Published online Mar 21, 2020. doi: 10.3748/wjg.v26.i11.1197
Peer-review started: December 6, 2019
First decision: February 16, 2020
Revised: February 21, 2020
Accepted: March 5, 2020
Article in press: March 5, 2020
Published online: March 21, 2020
Processing time: 106 Days and 1.3 Hours
BRIP1 is a helicase that partners with BRCA1 in the homologous recombination (HR) step in the repair of DNA inter-strand cross-link lesions. It is a rare cause of hereditary ovarian cancer in patients with no mutations of BRCA1 or BRCA2. The role of the protein in other cancers such as gastrointestinal (GI) carcinomas is less well characterized but given its role in DNA repair it could be a candidate tumor suppressor similarly to the two BRCA proteins.
To analyze the role of helicase BRIP1 (FANCJ) in GI cancers pathogenesis.
Publicly available data from genomic studies of esophageal, gastric, pancreatic, cholangiocarcinomas and colorectal cancers were interrogated to unveil the role of BRIP1 in these carcinomas and to discover associations of lesions in BRIP1 with other more common molecular defects in these cancers.
Molecular lesions in BRIP1 were rare (3.6% of all samples) in GI cancers and consisted almost exclusively of mutations and amplifications. Among mutations, 40% were possibly pathogenic according to the OncoKB database. A majority of BRIP1 mutated GI cancers were hyper-mutated due to concomitant mutations in mismatch repair or polymerase ε and δ1 genes. No associations were discovered between amplifications of BRIP1 and any mutated genes. In gastroesophageal cancers BRIP1 amplification commonly co-occurs with ERBB2 amplification.
Overall BRIP1 molecular defects do not seem to play a major role in GI cancers whereas mutations frequently occur in hypermutated carcinomas and co-occur with other HR genes mutations. Despite their rarity, BRIP1 defects may present an opportunity for therapeutic interventions similar to other HR defects.
Core tip: BRIP1 gene alterations are uncommon in gastrointestinal cancers. Mutations frequently occur in hypermutated carcinomas and co-occur with other homologous recombination genes mutations. Despite their rarity, BRIP1 defects may present an opportunity for therapeutic interventions similar to other homologous recombination defects.
- Citation: Voutsadakis IA. Landscape of BRIP1 molecular lesions in gastrointestinal cancers from published genomic studies. World J Gastroenterol 2020; 26(11): 1197-1207
- URL: https://www.wjgnet.com/1007-9327/full/v26/i11/1197.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i11.1197
BRIP1 [BRCA1 interacting protein C-terminal helicase 1, alternatively called FANCJ, Fanconi Anemia (FA) complementation group J or BACH1, BRCA1 Associated C-terminal Helicase 1] is a 1249 amino-acid protein with helicase function that participates in DNA homeostasis. The gene (Gene ID: 83990) is located at human chromosome 17q23.2 and consists of 20 exons, 19 of which (exons 2 to 20) are coding. BRIP1 protein plays a role in DNA repair through homologous recombination (HR) and interacts with BRCA1[1]. BRIP1 has also BRCA1 independent effects in DNA repair that depend on the helicase activity[2]. Besides BRCA1, BRIP1 interacts with mismatch repair (MMR) protein MLH1 and promotes signaling for apoptosis at sites with O6-methylated guanine adducts[3]. BRIP1 mutant cells that lose the ability for MLH1 interaction survive better when methyl-guanine methyltransferase MGMT is functional as MGMT has more time to process the defective site. BRIP1-MLH1 interaction may be as important as the interaction with BRCA1 in signaling from inter-strand cross-links and underlines the role of BRIP1 as a key player at the cross-roads of DNA repair though the FA pathway and the MMR as well as the HR pathway[4]. Besides inter-strand cross-links, a role of BRIP1 in repairing other abnormal DNA structures, such as G-quadruplex structures and hairpins, arising during DNA replication, under replication stress, has been recently established[5].
BRIP1 has been implicated in hereditary ovarian cancers that lack BRCA1 or BRCA2 mutations[6]. Up to 0.6%-0.9% of ovarian cancers may carry pathogenic variants in BRIP1, although the percentage may vary in different populations[7]. A role of BRIP1 in hereditary breast cancer has also been proposed but is debated[8,9]. Similarly, rare cases of prostate cancer with BRIP1 mutations reminiscent of prostate cancer in BRCA2 families have been reported[10,11]. Leukemia predisposition is part of FA and has been described with BRIP1 hereditary mutations, in common with other FA complementation group gene mutations[12]. The implication of BRIP1 as a tumor suppressor in other hereditary cancers or in sporadic cancers is even less clear.
This paper investigates the role of BRIP1 defects in gastrointestinal (GI) cancers exploring publicly available genomic data from The Cancer Genome Atlas (TCGA) available in the cBioportal of cancer genomics platform.
Studies performed by TCGA consortium (PanCancer Atlas) that were evaluated in the current investigation included esophageal adenocarcinoma (containing 182 samples), gastric adenocarcinoma (containing 440 samples), pancreatic adenocarcinoma (containing 184 samples), colorectal cancer (containing 594 samples), cholangio-carcinoma (with 36 samples)[13-17]. Analyses were performed in the cBioCancer Genomics Portal (cBioportal, http://www.cbioportal.org) platform[18,19]. cBioportal contains 172 non-overlapping genomic studies published by TCGA and by other investigators worldwide and empowers interrogation of each study or group of studies for genetic lesions in any gene of interest, in a user-friendly manner. The five studies selected for the current investigation cover the most updated available TCGA results of the most common GI cancers.
cBioportal currently provides assessment of the functional implications of mutations of interest using the mutation assessor and other relevant tools. The mutation assessor (mutationassessor.org) uses a multiple sequence alignment algorithm to assign a prediction score of functional significance to each mutation[20]. Data from the mutation assessor as reported in cBioportal were used for evaluation of putative functional repercussions of BRIP1 mutations and other mutations of interest. Data from the OncoKB database, a precision oncology database annotating the biologic and oncogenic significance of somatic cancer mutations were incorporated in the functional assessment of discussed mutations[21].
Survival of gastric cancer patients with high expression of BRIP1 mRNA vs those with low BRIP1 mRNA expression was compared using the online tool Kaplan Meier Plotter (kmplot.com)[22]. This online tool currently does not include other GI cancers.
Investigation of BRIP1 promoters was performed using the EPD database (http://epd.epfl.ch) and putative transcription factor binding sites were identified using the JASPAR CORE 2018 vertebrate database[23].
For further analyses that could not be performed directly in cBioportal, the list of genes and relevant mutated or amplified samples from each study of interest was transferred to an Excel sheet (Microsoft Corp., Redmond, WA) for performance of required calculations. Categorical and continuous data were compared with the Fisher’s exact test and the t test respectively. Correlations were explored with the Pearson correlation coefficient. All statistical comparisons were considered significant if P < 0.05. Correction for multiple comparisons was performed using the Benjamini-Hochberg false discovery rate correction procedure.
