Published online Jul 7, 2020. doi: 10.3748/wjg.v26.i25.3650
Peer-review started: February 10, 2020
First decision: March 15, 2020
Revised: April 3, 2020
Accepted: June 18, 2020
Article in press: June 18, 2020
Published online: July 7, 2020
Processing time: 141 Days and 11.3 Hours
The risk of gastric cancer increases in patients with Helicobacter pylori-associated chronic atrophic gastritis (CAG). X-ray examination can evaluate the condition of the stomach, and it can be used for gastric cancer mass screening. However, skilled doctors for interpretation of X-ray examination are decreasing due to the diverse of inspections.
To evaluate the effectiveness of stomach regions that are automatically estimated by a deep learning-based model for CAG detection.
We used 815 gastric X-ray images (GXIs) obtained from 815 subjects. The ground truth of this study was the diagnostic results in X-ray and endoscopic examinations. For a part of GXIs for training, the stomach regions are manually annotated. A model for automatic estimation of the stomach regions is trained with the GXIs. For the rest of them, the stomach regions are automatically estimated. Finally, a model for automatic CAG detection is trained with all GXIs for training.
In the case that the stomach regions were manually annotated for only 10 GXIs and 30 GXIs, the harmonic mean of sensitivity and specificity of CAG detection were 0.955 ± 0.002 and 0.963 ± 0.004, respectively.
By estimating stomach regions automatically, our method contributes to the reduction of the workload of manual annotation and the accurate detection of the CAG.
Core tip: To construct a computer-aided diagnosis system, a method to detect chronic atrophic gastritis from gastric X-ray images (GXIs) with a patch-based convolutional neural network is presented in this paper. The proposed method utilizes two GXI groups for training: a manual annotation group and an automatic annotation group. The manual annotation group consists of GXIs for which we manually annotate the stomach regions, and the automatic annotation group consists of GXIs for which we automatically estimate the stomach regions. By utilizing GXIs with the stomach regions for training, the proposed method enables chronic atrophic gastritis detection that automatically eliminates the negative effect of the outside regions.
- Citation: Kanai M, Togo R, Ogawa T, Haseyama M. Chronic atrophic gastritis detection with a convolutional neural network considering stomach regions. World J Gastroenterol 2020; 26(25): 3650-3659
- URL: https://www.wjgnet.com/1007-9327/full/v26/i25/3650.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i25.3650
Gastric cancer is the third leading cause of death in all types of malignancies behind lung cancer and colorectal cancer[1]. One of the major risk factors of gastric cancer is Helicobacter pylori (H. pylori) infection[2-4]. Chronic atrophic gastritis (CAG) induced by H. pylori leads to atrophic mucosa[5], which increases the risk of gastric cancer[6]. Moreover, it has been revealed that H. pylori eradication therapy is effective for the reduction of the risk of gastric cancer[7-9].
According to the International Agency for Research on Cancer, the number of new cases of gastric cancer in Eastern Asia accounts for more than half of those in the world. For the reduction of gastric cancer mortality, population-based screening for gastric cancer has been conducted through endoscopic and X-ray examinations in Japan[10]. Although the detection rate of early gastric cancer by endoscopic examination is higher than that by X-ray examination[11], mass screening using endoscopic examination has some problems, e.g., a limit to the number of patients who can be examined[12]. Therefore, in Japan, endoscopic examination is often performed for cases in which something unusual is found by X-ray examination[13]. However, the interpretation of gastric X-ray images (GXIs) requires sufficient experience and knowledge, and there is a shortage of doctors who are skilled in diagnosis[10]. The development of computer-aided diagnosis (CAD) systems is needed to help doctors who do not have sufficient experience and knowledge.
For realizing CAD systems, researchers have been exploring methods for CAG detection from GXIs[14-17]. In early works, attempts were made to describe the visual features of CAG with mathematical models[14,15]. For more accurate detection, in the papers[16,17], we have tried to introduce convolutional neural networks (CNNs)[18] since it has been reported that CNNs outperform methods with hand-crafted features in various tasks[19-21]. We adopted a CNN that was trained on patches obtained by dividing original images, i.e., a patch-based CNN, to preserve detailed textures of GXIs since they had high resolutions. In our previous investigation[17], we focused on the outside patches of the stomach since textures of these patches do not depend on the image-level ground truth (GT), i.e., CAG or non-CAG. In clinical settings, GXIs generally have only the image-level GT. Therefore, we introduced manual annotation of stomach regions for all GXIs used in training and assigned the patch-level class labels based on the image-level GT and the stomach regions. Although the previously reported method already achieved high detection performance (sensitivity: 0.986, specificity: 0.945), there remains a problem. In general, CNNs require a large number of labeled images for training to determine millions of parameters that can capture the semantic contents of images. However, manually annotating stomach regions for a large number of GXIs is time- and labor-consuming. In other words, the previous method can practically utilize only a small number of GXIs even in the case of numerous GXIs being available for training.
