Published online Jun 14, 2020. doi: 10.3748/wjg.v26.i22.3098
Peer-review started: January 6, 2020
First decision: January 19, 2020
Revised: March 30, 2020
Accepted: May 28, 2020
Article in press: May 28, 2020
Published online: June 14, 2020
Processing time: 160 Days and 7.7 Hours
Several studies have employed animal models to explore the association between microbiota and interleukin (IL) 10 signaling; however, limited information is available about the human microbiome.
To characterize the microbiome in patients with IL10RA mutations and to explore the association between gut dysbiosis and disease severity.
Fecal samples were collected from patients who were diagnosed with loss-of-function mutations in the IL10RA gene between January 2017 and July 2018 at the Children's Hospital of Fudan University. Age-matched volunteer children were recruited as healthy controls. Patients with Crohn's disease (CD) were used as disease controls to standardize the antibiotic exposure. Microbial DNA was extracted from the fecal samples. All analyses were based on the 16S rRNA gene sequencing data.
Seventeen patients with IL10RA mutations (IL10RA group), 17 patients with pediatric CD, and 26 healthy children were included. Both patients with IL10RA mutations and those with CD exhibited a reduced diversity of gut microbiome with increased variability. The relative abundance of Firmicutes was substantially increased in the IL10RA group (P = 0.02). On further comparison of the relative abundance of taxa between patients with IL10RA mutations and healthy children, 13 taxa showed significant differences. The IL10RA-specific dysbiosis indices exhibited a significant positive correlation with weighted pediatric CD activity index and simple endoscopic score for CD.
In patients with IL10RA mutations and early onset inflammatory bowel disease, gut dysbiosis shows a moderate association with disease severity.
Core tip: Understanding the role of microbes in sub-populations of inflammatory bowel disease patients is important. The focus on this relatively unique and uniform interleukin (IL)10RA group provides an excellent opportunity. In this study, clinical variables of IL10RA-deficient patients (such as disease course) were linked with changes in the stool microbiome, which implies potential clinical relevance of the changes in microbial populations.
- Citation: Xue AJ, Miao SJ, Sun H, Qiu XX, Wang SN, Wang L, Ye ZQ, Zheng CF, Huang ZH, Wang YH, Huang Y. Intestinal dysbiosis in pediatric Crohn's disease patients with IL10RA mutations. World J Gastroenterol 2020; 26(22): 3098-3109
- URL: https://www.wjgnet.com/1007-9327/full/v26/i22/3098.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i22.3098
Inflammatory bowel disease (IBD) includes Crohn's disease (CD), ulcerative colitis, and IBD unclassified. The pathogenetic mechanism of IBD is believed to involve inappropriate immune response to gut microbiota in genetically susceptible individuals. Recent studies have provided some insights into the role of complex host-microbiota interactions in the pathogenesis of IBD[1]. The next-generation sequencing approaches have helped unravel the genetic factors involved in the pathogenesis of infantile-onset IBD (age at diagnosis: < 2 years)[2]. The interleukin (IL)10 gene is one of the important genes that are known to affect the risk of IBD.
IL10 is an anti-inflammatory cytokine that inhibits intestinal inflammation. Patients with IL10 or IL10R deficiency can develop severe infantile colitis resembling CD[3]. Recent studies have revealed an association between host genetic variants and gut microbial changes; in addition, the underlying interactions were found to contribute to the onset and severity of IBD[4-7]. However, the role of microbiota in patients with infantile-onset IBD who have IL10 signaling defects is not clear. Several studies have employed animal models to explore the association between microbiota and IL10 signaling, because the link between aberrant IL10 signaling and IBD was first established in IL10-/- mice[8]. Colitis occurs in the presence of intestinal microbiota and changes in intestinal microbiota may also modulate the inflammatory response. For example, introduction of Lactobacillus plantarum 299v, Lactobacillus salivarius 433118, and Bifidobacterium infantis 35624 was shown to attenuate colitis[9-11], while introduction of Enterococcus faecalis, Escherichia coli, Helicobacter bilis, and Helicobacter hepaticus exacerbated the inflammation[12-14]. IBD (in animal models) occurs only in the presence of intestinal microbiota, as germ-free animals do not develop colitis. However, information pertaining to human microbiome is not well characterized.
Mutations in IL10RA, a gene that encodes one of the subunits of IL10R, have been identified as the most common causal mutations in infantile-onset IBD in China[15-17]. Thus, we conducted this study to characterize the microbiome in patients with IL10RA mutations and to explore the association between the disease severity and gut dysbiosis.
This was a single-center observational study. Pediatric patients (age: 0–18 years) who were initially diagnosed with loss-of-function mutations in the IL10RA gene at the Children's Hospital of Fudan University (China) between January 2017 and July 2018 were enrolled (IL10RA group). IL10RA gene mutations were identified by whole-exome sequencing or targeted gene panel and confirmed by Sanger sequencing, as described elsewhere[16,17]. Patients who did not undergo endoscopy at our center or who had been diagnosed with IL10RA gene mutations before transfer to our hospital were excluded. Moreover, we excluded patients with a history of ostomy and those with a history of extensive bowel resection. Two groups were used as controls. Age-matched volunteer children who did not receive any medical treatment or antibiotics and had no evidence of gastrointestinal disease or symptoms were recruited as healthy controls (HC group). Patients with a confirmed diagnosis of CD based on radiological, endoscopic, and histopathological evaluations after a minimum of 6-mo follow-up were enrolled as CD controls. CD patients who developed the disease at the age of less than 6 years were screened for relevant mutations by whole exosome sequencing; the sequencing results were negative for all these patients. Fecal samples were collected prior to bowel preparation for endoscopy. Samples were transported to the laboratory and stored at -80°C prior to further processing.
