Published online Dec 14, 2019. doi: 10.3748/wjg.v25.i46.6728
Peer-review started: September 24, 2019
First decision: November 4, 2019
Revised: November 18, 2019
Accepted: November 29, 2019
Article in press: November 29, 2019
Published online: December 14, 2019
Processing time: 81 Days and 4.2 Hours
Long non-coding RNA (lncRNA) is abnormally expressed in various malignant tumors. In recent years, it has been found that IncRNA HULC is increasingly expressed in pancreatic cancer tissues and is involved in the development and progression of pancreatic cancer. However, the clinical value of serum HULC in pancreatic cancer remains unclear, and there are few studies on how HULC regulates the biological function of pancreatic cancer cells.
To determine the value of lncRNA HULC in the diagnosis and prognosis of pancreatic cancer, and its possible biological potential.
Sixty patients with pancreatic cancer and sixty patients with benign pancreatic diseases admitted to Xiangya Hospital, Central South University were assigned to the pancreatic cancer group and the benign disease group, respectively, and another 60 healthy subjects were enrolled as the normal group during the same period. HULC-siRNA and NC-siRNA were transfected into pancreatic cancer cells. Quantitative real-time polymerase chain reaction was performed to determine the expression of HULC in tissues, serum, and cells. Western Blot was carried out to determine the expression of β-catenin, c-myc, and cyclin D1 in cells, and the cell counting kit-8, flow cytometry, and Transwell assay were conducted to determine the proliferation, apoptosis and invasion of cells.
Highly expressed in the tissues and serum of pancreatic cancer patients, HULC showed good clinical value in distinguishing between patients with pancreatic cancer, patients with benign pancreatic diseases and healthy subjects. HULC was related to pathological parameters including tumor size, T staging, M staging and vascular invasion, and the area-under-the-curve for evaluating these four parameters was 0.844, 0.834, 0.928 and 0.818, respectively. Patients with low expression of HULC had a significantly higher 3-year overall survival (OS) and 5-year OS than those with high expression. T staging, M staging, vascular invasion, and HULC were independent prognostic factors affecting the 3-year OS of patients with pancreatic cancer. Inhibition of HULC expression prevented the proliferation and invasion of pancreatic cancer cells, promoted apoptosis, and inhibited the expression of Wnt/β-catenin signaling pathway-related proteins, β-catenin, c-myc, and cyclin D1. The Wnt/β-catenin signaling pathway agonist (LiCl) restored proliferation, apoptosis, and invasion of pancreatic cancer cells with inhibited expression of HULC.
HULC is an effective marker for the diagnosis and prognosis of pancreatic cancer, which may affect the biological function of pancreatic cancer cells through the Wnt/β-catenin signaling pathway.
Core tip: In this study, we evaluated the role of HULC in the diagnosis and prognosis of pancreatic cancer and its possible biological potential. Firstly, we studied the expression of HULC in the serum and tissues of patients with pancreatic cancer. Secondly, we analyzed the relationship between HULC and the clinical pathological parameters and prognosis of patients. Finally, we determined the effects of inhibiting HULC expression on the proliferation, apoptosis and invasion of pancreatic cancer cells and its possible biological potential.
- Citation: Ou ZL, Luo Z, Lu YB. Long non-coding RNA HULC as a diagnostic and prognostic marker of pancreatic cancer. World J Gastroenterol 2019; 25(46): 6728-6742
- URL: https://www.wjgnet.com/1007-9327/full/v25/i46/6728.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i46.6728
Pancreatic cancer, a highly invasive and malignant tumor, is the main cause of cancer death in humans[1]. Surgical resection is effective in patients with early pancreatic cancer, but patients with pancreatic cancer have a low early diagnosis rate due to insidious symptoms in the early stage, and these patients are usually diagnosed at a late stage; thus, their surgical resection rate is relatively low, resulting in poor prognosis and a 5-year overall survival rate (OS) of approximately 6%[2]. Although significant advances in the diagnosis and treatment of tumors have been achieved due to the continuous development of molecular diagnosis and targeted therapy, pancreatic cancer, which is a highly invasive disease, still lacks effective biomarkers for prevention, diagnosis, metastasis and prognosis[3]. Therefore, determining the occurrence, development, and prognosis of pancreatic cancer, and the potential underlying mechanism is helpful for clinicians to assess a more appropriate therapeutic regimen for pancreatic cancer.
Carbohydrate antigen 19-9 (CA199) is an important serum index for the diagnosis of pancreatic cancer clinically, but it lacks certain specificity[4]. Long non-coding RNA (lncRNA) is a non-coding RNA longer than 200bp, whose genomic position is closely related to protein-coding genes[5]. It was reported that most lncRNAs have no RNA transcripts with protein-coding potential, but they are involved in various biological processes in tumors, and play an important role in the cycle, differentiation, proliferation and regulation of cells[6-8]. A growing body of research suggests that lncRNAs are abnormally expressed in various cancers, some of which play an important role in the development and progression of tumor inhibition, and have diagnostic and prognostic value for tumors[9,10]. LncRNA HULC is up-regulated in tumors such as liver cancer and gastric cancer, and it can regulate and control the development and progression of tumors through a variety of molecular mechanisms[11,12]. A previous study indicated that HULC is a promoting factor in pancreatic cancer, and may be a prognostic biomarker for pancreatic cancer[13]. However, the relationship between serum HULC and the diagnosis and prognosis of pancreatic cancer, and its possible molecular mechanism are still under investigation.
