanauksorn V, Bhattacharya S, Davidson BR, Dhillon AP, Kakkar AK, Williamson RC, Lemoine NR. Genomic heterogeneity in synchronous hepatocellular carcinomas. Gut. 1999;45:761-765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 57. | Xue R, Li R, Guo H, Guo L, Su Z, Ni X, Qi L, Zhang T, Li Q, Zhang Z, Xie XS, Bai F, Zhang N. Variable Intra-Tumor Genomic Heterogeneity of Multiple Lesions in Patients With Hepatocellular Carcinoma. Gastroenterology. 2016;150:998-1008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 173] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 58. | Furuta M, Ueno M, Fujimoto A, Hayami S, Yasukawa S, Kojima F, Arihiro K, Kawakami Y, Wardell CP, Shiraishi Y, Tanaka H, Nakano K, Maejima K, Sasaki-Oku A, Tokunaga N, Boroevich KA, Abe T, Aikata H, Ohdan H, Gotoh K, Kubo M, Tsunoda T, Miyano S, Chayama K, Yamaue H, Nakagawa H. Whole genome sequencing discriminates hepatocellular carcinoma with intrahepatic metastasis from multi-centric tumors. J Hepatol. 2017;66:363-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 85] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 59. | Zhai W, Lim TK, Zhang T, Phang ST, Tiang Z, Guan P, Ng MH, Lim JQ, Yao F, Li Z, Ng PY, Yan J, Goh BK, Chung AY, Choo SP, Khor CC, Soon WW, Sung KW, Foo RS, Chow PK. The spatial organization of intra-tumour heterogeneity and evolutionary trajectories of metastases in hepatocellular carcinoma. Nat Commun. 2017;8:4565. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 106] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 60. | Shi JY, Xing Q, Duan M, Wang ZC, Yang LX, Zhao YJ, Wang XY, Liu Y, Deng M, Ding ZB, Ke AW, Zhou J, Fan J, Cao Y, Wang J, Xi R, Gao Q. Inferring the progression of multifocal liver cancer from spatial and temporal genomic heterogeneity. Oncotarget. 2016;7:2867-2877. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 61. | Lin DC, Mayakonda A, Dinh HQ, Huang P, Lin L, Liu X, Ding LW, Wang J, Berman BP, Song EW, Yin D, Koeffler HP. Genomic and Epigenomic Heterogeneity of Hepatocellular Carcinoma. Cancer Res. 2017;77:2255-2265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 161] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 62. | Xu LX, He MH, Dai ZH, Yu J, Wang JG, Li XC, Jiang BB, Ke ZF, Su TH, Peng ZW, Guo Y, Chen ZB, Chen SL, Peng S, Kuang M. Genomic and transcriptional heterogeneity of multifocal hepatocellular carcinoma. Ann Oncol. 2019;30:990-997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 103] [Article Influence: 14.7] [Reference Citation Analysis (10)] |
| 63. | Torrecilla S, Sia D, Harrington AN, Zhang Z, Cabellos L, Cornella H, Moeini A, Camprecios G, Leow WQ, Fiel MI, Hao K, Bassaganyas L, Mahajan M, Thung SN, Villanueva A, Florman S, Schwartz ME, Llovet JM. Trunk mutational events present minimal intra- and inter-tumoral heterogeneity in hepatocellular carcinoma. J Hepatol. 2017;67:1222-1231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 132] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 64. | Losic B, Craig AJ, Villacorta-Martin C, Martins-Filho SN, Akers N, Chen X, Ahsen ME, von Felden J, Labgaa I, DʹAvola D, Allette K, Lira SA, Furtado GC, Garcia-Lezana T, Restrepo P, Stueck A, Ward SC, Fiel MI, Hiotis SP, Gunasekaran G, Sia D, Schadt EE, Sebra R, Schwartz M, Llovet JM, Thung S, Stolovitzky G, Villanueva A. Intratumoral heterogeneity and clonal evolution in liver cancer. Nat Commun. 2020;11:291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 276] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 65. | Ding X, He M, Chan AWH, Song QX, Sze SC, Chen H, Man MKH, Man K, Chan SL, Lai PBS, Wang X, Wong N. Genomic and Epigenomic Features of Primary and Recurrent Hepatocellular Carcinomas. Gastroenterology. 2019;157:1630-1645.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 147] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 66. | McGranahan N, Swanton C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell. 2017;168:613-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1799] [Cited by in RCA: 1980] [Article Influence: 220.0] [Reference Citation Analysis (0)] |