Clinical data pertaining to the following variables were obtained from the medical records: Age, sex, weight, age at onset, medical history, diet, disease behavior, laboratory results, clinical diagnosis, treatment, and endoscopic findings. Laboratory results included hemoglobin, C-reactive protein (CRP), and IL6 levels. Clinical activity was assessed at sample collection using Simple Endoscopic Score for CD (SES-CD), weighted pediatric CD activity index (wPCDAI), and Mucosal-Inflammation Non-Invasive (MINI) index [a newly developed noninvasive index that incorporates fecal calprotectin, CRP, erythrocyte sedimentation rate (ESR), and the stool item from the PCDAI for pediatric CD[18]].
Microbial DNA was extracted using the FastDNA SPIN Kit (MP Biomedicals, Santa Ana, CA, United States) according to the manufacturer's instructions. DNA concentration and purity were measured using the NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, United States), and DNA quality was checked by 1% agarose gel electrophoresis. V3-V4 hypervariable regions of the bacteria 16S rRNA gene were amplified using the thermocycler PCR system (GeneAmp 9700, ABI, United States). Purified amplicons were pooled in equimolar and paired-end sequenced on the Illumina MiSeq Platform (Illumina, San Diego, United States) according to standard protocols of the Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China).
Data analyses were performed using Stata 13.1 for Windows (Stata Corp LP, TX, United States), Prism 6 version 6.02 (GraphPad Software, lnc, San Diego, CA, United States), and MedCalc Statistical Software version 19.0.4 (MedCalc Software bvba, Ostend, Belgium). Categorical variables are presented as frequencies (percentages). Continuous variables are presented as the mean (standard deviation). Fisher’s exact test was used to compare categorical variables when cell sizes were less than 1. The Mann-Whitney U test was used to compare continuous variables between two groups, while the Kruskal-Wallis method was used to compare three or more groups. Spearman correlation analysis was performed to assess correlation between variables. The conservative Bonferroni correction was adopted for multiple tests. Two-tailed P values < 0.05 were considered indicative of statistical significance. For bioinformatics analyses, the α-diversity metrics and β-diversity were calculated using unweighted unifrac distances and represented in principal co-ordinates analysis (PCoA) using the open-access online Majorbio I-Sanger Cloud Platform (http://www.i-sanger.com).
A total of 32 IL10RA-deficient patients were admitted to our hospital during the study reference period. Fifteen patients were excluded for the following reasons: Six patients were undergoing follow-up for disease revaluation; of these, four had undergone allo-hematopoietic stem cell transplantation while the other two were in remission after treatment with thalidomide. Four patients were excluded because of a history of ostomy. Five patients had confirmed IL10RA gene mutations and had undergone endoscopy prior to referral to our hospital. Our study finally included 17 IL10RA-deficient patients, defined as the IL10RA group (Supplementary Table 1). In addition, 17 patients with CD and 26 healthy children were also enrolled. The detailed clinical characteristics and medication history are shown in Table 1.
| IL10RA | CD | HC | P value1 | |