Therefore, we investigated the clinical value of HULC in pancreatic cancer and its possible molecular mechanism by detecting the expression of HULC in patients with pancreatic cancer. The aim of this study was to identify a reliable tumor marker for diagnosis and prognosis evaluation and a potential drug target for pancreatic cancer.
Sixty patients with pancreatic cancer and sixty patients with benign pancreatic diseases admitted to Xiangya Hospital, Central South University from January 2012 to March 2014 were assigned to the pancreatic cancer group and the benign disease group, respectively. The pancreatic cancer group consisted of 43 males and 17 females with an average age of 54.9 ± 9.6 years, and the benign disease group consisted of 40 males and 20 females with an average age of 53.2 ± 8.7 years. Additionally, 60 healthy subjects (39 males and 21 females with an average age of 51.3 ± 8.4 years) were enrolled as the normal group during the same period. There was no significant difference in age and sex between the three groups. Inclusion criteria were as follows: Patients diagnosed with pancreatic cancer based on pathology, cytology, and imaging[14], and patients who had not received relevant anti-tumor treatment and who had signed an informed consent form. The experimental process was approved by the Ethics Committee of Xiangya Hospital, Central South University and it was in conformity with the Declaration of Helsinki. Exclusion criteria were as follows: Patients with comorbidities such as liver cirrhosis or blood coagulation dysfunction; patients without detailed general clinical data; patients unwilling to cooperate during follow-up; patients whose expected survival time was less than 1 month, and patients not included in the follow-up period.
The following instruments and reagents were obtained: An ABI Stepone Plusreal-time fluorescence ratio PCR instrument, Lipofectamine™ 2000 transfection kit, Trizol extraction kit, and Annexin V/PI apoptosis detection kit (Invitrogen, Carlsbad, CA, United States); BxPC3, PANC-1, AsPC-1, and CFPAC-1 cells, and hTERT-HPNE normal human pancreatic duct epithelial cells (Shanghai Aolu Biotechnology Co., Ltd., China); SYBR green PCR master mix (Applied Biosystems, Waltham, MA, United States); cell counting kit-8 (AmyJet Scientific Inc., Wuhan, China); 10% fetal bovine serum, mixed penicillin-streptomycin solution (100 × double-antibody), Dulbecco's modified Eagle’s medium (DMEM), and Transwell kit (GibcoTM BRL, Gaithersburg, MD, United States). β-catenin polyclonal goat immunoglobulin G (IgG), cyclin D1 polyclonal goat IgG, c-myc polyclonal goat IgG, β-actin, horseradish peroxidase (HRP)-labeled goat anti-mouse secondary antibody (R&D Systems, Minneapolis, MN, United States); electrochemiluminescence (ECL) kit, bicinchoninic acid (BCA) kit, and Multiskan™ GO full-wavelength enzyme mark instrument (Thermo Fisher Scientific-CN); FACS Canto flow cytometry (Becton Dickinson, Franklin Lakes, NJ, United States); DR5000 ultraviolet visible spectrophotometry (BioRad, Hercules, United States), and Wnt pathway agonist LiCl (Sigma-Aldrich, China). Primer sequences were all synthesized by Shanghai Sangon Biotech Co., Ltd., China.
Tumor tissues and corresponding tumor-adjacent tissues were sampled from patients with pancreatic cancer and stored in liquid nitrogen. Elbow vein blood (5 mL) was obtained from patients in the three groups, and centrifuged at 3000 × g for 10 min to extract serum. The serum was stored for later analysis.
Pancreatic cancer cell lines were placed in DMEM containing 10% fetal bovine serum and mixed penicillin-streptomycin solution, cultured in a cell incubator with saturated humidity and 5% CO2 at a constant temperature of 37 °C, then transfected with HULC-siRNA (siRNA-HULC) and control plasmid NC-siRNA (siRNA-NC), respectively, according to the Lipofectamine™2000 transfection kit instructions. Primers were transfected into cells with the greatest difference in HULC expression, and after 6 h, the cells were cultured in medium containing 10% fetal bovine serum. Quantitative real-time polymerase chain reaction (qRT-PCR) was used to identify the efficiency of cell transfection.
Total RNA in serum, tissues and cells was extracted using Trizol extraction kits. The concentration and purity of the extracted RNA were determined using ultraviolet spectrophotometry, and the integrity of RNA was determined using 1% agarose gel electrophoresis. RNA was reverse transcribed into cDNA according to the reverse transcription kit instructions, and the reverse transcribed samples were stored at -20 °C for later analysis. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was included as an internal reference. The upstream and downstream of HULC were 5’-ACCTCCAGAACTGTGATCCAAAATG-3’, and 5’-TCTTGCTTGATGCTTTGGTCTG-3’, respectively, and those of GAPDH were 5’-CAGCCAGGAGAAATCAAACAG-3’, and 5’-GACTGAGTACCTGAACCGGC-3’, respectively. The reaction was performed on a real-time fluorescence ratio PCR instrument, and the amplification was performed under pre-denaturation at 95 °C for 30 s, followed by 40 cycles of denaturation at 95 °C for 5 s and annealing and elongation at 60 °C for 30 s. Data were obtained from three independent experiments, and the relative expression of genes was determined after calculation using the 2-△△CT method.