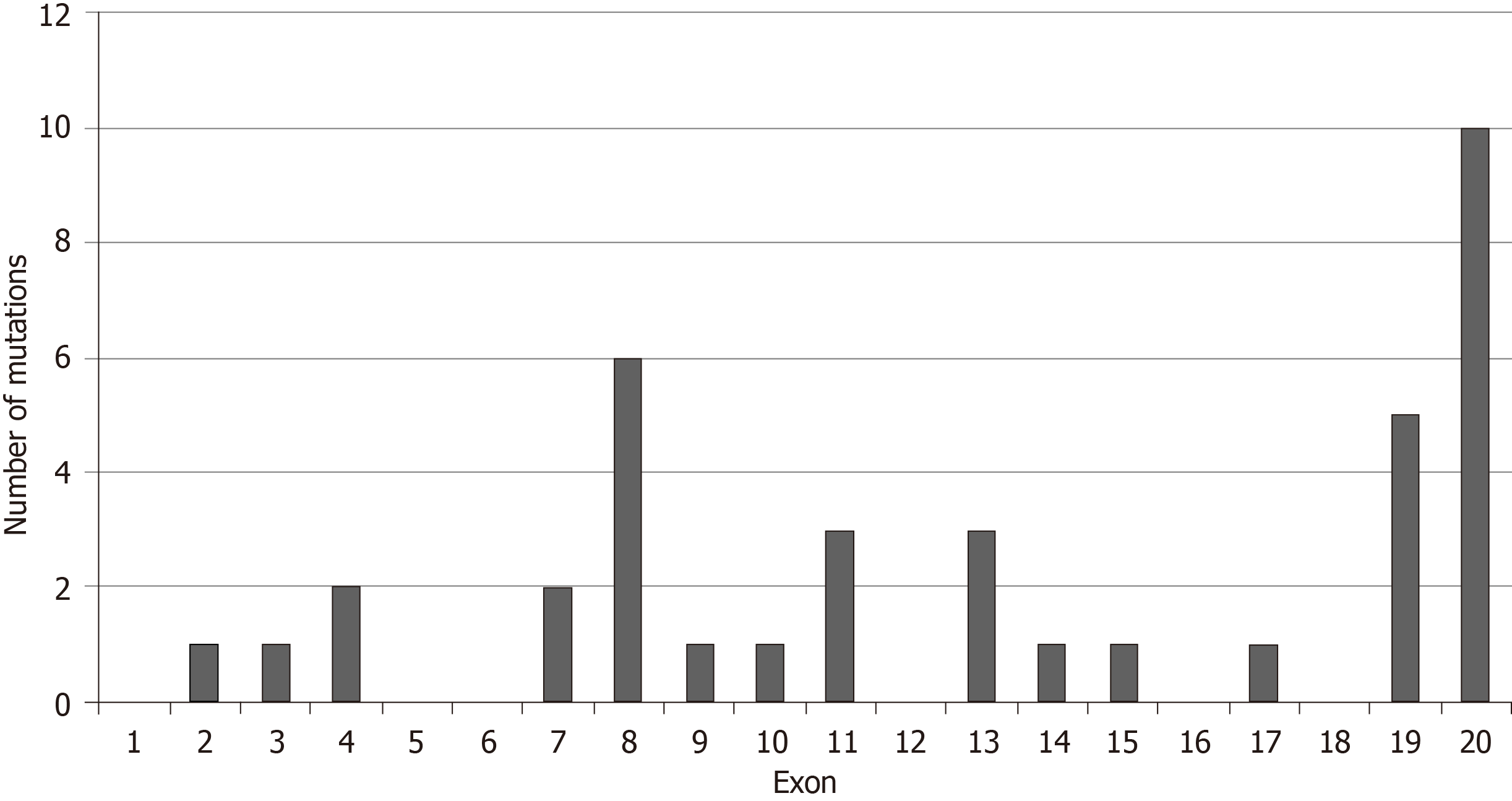
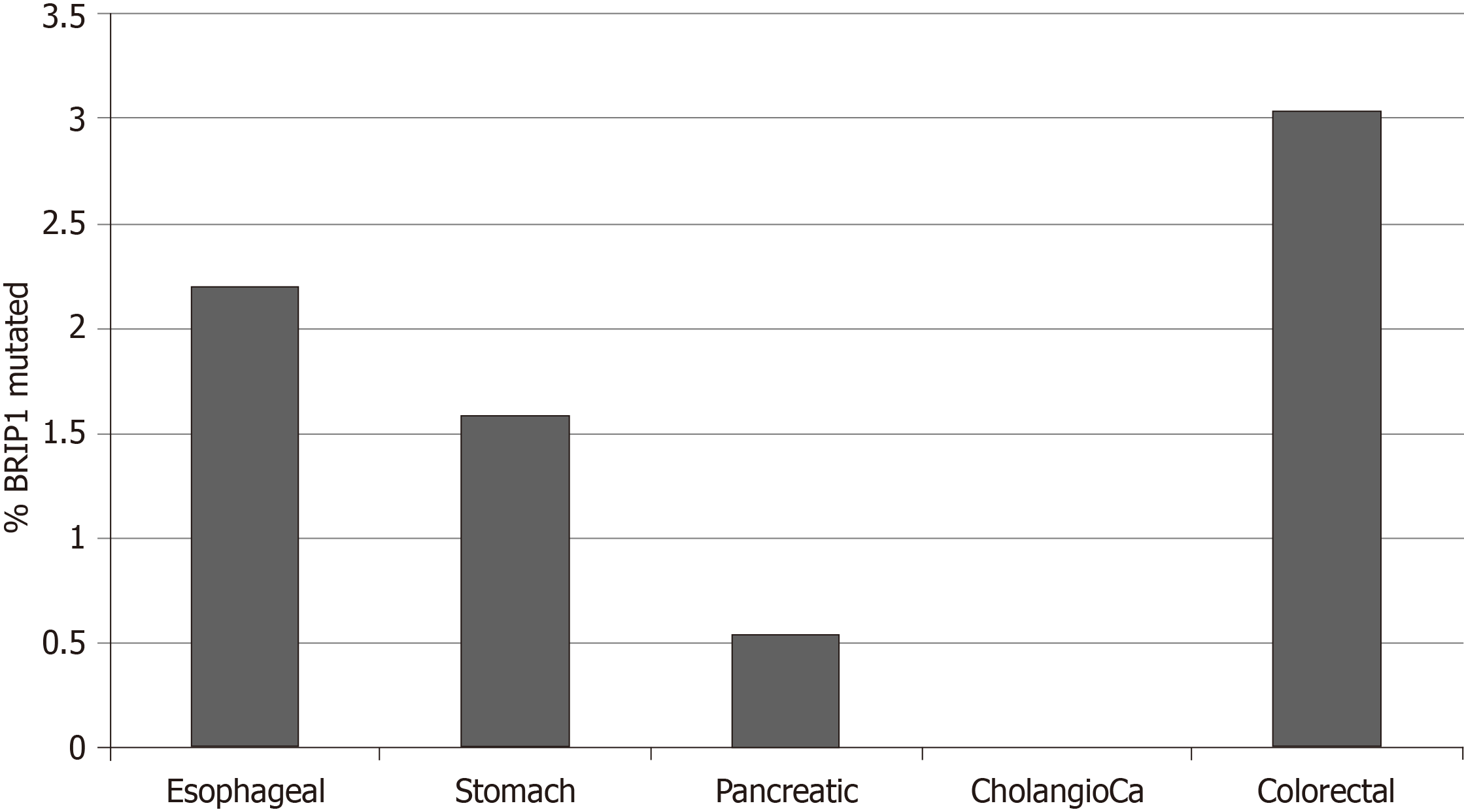
The frequency of BRIP1 mutations was low in the GI cancers examined. Among the 1436 samples included in the 5 interrogated studies, 30 samples (2.1%) had one or more BRIP1 mutations. There was a total of 38 BRIP1 mutations in these 30 samples. The distribution of mutations in the exons of BRIP1 is shown in Figure 1. Six of 38 mutations (15.8%) were listed as likely oncogenic in the OncoKB database (Table 1). These six mutations occur in different aminoacid residues in different exons besides a mutation at aminoacid I504 recurring in two samples and resulting in frame shift and protein truncation shortly thereafter. The remaining four likely oncogenic BRIP1 mutations are nonsense mutations. The incidence of mutated BRIP1 samples in each of the 5 studies was as follows: esophageal cancer 2.2% (4 of 182 cases), gastric cancer 1.6% (7 of 440 cases), pancreatic cancer 0.5% (1 of 184 cases), no mutations observed in the 36 samples of the cholangiocarcinoma study, colorectal cancer 3% (18 of 594 cases) (Figure 2).