| 67. | Friemel J, Rechsteiner M, Frick L, Böhm F, Struckmann K, Egger M, Moch H, Heikenwalder M, Weber A. Intratumor heterogeneity in hepatocellular carcinoma. Clin Cancer Res. 2015;21:1951-1961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 237] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 68. | An FQ, Matsuda M, Fujii H, Tang RF, Amemiya H, Dai YM, Matsumoto Y. Tumor heterogeneity in small hepatocellular carcinoma: analysis of tumor cell proliferation, expression and mutation of p53 AND beta-catenin. Int J Cancer. 2001;93:468-474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 61] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 69. | Ling S, Hu Z, Yang Z, Yang F, Li Y, Lin P, Chen K, Dong L, Cao L, Tao Y, Hao L, Chen Q, Gong Q, Wu D, Li W, Zhao W, Tian X, Hao C, Hungate EA, Catenacci DV, Hudson RR, Li WH, Lu X, Wu CI. Extremely high genetic diversity in a single tumor points to prevalence of non-Darwinian cell evolution. Proc Natl Acad Sci USA. 2015;112:E6496-E6505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 249] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 70. | Sottoriva A, Kang H, Ma Z, Graham TA, Salomon MP, Zhao J, Marjoram P, Siegmund K, Press MF, Shibata D, Curtis C. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015;47:209-216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 711] [Cited by in RCA: 791] [Article Influence: 71.9] [Reference Citation Analysis (0)] |
| 71. | Liu J, Dang H, Wang XW. The significance of intertumor and intratumor heterogeneity in liver cancer. Exp Mol Med. 2018;50:e416. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 174] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 72. | Zhang Q, Lou Y, Yang J, Wang J, Feng J, Zhao Y, Wang L, Huang X, Fu Q, Ye M, Zhang X, Chen Y, Ma C, Ge H, Wang J, Wu J, Wei T, Chen Q, Wu J, Yu C, Xiao Y, Feng X, Guo G, Liang T, Bai X. Integrated multiomic analysis reveals comprehensive tumour heterogeneity and novel immunophenotypic classification in hepatocellular carcinomas. Gut. 2019;68:2019-2031. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 266] [Article Influence: 38.0] [Reference Citation Analysis (13)] |
| 73. | Buczak K, Ori A, Kirkpatrick JM, Holzer K, Dauch D, Roessler S, Endris V, Lasitschka F, Parca L, Schmidt A, Zender L, Schirmacher P, Krijgsveld J, Singer S, Beck M. Spatial Tissue Proteomics Quantifies Inter- and Intratumor Heterogeneity in Hepatocellular Carcinoma (HCC). Mol Cell Proteomics. 2018;17:810-825. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 65] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 74. | Kurebayashi Y, Ojima H, Tsujikawa H, Kubota N, Maehara J, Abe Y, Kitago M, Shinoda M, Kitagawa Y, Sakamoto M. Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology. 2018;68:1025-1041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 349] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 75. | Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, Hackl H, Trajanoski Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017;18:248-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1446] [Cited by in RCA: 3475] [Article Influence: 386.1] [Reference Citation Analysis (0)] |
| 76. | Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1271] [Cited by in RCA: 3287] [Article Influence: 365.2] [Reference Citation Analysis (0)] |
| 77. | Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, Diehn M, West RB, Plevritis SK, Alizadeh AA. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938-945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2244] [Cited by in RCA: 2468] [Article Influence: 224.4] [Reference Citation Analysis (0)] |
| 78. | Racle J, de Jonge K, Baumgaertner P, Speiser DE, Gfeller D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife. 2017;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 394] [Cited by in RCA: 991] [Article Influence: 110.1] [Reference Citation Analysis (0)] |