Lysed cells were collected and transferred into a centrifuge tube, and centrifuged at 12000 × g for 10 min at 4 °C to collect the supernatant containing the protein samples. The protein concentrations in these samples were determined using the BAC method, and the samples were diluted with lysis buffer to prepare 20 mg/mL protein. In addition, 8.00% separation gel and 5.00% spacer gel were also prepared. The samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to a polyvinylidene fluoride membrane. The samples were added to β-catenin, cyclin D1, and c-myc primary antibodies (1:1000), and internal reference β-actin (1:3000), sealed overnight at 4 °C, and then added to HRP-labeled goat anti-mouse secondary antibody (1:5000), incubated at 37 °C for 1 h, and rinsed with Tris-buffered saline Tween-20 three times, for 5 min each time. The samples were then developed in a darkroom to remove excess liquid on the membrane, and prepared for ECL. The protein bands of the samples were scanned, and their grey values were analyzed using Quantity One (Molecular Devices Corp, The Bay Area, CA, United States).
The proliferation of cells was determined using the cell counting kit-8 (CCK-8) assay as follows: Cells transfected for 48 h were seeded into a 96-well plate at 2 × 103 cells/well, and then determined by adding 100 mL of CCK-8 dilute solution to the plate at 24 h, 48 h, 72 h, and 96 h, respectively. The plate was incubated for 2 h in 5% CO2 and the optical density (OD) of each well was measured at a wavelength of 450 nm using an enzyme mark instrument and repeated three times.
Cells transfected for 48 h were digested with 0.25% trypsin, washed with phosphate buffer saline twice, and then resuspended in 100 μL AnnexinV binding buffer to prepare a 1 × 106 cells/mL suspension. The suspension was incubated at 4 °C for 15 min with 5 μL Annexin-V/FITC solution, and then incubated at 4 °C for 5 min with 5 μL PI staining solution. Flow cytometry was performed three times and the average value was obtained.
A Transwell chamber coated with Matrigel glue was left to stand at 37 °C for 30 min, and then serum-free DMEM was added to resuspend the cells at a cell density of 4 × 105 cells/mL. 200 μL cell suspension was added to the upper chamber, and 800 μL DMEM (10% fetal bovine serum) was added to the lower chamber. The chamber was cultured for 24-48 h, the cells were immobilized with 4% paraformaldehyde, and stained with 0.1% crystal violet, and the cells in the upper chamber were scraped off. Ten high power fields were randomly selected under an optical microscope and the cells in the basement membrane of the chamber were counted. These cells represented the cell invasion ability.
The patients were followed up for 5 years by telephone, WeChat, or clinic medical records once every three months to record their survival. OS refers to the time from the beginning of treatment to death or the last follow-up.