| Sample ID | Cancer type | Protein change | Mutation type | Copy number | Allele frequency | Number of mutations | Exon |
| TCGA-A6-3807-01 | Colon adenocarcinoma | S1117* | Nonsense_muta-tion | Diploid | 0.21 | 90 | 20 |
| TCGA-DT-5265-01 | Rectal adenocarcinoma | Q227* | Nonsense_muta-tion | Shallow del | 0.59 | 81 | 7 |
| TCGA-AA-3496-01 | Colon adenocarcinoma | I504Nfs*7 | Frame_Shift_Ins | Gain | 0.43 | 145 | 11 |
| TCGA-AZ-4315-01 | Colon adenocarcinoma | E357* | Nonsense_muta-tion | Diploid | 0.32 | 6317 | 8 |
| TCGA-L5-A4OE-01 | Esophageal adenocarcinoma | Q126* | Nonsense_muta-tion | Gain | 0.35 | 267 | 4 |
| TCGA-CG-5721-01 | Gastric adenocarcinoma | I504Sfs*22 | Frame_Shift_Del | Diploid | 0.22 | 3725 | 11 |
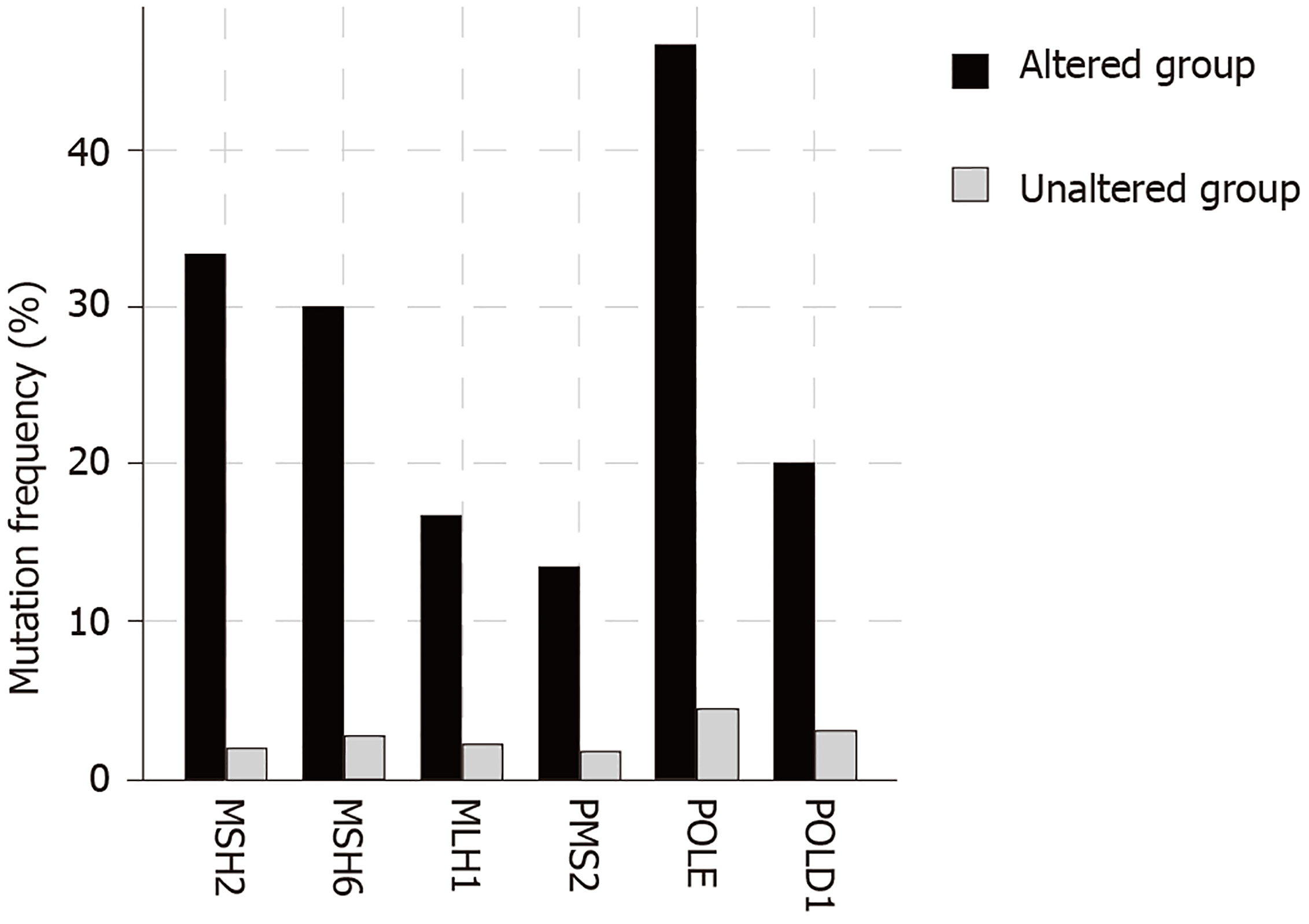
The total number of mutations in BRIP1 mutant samples varied widely ranging between 78 and 11438. The mean and median number of mutations were high (2993.2 and 1747.5 respectively) and 17 of 30 samples with BRIP1 mutations (56.7%) had more than 1000 mutations each. Such a heavy mutation burden is usually observed in cancers with microsatellite instability (MSI) due to mutations in genes that encode for MMR proteins that include MSH2, MSH6, MLH1 and PMS2 or alternatively in cancers with mutations in polymerases ε and δ1 (POLE and POLD1 respectively). Indeed, 18 of the 30 BRIP1 mutated samples (60%) contained one or more co-occurring mutations in one of these six genes. The mean number of mutations in the 18 samples with at least one co-occurring MSI/POLE/POLD1 mutations was 4813 while the mean number of mutations in the 12 samples without any co-occurring MSI/POLE/POLD1 mutations was 262.7. Seventeen of the 18 samples with at least one MSI/ POLE/POLD1 mutation had over 1000 total mutations, while none of the 12 samples with BRIP1 mutations but no MSI/POLE/POLD1 mutation had over 1000 total mutations. Two samples, including the single sample with a BRIP1 mutation in the pancreatic cancer study that contained the higher number of total mutations, an extraordinary 11438, contained mutations in all six MSI/POLE/POLD1 genes. The percentage of mutations in each of the six genes in BRIP1-mutated samples was significantly higher than this percentage in the samples of the 5 studies without BRIP1 mutations (Figure 3). POLE mutations were observed in 14 of the 30 BRIP1 mutant samples (46.7%). Nine of these POLE mutations were deemed likely oncogenic by the OncoKB database, including 4 samples with the known POLE hotspot mutations V411L and 2 samples with P286R/L hotspot mutations. The 5 studies contained 74 samples (5.1%) with mutations in POLE and among those 5 and 4 were the hotspot mutations V411L and P286R/L. Thus, a significant proportion of these characterized deleterious mutations co-occur with BRIP1 mutations. Overall these data suggest that BRIP1 mutations do not cause increased tumor burden but are commonly observed in samples with underlying MSI/POLE/POLD1 mutations and thus a substantial subset of GI cancers with somatically mutated BRIP1 have a high tumor mutation burden.