In this paper, we propose a novel CAG detection method that requires manual annotation of stomach regions for only a small number of GXIs. The main contribution of this paper is the effective use of stomach regions that are manually annotated for a part of GXIs used in training. We assume that distinguishing the inside and outside patches of the stomach is much easier for patch-based CNNs than distinguishing whether the patches are extracted from CAG images or non-CAG images. Therefore, we newly introduce the automatic estimation of stomach regions for non-annotated GXIs. Herewith, we can reduce the workload of manual annotation and train a patch-based CNN that considers stomach regions with all GXIs even when we manually annotate stomach regions for some of the GXIs used in training.
The proposed method that requires manual annotation of stomach regions for only a small number of GXIs to detect CAG is presented in this section. This study was reviewed and approved by the institutional review board. Patients were not required to give informed consent to this study since the analysis used anonymous data that were obtained after each patient agreed to inspections by written consent. In this study, Kanai M from Graduate School of Information Science and Technology, Hokkaido University, Togo R from Education and Research Center for Mathematical and Data Science, Hokkaido University, Ogawa T and Haseyama M from Faculty of Information Science and Technology, Hokkaido University, took charge of the statistical analysis since they have an advanced knowledge of statistical analysis.
The GXIs used in this study were obtained from 815 subjects. Each subject underwent a gastric X-ray examination and an endoscopic examination at The University of Tokyo Hospital in 2010, and GXIs that had the same diagnostic results in both examinations were used in this study. Criteria for exclusion were the usage history of gastric acid suppressants, a history of H. pylori eradication therapy, and insufficient image data. The ground truth for this study was the diagnostic results in X-ray and endoscopic examinations. In the X-ray evaluation, subjects were classified into four categories, i.e., “normal”, “mild”, “moderate” and “severe”, based on atrophic levels[22]. It should be noted that the stomach with non-CAG has straight and fine fold distributions and fine mucosal surfaces, and the stomach with CAG has non-straight and snaked folds and coarse mucosal surfaces. X-ray examination can visualize these atrophic characteristics by barium contrast medium. We show that these differences can be trained on a chronic atrophic gastritis detection model with a small number of training images in this paper. We regarded subjects whose diagnosis results were “normal” as non-CAG subjects and the other subjects as CAG subjects. In contrast, in the endoscopic examination, subjects were classified into seven categories, i.e., no atrophic change (C0), three closed types of atrophic gastritis (C1, C2, C3) and three open types of atrophic gastritis (O1, O2, O3) based on the Kimura-Takemoto seven-grade classification[23]. Since C1 is defined as the atrophic borderline, we excluded subjects whose diagnosis results were C1 from the dataset. We regarded subjects whose diagnosis results were C0 as non-CAG subjects and the other subjects as CAG subjects. As a result, we regarded 240 subjects as CAG subjects and 575 subjects as non-CAG subjects.
According to the specific condition of GXIs, the size of GXIs used in this study was 2048 × 2048 pixels with an 8-bit grayscale. Technique of fluoroscopy was a digital radiography system. Exposure was controlled by an automatic exposure control mechanism. To realize the learning of our model at the patch level, GXIs taken from the double-contrast frontal view of the stomach in the supine position were used in this study.
For more accurate detection, we focus on the outside patches of the stomach since textures of these patches do not depend on the image-level GT at all. Although stomach regions can be easily determined without highly dedicated knowledge, manually annotating stomach regions for a large number of GXIs is not practical. In order to overcome this problem, we split GXIs for training into the following two groups.
(1) Manual annotation group (MAG): This group consists of GXIs for which we manually annotate the stomach regions. It is ideal for the number of GXIs in this group to be small since annotation for a large number of GXIs is time-and labor-consuming; (2) Automatic annotation group (AAG): This group consists of GXIs for which we automatically estimate the stomach regions with a CNN. By estimating the stomach regions automatically, a large number of GXIs can be used for training without the large burden of manual annotation.