SPSS 20.0 (SPSS, Inc., Chicago, IL, United States) was used for statistical analysis. Data in normal distribution were expressed by the mean ± standard deviation (mean ± SD). Comparisons between groups related to measurement data were carried out using the independent-samples T test, and comparisons of data at multiple time points were performed by the repeated measures analysis of variance. Back-test was performed using the Bonferroni method. Comparisons between multiple groups in means were carried out using one-way ANOVA, and post hoc comparison was performed using the LSD-t test. Receiver operating characteristic (ROC) curves were adopted to assess diagnostic value, and Pearson’s test was used to analyze correlations. The Kaplan-Meier method was used to draw the 3-year OS and 5-year OS curves of patients, and the log-rank test was used to compare these curves. Cox regression was adopted for univariate and multivariate analyses of relevant factors regarding the prognosis of patients. aP < 0.05 indicated a significant difference.
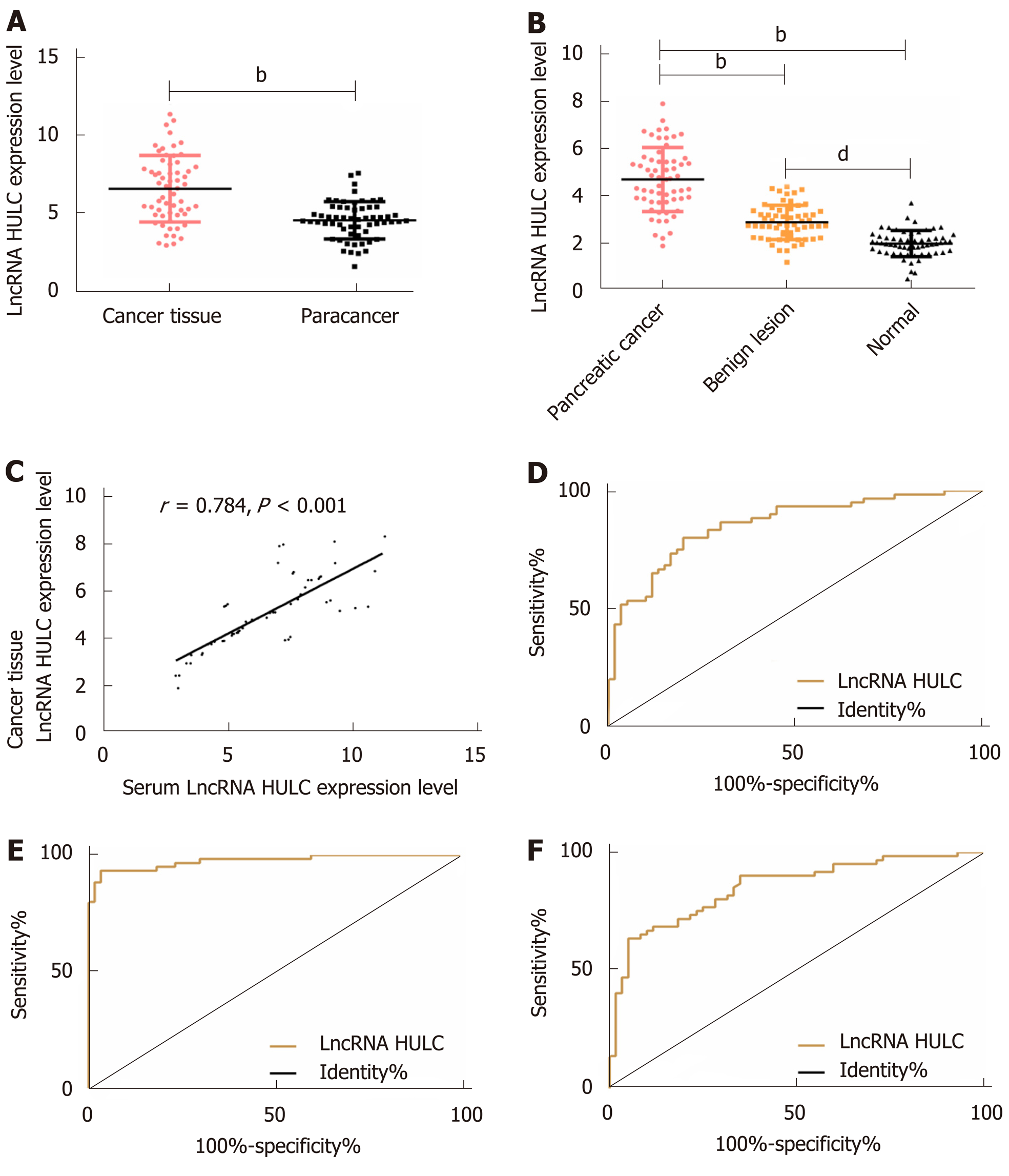
The qRT-PCR results revealed that there were significant differences among the three groups in the expression of HULC in serum (bP < 0.01): The pancreatic cancer group showed significantly higher expression of serum HULC than the benign disease group and the normal group (bP < 0.01), and the benign group showed significantly higher expression of HULC than the normal group (bP < 0.01). Pearson’s test results showed that there was a positive correlation between tissues and serum in the pancreatic cancer group in terms of expression of HULC (r = 0.784, bP < 0.01). ROC curves showed that the area-under-the-curve (AUC), sensitivity, and specificity of HULC in distinguishing patients with pancreatic cancer from those with benign pancreatic diseases were 0.856, 80.00%, and 80.00%, respectively; these values for distinguishing patients with pancreatic cancer from healthy subjects were 0.975, 93.33%, and 96.67%, respectively, and those for distinguishing patients with benign diseases from healthy subjects were 0.850, 63.33%, and 95.00%, respectively (Figure 1 and Table 1).
| Parameter | AUC | 95%CI | SE | Cut-off | Sensitivity (%) | Specificity (%) |
| Pancreatic cancer group-benign disease group | 0.856 | 0.788-0.923 | 0.034 | > 3.690 | 80.00 | 80.00 |
| Pancreatic cancer group-normal group | 0.975 | 0.950-1.000 | 0.012 | > 2.915 | 93.33 | 96.67 |
| Benign disease group-normal group | 0.850 | 0.781-0.919 | 0.035 | > 2.659 | 63.33 | 95.00 |
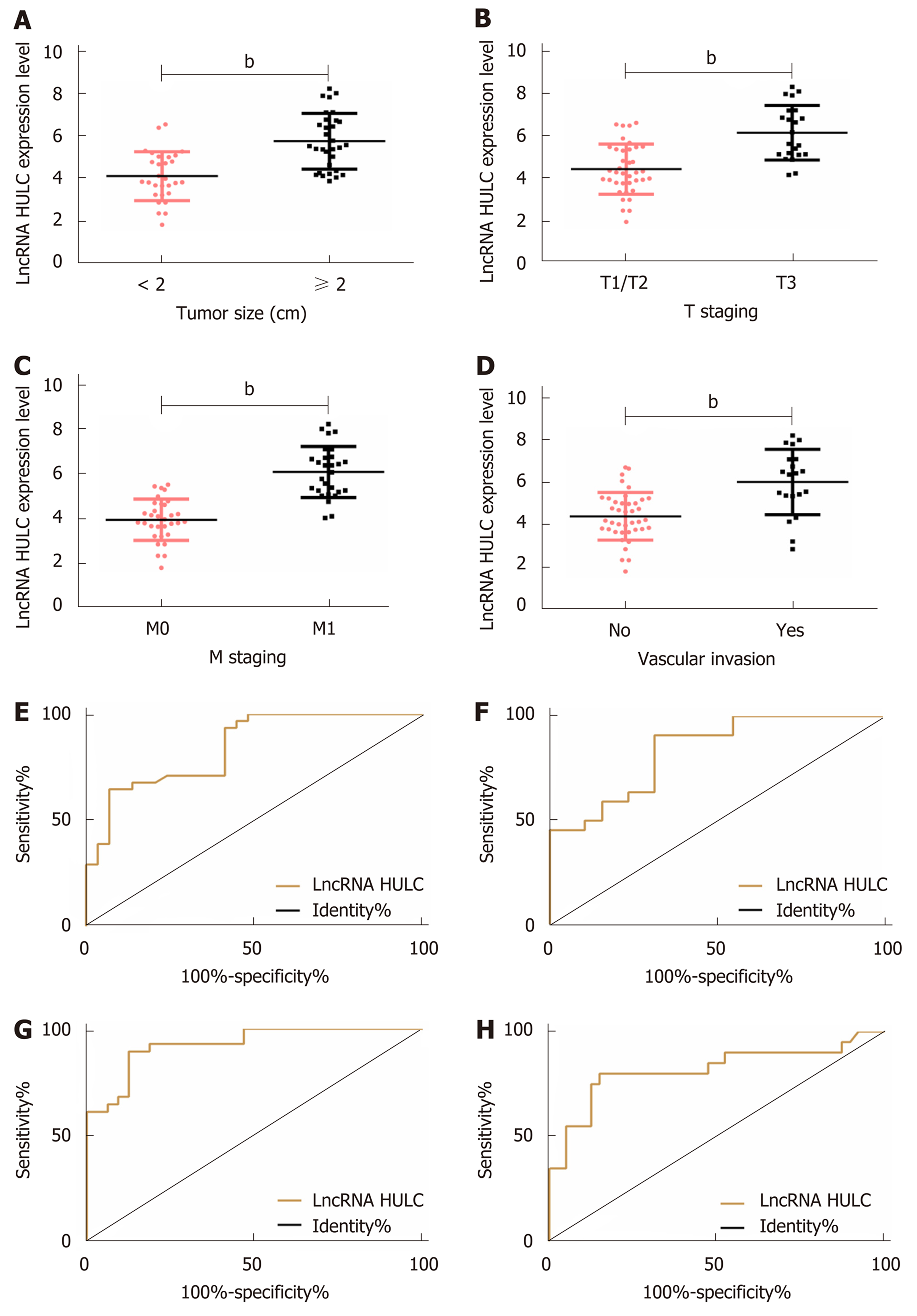
The relationships between serum HULC and pathological parameters in patients with pancreatic cancer were evaluated, and the results revealed that HULC was related to tumor size, T staging, M staging and vascular invasion (all P < 0.01). In addition, ROC curves showed that the AUC of HULC in determining tumor size, T staging, M staging, and vascular invasion was 0.844, 0.834, 0.928 and 0.818, respectively (Tables 2 and 3, Figure 2).