| Number of samples | 17 | 17 | 26 | |
| Demographics | ||||
| Age (mean ± SD, yr) | 1.00 ± 0.68 | 8.92 ± 4.80 | 1.61 ± 1.71 | < 0.001 |
| Sex (M/F) | 10/7 | 11/6 | 17/9 | 0.724 |
| Weight and BMI | ||||
| Weight (mean ± SD, kg) | 7.31 ± 2.58 | 27.18 ± 13.04 | NA | < 0.001 |
| BMI (mean ± SD) | 14.53 ± 2.20 | 15.25 ± 2.44 | NA | 0.6175 |
| Disease activity | ||||
| CRP (mean ± SD, mg/L) | 40.65 ± 28.13 | 34.06 ± 31.92 | NA | 0.4486 |
| wPCDAI (mean ± SD) | 52.21 ± 17.65 | 40.88 ± 20.02 | NA | 0.1253 |
| SES-CD (mean ± SD) | 20.56 ± 7.56 | 12.94 ± 8.60 | NA | 0.0158 |
| MINI index (mean ± SD) | 19.88 ± 4.96 | 16.47 ± 2.92 | NA | 0.0027 |
| Disease duration and anemia | ||||
| Disease duration (mean ± SD, mo) | 9.93 ± 7.75 | 7.10 ± 9.30 | NA | 0.0706 |
| Disease location | ||||
| Ileum only (%) | 0 (0) | 3 (18) | NA | < 0.001 |
| Colon only (%) | 17 (100) | 3 (18) | NA | |
| Both (%) | 0 (0) | 11 (64) | NA | |
| Disease behaviour | ||||
| Montreal classification B1 (%) | 11 (65) | 15 (88) | NA | 0.106 |
| Montreal classification B2 (%) | 5 (29) | 2 (12) | NA | 0.203 |
| Montreal classification B3 (%) | 4 (24) | 0 (0) | NA | 0.033 |
| Montreal classification P (%) | 16 (94) | 4 (24) | NA | < 0.001 |
| Serology | ||||
| IL6 (mean ± SD, pg/mL) | 122.47 ± 152.22 | 37.50 ± 25.08 | NA | 0.0211 |
| Birth and breast feeding | ||||
| Cesarian section (%) | 11 (65) | 8 (47) | NA | 0.300 |
| Breast feeding (%) | 14 (82) | 13 (76) | NA | 0.671 |
| IBD medication | ||||
| Mesalazine (%) | 11 (65) | 11 (65) | NA | 1.000 |
| Steroids (%) | 1 (6) | 2 (12) | NA | 0.542 |
| Thalidomide (%) | 4 (24) | 0 (0) | NA | 0.014 |
| Other medication | ||||
| Antibiotics (%) | 13 (76) | 13 (76) | NA | 1.000 |
| Proton pump inhibitors (%) | 1 (6) | 9 (53) | NA | 0.001 |
| Marzulene-S (%) | 1 (6) | 1 (6) | NA | 1.000 |
| Self-reported diets | ||||
| Amino acid formula (%) | 2 (12) | 0 (0) | NA | 0.089 |
| Extensive hydrolyzed formula (%) | 3 (18) | 1 (6) | NA | 0.277 |
| Other diet (%) | 12 (71) | 10 (59) | NA | 0.473 |
Among patients in the IL10RA and CD groups, 76% were exposed to antibiotics within the last month before sample collection (Table 1). The average age at diagnosis in the IL10RA group was significantly lower than that in the CD group (P < 0.001). In addition, patients in the IL10RA group showed a significantly lower level of hemoglobin (P = 0.0035), higher level of IL6 (P = 0.0192), and a more severe phenotype [as reflected by a higher SES-CD score (P = 0.0157) and MINI index (P = 0.0025)] as compared to those in the CD group. Colon and the perianal region appeared to be more commonly affected in the IL10RA group (Table 1). We then performed receiver operating characteristic (ROC) curve analysis to evaluate the ability of these clinical variables in discriminating the IL10RA group from the CD group. Of the clinical variables that showed significant differences between the IL10RA and CD groups, the age at initial admission showed the best predictive ability [area under the curve (AUC) = 0.925] with a sensitivity of 94.12% and specificity of 88.24%. Other predictive factors were MINI (AUC = 0.861), SES-CD (AUC = 0.786), hemoglobin (AUC = 0.780), and IL6 (AUC = 0.757); this showed that IL10RA-deficient patients had more severe disease than patients in the CD group (Supplementary Figure 1).
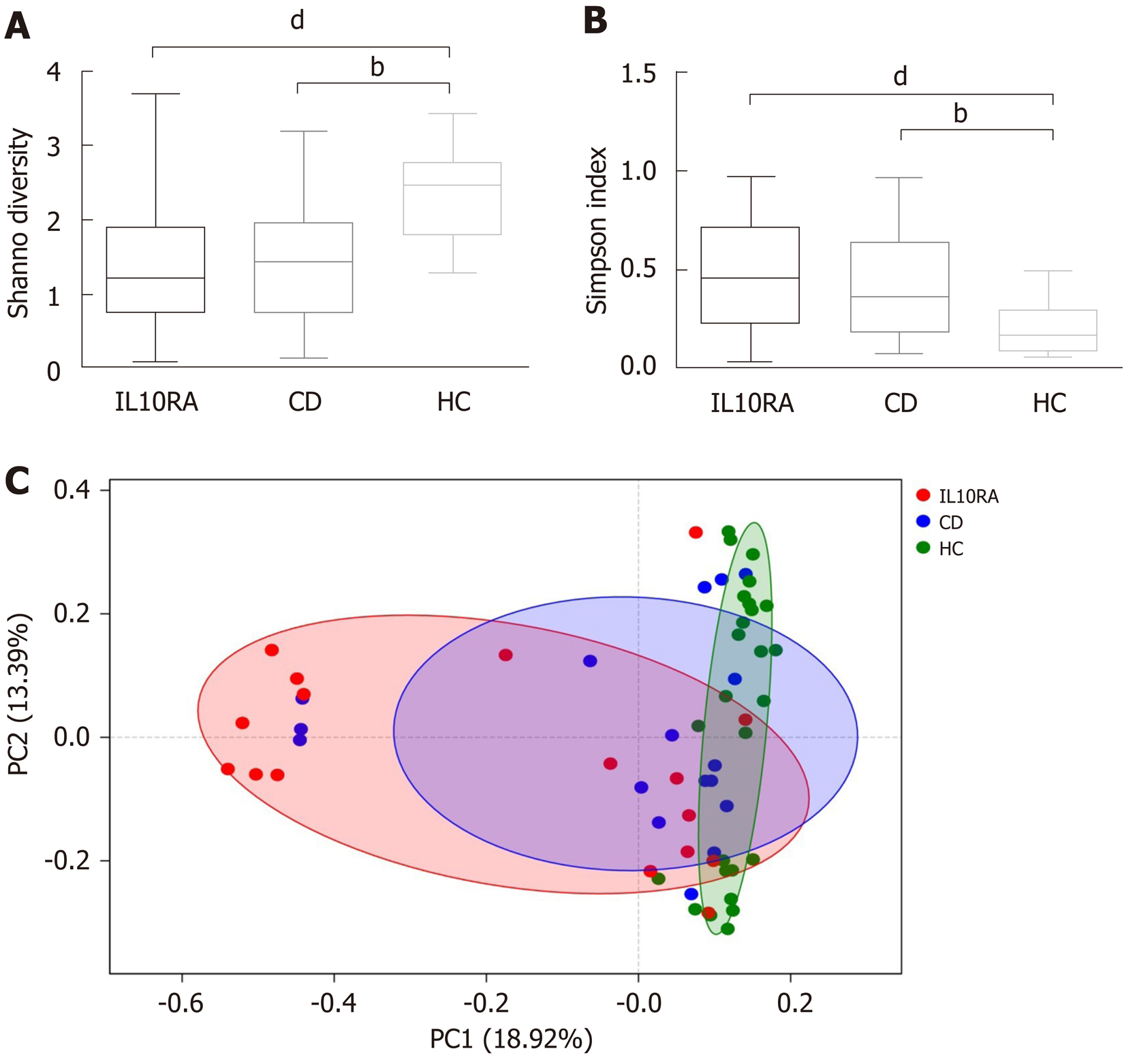