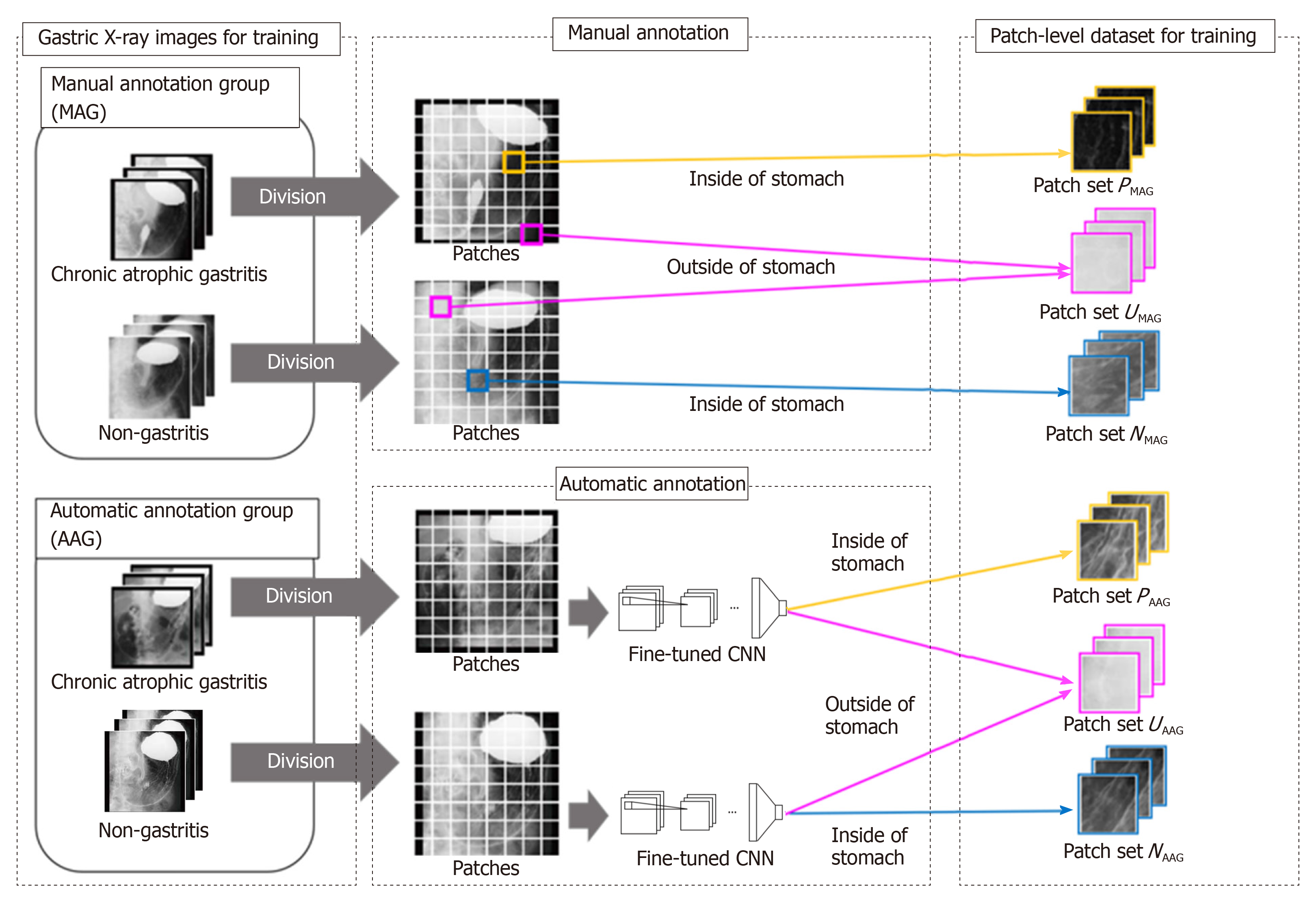
An overview of the preparation of the dataset is shown in Figure 1. The preparation of the dataset for training consists of two steps. In the first step, we annotate the stomach regions for GXIs in the MAG manually and select patches for training from GXIs in the MAG. In the second step, we estimate the stomach regions for GXIs in the AAG automatically and select patches for training from GXIs in the AAG.
First step - Patch selection for training from the MAG: Each GXI in the MAG is divided into patches. To separate the inside and outside patches of the stomach, we categorize the patches with the following three kinds of patch-level class labels (P, N, U): P: inside patches of the stomach in CAG images, i.e., positive patches; N: inside patches of the stomach in non-CAG images, i.e., negative patches; U: outside patches of the stomach in both CAG and non-CAG images, i.e., unrelated patches.
Note that if more than 80% regions within patches are included in the inside of the stomach, they were annotated as P or N. Furthermore, if less than 1% regions within patches are included in the inside of the stomach, they were annotated as U. Otherwise, we discard such patches from the training dataset. We denote a set of patches with each patch-level class label (P, N, U) as PMAG, NMAG, and UMAG. By setting the class label U, we can train a patch-based CNN that can distinguish the inside and outside patches of the stomach.