| Clinicopathological parameters | n | HULC | t | P value |
| Gender | 0.016 | 0.987 | ||
| Male | 43 | 4.994 ± 1.520 | ||
| Female | 17 | 4.987 ± 1.451 | ||
| Age (yr) | 0.969 | 0.336 | ||
| < 60 | 35 | 4.834 ± 1.498 | ||
| ≥ 60 | 25 | 5.212 ± 1.477 | ||
| Tumor size (cm) | 3.923 | < 0.001b | ||
| < 2 | 29 | 4.293 ± 1.368 | ||
| ≥ 2 | 31 | 5.900 ± 0.988 | ||
| T staging | 5.248 | < 0.001b | ||
| T1/T2 | 38 | 4.354 ± 1.196 | ||
| T3 | 22 | 6.093 ± 1.304 | ||
| N staging | 1.126 | 0.265 | ||
| N0 | 39 | 4.743 ± 1.497 | ||
| N1 | 21 | 5.286 ± 1.281 | ||
| M staging | 5.120 | < 0.001b | ||
| M0 | 32 | 4.221 ± 1.202 | ||
| M1 | 28 | 5.872 ± 1.295 | ||
| Differentiation | 0.829 | 0.411 | ||
| High/medium | 49 | 4.916 ± 1.420 | ||
| Low | 11 | 5.329 ± 1.802 | ||
| Nerve involvement | 0.401 | 0.690 | ||
| No | 55 | 4.971 ± 1.386 | ||
| Yes | 5 | 5.283 ± 2.876 | ||
| Vascular invasion | 3.660 | < 0.001b | ||
| No | 40 | 4.540 ± 1.324 | ||
| Yes | 20 | 5.896 ± 1.412 | ||
| Smoking history | 0.101 | 0.920 | ||
| No | 36 | 4.976 ± 1.405 | ||
| Yes | 24 | 5.016 ± 1.637 | ||
| History of alcoholism | 0.494 | 0.623 | ||
| No | 42 | 4.929 ± 1.64 | ||
| Yes | 18 | 5.138 ± 1.084 |
| Pathological parameters | AUC | 95%CI | SE | Cut-off | Sensitivity (%) | Specificity (%) |
| Tumor size (cm) | 0.844 | 0.746-0.941 | 0.050 | > 5.408 | 93.10 | 64.52 |
| T staging | 0.834 | 0.732-0.935 | 0.052 | > 4.819 | 90.91 | 68.42 |
| M staging | 0.928 | 0.865-0.990 | 0.032 | > 5.094 | 89.29 | 87.50 |
| Vascular invasion | 0.818 | 0.687-0.949 | 0.067 | > 5.408 | 80.00 | 85.00 |
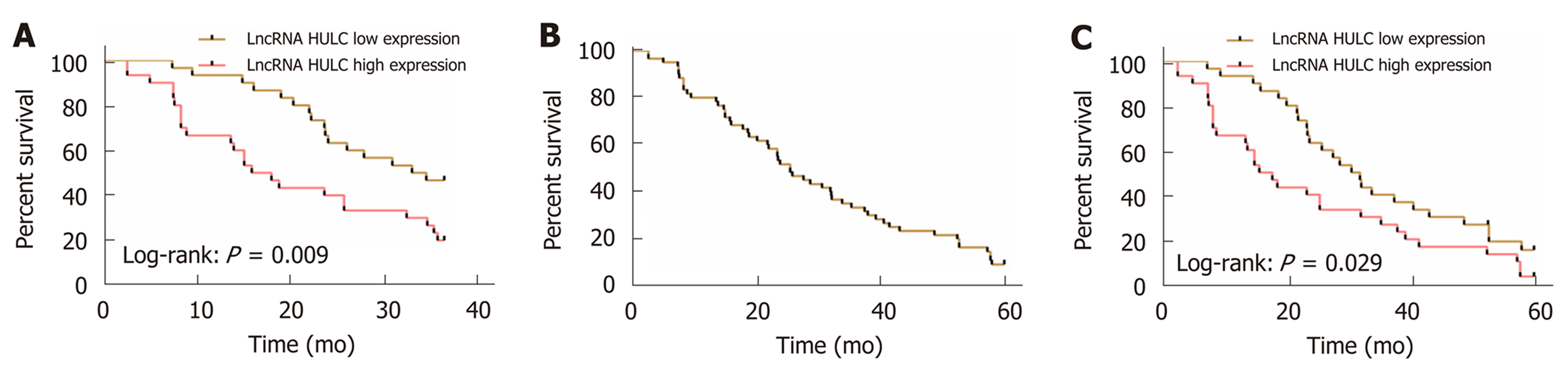
The patients with pancreatic cancer were successfully followed up for 3 or 5 years, and it was found that the 3-year OS and 5-year OS of these patients were 35.00% (21/60), and 10.00% (6/60), respectively. The median serum HULC (4.661) was taken as the cut-point. The 3-year OS and 5-year OS of patients with low expression of HULC were significantly higher than those of patients with high expression (aP < 0.05). Multivariate Cox regression revealed that T staging (aP < 0.01), M staging (aP < 0.05), vascular invasion (aP < 0.05) and HULC (bP < 0.01) were independent prognostic factors affecting the 3-year OS of patients with pancreatic cancer (Figure 3, Tables 4 and 5).