Both patients with IL10RA deficiency and patients in the CD group exhibited a reduced microbial diversity. The Shannon index values for the IL10RA, CD, and HC groups were 1.39 ± 0.85, 1.45 ± 0.87, and 2.36 ± 0.61, respectively (Figure 1A). This result was consistent with the Simpson index as an indicator of microbial diversity. The average Simpson index values were 0.47 ± 0.27, 0.44 ± 0.29, and 0.19 ± 0.11, respectively (Figure 1B). The IL10RA group showed a significantly reduced microbial diversity (P < 0.0001) as compared to the CD group (P < 0.001). As for beta diversity measured by the unweighted unifrac distance of the OTU community structure, the microbiome of the HC group clustered together and shared similar microbial profiles; however, the microbial composition in the IL10RA group was scattered and showed greater heterogeneity than that in the CD group (Figure 1C).
At the phylum level, Firmicutes, Actinobacteria, Proteobacteria, and Bacteroidetes were the predominant phyla in all groups (Figure 2A). The relative abundance of Firmicutes and Actinobacteria showed significant differences among groups (Figure 2B). After FDR correction for multiple tests, the relative abundance of Firmicutes in the IL10RA group was substantially greater than that in the HC group (P = 0.02). On further comparison of the relative abundance between the IL10RA and HC groups, significant differences were observed with respect to 13 taxa, all of which belonged to phylum Firmicutesor Actinobacteria (Table 2). Some microbial changes which were reported to be associated with risk variants or mutations of other CD candidate genes are listed in Table 2[4,6,7,19-23].
| Ref. | Gene | Human/animal | Major findings |
| [6,19] | IL10 | Il10-/- mice | Decrease in diversity and richness |
| ↑Proteobacteria and Escherichia coli (during onset of inflammation) | |||
| ↓Bacteroidetes and Firmicutes | |||
| Current study | IL10RA | Human cohort | Decrease in diversity and increase in variability |
| ↑Firmicutes, Enterococcaceae, Enterococcus, Lactobacillales, Bacilli, and Micrococcales | |||
| ↓Bifidobacteriales, Bifidobacteriaceae, Bifidobacterium, Veillonellaceae, Clostridiales, Clostridia, Selenomonadales, and Negativicutes | |||
| [4,7,20] | NOD2 | Human cohort | ↓Roseburia, Faecalibacterium prausnitzii, Bacteroides and Bacteroidia |
| ↑Eubacteriaceae and Enterobacteriaceae | |||
| [21] | NOD2 | Nod2-/- mice | Decrease in diversity and richness |
| ↑Bacteroides, Bacteroidaceae, and B.acidifaciens | |||
| ↓Proteobacteria, Helicobacter hepaticus, and Desulfovibro spp | |||
| [22] | ATG16L1 | Human cohort | ↑Fusobacteriaceae |
| [4] | CARD9 | Human cohort | ↓Firmicutes |
| [23] | CARD9 | Card9-/- mice | Decreased stability |
| ↓Adlercreutzia, Actinobacteria, and Lactobacillus reuteri |
Subsequently, we used a random forest classifier and performed linear discriminant analysis (LDA) to determine the effect size of taxa on the dysbiosis in each group (Figure 2C). Of the identified taxa in the IL10RA group, Lactobacillales, Bacilli, Enterococcaceae, Enterococcus, and Firmicutes were enriched in abundance with variable importance (LDA) scores of greater than 5 (Figure 2C). The LDA scores of Clostridia, Clostridiales, Bifidobacterium, Bifidobacteriales, Bifidobacteriaceae, and Actinobacteria were greater than 5 in the HC group. The top five taxa contributing to dysbiosis in the CD group were Veillonellaceae, Megamonas, Micrococcaceae, Rothia, and Micrococcales (Figure 2C).