Among the six samples with likely oncogenic BRIP1 mutations four samples had lower total mutation number (between 81 and 267, Table 1) and three of them had no concomitant MSI/POLE/POLD1 mutations while the fourth, a colorectal cancer sample with a BRIP1 frameshift mutation at I507 had a mutation in MLH1 at L697. These data suggest that likely oncogenic BRIP1 mutations could contribute to cancer pathogenesis without producing hypermutability. Other BRIP1 mutations with unknown significance may be passengers in hyper-mutated cancers.
In colorectal cancer two thirds of BRIP1-mutated samples (12 of 18) contained also mutations in one or more of the commonly mutated genes of the KRAS/BRAF pathway (KRAS/NRAS/BRAF/PTEN/PIK3CA). There was a significant co-occurrence of BRIP1 mutations with mutations in BRAF and PTEN. However, all 12 samples with BRIP1 mutations co-occurring with the five genes of the KRAS/BRAF pathway were hypermutated and contained mutations in either POLE or POLD1 or both. Thus, the presence of BRIP1 mutations in samples with mutations in genes of the KRAS/BRAF pathway may be co-incidental due to the high mutations burden of hypermutated cancers.
Proteins directly interacting with BRIP1 during the DNA repair function include BRCA1 and MLH1. Thus, mutations of these proteins, especially in their BRIP1-interacting domains, or deletions of BRCA1 and MLH1 even in the absence of BRIP1 lesions per se may result in interference with normal function of BRIP1. BRCA1 interacts with BRIP1 through its BRCT domain (aminoacids 1662-1723 and 1757-1842). Mutations in BRCT domain of BRCA1 were observed in only one sample of the total 1436 samples in the 5 studies of GI cancers. Deletions of BRCA1 were also rare, observed in 3 samples. MLH1 interacts with BRIP1 through its carboxyterminal domain (aminoacids 478-744). Mutations in this part of MLH1 are rare, occurring in 10 samples among the 1436 total samples of the 5 GI cancers studies. Deletion of MLH1 occurred in a single sample.
Several other genes of the FA pathway were found to have low mutation frequencies in the 5 studies examined. BRCA2 was the only gene that had a mutation percentage above 3%, specifically 6%. Despite low mutation frequencies, mutations in several of these genes such as BRCA1, BRCA2, FANCI, FANCD2, PALB2, FANCC and RAD51C were all observed to statistically significantly co-occur with BRIP1 mutations (P < 0.001, Q < 0.001).
Comparison of BRIP1 mutations in GI cancers with BRIP1 mutations in breast and ovarian cancer disclosed that in breast cancers BRIP1 mutations are uncommon (10 of 996 samples in the TCGA study of breast cancer, 1%) and contained concomitant MSI/POLE/POLD1 mutations in 3 samples[24]. Similar with GI cancers, mutations of BRIP1 in breast cancers are widely spread in different exons. In the TCGA study of ovarian cancer the 4 of 5 BRIP1 mutated samples were observed in the absence of MSI or POLE/POLD1 mutations and 3 of the 4 samples were concentrated in the DEAD-2 domain (aminoacids 248 to 415)[25].
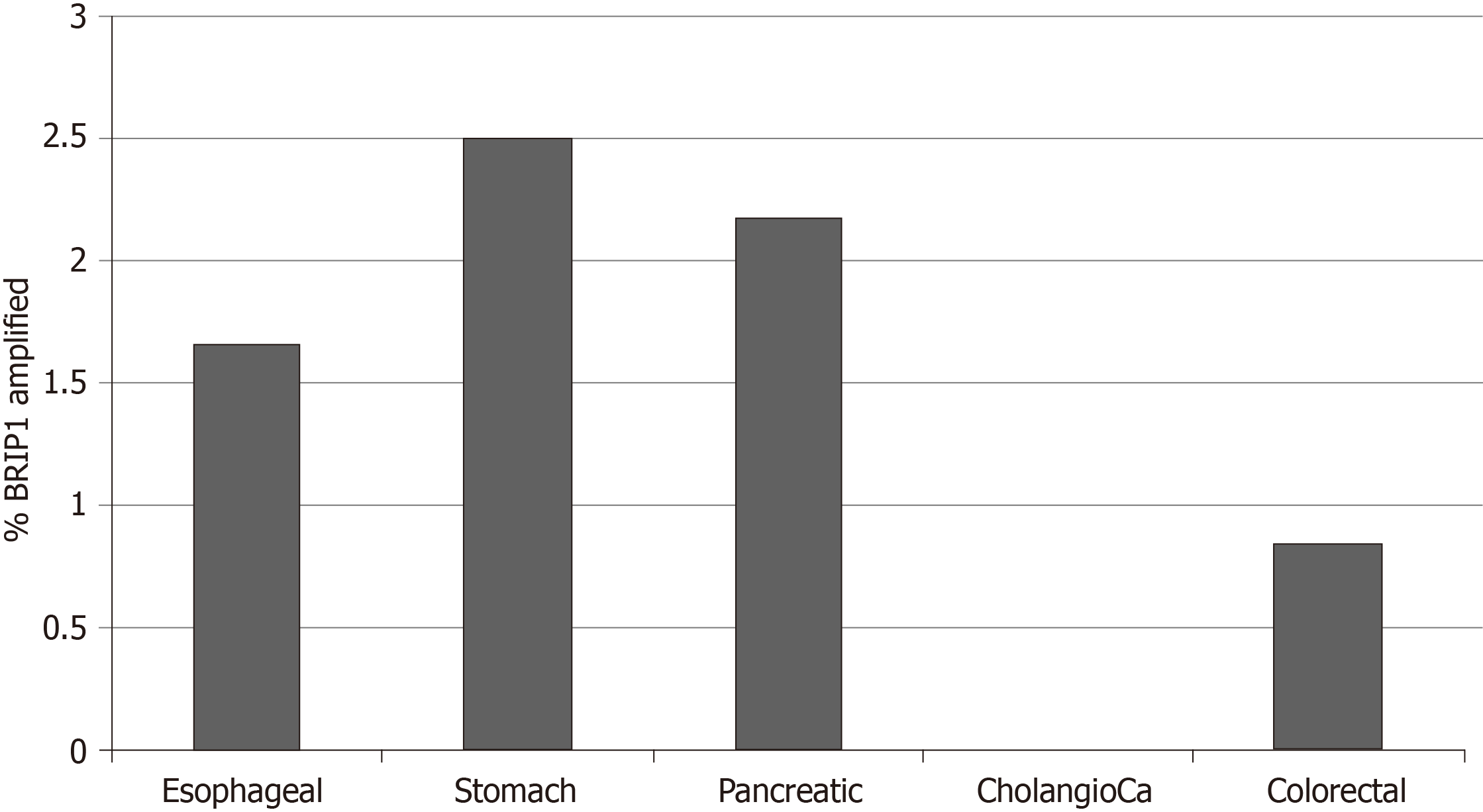
Copy number alterations of BRIP1 were also uncommon in the studies of the GI cancers examined in this analysis and included 23 BRIP1 amplified samples (1.6%) and a single deleted sample which occurred in an esophageal cancer. Percentages of amplified samples in the various cancers are presented in Figure 4. One thousand four hundred twenty-eight genes were co-amplified significantly more often in BRIP1 amplified samples than in BRIP1 non-amplified. Most significant correlations, including the entire list of the top 100 most significantly co-amplified genes were neighboring genes at 17q22-17q24 loci. In gastroesophageal adenocarcinomas, ERBB2 gene located at 17q12 is commonly amplified in about 15% of cases. Co-amplification of ERBB2 was observed in 10 of 14 (71.4%) of BRIP1 amplified gastroesophageal cancer cases (P < 0.001), suggesting that the two genes may be parts of the same amplicon in these cases. As a comparison in breast cancer, where ERBB2 is also commonly amplified, amplification of the two genes co-occurs with statistical significance (P < 0.001) and 36 of the 82 cases (43.9%) with BRIP1 amplifications contained concomitant amplification of ERBB2.