| Factors | Variables | Assignment |
| Gender | X1 | Male = 1, female = 2 |
| Age (yr) | X2 | < 60 = 1, ≥ 60 = 2 |
| Tumor size (cm) | X3 | < 2 = 1, ≥ 2 = 2 |
| T staging | X4 | T1/T2 = 1, T3 = 2 |
| N staging | X5 | N0 = 1, N1 = 2 |
| M staging | X6 | M0 = 1, M1 = 2 |
| Differentiation | X7 | High/medium = 1, low = 2 |
| Nerve involvement | X8 | No = 1, yes = 2 |
| Vascular invasion | X9 | No = 1, yes = 2 |
| Smoking history | X10 | No = 1, yes = 2 |
| History of alcoholism | X11 | No = 1, yes = 2 |
| HULC | X12 | < 4.661 = 1, ≥ 4.661 = 2 |
| Factors | Single factor | Multiple factors | ||
| HR (95CI%) | P value | HR (95CI%) | P value | |
| Gender | 1.195 (0.348-4.176) | 0.773 | ||
| Age (yr) | 1.549 (0.519-4.628) | 0.438 | ||
| Tumor size (cm) | 1.406 (1.092-1.812) | 0.043a | 1.138 (0.857-1.514) | 0.372 |
| T staging | 6.459 (1.408-29.575) | 0.017a | 12.852 (2.297-71.238) | 0.005b |
| N staging | 0.564 (0.201-1.580) | 0.276 | ||
| M staging | 0.292 (0.124-0.689) | 0.005b | 0.357 (0.158-0.735) | 0.013a |
| Differentiation | 0.553 (0.157-2.042) | 0.372 | ||
| Nerve involvement | 1.008 (0.991-1.022) | 0.368 | ||