We detected a strong correlation between SES-CD and wPCDAI (r = 0.71, P = 0.0026) within the IL10RA group. However, SES-CD did not show any correlation with wPCDAI or MINI index in the CD group (P > 0.05). On combining these two groups together, we found a significant correlation of SES-CD with both wPCDAI (Spearman r = 0.58, P = 0.0004) and MINI index (r = 0.52, P = 0.0020).
IL10RA-specific dysbiosis indices were calculated based on the relative abundance of five taxa at the order level (Lactobacillales, Micrococcales, Veillonellaceae, Clostridiales, and Selenomonadales), according to a previously defined method[24]. The dysbiosis indices were associated with the Shannon indices in both the IL10RA (r = -0.66, P = 0.0052) and CD groups (r = -0.66, P = 0.0046); this suggests that the abundance of these five taxa largely captured the dysbiosis. We found a significant correlation of wPCDAI and SES-CD scores with the dysbiosis indices (Table 3). Hemoglobin level and disease duration showed an inverse correlation with the dysbiosis indices. No significant association was found between the dysbiosis index and the MINI index and the level of IL6 or CRP. In our exploratory analysis, the dysbiosis index seemed to fit better with SES-CD score, hemoglobin, and disease duration within the IL10RA group as compared to that in the CD group (as reflected by higher values of the correlation coefficient; Table 3).
| Variable | Combined, r1 (P value) | IL10RA, r (P value) | CD, r (P value) |
| wPCDAI | 0.52 (0.002) | 0.51 (0.040) | 0.54 (0.027) |
| SES-CD | 0.48 (0.005) | 0.51 (0.044) | 0.35 (0.174) |
| MINI index | 0.26 (0.131) | 0.32 (0.213) | 0.17 (0.510) |
| CRP, mg/L | 0.11 (0.532) | 0.16 (0.526) | 0.18 (0.493) |
| IL6, pg/mL | 0.17 (0.346) | 0.22 (0.417) | 0.01 (0.961) |
| Hb, g/L | -0.54 (0.0009) | -0.56 (0.022) | -0.51 (0.037) |
| Disease duration (mo) | -0.35 (0.045) | -0.50 (0.043) | -0.40 (0.109) |
In this observational study, we observed reduced diversity and increased variability of gut microbiome in patients with IL10RA mutations based on the 16S rRNA sequencing data. Patients with IL10RA mutations had early disease onset and experienced more severe colitis. We also explored the association between intestinal dysbiosis and the disease severity in these patients.
In previous studies, microbial diversity exhibited a negative correlation with severity of IBD; however, the mutation status was not factored in these studies[4,7,20,23]. In our study, patients in both the IL10RA and CD groups showed a reduced diversity compared with the healthy children. The lack of significant difference between these two groups with respect to diversity indices was likely attributable to similar antibiotic exposure (76%). Given the average age and BMI of the IL10RA-deficient patients, this might also be due to the limitation of the synthetic diversity descriptor, which potentially masks the multi-factor impact on microbiome[25]. Another interesting finding was the variability in different groups. We speculate that the resilience of the gut microbiota varied in individuals with different diseases. Patients exposed to intrinsic factors (i.e., IL10RA deficiency) with an immature gut microbiome may harbor the microbiome that is most vulnerable to environmental disturbances.
Gevers et al[24] reported the microbial dysbiosis in new-onset pediatric CD: Increased abundance of Enterobacteriaceae, Pasteurellacaea, Veillonellaceae, and Fusobacteriaceae and decreased abundance of Erysipelotrichales, Bacteroidales, and Clostridiales. A systematic review showed that patients with active CD have lower abundance of Clostridium leptum, Faecalibacterium prausnitzii, and Bifidobacterium[26]. Consistent with previous studies, we observed increased Veillonellaceae in the CD group; Clostridiales and Bifidobacterium were decreased in the IL10RA group regardless of the antibiotic exposure. Gevers et al[24] found that exposure to antibiotics amplified the dysbiosis; however, exclusion of samples from subjects with antibiotic exposure did not change the key players. In a study by Knights et al[20], recent antibiotic usage was inversely associated with Firmicutes, Blautia, Ruminococcac, Tenericutes, and Lachnospiraceae; however, Proteobacteria and Bacilli showed a positive correlation with the medication history. Our study showed increased abundance of Bacilli in the IL10RA group. However, the abundance of Firmicutes was also increased in the IL10RA group.