Second step - Patch selection for training from the AAG: For estimating the stomach regions for GXIs in the AAG, we introduce a fine-tuning technique that is effective when only a small number of images for training are available[24]. First, we prepare a CNN whose weights are transferred from a CNN pre-trained for image classification with a large number of labeled natural images. The number of nodes on the last fully connected layer in the CNN is altered to the number of the patch-level class labels (P, N, U). Due to this alteration, we initialize weights of the last fully connected layer with random values sampled from a uniform distribution. Next, the CNN is fine-tuned with the patches obtained in the first step to calculate the probabilities pc (c belong to {P,N,U}, ∑cpc = 1) of belonging to the patch-level class label c. By transferring the weights from the pre-trained CNN, accurate prediction of the patch-level class labels is realized even when the number of GXIs in the MAG is small. Second, we estimate the stomach regions for GXIs in the AAG. We divide each GXI in the AAG into patches. By inputting the patches into the fine-tuned CNN, we calculate the probabilities pc. We can regard pP + pN and pU as the probability of the inside of the stomach and the probability of the outside of the stomach, respectively. Therefore, we handle the patches that satisfy pP + pN ≥ α (0.5 < α ≤ 1) and the patches that satisfy pU ≥ α as the inside and outside patches of the stomach, respectively. Finally, we assign three kinds of patch-level class labels (P, N, U) for the patches based on the estimated results and the image-level GT. We denote a set of selected patches with patch-level class labels (P, N, U) as PAAG, NAAG and UAAG respectively. By estimating the stomach regions for the AAG, we can add PAAG, NAAG, and UAAG to the dataset for training even in the case of manual annotation of the stomach regions only for the MAG.
With all selected patches (P = PMAG + PAAG, N = NMAG + NAAG, U = UMAG + UAAG), we retrain the fine-tuned CNN to predict the patch-level class labels. For a GXI Xtest whose stomach regions and GT are unknown, we estimate an image-level class label ytestbelong to {1,0} that indicates CAG or non-CAG. First, we divide the target image Xtest into patches. We denote the patch-level class label predicted by the retrained CNN as cpred belong to {P,N,U}. In order to eliminate the influence of patches outside the stomach, we select patches that satisfy cpred = P or cpred = N for estimating the image-level class label. We calculate the ratio R with the selected patches as follows: R = MP/(MP + MN), where MP and MN are the numbers of patches that satisfy cpred = P and cpred = N, respectively. Finally, the image-level estimation result ytest for the target image Xtest is obtained as follows: ytest = 1 if R < β, otherwise, ytest = 0, where β is a predefined threshold. By selecting patches that satisfy cpred = P or cpred = N, estimating the image-level class label without the negative effect of regions outside the stomach is feasible.
A total of 815 GXIs including 200 images (100 CAG and 100 non-CAG images) for training and 615 images (140 CAG and 475 non-CAG images) for evaluation were used in this experiment. We set the number of GXIs in the MAG NMAG to {10, 20,…, 50}. Note that the numbers of CAG and non-CAG images in the MAG were both set to NMAG/2. We randomly sampled GXIs for the MAG from those for training, and the rest of them were used for the AAG. For an accurate evaluation, random sampling and calculating the performance of CAG detection were repeated five times at each NMAG. It took approximately 24 h to perform each trial. Note that all networks were computed on a single NVIDIA GeForce RTX 2080 Ti GPU. In this study, we extracted patches of 299 × 299 pixels in size from GXIs at intervals of 50 pixels. The following parameters were used for training of the CNN model: Batch size = 32, learning rate = 0.0001, momentum = 0.9, and the number of epochs = 50. The threshold for estimating stomach regions α was set to 0.9.
The TensorFlow framework[25] was utilized for training CNNs. We utilized the Inception-v3[26] model with weights trained on ImageNet[27] for fine-tuning. To confirm the effectiveness of utilizing not only the MAG but also the AAG for training, we compared the proposed method utilizing only the MAG with the proposed method utilizing the MAG and AAG. Hereinafter, (MAG only) denotes the proposed method utilizing only the MAG, and (MAG + AAG) denotes the proposed method utilizing both the MAG and AAG.
The performance was measured by the following harmonic mean (HM) of sensitivity and specificity: HM = (2 × Sensitivity × Specificity)/( Sensitivity + Specificity).
Note that HM was obtained at threshold β providing the highest HM.