| Vascular invasion | 6.108 (1.385-24.954) | 0.018a | 2.601 (1.032-6.538) | 0.041a |
| Smoking history | 2.270 (0.901-1.792) | 0.175 | ||
| History of alcoholism | 2.748 (0.705-10.701) | 0.143 | ||
| HULC | 8.356 (2.128-3.846) | 0.002b | 5.817 (2.157-15.756) | 0.003b |
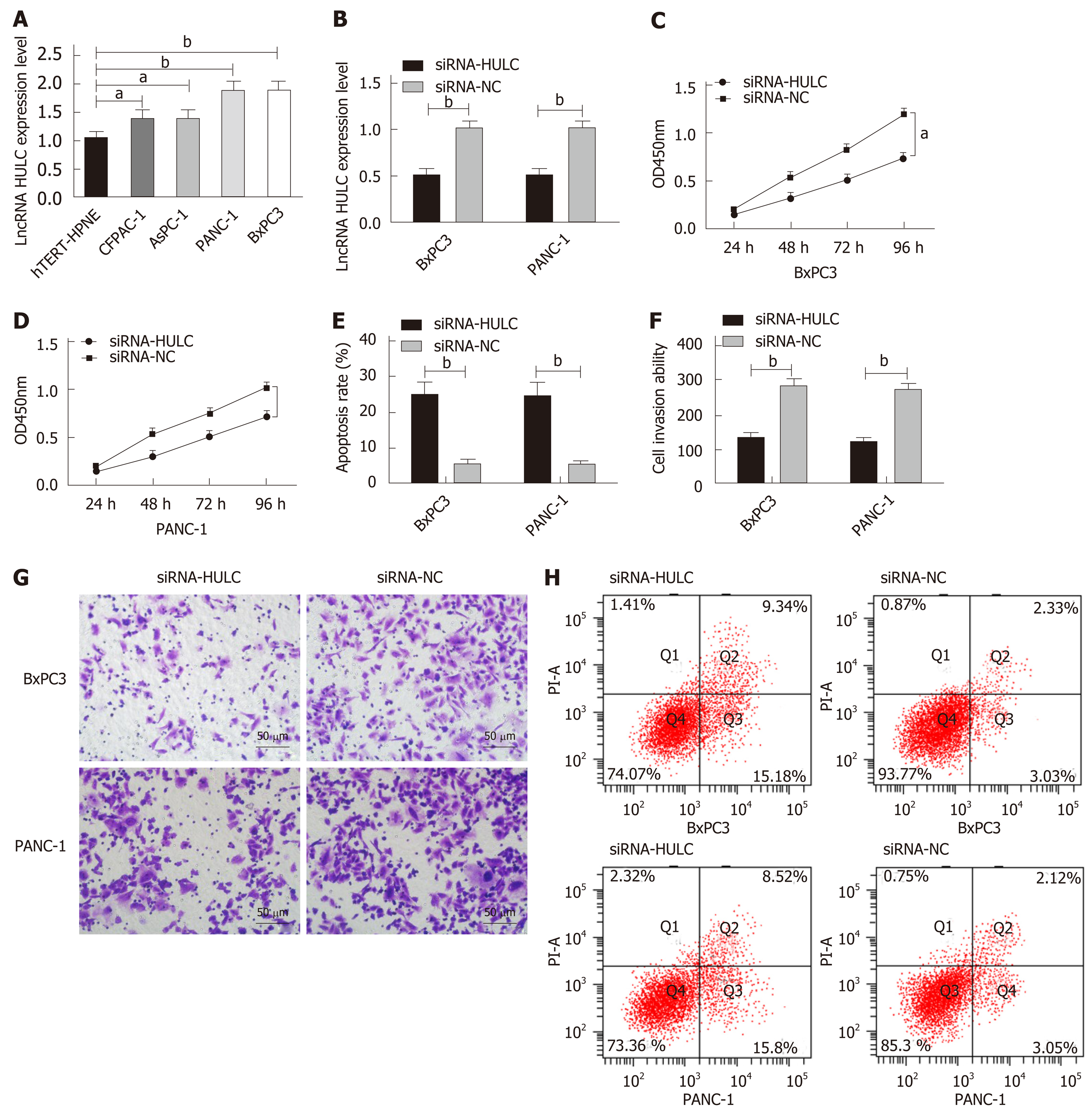
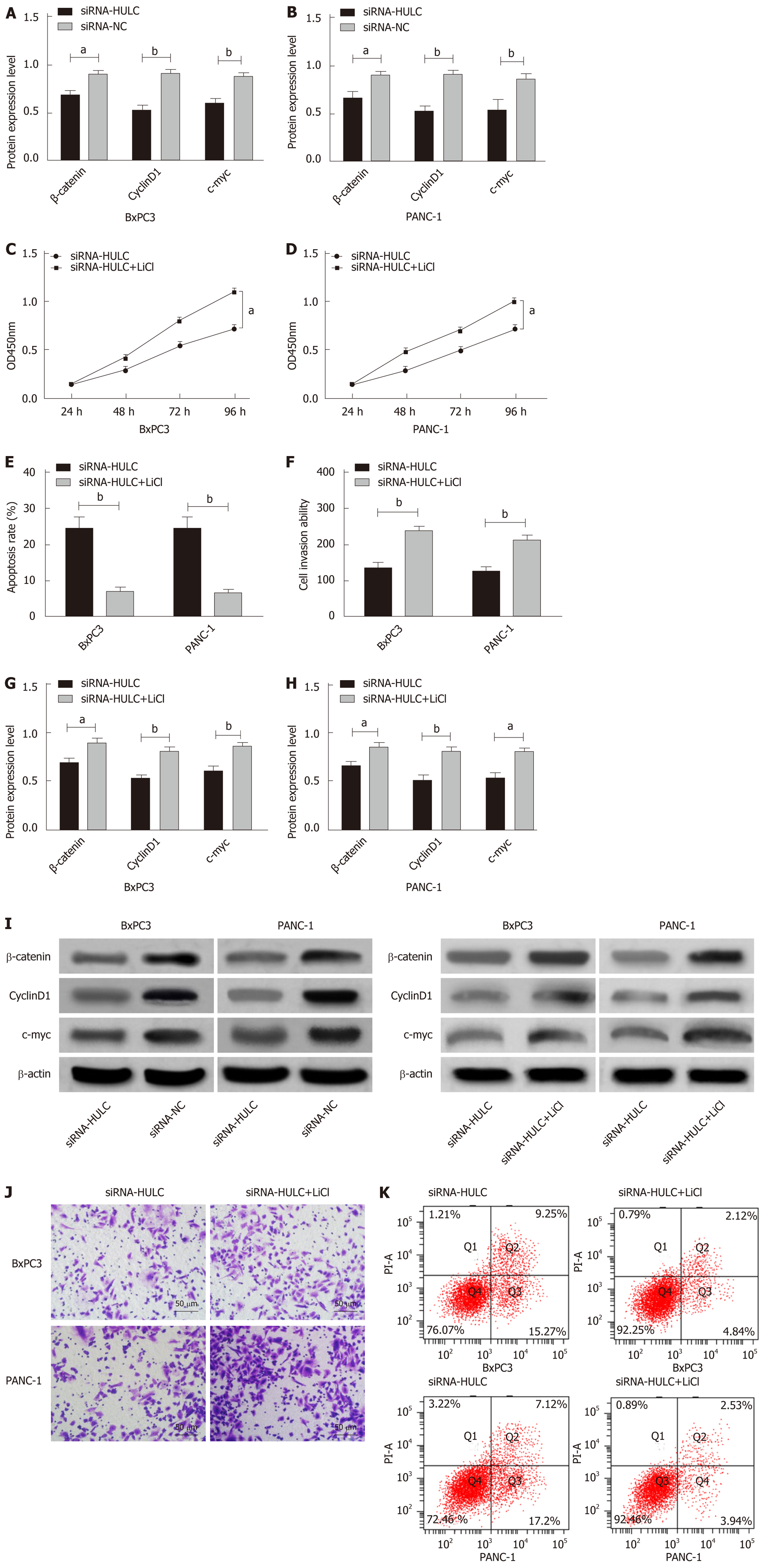
qRT-PCR was performed to determine the expression of HULC in cell lines, and it was found that compared with hTERT-HPNE normal human pancreatic duct epithelial cells, BxPC3, PANC-1, AsPC-1, and CFPAC-1 cells showed significantly increased expression of HULC (all aP < 0.05). BxPC3 and PANC-1 cells with the greatest expression difference were selected for transfection, and after transfection, the siRNA-HULC group showed significantly lower expression of HULC than the siRNA-NC group (bP < 0.01). Compared with the siRNA-NC group, the siRNA-HULC group showed significantly decreased proliferation ability (aP < 0.05), significantly lower apoptosis rate and significantly lower cell invasion ability according to the CCK-8 assay, flow cytometry, and Transwell experiment, respectively (bP < 0.01) (Figure 4).