Recent studies have shown the effects of Mendelian disorders on the intestinal microbiome, the function of the intestinal mucosa, and the immune response in the gut[4,6,7,19-23]. In parallel, studies have also identified the role of microbiota in initiating and exacerbating the disease[12-14,19]. Besides genetic defects in IL10 and its receptor, a set of causal variants and causative genes have also been identified in IBD. Variants of genes that affect the risk of IBD and have been associated with altered composition of the microbiome are listed in Table 2. These genes are involved in the intestinal immune response to microbes. For instance, impaired function of nucleotide-binding oligomerization domain-containing protein 2 (NOD2) in sensing the bacterial lipopolysaccharide may cause an increase in bacteria that produce these products (e.g., Escherichia species and Bacteroides vulgatus)[4,7,20,21]. Caspase recruitment domain family member 9 (CARD9) was shown to affect the composition of the gut microbiota by altering the production of microbial metabolites[23]. Mutations in ATG16L1 were found to decrease the secretion of antimicrobial peptides by Paneth cells and to impair the elimination of specific bacteria through phagocytosis[22]. In addition, polymorphisms in MHC class II genes affect the production of IgA in response to microbes[27]. In NHE3-deficient mice, altered electrolyte transport and mucosal pH may represent a key mechanism of reduced colonic microbial diversity[27]. We found no mutual microbial changes between patients with IL10RA mutations and those with other reported risk variants in IBD; this indicates that IL10 signaling defects may impact the microbiota via other pathways. Defects in STAT3, the signaling molecule downstream of IL10 receptors, have been recently implicated in skin microbial imbalance. In addition, failure of the MyD88-Stat3 signaling in Treg cells was shown to result in dysbiosis[28,29].
IBD is a heterogenous disease. Owing to considerable dissociation between clinical symptoms and mucosal inflammation in CD, development of therapeutic strategies targeting different sub-groups of patients based on age, disease severity, and disease location is a key challenge. Versions of PCDAI exhibited only a fair correlation with SES-CD (r = 0.33-0.45)[18]. Few studies have performed parallel scoring of the dysbiosis index, SES-CD, and wPCDAI. The MINI index was also validated in our cohort, but it showed no superiority in the accordance of SES-CD. We found that the correlation between SES-CD and wPCDAI was stronger in the IL10RA group.
This observational study was a pilot effort to characterize IL10RA-specific microbial alterations and has several limitations. First, the descriptive nature of the study does not permit any causal inferences. Prospective trials enrolling larger treatment-naive populations at high risk with longitudinal follow-up would provide insights into the role of microbes in the onset of inflammation. Second, 16S rRNA sequencing has its limitations; shotgun metagenomic sequencing with a higher taxonomic resolution may capture microbial shifts in full complexity. Further investigations may be warranted to identify the shifts in functional or metabolic capabilities of the microbiome. Third, comparison between CD patients and those with IL10RA mutations was not corrected for other factors; patients with IL10RA mutations are young and typically have a greater propensity for colonic disease.
In summary, the advent of new methodologies can facilitate a better understanding of the interactions between genetic factors and the gut microbiome. To the best of our knowledge, this is the first report of microbial dysbiosis in this sub-population of IBD patients with IL10RA mutations; our findings may facilitate further attempts to develop microbial therapeutics. Gut dysbiosis in patients with IL10RA mutations showed a moderate association with disease severity in this study. Further studies should focus on the precise role of the microbiota in the etiology of IBD in terms of host genetic susceptibility; this constitutes an attractive target for a given host genome.
Several studies have employed animal models to explore the association between microbiota and interleukin (IL)10 signaling; however, limited information is available about the human microbiome. To the best of our knowledge, this is the first report of microbial dysbiosis in this sub-population of inflammatory bowel diseases (IBD) patients with IL10RA mutations.
Patients with IL10RA mutations had early disease onset and experienced more severe colitis. Recent studies have revealed an association between host genetic variants and gut microbial changes; in addition, the underlying interactions were found to contribute to the onset and severity of IBD. However, the role of microbiota in patients with infantile-onset IBD who have IL10 signaling defects is not clear. Our findings may facilitate further attempts to develop microbial therapeutics in these patients.
We aimed to characterize the microbiome in patients with IL10RA mutations and to explore the association between gut dysbiosis and disease severity. We observed a reduced diversity and increased variability of gut microbiome in patients with IL10RA mutations. We also explored the association between intestinal dysbiosis and the disease severity in these patients. Further studies should focus on the precise role of the microbiota in the etiology of IBD in terms of host genetic susceptibility; this constitutes an attractive target for a given host genome.
Fecal samples were collected from patients who were diagnosed with loss-of-function mutations in the IL10RA gene. Age-matched volunteer children were recruited as healthy controls. Patients with Crohn's disease (CD) were used as disease controls to standardize the antibiotic exposure. Microbial DNA was extracted from the fecal samples. All analyses were based on the 16S rRNA gene sequencing data.
Seventeen patients with IL10RA mutations, 17 patients with pediatric CD, and 26 healthy children were included. Both patients with IL10RA mutations and those with CD exhibited a reduced diversity of gut microbiome with increased variability. The relative abundance of Firmicutes was substantially increased in the IL10RA group. On further comparison of the relative abundance of taxa between patients with IL10RA mutations and healthy children, 13 taxa showed significant differences. The IL10RA-specific dysbiosis indices exhibited a significant positive correlation with weighted pediatric CD activity index and simple endoscopic score for CD. This observational study was a pilot effort to characterize IL10RA-specific microbial alterations and does not permit any causal inferences.
In patients with IL10RA mutations and early onset IBD, gut dysbiosis showed a moderate association with disease severity. In this study, clinical variables of IL10RA-deficient patients (such as disease course) were linked with changes in the stool microbiome, which implies potential clinical relevance of the changes in microbial populations.