No significant correlations of BRIP1 amplification with mutations in any gene were found in the GI cancers. For example, co-occurrence of BRIP1 amplification with the most commonly mutated tumor suppressor TP53 was observed in 45.8% of BRIP1 amplified samples while 55.6% of BRIP1 non-amplified samples had TP53 mutations (P = 0.22). Similarly, co-occurrence of BRIP1 amplification with the most commonly mutated oncogene KRAS was seen in 16.7% of BRIP1 amplified samples, while 26.6% of BRIP1 non-amplified samples had KRAS mutations (P = 0.19).
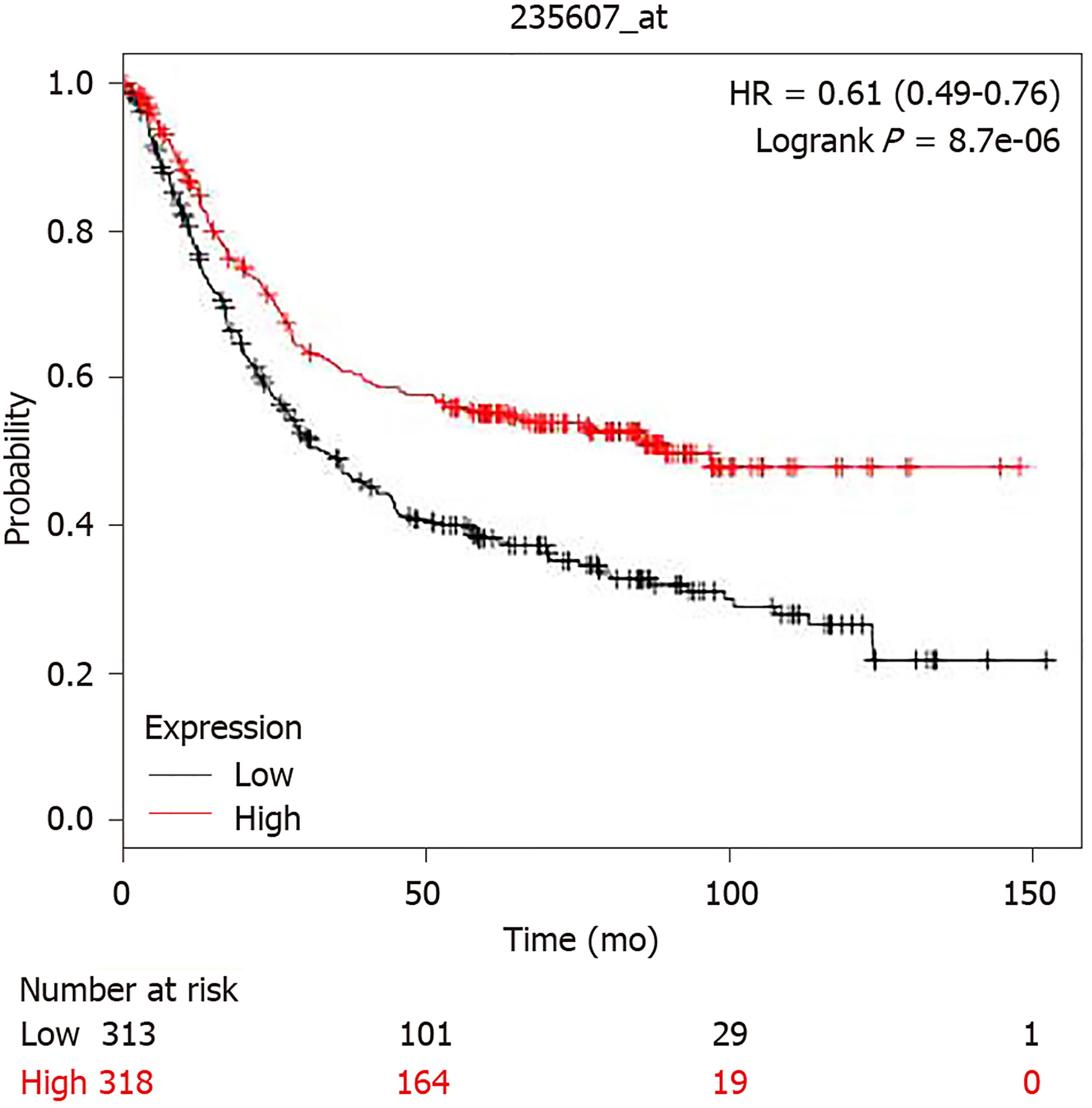
The promoter region of BRIP1 gene (from -499 to 100 from Transcription Start Site) contains 5 binding motif sequences for E2F1 transcription factor at -468, -467, -227, -72 and -71. However, despite this promoter binding potential, E2F and BRIP1 overexpression does not correlate in colorectal cancer (P = 0.13), suggesting that E2F1 activity does not lead to over-expression of its potential target BRIP1. E2F1 was proposed as a part of a panel of genes together with MYBL2 and FOXM1 that may predict tumor aneuploidy[26]. Consistent with the lack of increased BRIP1 expression in tumors with E2F1 over-expression, aneuploidy scores in BRIP1 amplified GI tumors were variable, suggesting that, despite the roles of BRIP1 in DNA repair mechanisms, no direct influence of its abundance with ploidy is evident. However, despite lack of clear association with aneuploidy, increased expression of BRIP1 mRNA (above the median) was associated with improved survival in patients with gastric carcinomas compared with patients whose cancers expressed lower BRIP1 (below the median in the series, Figure 5). Similar results were observed when only patients with localized gastric cancers were included in the survival analysis.
Another potential transcription factor of interest in the regulation of BRIP1 is AP1 (a heterodimer of FOS and JUN) because it is often activated downstream of KRAS/BRAF/MAPK pathway, which is often dysregulated in GI cancers. However, no binding sites of AP1 were present in the BRIP1 promoter.
BRIP1 (alternatively called FANCJ or BACH1) is a protein involved in DNA repair and named for both its interaction with BRCA1 and its being a FA pathway member. It belongs to a family of iron-sulfur helicases together with RTEL1, DDX11 and XPD[27]. As a BRCA1 collaborator, BRIP1 participates in DNA repair of inter-strand cross-links through HR downstream of the core FA complex and following ID2 complex (consisting of proteins FANCI and FANCD2) mono-ubiquitination[28]. Other roles of BRIP1 in DNA lesions metabolism have been revealed more recently. BRIP1 participates in protection of DNA from degradation at a stalled fork[29]. In addition, FANCJ directly interacts with MMR protein MLH1 and participates in bridging MMR complexes with the HR machinery for replication restart after inter-strand cross-links repair[30]. Moreover, a direct role of BRIP1 in resolution of G-quadruplex structures and hairpins arising during replication on single strand DNA, especially in microsatellite sites, has been revealed[31]. Consistent with this last role, cells from FANCJ FA patients show MSI, in contrast to other complementation groups[32].
The current study took advantage of published genomic data by the TCGA and the cBioportal platform as well as other online tools to investigate the role of BRIP1 in common GI cancers. Main findings include the low frequency of BRIP1 defects in GI cancers and a significant association of BRIP1 mutations with defects of MSI/polymerase ε and δ1 genes and the mutator phenotype. In view of the role of BRIP1 helicase in resolution of abnormal DNA structures often affecting microsatellite sites the association is intriguing and may promote MSI. Consistent with this hypothesis, samples with BRIP1 mutations in the five studies had a mean of 4813 mutations while the mean number of mutations in the 83 samples of the colorectal TCGA study, for example, with one or more MSI/POLE/POLD1 mutations was 1734. An alternative hypothesis is that samples with more functionally robust MSI/POLE/POLD1 mutations, producing higher total mutation burden, would contain more commonly passenger BRIP1 mutations.