Our experimental results are shown in this section. Examples of GXIs for evaluation are shown in Figure 2. First, we evaluate the performance of the fine-tuned CNN to select patches from the AAG. Note that the fine-tuned CNN doesn’t have to distinguish with high accuracy whether the patches are extracted from CAG images or non-CAG images since we utilize the fine-tuned CNN only for estimating the stomach regions. Figure 3 shows the visualization results obtained by applying the fine-tuned CNN to the CAG image shown in Figure 2A.
Specifically, the estimated patches of the inside and outside of the stomach and visualization of pP and pN (i.e., calculated probabilities of belonging to the patch-level class labels P and N) are shown in Figure 3. It is notable that the inside and outside regions of the stomach partially overlapped since GXIs were divided into patches with the overlap in this experiment. As shown in Figure 3, regions whose probabilities pP and pN are high tend to increase and decrease, respectively, as NMAG (i.e., the number of GXIs in the MAG) increases. In contrast, the stomach regions were estimated with high accuracy and the estimated stomach regions do not depend on the change of NMAG. Therefore, it is worth utilizing the fine-tuned CNN for estimating the stomach regions.
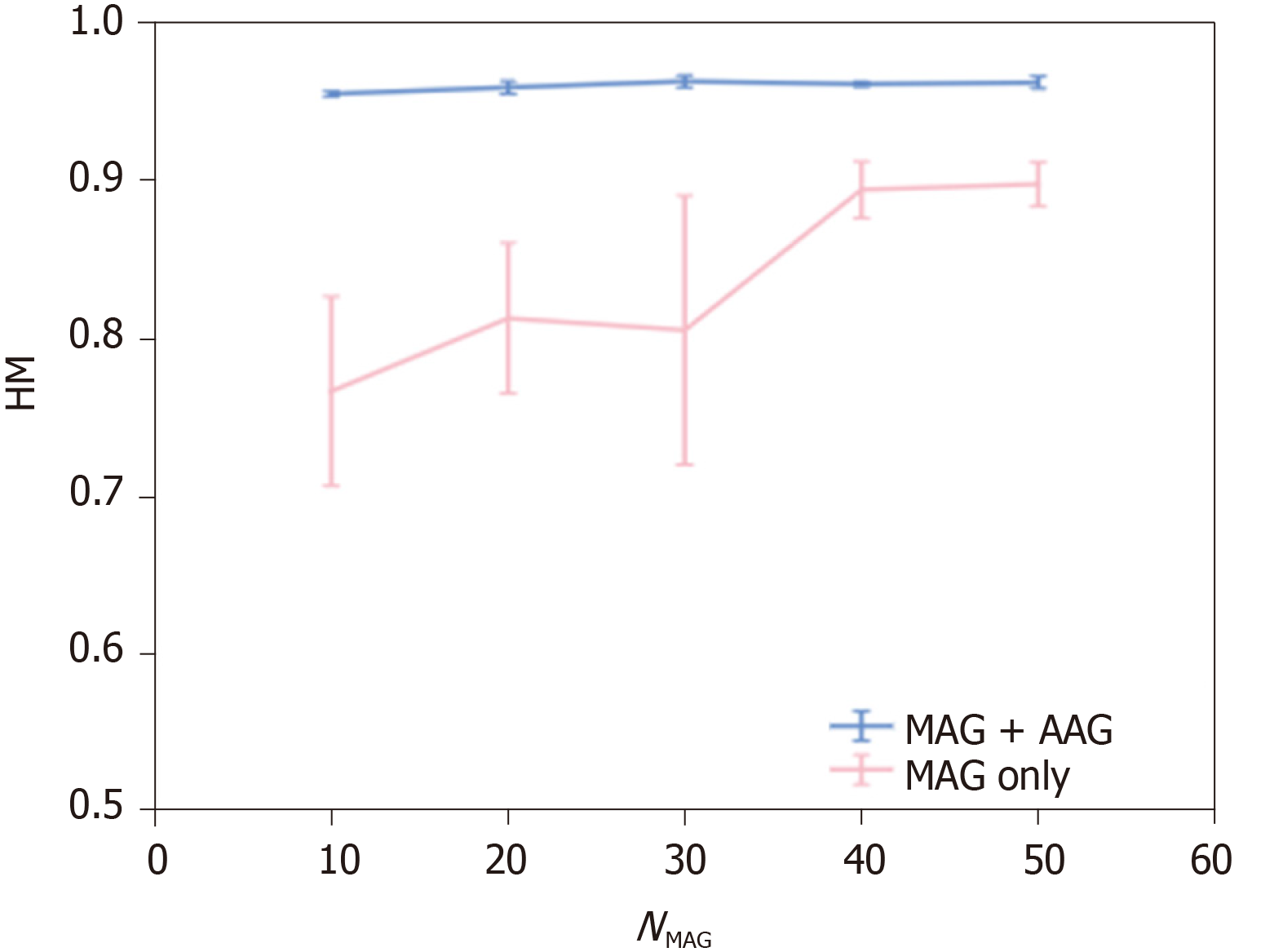
Next, we evaluate the performance of CAG detection. To confirm the effectiveness of considering stomach regions, we evaluated the detection performance of a baseline method that did not consider the stomach regions by setting patch-level class labels to the same as the image-level GT. As a result of utilizing 200 GXIs for the training, HM of the baseline method was 0.945. We show the detection performance of methods that considered the stomach regions. The detection performance of (MAG only) and that of (MAG + AAG) are shown in Figure 4. In Figure 4, HMs are shown as means ± SD of five trials. As shown in Figure 4, the mean of HM by (MAG + AAG) is higher than that by (MAG only) at each NMAG. The standard deviation of HM by (MAG + AAG) is smaller than that by (MAG only) at each NMAG. Therefore, the effectiveness of utilizing not only the MAG but also the AAG for the training is confirmed when the stomach regions are manually annotated for the same number of GXIs. Besides, to confirm the effect of reducing the workload of the manual annotation on detection performance, we evaluated the detection performance of a method that manually annotated the stomach regions for all GXIs used in the training (i.e., NMAG = 200). As a result, the HM of this method was 0.965. The negative effect of reducing the workload of manual annotation is small since the mean the HM of (MAG + AAG) approaches the HM of this method even when NMAG is small.
This study demonstrated that highly accurate detection of CAG was feasible even when we manually annotated the stomach regions for a small number of GXIs. Figure 4 (MAG only) indicates that a larger number of GXIs with annotation of the stomach regions are required to realize highly accurate detection. In contrast, a highly accurate estimation of the stomach regions is feasible even when the number of manually annotated images is limited. Therefore, highly accurate detection of CAG and reduction of the labor required for manually annotating the stomach regions are simultaneously realized by the proposed method when a large number of GXIs with the image-level GT are available for training.