16S rRNA sequencing has its limitations; shotgun metagenomic sequencing with a higher taxonomic resolution may capture microbial shifts in full complexity. Further investigations may be warranted to identify the shifts in functional or metabolic capabilities of the microbiome. Prospective trials enrolling larger treatment-naive populations at high risk with longitudinal follow-up would provide insights into the role of microbes in the onset of inflammation.
We would like to thank all the patients and families who participated in the study. Many thanks to all the reviewers who have provided helpful suggestions and corrections.
| 1. | Chu H, Khosravi A, Kusumawardhani IP, Kwon AH, Vasconcelos AC, Cunha LD, Mayer AE, Shen Y, Wu WL, Kambal A, Targan SR, Xavier RJ, Ernst PB, Green DR, McGovern DP, Virgin HW, Mazmanian SK. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science. 2016;352:1116-1120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 508] [Article Influence: 50.8] [Reference Citation Analysis (1)] |
| 2. | Uhlig HH, Muise AM. Clinical Genomics in Inflammatory Bowel Disease. Trends Genet. 2017;33:629-641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 83] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 3. | Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schäffer AA, Noyan F, Perro M, Diestelhorst J, Allroth A, Murugan D, Hätscher N, Pfeifer D, Sykora KW, Sauer M, Kreipe H, Lacher M, Nustede R, Woellner C, Baumann U, Salzer U, Koletzko S, Shah N, Segal AW, Sauerbrey A, Buderus S, Snapper SB, Grimbacher B, Klein C. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033-2045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1149] [Cited by in RCA: 1119] [Article Influence: 65.8] [Reference Citation Analysis (0)] |
| 4. | Aschard H, Laville V, Tchetgen ET, Knights D, Imhann F, Seksik P, Zaitlen N, Silverberg MS, Cosnes J, Weersma RK, Xavier R, Beaugerie L, Skurnik D, Sokol H. Genetic effects on the commensal microbiota in inflammatory bowel disease patients. PLoS Genet. 2019;15:e1008018. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 5. | Cohen LJ, Cho JH, Gevers D, Chu H. Genetic Factors and the Intestinal Microbiome Guide Development of Microbe-Based Therapies for Inflammatory Bowel Diseases. Gastroenterology. 2019;156:2174-2189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 145] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 6. | Pellicciotta M, Rigoni R, Falcone EL, Holland SM, Villa A, Cassani B. The microbiome and immunodeficiencies: Lessons from rare diseases. J Autoimmun. 2019;98:132-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 7. | Imhann F, Vich Vila A, Bonder MJ, Fu J, Gevers D, Visschedijk MC, Spekhorst LM, Alberts R, Franke L, van Dullemen HM, Ter Steege RWF, Huttenhower C, Dijkstra G, Xavier RJ, Festen EAM, Wijmenga C, Zhernakova A, Weersma RK. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut. 2018;67:108-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 420] [Cited by in RCA: 589] [Article Influence: 73.6] [Reference Citation Analysis (1)] |
| 8. | Madsen KL. Inflammatory bowel disease: lessons from the IL-10 gene-deficient mouse. Clin Invest Med. 2001;24:250-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 63] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 9. | Schultz M, Veltkamp C, Dieleman LA, Grenther WB, Wyrick PB, Tonkonogy SL, Sartor RB. Lactobacillus plantarum 299V in the treatment and prevention of spontaneous colitis in interleukin-10-deficient mice. Inflamm Bowel Dis. 2002;8:71-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 246] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 10. | McCarthy J, O'Mahony L, O'Callaghan L, Sheil B, Vaughan EE, Fitzsimons N, Fitzgibbon J, O'Sullivan GC, Kiely B, Collins JK, Shanahan F. Double blind, placebo controlled trial of two probiotic strains in interleukin 10 knockout mice and mechanistic link with cytokine balance. Gut. 2003;52:975-980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 342] [Cited by in RCA: 303] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 11. | Madsen KL, Doyle JS, Tavernini MM, Jewell LD, Rennie RP, Fedorak RN. Antibiotic therapy attenuates colitis in interleukin 10 gene-deficient mice. Gastroenterology. 2000;118:1094-1105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 180] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 12. | Kim SC, Tonkonogy SL, Albright CA, Tsang J, Balish EJ, Braun J, Huycke MM, Sartor RB. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology. 2005;128:891-906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 346] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 13. | Balish E, Warner T. Enterococcus faecalis induces inflammatory bowel disease in interleukin-10 knockout mice. Am J Pathol. 2002;160:2253-2257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 241] [Cited by in RCA: 255] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 14. | Burich A, Hershberg R, Waggie K, Zeng W, Brabb T, Westrich G, Viney JL, Maggio-Price L. Helicobacter-induced inflammatory bowel disease in IL-10- and T cell-deficient mice. Am J Physiol Gastrointest Liver Physiol. 