In pancreatic cancer, where MSI and POLE/POLD1 mutations are rare, BRIP1 mutations are very rare. Specifically, only one mutation was detected in the TCGA pancreatic cancer study. Another more extensive genomic study that included 359 pancreatic adenocarcinoma samples found no BRIP1 mutations in any of them[33].
The partner of BRIP1, BRCA1 is an important player in HR and, in this capacity, it needs to interact with chromatin. BRIP1 stabilizes this interaction. In contrast, oncogenic KRAS promotes down-regulation of BRIP1 and BRCA1 dissociation from chromatin leading to cell senescence[34]. Activating mutations in KRAS or other proteins of the pathway are common in GI cancers and thus may affect DNA repair through impairment of the BRCA1/BRIP1 function. This may imply that KRAS and BRCA1/BRIP1 lesions would be redundant and mutually exclusive. In this study no such mutual exclusivity between BRIP1 mutations and KRAS mutations was observed and in fact a co-occurrence of BRIP1 mutations with mutations of other genes of KRAS pathways was present instead. This may be due to the common association of both BRIP1 and KRAS pathway mutations with MSI/hypermutable cancers or alternatively due to lack of functional repercussions for some of these BRIP1 mutations.
Gastric cancers with BRIP1 mRNA expression above the mean seem to have a better prognosis than counterparts with lower BRIP1 mRNA expression. This may suggest that cancers that up-regulate BRIP1 could have a less aggressive course due to a better ability to repair DNA lesions and possibly decreased genomic lesions accumulation[35].
Despite the fact that the BRIP1 gene promoter area upstream of its transcription start site contains several putative binding motifs for transcription factor E2F1 and the fact that E2F factors have been confirmed to bind and up-regulate BRIP1 in vitro[36], no correlation of the expression of the two genes at the mRNA level in GI cancers was observed in the current interrogation of TCGA studies. This may imply, among other plausible explanations, that other transcription factors are involved in the regulation of BRIP1 obscuring the effect of E2F factors or that increased mRNA expression of E2F does not translate into increased expression of the proteins or increased transcription function. Another candidate transcription factor, AP1, often activated downstream of oncogenic KRAS, was ruled out as a direct regulator of BRIP1 as it possesses no binding sites in BRIP1 promoter.
Overall this study suggests that neutralization of BRIP1 as a tumor suppressor seems to play a minor role in GI cancers pathogenesis. However, a contribution as a defect with cumulative influence in cancers with the mutator phenotype is plausible and may be selected by promoting survival in cells with MMR or polymerase mutations, for example if it would contribute to defects in antigen presentation machinery in hypermutated cancers[37]. The association of BRIP1 with the mutator phenotype is intriguing in the current era of immunotherapy of cancer. If a contribution of BRIP1 to an expansion of instability in hypermutated cancers is confirmed, mutations in the gene may become an additional potential predictive marker of response to immunotherapies. In addition, it may suggest potential avenues for combination therapies, for example with immune checkpoint inhibitors and PARP inhibitors. Indeed, such combinations are in development[38].
Gastrointestinal (GI) cancers are, as a group, very common and their pathogenesis has been progressively elucidated over the last 30 years. However, the role of genetic lesions in homologous recombination (HR) DNA repair remains less well characterized in these cancers.
BRIP1 is a helicase with a role in HR as well as other key functions in DNA metabolism. Its specific role in GI cancers has rarely been reported. Further elucidation of molecular lesions in this gene may pave the way for targeted therapeutic interventions.
To analyze molecular defects of helicase BRIP1 (FANCJ) in GIcancers pathogenesis.
GIcancer studies from The Cancer Genome Atlas (TCGA) were analyzed using the cBioportal platform and other precision medicine databases. TCGA studies were interrogated for BRIP1 mutations and copy number alterations. Associations with other key lesions in GIcancers as well as with the total tumor mutation burden in these cancers were analyzed. Additional analyses that could not be performed directly in the cBioportal platform were performed in Excel (Microsoft Corp., Redmond, WA) after transfer of the relevant data. Appropriate statistical tests (the Fisher’s exact test and the t test respectively) were used for analysis of categorical and continuous data.
Molecular lesions in BRIP1 are observed in 3.6% of GI cancers and consisted almost exclusively of mutations and amplifications. Two fifths of all BRIP1 mutations are considered possibly pathogenic. Most BRIP1 mutated GI cancers have concomitant mutations in MMR genes or one of the replication polymerases, polymerase ε and δ1 genes. No associations were discovered between amplifications of BRIP1 and any mutated genes. BRIP1 amplification commonly co-occurs with ERBB2 amplification, a comparatively common amplification in gastroesophageal cancers.
BRIP1 gene lesions are not major pathogenic players in GI cancers. Association with microsatellite unstable cancers and ERBB2 amplifications in gastroesophageal cancers is worth noting.
Molecular defects in helicase BRIP1, albeit rare, may provide opportunities for novel therapies in GI cancers. Their association with the mutator phenotype is intriguing in the current era of immunotherapy of cancer. BRIP1 defects may contribute to an expansion of instability in hypermutated cancers. Thus, BRIP1 mutations could be an additional potential predictive marker of response to immunotherapies. A role of combination therapies, including immunotherapies with targeted therapies active in cancers with HR defects such as PARP inhibitors, in BRIP defective GI cancers is worth exploring.