| 79. | Dong LQ, Peng LH, Ma LJ, Liu DB, Zhang S, Luo SZ, Rao JH, Zhu HW, Yang SX, Xi SJ, Chen M, Xie FF, Li FQ, Li WH, Ye C, Lin LY, Wang YJ, Wang XY, Gao DM, Zhou H, Yang HM, Wang J, Zhu SD, Wang XD, Cao Y, Zhou J, Fan J, Wu K, Gao Q. Heterogeneous immunogenomic features and distinct escape mechanisms in multifocal hepatocellular carcinoma. J Hepatol. 2020;72:896-908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 150] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 80. | Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, Kang B, Hu R, Huang JY, Zhang Q, Liu Z, Dong M, Hu X, Ouyang W, Peng J, Zhang Z. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell. 2017;169:1342-1356.e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1011] [Cited by in RCA: 1601] [Article Influence: 177.9] [Reference Citation Analysis (6)] |
| 81. | Zhang Q, He Y, Luo N, Patel SJ, Han Y, Gao R, Modak M, Carotta S, Haslinger C, Kind D, Peet GW, Zhong G, Lu S, Zhu W, Mao Y, Xiao M, Bergmann M, Hu X, Kerkar SP, Vogt AB, Pflanz S, Liu K, Peng J, Ren X, Zhang Z. Landscape and Dynamics of Single Immune Cells in Hepatocellular Carcinoma. Cell. 2019;179:829-845.e20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 763] [Cited by in RCA: 1118] [Article Influence: 159.7] [Reference Citation Analysis (0)] |
| 82. | Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R, Rosenthal R, Salm M, Horswell S, Escudero M, Matthews N, Rowan A, Chambers T, Moore DA, Turajlic S, Xu H, Lee SM, Forster MD, Ahmad T, Hiley CT, Abbosh C, Falzon M, Borg E, Marafioti T, Lawrence D, Hayward M, Kolvekar S, Panagiotopoulos N, Janes SM, Thakrar R, Ahmed A, Blackhall F, Summers Y, Shah R, Joseph L, Quinn AM, Crosbie PA, Naidu B, Middleton G, Langman G, Trotter S, Nicolson M, Remmen H, Kerr K, Chetty M, Gomersall L, Fennell DA, Nakas A, Rathinam S, Anand G, Khan S, Russell P, Ezhil V, Ismail B, Irvin-Sellers M, Prakash V, Lester JF, Kornaszewska M, Attanoos R, Adams H, Davies H, Dentro S, Taniere P, O'Sullivan B, Lowe HL, Hartley JA, Iles N, Bell H, Ngai Y, Shaw JA, Herrero J, Szallasi Z, Schwarz RF, Stewart A, Quezada SA, Le Quesne J, Van Loo P, Dive C, Hackshaw A, Swanton C; TRACERx Consortium. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med. 2017;376:2109-2121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1581] [Cited by in RCA: 1721] [Article Influence: 191.2] [Reference Citation Analysis (0)] |
| 83. | Martins-Filho SN, Alves VAF, Wakamatsu A, Maeda M, Craig AJ, Assato AK, Villacorta-Martin C, D'Avola D, Labgaa I, Carrilho FJ, Thung SN, Villanueva A. A phenotypical map of disseminated hepatocellular carcinoma suggests clonal constraints in metastatic sites. Histopathology. 2019;74:718-730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 84. | Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, Petritsch C, Ji HP, Maley CC. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med. 2016;22:105-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 490] [Cited by in RCA: 606] [Article Influence: 55.1] [Reference Citation Analysis (0)] |
| 85. | Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, Seth S, Chow CW, Cao Y, Gumbs C, Gold KA, Kalhor N, Little L, Mahadeshwar H, Moran C, Protopopov A, Sun H, Tang J, Wu X, Ye Y, William WN, Lee JJ, Heymach JV, Hong WK, Swisher S, Wistuba II, Futreal PA. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346:256-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 667] [Cited by in RCA: 780] [Article Influence: 65.0] [Reference Citation Analysis (0)] |
| 86. | Harbst K, Lauss M, Cirenajwis H, Isaksson K, Rosengren F, Törngren T, Kvist A, Johansson MC, Vallon-Christersson J, Baldetorp B, Borg Å, Olsson H, Ingvar C, Carneiro A, Jönsson G. Multiregion Whole-Exome Sequencing Uncovers the Genetic Evolution and Mutational Heterogeneity of Early-Stage Metastatic Melanoma. Cancer Res. 2016;76:4765-4774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 82] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 87. | Juric D, Castel P, Griffith M, Griffith OL, Won HH, Ellis H, Ebbesen SH, Ainscough BJ, Ramu A, Iyer G, Shah RH, Huynh T, Mino-Kenudson M, Sgroi D, Isakoff S, Thabet A, Elamine L, Solit DB, Lowe SW, Quadt C, Peters M, Derti A, Schegel R, Huang A, Mardis ER, Berger MF, Baselga J, Scaltriti M. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kα inhibitor. Nature. 