The proposed method can be applied to other tasks in the field of medical image analysis since regions outside a target organ in medical images adversely affect the performance of the tasks. With only a simple annotation of regions of the target organ for a small number of medical images, the proposed method will enable accurate analysis that excludes the effect of regions outside the target organ.
In general, endoscopic examination is superior to an X-ray examination for the evaluation of CAG in imaging inspections[28]. The endoscopic examination has been recommended for gastric cancer mass screening programs in East Asian countries in recent years. For example, South Korea has started the endoscopic examination-based gastric cancer screening program since 2002, and the proportion of individuals who underwent endoscopic examination greatly increased from 31.15% in 2002 to 72.55% in 2011[28]. Also, Japan has started the endoscopic examination-based gastric cancer mass screening program in addition to an X-ray examination since 2016. However, there remains the problem that the number of individuals who can be examined in a day is limited. Hence, X-ray examination still plays an important role in gastric cancer mass screening.
To realize effective gastric cancer mass screening, it is crucial to narrow down individuals who need endoscopic examination by evaluating the condition of the stomach. Then CAD systems that can provide additional information to doctors will be helpful. Particularly, our approach presented in this paper realized the construction of machine learning-based CAG detection with a small number of training images. This suggests that the CAG detection method can be trained with data from a small-scale or medium-scale hospital without a large number of medical images for training.
This study has a few limitations. First, GXIs taken from only a single angle were analyzed in this study. In general X-ray examinations, GXIs are taken from multiple angles for each patient to examine the inside of the stomach thoroughly. Therefore, the detection performance will be improved by applying the proposed method to GXIs taken from multiple angles. Furthermore, the GXIs analyzed in this study were obtained in a single medical facility. To verify versatility, the proposed method should be applied to GXIs obtained in various medical facilities.
In this paper, a method for CAG detection from GXIs is presented. In the proposed method, we manually annotate the stomach regions for some of the GXIs used in training and automatically estimate the stomach regions for the rest of the GXIs. By using GXIs with the stomach regions for training, the proposed method realizes accurate CAG detection that automatically excludes the effect of regions outside the stomach. Experimental results showed the effectiveness of the proposed method.
It has been reported that chronic atrophic gastritis (CAG) induce by Helicobacter pylori infection increases the risk of gastric cancer. X-ray examination can evaluate the condition of the stomach for mass screening. On the other hand, there remains a problem that skilled doctors are decreasing.
Researches for the detection of CAG have been conducted, especially, deep learning-based techniques have achieved high recognition performance in general image datasets. However, early works need a large number of labeled images for training.
The study aimed to evaluate the effectiveness of a deep learning technique with a small number of training images with the stomach region annotation.