| 1. | Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DC, Sgroi DC, Lane WS, Haber DA, Livingston DM. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 489] [Cited by in RCA: 532] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 2. | Bridge WL, Vandenberg CJ, Franklin RJ, Hiom K. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat Genet. 2005;37:953-957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 169] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 3. | Xie J, Guillemette S, Peng M, Gilbert C, Buermeyer A, Cantor SB. An MLH1 mutation links BACH1/FANCJ to colon cancer, signaling, and insight toward directed therapy. Cancer Prev Res (Phila). 2010;3:1409-1416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 4. | Peng M, Litman R, Xie J, Sharma S, Brosh RM, Cantor SB. The FANCJ/MutLalpha interaction is required for correction of the cross-link response in FA-J cells. EMBO J. 2007;26:3238-3249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 152] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 5. | Bharti SK, Awate S, Banerjee T, Brosh RM. Getting Ready for the Dance: FANCJ Irons Out DNA Wrinkles. Genes (Basel). 2016;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 6. | Ramus SJ, Song H, Dicks E, Tyrer JP, Rosenthal AN, Intermaggio MP, Fraser L, Gentry-Maharaj A, Hayward J, Philpott S, Anderson C, Edlund CK, Conti D, Harrington P, Barrowdale D, Bowtell DD, Alsop K, Mitchell G; AOCS Study Group, Cicek MS, Cunningham JM, Fridley BL, Alsop J, Jimenez-Linan M, Poblete S, Lele S, Sucheston-Campbell L, Moysich KB, Sieh W, McGuire V, Lester J, Bogdanova N, Dürst M, Hillemanns P; Ovarian Cancer Association Consortium, Odunsi K, Whittemore AS, Karlan BY, Dörk T, Goode EL, Menon U, Jacobs IJ, Antoniou AC, Pharoah PD, Gayther SA. Germline Mutations in the BRIP1, BARD1, PALB2, and NBN Genes in Women With Ovarian Cancer. J Natl Cancer Inst. 2015;107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 318] [Cited by in RCA: 305] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 7. | Weitzel JN, Neuhausen SL, Adamson A, Tao S, Ricker C, Maoz A, Rosenblatt M, Nehoray B, Sand S, Steele L, Unzeitig G, Feldman N, Blanco AM, Hu D, Huntsman S, Castillo D, Haiman C, Slavin T, Ziv E. Pathogenic and likely pathogenic variants in PALB2, CHEK2, and other known breast cancer susceptibility genes among 1054 BRCA-negative Hispanics with breast cancer. Cancer. 2019;125:2829-2836. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 8. | Velázquez C, Esteban-Cardeñosa EM, Lastra E, Abella LE, de la Cruz V, Lobatón CD, Durán M, Infante M. Unraveling the molecular effect of a rare missense mutation in BRIP1 associated with inherited breast cancer. Mol Carcinog. 2019;58:156-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 9. | Venkateshwari A, Clark DW, Nallari P, Vinod C, Kumarasamy T, Reddy G, Jyothy A, Kumar MV, Ramaiyer R, Palle K. BRIP1/FANCJ Mutation Analysis in a Family with History of Male and Female Breast Cancer in India. J Breast Cancer. 2017;20:104-107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 10. | Pilié PG, Johnson AM, Hanson KL, Dayno ME, Kapron AL, Stoffel EM, Cooney KA. Germline genetic variants in men with prostate cancer and one or more additional cancers. Cancer. 2017;123:3925-3932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 45] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 11. | Kote-Jarai Z, Jugurnauth S, Mulholland S, Leongamornlert DA, Guy M, Edwards S, Tymrakiewitcz M, O'Brien L, Hall A, Wilkinson R, Al Olama AA, Morrison J, Muir K, Neal D, Donovan J, Hamdy F, Easton DF, Eeles R; UKGPCS Collaborators; British Association of Urological Surgeons' Section of Oncology. A recurrent truncating germline mutation in the BRIP1/FANCJ gene and susceptibility to prostate cancer. Br J Cancer. 2009;100:426-430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 54] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 12. | Steinberg-Shemer O, Goldberg TA, Yacobovich J, Levin C, Koren A, Revel-Vilk S, Ben-Ami T, Kuperman AA, Shkalim Zemer V, Toren A, Kapelushnik J, Ben-Barak A, Miskin H, Krasnov T, Noy-Lotan S, Dgany O, Tamary H. Characterization and genotype-phenotype correlation of patients with Fanconi anemia in a multi-ethnic population. Haematologica. 2019;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 13. | Cancer Genome Atlas Research Network; Analysis Working Group: Asan University; BC Cancer Agency; Brigham and Women’s Hospital; Broad Institute; Brown University; Case Western Reserve University; Dana-Farber Cancer Institute; Duke University; Greater Poland Cancer Centre; Harvard Medical School; Institute for Systems Biology; KU Leuven; Mayo Clinic; Memorial Sloan Kettering Cancer Center; National Cancer Institute; Nationwide Children’s Hospital; Stanford University; University of Alabama; University of Michigan; University of North Carolina; University of Pittsburgh; University of Rochester; University of Southern California; University of Texas MD Anderson Cancer Center; University of Washington; Van Andel Research Institute; Vanderbilt University; Washington University; Genome Sequencing Center: Broad Institute; Washington University in St. Louis; Genome Characterization Centers: BC Cancer Agency; Broad Institute; Harvard Medical School; Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins University; University of North Carolina; University of Southern California Epigenome Center; University of Texas MD Anderson Cancer Center; Van Andel Research Institute; Genome Data Analysis Centers: Broad Institute; Brown University:; Harvard Medical School; Institute for Systems Biology; Memorial Sloan Kettering Cancer Center; University of California Santa Cruz; University of Texas MD Anderson Cancer Center; Biospecimen Core Resource: International Genomics Consortium; Research Institute at Nationwide Children’s Hospital; Tissue Source Sites: Analytic Biologic Services; Asan Medical Center; Asterand Bioscience; Barretos Cancer Hospital; BioreclamationIVT; Botkin Municipal Clinic; Chonnam National University Medical School; Christiana Care Health System; Cureline; Duke University; Emory University; Erasmus University; Indiana University School of Medicine; Institute of Oncology of Moldova; International Genomics Consortium; Invidumed; Israelitisches Krankenhaus Hamburg; Keimyung University School of Medicine; Memorial Sloan Kettering Cancer Center; National Cancer Center Goyang; Ontario Tumour Bank; Peter MacCallum Cancer Centre; Pusan National University Medical School; Ribeirão Preto Medical School; St. Joseph’s Hospital &Medical Center; St. Petersburg Academic University; Tayside Tissue Bank; University of Dundee; University of Kansas Medical Center; University of Michigan; University of North Carolina at Chapel Hill; University of Pittsburgh School of Medicine; University of Texas MD Anderson Cancer Center; Disease Working Group: Duke University; Memorial Sloan Kettering Cancer Center; National Cancer Institute; University of Texas MD Anderson Cancer Center; Yonsei University College of Medicine; Data Coordination Center: CSRA Inc; Project Team: National Institutes of Health. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169-175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1458] [Cited by in RCA: 1409] [Article Influence: 156.6] [Reference Citation Analysis (0)] |
| 14. | Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5015] [Cited by in RCA: 5095] [Article Influence: 424.6] [Reference Citation Analysis (4)] |
| 15. | Cancer Genome Atlas Research Network. Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2017;32:185-203.e13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1247] [Cited by in RCA: 1502] [Article Influence: 166.9] [Reference Citation Analysis (13)] |
| 16. | Farshidfar F, Zheng S, Gingras MC, Newton Y, Shih J, Robertson AG, Hinoue T, Hoadley KA, Gibb EA, Roszik J, Covington KR, Wu CC, Shinbrot E, Stransky N, Hegde A, Yang JD, Reznik E, Sadeghi S, Pedamallu CS, Ojesina AI, Hess JM, Auman JT, Rhie SK, Bowlby R, Borad MJ; Cancer Genome Atlas Network, Zhu AX, Stuart JM, Sander C, Akbani R, Cherniack AD, Deshpande V, Mounajjed T, Foo WC, Torbenson MS, Kleiner DE, Laird PW, Wheeler DA, McRee AJ, Bathe OF, Andersen JB, Bardeesy N, Roberts LR, Kwong LN. Integrative Genomic Analysis of Cholangiocarcinoma Identifies Distinct IDH-Mutant Molecular Profiles. Cell Rep. 2017;18:2780-2794. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 333] [Cited by in RCA: 401] [Article Influence: 44.6] [Reference Citation Analysis (1)] |