2015;518:240-244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 379] [Cited by in RCA: 490] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 88. | Kwak EL, Ahronian LG, Siravegna G, Mussolin B, Borger DR, Godfrey JT, Jessop NA, Clark JW, Blaszkowsky LS, Ryan DP, Lennerz JK, Iafrate AJ, Bardelli A, Hong TS, Corcoran RB. Molecular Heterogeneity and Receptor Coamplification Drive Resistance to Targeted Therapy in MET-Amplified Esophagogastric Cancer. Cancer Discov. 2015;5:1271-1281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 149] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 89. | Russo M, Siravegna G, Blaszkowsky LS, Corti G, Crisafulli G, Ahronian LG, Mussolin B, Kwak EL, Buscarino M, Lazzari L, Valtorta E, Truini M, Jessop NA, Robinson HE, Hong TS, Mino-Kenudson M, Di Nicolantonio F, Thabet A, Sartore-Bianchi A, Siena S, Iafrate AJ, Bardelli A, Corcoran RB. Tumor Heterogeneity and Lesion-Specific Response to Targeted Therapy in Colorectal Cancer. Cancer Discov. 2016;6:147-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 340] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 90. | Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15:599-616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1487] [Cited by in RCA: 1463] [Article Influence: 182.9] [Reference Citation Analysis (0)] |
| 91. | Zhu AX, Finn RS, Edeline J, Cattan S, Ogasawara S, Palmer D, Verslype C, Zagonel V, Fartoux L, Vogel A, Sarker D, Verset G, Chan SL, Knox J, Daniele B, Webber AL, Ebbinghaus SW, Ma J, Siegel AB, Cheng AL, Kudo M; KEYNOTE-224 investigators. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial. Lancet Oncol. 2018;19:940-952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1184] [Cited by in RCA: 1983] [Article Influence: 247.9] [Reference Citation Analysis (0)] |
| 92. | Li J, Byrne KT, Yan F, Yamazoe T, Chen Z, Baslan T, Richman LP, Lin JH, Sun YH, Rech AJ, Balli D, Hay CA, Sela Y, Merrell AJ, Liudahl SM, Gordon N, Norgard RJ, Yuan S, Yu S, Chao T, Ye S, Eisinger-Mathason TSK, Faryabi RB, Tobias JW, Lowe SW, Coussens LM, Wherry EJ, Vonderheide RH, Stanger BZ. Tumor Cell-Intrinsic Factors Underlie Heterogeneity of Immune Cell Infiltration and Response to Immunotherapy. Immunity. 2018;49:178-193.e7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 659] [Cited by in RCA: 582] [Article Influence: 72.8] [Reference Citation Analysis (0)] |
| 93. | Wolf Y, Bartok O, Patkar S, Eli GB, Cohen S, Litchfield K, Levy R, Jiménez-Sánchez A, Trabish S, Lee JS, Karathia H, Barnea E, Day CP, Cinnamon E, Stein I, Solomon A, Bitton L, Pérez-Guijarro E, Dubovik T, Shen-Orr SS, Miller ML, Merlino G, Levin Y, Pikarsky E, Eisenbach L, Admon A, Swanton C, Ruppin E, Samuels Y. UVB-Induced Tumor Heterogeneity Diminishes Immune Response in Melanoma. Cell. 2019;179:219-235.e21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 245] [Cited by in RCA: 292] [Article Influence: 41.7] [Reference Citation Analysis (0)] |
| 94. | Corcoran RB, Chabner BA. Application of Cell-free DNA Analysis to Cancer Treatment. N Engl J Med. 2018;379:1754-1765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 481] [Cited by in RCA: 712] [Article Influence: 89.0] [Reference Citation Analysis (0)] |
| 95. | Georgiadis A, Durham JN, Keefer LA, Bartlett BR, Zielonka M, Murphy D, White JR, Lu S, Verner EL, Ruan F, Riley D, Anders RA, Gedvilaite E, Angiuoli S, Jones S, Velculescu VE, Le DT, Diaz LA, Sausen M. Noninvasive Detection of Microsatellite Instability and High Tumor Mutation Burden in Cancer Patients Treated with PD-1 Blockade. Clin Cancer Res. 2019;25:7024-7034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 119] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 96. | Okajima W, Komatsu S, Ichikawa D, Miyamae M, Ohashi T, Imamura T, Kiuchi J, Nishibeppu K, Arita T, Konishi H, Shiozaki A, Morimura R, Ikoma H, Okamoto K, Otsuji E. Liquid biopsy in patients with hepatocellular carcinoma: Circulating tumor cells and cell-free nucleic acids. World J Gastroenterol. 2017;23:5650-5668. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 71] [Cited by in RCA: 78] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 97. | Cai ZX, Chen G, Zeng YY, Dong XQ, Lin MJ, Huang XH, Zhang D, Liu XL, Liu JF. Circulating tumor DNA profiling reveals clonal evolution and real-time disease progression in advanced hepatocellular carcinoma. Int J Cancer. 