A total of 815 gastric X-ray images (GXIs) were used in our analysis. The ground truth of this study was the diagnostic results in X-ray and endoscopic examinations. For a part of GXIs for training, the stomach regions are manually annotated. A deep learning model is trained with the stomach region annotations. For the rest of them, the stomach regions are automatically estimated by the learned model. Finally, a model for automatic CAG detection is trained with all GXIs for training.
In the case that the stomach regions were manually annotated for only 10 GXIs and 30 GXIs, the harmonic mean of sensitivity and specificity of CAG detection were 0.955 ± 0.002 and 0.963 ± 0.004, respectively.
By estimating stomach regions automatically, our method contributes to the reduction of the workload of manual annotation and the accurate detection of the CAG.
Our CAG detection method can be trained with data from a small-scale or medium-scale hospital without medical data sharing that having the risk of leakage of personal information.
Experimental data were provided by the University of Tokyo Hospital in Japan. We express our thanks to Katsuhiro Mabe of the Junpukai Health Maintenance Center, and Nobutake Yamamichi of The University of Tokyo.
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 56684] [Article Influence: 7085.5] [Reference Citation Analysis (135)] |
| 2. | Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784-789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3126] [Cited by in RCA: 3248] [Article Influence: 129.9] [Reference Citation Analysis (1)] |
| 3. | Ohata H, Kitauchi S, Yoshimura N, Mugitani K, Iwane M, Nakamura H, Yoshikawa A, Yanaoka K, Arii K, Tamai H, Shimizu Y, Takeshita T, Mohara O, Ichinose M. Progression of chronic atrophic gastritis associated with Helicobacter pylori infection increases risk of gastric cancer. Int J Cancer. 2004;109:138-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 354] [Cited by in RCA: 378] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 4. | Vannella L, Lahner E, Annibale B. Risk for gastric neoplasias in patients with chronic atrophic gastritis: a critical reappraisal. World J Gastroenterol. 2012;18:1279-1285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 111] [Cited by in RCA: 113] [Article Influence: 8.1] [Reference Citation Analysis (5)] |
| 5. | Bavi R, Kumar R, Rampogu S, Son M, Park C, Baek A, Kim HH, Suh JK, Park SJ, Lee KW. Molecular interactions of UvrB protein and DNA from Helicobacter pylori: Insight into a molecular modeling approach. Comput Biol Med. 2016;75:181-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 6. | Kuipers EJ, Lundell L, Klinkenberg-Knol EC, Havu N, Festen HP, Liedman B, Lamers CB, Jansen JB, Dalenback J, Snel P, Nelis GF, Meuwissen SG. Atrophic gastritis and Helicobacter pylori infection in patients with reflux esophagitis treated with omeprazole or fundoplication. N Engl J Med. 1996;334:1018-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 514] [Cited by in RCA: 495] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 7. | Ma JL, Zhang L, Brown LM, Li JY, Shen L, Pan KF, Liu WD, Hu Y, Han ZX, Crystal-Mansour S, Pee D, Blot WJ, Fraumeni JF, You WC, Gail MH. Fifteen-year effects of Helicobacter pylori, garlic, and vitamin treatments on gastric cancer incidence and mortality. J Natl Cancer Inst. 2012;104:488-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 314] [Cited by in RCA: 360] [Article Influence: 25.7] [Reference Citation Analysis (1)] |
| 8. | Li WQ, Ma JL, Zhang L, Brown LM, Li JY, Shen L, Pan KF, Liu WD, Hu Y, Han ZX, Crystal-Mansour S, Pee D, Blot WJ, Fraumeni JF, You WC, Gail MH. Effects of Helicobacter pylori treatment on gastric cancer incidence and mortality in subgroups. J Natl Cancer Inst. 2014;106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 107] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 9. | IARC Helicobacter pylori Working Group. Helicobacter pylori eradication as strategy for preventing gastric cancer. Lyon, Fr Int Agency Res Cancer (IARC Work Gr Reports, No 8). 2014. |
| 10. | Sugano K. Screening of gastric cancer in Asia. Best Pract Res Clin Gastroenterol. 2015;29:895-905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 160] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 11. | Tashiro A, Sano M, Kinameri K, Fujita K, Takeuchi Y. Comparing mass screening techniques for gastric cancer in Japan. World J Gastroenterol. 2006;12:4873-4874. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 58] [Reference Citation Analysis (0)] |