| 17. | Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330-337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6773] [Cited by in RCA: 6866] [Article Influence: 490.4] [Reference Citation Analysis (10)] |
| 18. | Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9144] [Cited by in RCA: 13506] [Article Influence: 964.7] [Reference Citation Analysis (0)] |
| 19. | Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8187] [Cited by in RCA: 11734] [Article Influence: 902.6] [Reference Citation Analysis (0)] |
| 20. | Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1298] [Cited by in RCA: 1583] [Article Influence: 105.5] [Reference Citation Analysis (0)] |
| 21. | Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, Rudolph JE, Yaeger R, Soumerai T, Nissan MH, Chang MT, Chandarlapaty S, Traina TA, Paik PK, Ho AL, Hantash FM, Grupe A, Baxi SS, Callahan MK, Snyder A, Chi P, Danila D, Gounder M, Harding JJ, Hellmann MD, Iyer G, Janjigian Y, Kaley T, Levine DA, Lowery M, Omuro A, Postow MA, Rathkopf D, Shoushtari AN, Shukla N, Voss M, Paraiso E, Zehir A, Berger MF, Taylor BS, Saltz LB, Riely GJ, Ladanyi M, Hyman DM, Baselga J, Sabbatini P, Solit DB, Schultz N. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol. 2017;2017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1079] [Cited by in RCA: 1378] [Article Influence: 153.1] [Reference Citation Analysis (0)] |
| 22. | Szász AM, Lánczky A, Nagy Á, Förster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A, Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322-49333. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 582] [Cited by in RCA: 765] [Article Influence: 95.6] [Reference Citation Analysis (0)] |
| 23. | Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R, Bessy A, Chèneby J, Kulkarni SR, Tan G, Baranasic D, Arenillas DJ, Sandelin A, Vandepoele K, Lenhard B, Ballester B, Wasserman WW, Parcy F, Mathelier A. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46:D1284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 331] [Cited by in RCA: 281] [Article Influence: 35.1] [Reference Citation Analysis (0)] |
| 24. | Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61-70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9574] [Cited by in RCA: 9735] [Article Influence: 695.4] [Reference Citation Analysis (0)] |
| 25. | Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609-615. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6439] [Cited by in RCA: 6333] [Article Influence: 422.2] [Reference Citation Analysis (0)] |
| 26. | Pfister K, Pipka JL, Chiang C, Liu Y, Clark RA, Keller R, Skoglund P, Guertin MJ, Hall IM, Stukenberg PT. Identification of Drivers of Aneuploidy in Breast Tumors. Cell Rep. 2018;23:2758-2769. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 27. | Estep KN, Brosh RM. RecQ and Fe-S helicases have unique roles in DNA metabolism dictated by their unwinding directionality, substrate specificity, and protein interactions. Biochem Soc Trans. 2018;46:77-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 28. | Niraj J, Färkkilä A, D'Andrea AD. The Fanconi Anemia Pathway in Cancer. Annu Rev Cancer Biol. 2019;3:457-478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 285] [Article Influence: 40.7] [Reference Citation Analysis (0)] |
| 29. | Peng M, Cong K, Panzarino NJ, Nayak S, Calvo J, Deng B, Zhu LJ, Morocz M, Hegedus L, Haracska L, Cantor SB. Opposing Roles of FANCJ and HLTF Protect Forks and Restrain Replication during Stress. Cell Rep. 2018;24:3251-3261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 62] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 30. | Cantor SB, Xie J. Assessing the link between BACH1/FANCJ and MLH1 in DNA crosslink repair. Environ Mol Mutagen. 2010;51:500-507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 31. | Barthelemy J, Hanenberg H, Leffak M. FANCJ is essential to maintain microsatellite structure genome-wide during replication stress. Nucleic Acids Res. 2016;44:6803-6816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 32. | Matsuzaki K, Borel V, Adelman CA, Schindler D, Boulton SJ. FANCJ suppresses microsatellite instability and lymphomagenesis independent of the Fanconi anemia pathway. Genes Dev. 2015;29:2532-2546. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 33. | Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC, Nourse C, Murtaugh LC, Harliwong I, Idrisoglu S, Manning S, Nourbakhsh E, Wani S, Fink L, Holmes O, Chin V, Anderson MJ, Kazakoff S, Leonard C, Newell F, Waddell N, Wood S, Xu Q, Wilson PJ, Cloonan N, Kassahn KS, Taylor D, Quek K, Robertson A, Pantano L, Mincarelli L, Sanchez LN, Evers L, Wu J, Pinese M, Cowley MJ, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chantrill LA, Mawson A, Humphris J, Chou A, Pajic M, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Lovell JA, Merrett ND, Toon CW, Epari K, Nguyen NQ, Barbour A, Zeps N, Moran-Jones K, Jamieson NB, Graham JS, Duthie F, Oien K, Hair J, Grützmann R, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Rusev B, Capelli P, Salvia R, Tortora G, Mukhopadhyay D, Petersen GM; Australian Pancreatic Cancer Genome Initiative, Munzy DM, Fisher WE, Karim SA, Eshleman JR, Hruban RH, Pilarsky C, Morton JP, Sansom OJ, Scarpa A, Musgrove EA, Bailey UM, Hofmann O, Sutherland RL, Wheeler DA, Gill AJ, Gibbs RA, Pearson JV, Waddell N, Biankin AV, Grimmond SMBailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC, Nourse C, Murtaugh LC, Harliwong I, Idrisoglu S, Manning S, Nourbakhsh E, Wani S, Fink L, Holmes O, Chin V, Anderson MJ, Kazakoff S, Leonard C, Newell F, Waddell N, Wood S, Xu Q, Wilson PJ, Cloonan N, Kassahn KS, Taylor D, Quek K, Robertson A, Pantano L, Mincarelli L, Sanchez LN, Evers L, Wu J, Pinese M, Cowley MJ, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chantrill LA, Mawson A, Humphris J, Chou A, Pajic M, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Lovell JA, Merrett ND, Toon CW, Epari K, Nguyen NQ, Barbour A, Zeps N, Moran-Jones K, Jamieson NB, Graham JS, Duthie F, Oien K, Hair J, Grützmann R, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Rusev B, Capelli P, Salvia R, Tortora G, Mukhopadhyay D, Petersen GM; Australian Pancreatic Cancer Genome Initiative, Munzy DM, Fisher WE, Karim SA, Eshleman JR, Hruban RH, Pilarsky C, Morton JP, Sansom OJ, Scarpa A, Musgrove EA, Bailey UM, Hofmann O, Sutherland RL, Wheeler DA, Gill AJ, Gibbs RA, Pearson JV, Waddell N, Biankin AV, Grimmond SM. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2848] [Cited by in RCA: 2725] [Article Influence: 272.5] [Reference Citation Analysis (7)] |
| 34. | Tu Z, Aird KM, Bitler BG, Nicodemus JP, Beeharry N, Xia B, Yen TJ, Zhang R. Oncogenic RAS regulates BRIP1 expression to induce dissociation of BRCA1 from chromatin, inhibit DNA repair, and promote senescence. Dev Cell. 2011;21:1077-1091. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 77] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 35. | Nakanishi R, Kitao H, Fujinaka Y, Yamashita N, Iimori M, Tokunaga E, Yamashita N, Morita M, Kakeji Y, Maehara Y. FANCJ expression predicts the response to 5-fluorouracil-based chemotherapy in MLH1-proficient colorectal cancer. Ann Surg Oncol. 2012;19:3627-3635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 36. | Eelen G, Vanden Bempt I, Verlinden L, Drijkoningen M, Smeets A, Neven P, Christiaens MR, Marchal K, Bouillon R, Verstuyf A. Expression of the BRCA1-interacting protein Brip1/BACH1/FANCJ is driven by E2F and correlates with human breast cancer malignancy. Oncogene. 2008;27:4233-4241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 35] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 37. | Voutsadakis IA. Polymerase epsilon mutations and concomitant β2-microglobulin mutations in cancer. Gene. 2018;647:31-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (1)] |
| 38. | Zimmer AS, Nichols E, Cimino-Mathews A, Peer C, Cao L, Lee MJ, Kohn EC, Annunziata CM, Lipkowitz S, Trepel JB, Sharma R, Mikkilineni L, Gatti-Mays M, Figg WD, Houston ND, Lee JM. A phase I study of the PD-L1 inhibitor, durvalumab, in combination with a PARP inhibitor, olaparib, and a VEGFR1-3 inhibitor, cediranib, in recurrent women's cancers with biomarker analyses. J Immunother Cancer. 2019;7:197. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 93] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Canada
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Chiu KW, Cerwenka H, Hashimoto N, Slomiany BL S-Editor: Dou Y L-Editor: A E-Editor: Liu JH