2017;141:977-985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 72] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 98. | Huang A, Zhang X, Zhou SL, Cao Y, Huang XW, Fan J, Yang XR, Zhou J. Detecting Circulating Tumor DNA in Hepatocellular Carcinoma Patients Using Droplet Digital PCR Is Feasible and Reflects Intratumoral Heterogeneity. J Cancer. 2016;7:1907-1914. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 87] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 99. | Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15:81-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1349] [Cited by in RCA: 2553] [Article Influence: 283.7] [Reference Citation Analysis (3)] |
| 100. | Hectors SJ, Wagner M, Bane O, Besa C, Lewis S, Remark R, Chen N, Fiel MI, Zhu H, Gnjatic S, Merad M, Hoshida Y, Taouli B. Quantification of hepatocellular carcinoma heterogeneity with multiparametric magnetic resonance imaging. Sci Rep. 2017;7:2452. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 80] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 101. | Kiryu S, Akai H, Nojima M, Hasegawa K, Shinkawa H, Kokudo N, Yasaka K, Ohtomo K. Impact of hepatocellular carcinoma heterogeneity on computed tomography as a prognostic indicator. Sci Rep. 2017;7:12689. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 102. | Sadeghi-Naini A, Sannachi L, Tadayyon H, Tran WT, Slodkowska E, Trudeau M, Gandhi S, Pritchard K, Kolios MC, Czarnota GJ. Chemotherapy-Response Monitoring of Breast Cancer Patients Using Quantitative Ultrasound-Based Intra-Tumour Heterogeneities. Sci Rep. 2017;7:10352. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 45] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 103. | Dong X, Wu P, Sun X, Li W, Wan H, Yu J, Xing L. Intra-tumour 18F-FDG uptake heterogeneity decreases the reliability on target volume definition with positron emission tomography/computed tomography imaging. J Med Imaging Radiat Oncol. 2015;59:338-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 104. | van Baardwijk A, Bosmans G, van Suylen RJ, van Kroonenburgh M, Hochstenbag M, Geskes G, Lambin P, De Ruysscher D. Correlation of intra-tumour heterogeneity on 18F-FDG PET with pathologic features in non-small cell lung cancer: a feasibility study. Radiother Oncol. 2008;87:55-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 60] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 105. | Willaime JM, Turkheimer FE, Kenny LM, Aboagye EO. Quantification of intra-tumour cell proliferation heterogeneity using imaging descriptors of 18F fluorothymidine-positron emission tomography. Phys Med Biol. 2013;58:187-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 61] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 106. | Rimola J. Heterogeneity of Hepatocellular Carcinoma on Imaging. Semin Liver Dis. 2020;40:61-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 107. | Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, Sanduleanu S, Larue RTHM, Even AJG, Jochems A, van Wijk Y, Woodruff H, van Soest J, Lustberg T, Roelofs E, van Elmpt W, Dekker A, Mottaghy FM, Wildberger JE, Walsh S. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1825] [Cited by in RCA: 3956] [Article Influence: 439.6] [Reference Citation Analysis (0)] |
| 108. | Saini A, Breen I, Pershad Y, Naidu S, Knuttinen MG, Alzubaidi S, Sheth R, Albadawi H, Kuo M, Oklu R. Radiogenomics and Radiomics in Liver Cancers. Diagnostics (Basel). 2018;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (1)] |
| 109. | O'Connor JP, Rose CJ, Waterton JC, Carano RA, Parker GJ, Jackson A. Imaging intratumor heterogeneity: role in therapy response, resistance, and clinical outcome. Clin Cancer Res. 2015;21:249-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 462] [Cited by in RCA: 512] [Article Influence: 46.5] [Reference Citation Analysis (0)] |
| 110. | Lee S, Kang TW, Song KD, Lee MW, Rhim H, Lim HK, Kim SY, Sinn DH, Kim JM, Kim K, Ha SY. Effect of Microvascular Invasion Risk on Early Recurrence of Hepatocellular Carcinoma After Surgery and Radiofrequency Ablation. Ann Surg. 2019;Online ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 248] [Article Influence: 49.6] [Reference Citation Analysis (0)] |