| 12. | Kato M, Asaka M. Recent development of gastric cancer prevention. Jpn J Clin Oncol. 2012;42:987-994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 13. | Yuan Y. A survey and evaluation of population-based screening for gastric cancer. Cancer Biol Med. 2013;10:72-80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 14. | Abe K, Miura D, Minami M. Features for discriminating helicobacter pylori infection from gastric X-ray images. In: Proc Int Conf Signal Processing Systems (ICSPS) 2016: 31-35. [DOI] [Full Text] |
| 15. | Ishihara K, Ogawa T, Haseyama M. Helicobacter Pylori infection detection from gastric X-ray images based on feature fusion and decision fusion. Comput Biol Med. 2017;84:69-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 16. | Ishihara K, Ogawa T, Haseyama M. Detection of gastric cancer risk from X-ray images via patch-based convolutional neural network. In: Proc IEEE Int Conf Image Processing (ICIP) 2017: 2055-2259. [DOI] [Full Text] |
| 17. | Kanai M, Togo R, Ogawa T, Haseyama M. Gastritis detection from gastric X-ray images via fine-tuning of patch-based deep convolutional neural network. In: Proc IEEE Int Conf Image Processing (ICIP) 2019: 1371-1375. [DOI] [Full Text] |
| 18. | LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. In: Proc the IEEE 1998: 2278-2324. [DOI] [Full Text] |
| 19. | Krizhevsky A, Sulskever Ii, Hinton GE. ImageNet Classification with Deep Convolutional Neural Networks. In: Proc Int Conf Adv Neural Inf Process Syst 2012: 1-9. [DOI] [Full Text] |
| 20. | Mai L, Jin H, Liu F. Composition-preserving deep photo aesthetics assessment. In: Proc IEEE Conf Computer Vision and Pattern Recognition (CVPR) 2016: 497-506. [DOI] [Full Text] |
| 21. | Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JAWM, van Ginneken B, Sánchez CI. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5573] [Cited by in RCA: 5359] [Article Influence: 595.4] [Reference Citation Analysis (1)] |
| 22. | Yamamichi N, Hirano C, Ichinose M, Takahashi Y, Minatsuki C, Matsuda R, Nakayama C, Shimamoto T, Kodashima S, Ono S, Tsuji Y, Niimi K, Sakaguchi Y, Kataoka Y, Saito I, Asada-Hirayama I, Takeuchi C, Yakabi S, Kaikimoto H, Matsumoto Y, Yamaguchi D, Kageyama-Yahara N, Fujishiro M, Wada R, Mitsushima T, Koike K. Atrophic gastritis and enlarged gastric folds diagnosed by double-contrast upper gastrointestinal barium X-ray radiography are useful to predict future gastric cancer development based on the 3-year prospective observation. Gastric Cancer. 2016;19:1016-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 23. | Quach DT, Hiyama T. Assessment of Endoscopic Gastric Atrophy according to the Kimura-Takemoto Classification and Its Potential Application in Daily Practice. Clin Endosc. 2019;52:321-327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 24. | Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, Jianming Liang. Convolutional Neural Networks for Medical Image Analysis: Full Training or Fine Tuning? IEEE Trans Med Imaging. 2016;35:1299-1312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1757] [Cited by in RCA: 1112] [Article Influence: 111.2] [Reference Citation Analysis (0)] |
| 25. | Mart’ A, Ashish A, Paul B, Eugene B, Zhifeng C, Craig C, Greg SC, Andy D, Jeffrey D, Matthieu D, Sanjay G, Ian G, Andrew H, Geoffrey I, Michael I, Yangqing J, Rafal J, Lukasz K, Manjunath K, Josh L, Dan M, Rajat M, Sherry M, Derek M, Chris O, Mike S, Jonathon S, Benoit S, Ilya S, Kunal T, Paul T, Vincent V, Vijay V, Fernanda V, Oriol V, Pete W, Martin W, Martin W, Yuan Y, Xiaoqiang Z. TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems, 2015. |
| 26. | Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. In: Proc IEEE Conf Computer Vision and Pattern Recognition (CVPR) 2016: 2818-2826. |
| 27. | Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, Berg C A, Fei-Fei L. ImageNet large scale visual recognition challenge. Int J Comput Vis. 2015;115:211–52. [RCA] [DOI] [Full Text] [Cited by in Crossref: 16940] [Cited by in RCA: 6807] [Article Influence: 618.8] [Reference Citation Analysis (0)] |
| 28. | Lee S, Jun JK, Suh M, Park B, Noh DK, Jung KW, Choi KS. Gastric cancer screening uptake trends in Korea: results for the National Cancer Screening Program from 2002 to 2011: a prospective cross-sectional study. Medicine (Baltimore). 2015;94:e533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 77] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Japan
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Abraham P, Cheng H, Vynios D S-Editor: Zhang L L-Editor: A E-Editor: Ma YJ