2001;281:G764-G778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 144] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 15. | Zheng C, Huang Y, Hu W, Shi J, Ye Z, Qian X, Huang Z, Xue A, Wang Y, Lu J, Tang Z, Wu J, Wang L, Peng K, Zhou Y, Miao S, Sun H. Phenotypic Characterization of Very Early-Onset Inflammatory Bowel Disease with Interleukin-10 Signaling Deficiency: Based on a Large Cohort Study. Inflamm Bowel Dis. 2019;25:756-766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 16. | Ye Z, Zhou Y, Huang Y, Wang Y, Lu J, Tang Z, Miao S, Dong K, Jiang Z. Phenotype and Management of Infantile-onset Inflammatory Bowel Disease: Experience from a Tertiary Care Center in China. Inflamm Bowel Dis. 2017;23:2154-2164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 42] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 17. | Huang Z, Peng K, Li X, Zhao R, You J, Cheng X, Wang Z, Wang Y, Wu B, Wang H, Zeng H, Yu Z, Zheng C, Wang Y, Huang Y. Mutations in Interleukin-10 Receptor and Clinical Phenotypes in Patients with Very Early Onset Inflammatory Bowel Disease: A Chinese VEO-IBD Collaboration Group Survey. Inflamm Bowel Dis. 2017;23:578-590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 51] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 18. | Cozijnsen MA, Ben Shoham A, Kang B, Choe BH, Choe YH, Jongsma MME, Russell RK, Ruemmele FM, Escher JC, de Ridder L, Koletzko S, Martín-de-Carpi J, Hyams J, Walters T, Griffiths A, Turner D. Development and Validation of the Mucosal Inflammation Noninvasive Index For Pediatric Crohn's Disease. Clin Gastroenterol Hepatol. 2020;18:133-140.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 19. | Maharshak N, Packey CD, Ellermann M, Manick S, Siddle JP, Huh EY, Plevy S, Sartor RB, Carroll IM. Altered enteric microbiota ecology in interleukin 10-deficient mice during development and progression of intestinal inflammation. Gut Microbes. 2013;4:316-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 143] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 20. | Knights D, Silverberg MS, Weersma RK, Gevers D, Dijkstra G, Huang H, Tyler AD, van Sommeren S, Imhann F, Stempak JM, Huang H, Vangay P, Al-Ghalith GA, Russell C, Sauk J, Knight J, Daly MJ, Huttenhower C, Xavier RJ. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 2014;6:107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 302] [Cited by in RCA: 299] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 21. | Mondot S, Barreau F, Al Nabhani Z, Dussaillant M, Le Roux K, Doré J, Leclerc M, Hugot JP, Lepage P. Altered gut microbiota composition in immune-impaired Nod2(-/-) mice. Gut. 2012;61:634-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 68] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 22. | Sadaghian Sadabad M, Regeling A, de Goffau MC, Blokzijl T, Weersma RK, Penders J, Faber KN, Harmsen HJ, Dijkstra G. The ATG16L1-T300A allele impairs clearance of pathosymbionts in the inflamed ileal mucosa of Crohn's disease patients. Gut. 2015;64:1546-1552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 23. | Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM, Brot L, Taleb S, Couturier-Maillard A, Nion-Larmurier I, Merabtene F, Seksik P, Bourrier A, Cosnes J, Ryffel B, Beaugerie L, Launay JM, Langella P, Xavier RJ, Sokol H. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22:598-605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1083] [Cited by in RCA: 1153] [Article Influence: 115.3] [Reference Citation Analysis (0)] |
| 24. | Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, Morgan XC, Kostic AD, Luo C, González A, McDonald D, Haberman Y, Walters T, Baker S, Rosh J, Stephens M, Heyman M, Markowitz J, Baldassano R, Griffiths A, Sylvester F, Mack D, Kim S, Crandall W, Hyams J, Huttenhower C, Knight R, Xavier RJ. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1945] [Cited by in RCA: 2470] [Article Influence: 205.8] [Reference Citation Analysis (1)] |
| 25. | Rashidi A, Kaiser T, Holtan SG, Weisdorf DJ, Khoruts A, Staley C. Pre-transplant recovery of microbiome diversity without recovery of the original microbiome. Bone Marrow Transplant. 2019;54:1115-1117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 26. | Prosberg M, Bendtsen F, Vind I, Petersen AM, Gluud LL. The association between the gut microbiota and the inflammatory bowel disease activity: a systematic review and meta-analysis. Scand J Gastroenterol. 2016;51:1407-1415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 155] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 27. | Bolnick DI, Snowberg LK, Caporaso JG, Lauber C, Knight R, Stutz WE. Major Histocompatibility Complex class IIb polymorphism influences gut microbiota composition and diversity. Mol Ecol. 2014;23:4831-4845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 135] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 28. | Larmonier CB, Laubitz D, Hill FM, Shehab KW, Lipinski L, Midura-Kiela MT, McFadden RM, Ramalingam R, Hassan KA, Golebiewski M, Besselsen DG, Ghishan FK, Kiela PR. Reduced colonic microbial diversity is associated with colitis in NHE3-deficient mice. Am J Physiol Gastrointest Liver Physiol. 2013;305:G667-G677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 65] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 29. | Smeekens SP, Huttenhower C, Riza A, van de Veerdonk FL, Zeeuwen PL, Schalkwijk J, van der Meer JW, Xavier RJ, Netea MG, Gevers D. Skin microbiome imbalance in patients with STAT1/STAT3 defects impairs innate host defense responses. J Innate Immun. 2014;6:253-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 77] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:
P-Reviewer: Schwarz SM S-Editor: Ma YJ L-Editor: Wang TQ E-Editor: Zhang YL