| 111. | Lei Z, Li J, Wu D, Xia Y, Wang Q, Si A, Wang K, Wan X, Lau WY, Wu M, Shen F. Nomogram for Preoperative Estimation of Microvascular Invasion Risk in Hepatitis B Virus-Related Hepatocellular Carcinoma Within the Milan Criteria. JAMA Surg. 2016;151:356-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 477] [Article Influence: 47.7] [Reference Citation Analysis (0)] |
| 112. | Xu X, Zhang HL, Liu QP, Sun SW, Zhang J, Zhu FP, Yang G, Yan X, Zhang YD, Liu XS. Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J Hepatol. 2019;70:1133-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 546] [Article Influence: 78.0] [Reference Citation Analysis (1)] |
| 113. | Abdel-Rahman O. Assessment of the discriminating value of the 8th AJCC stage grouping for hepatocellular carcinoma. HPB (Oxford). 2018;20:41-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 114. | Minagawa M, Ikai I, Matsuyama Y, Yamaoka Y, Makuuchi M. Staging of hepatocellular carcinoma: assessment of the Japanese TNM and AJCC/UICC TNM systems in a cohort of 13,772 patients in Japan. Ann Surg. 2007;245:909-922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 286] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 115. | Sankin A, Hakimi AA, Mikkilineni N, Ostrovnaya I, Silk MT, Liang Y, Mano R, Chevinsky M, Motzer RJ, Solomon SB, Cheng EH, Durack JC, Coleman JA, Russo P, Hsieh JJ. The impact of genetic heterogeneity on biomarker development in kidney cancer assessed by multiregional sampling. Cancer Med. 2014;3:1485-1492. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 106] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 116. | Huang A, Zhao X, Yang XR, Li FQ, Zhou XL, Wu K, Zhang X, Sun QM, Cao Y, Zhu HM, Wang XD, Yang HM, Wang J, Tang ZY, Hou Y, Fan J, Zhou J. Circumventing intratumoral heterogeneity to identify potential therapeutic targets in hepatocellular carcinoma. J Hepatol. 2017;67:293-301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 84] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 117. | Shen YC, Hsu CL, Jeng YM, Ho MC, Ho CM, Yeh CP, Yeh CY, Hsu MC, Hu RH, Cheng AL. Reliability of a single-region sample to evaluate tumor immune microenvironment in hepatocellular carcinoma. J Hepatol. 2020;72:489-497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 118. | Kudo M, Ikeda M, Motomura K, Okusaka T, Kato N, Dutcus CE, Hisai T, Suzuki M, Ikezawa H, Iwata T, Kumada H, Kobayashi M. A phase Ib study of lenvatinib (LEN) plus nivolumab (NIV) in patients (pts) with unresectable hepatocellular carcinoma (uHCC): Study 117. J Clin Oncol. 2020;38:513-513. [RCA] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 119. | Llovet JM, Kudo M, Cheng AL, Finn RS, Galle PR, Kaneko S, Meyer T, Qin S, Dutcus CE, Chen E, Dubrovsky L, Zhu AX. Lenvatinib (len) plus pembrolizumab (pembro) for the first-line treatment of patients (pts) with advanced hepatocellular carcinoma (HCC): Phase 3 LEAP-002 study. J Clin Oncol. 2019;37:TPS4152-TPS4152. [RCA] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 77] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 120. | Nuciforo S, Fofana I, Matter MS, Blumer T, Calabrese D, Boldanova T, Piscuoglio S, Wieland S, Ringnalda F, Schwank G, Terracciano LM, Ng CKY, Heim MH. Organoid Models of Human Liver Cancers Derived from Tumor Needle Biopsies. Cell Rep. 2018;24:1363-1376. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 199] [Cited by in RCA: 351] [Article Influence: 50.1] [Reference Citation Analysis (0)] |
| 121. | Gao Q, Wang ZC, Duan M, Lin YH, Zhou XY, Worthley DL, Wang XY, Niu G, Xia Y, Deng M, Liu LZ, Shi JY, Yang LX, Zhang S, Ding ZB, Zhou J, Liang CM, Cao Y, Xiong L, Xi R, Shi YY, Fan J. Cell Culture System for Analysis of Genetic Heterogeneity Within Hepatocellular Carcinomas and Response to Pharmacologic Agents. Gastroenterology. 2017;152:232-242.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 110] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:
P-Reviewer: Capasso R, Grassi G, Link A S-Editor: Gong ZM L-Editor: Wang TQ E-Editor: Zhang YL