Western Blot assay results showed that inhibiting the expression of HULC lowered the expression of Wnt/β-catenin signaling pathway-related proteins, β-catenin, cyclin D1 and c-myc, in BxPC3 and PANC-1 cells (all aP < 0.05). In order to further evaluate the role of HULC in regulating the Wnt/β-catenin pathway, the Wnt/β-catenin signaling pathway agonist (LiCl) was added to BxPC3 and PANC-1 cells to inhibit the expression of HULC, and demonstrated that LiCl restored the proliferation, invasion and apoptosis abilities of BxPC3 and PANC-1 cells with inhibited expression of HULC (all aP < 0.05), and caused a significant increase in the expression of β-catenin, cyclin D1 and c-myc proteins in BxPC3 and PANC-1 cells (all aP < 0.05) (Figure 5).
Pancreatic cancer is a highly malignant tumor with high morbidity and mortality[15]. It has no obvious characteristics in the early stage; thus, its diagnostic rate is relatively low, and most patients have advanced stage when diagnosed resulting in a low resection rate, recurrence, metastasis, and poor prognosis[16].
HULC is a lncRNA. Previous studies have confirmed that HULC is closely related to various malignant tumors such as hepatocellular carcinoma and leukemia[17-19]. This study determined the expression of HULC in tissues and serum of patients with pancreatic cancer, and it was found that cancer tissues and serum showed significantly higher expression of HULC than corresponding tumor-adjacent tissues, which indicated that HULC may be a target for the diagnosis and treatment of pancreatic cancer. We then analyzed the relationship between serum HULC and pathological features of pancreatic cancer, and found that HULC was related to tumor size, T staging, M staging and vascular invasion. A study by Peng et al[20] revealed that lncRNA HULC showed significantly up-regulated expression in pancreatic cancer tissues, and was closely related to advanced lymph node metastasis, tumor size, and vascular invasion, which were similar to our findings. In addition, we selected serum rather than pathological tissue as the test sample, as serum was easier to obtain and would cause less trauma. Imaging is the most important detection method for evaluating and identifying pathological parameters of pancreatic cancer such as TNM staging and lymph node metastasis, but the final diagnosis still depends on pathological examination[21,22]. Some existing serological indices such as CA199 lack specificity in diagnosing pancreatic cancer[23]. Based on the ROC curves, we found that HULC had good diagnostic value in distinguishing between patients with pancreatic cancer, patients with benign pancreatic diseases and healthy subjects. We also found that serum HULC had good diagnostic value in tumor size, T staging, M staging and vascular invasion. We followed up patients with pancreatic cancer for 3 or 5 years, and found that the 3-year OS and 5-year OS were 35.00% and 10.00%, respectively. Considering that pancreatic cancer is one of the malignant tumors with the lowest 5-year survival rate in humans, we conducted Cox regression analysis of the 3-year OS in patients with pancreatic cancer, and found that T staging, M staging, vascular invasion, and serum HULC were independent prognostic factors affecting the 3-year OS of these patients. Previous studies have revealed that T staging, M staging and vascular invasion are independent prognostic factors affecting the prognosis of patients with pancreatic cancer[24,25], but this is the first time that serum HULC has been confirmed to be a prognostic factor of pancreatic cancer. We preliminarily confirmed that HULC is expected to be a biomarker for the diagnosis and prognosis of patients with pancreatic cancer.
In order to determine the possible biological potential of HULC in the progression of pancreatic cancer, we transfected HULC into cell lines and found that the proliferation and invasion abilities of cells transfected with HULC were significantly inhibited and the apoptosis rate was significantly increased, which suggested that downregulation of HULC expression could inhibit the proliferation and invasion of pancreatic cancer cells and promote apoptosis. However, it is not clear which pathway is involved in HULC regulation. The Wnt/β-catenin signal transduction pathway can regulate cellular processes in tumors such as proliferation, invasion and other signaling pathways by regulating β-catenin ability[26,27]. A previous study indicated that lncRNA CUDR can control the malignant differentiation of human liver stem cells by regulating the CUDR-HULC/CUDR-β-catenin signal transduction pathway[28], and a study by Zhao et al[29] showed that lncRNA HNF1A-AS1 can accelerate proliferation and metastasis of osteosarcoma cells by activating the Wnt/β-catenin pathway. In order to understand the specific regulation mechanism of HULC in pancreatic cancer progression, we performed Western Blot assay to determine the Wnt/β-catenin signaling pathway-related proteins following inhibited expression of HULC, to assess whether down-regulating the expression of HULC affects the Wnt/β-catenin signaling pathway and to evaluate its role in tumor occurrence and development. The results showed that inhibiting the expression of HULC lowered the expression of β-catenin, cyclin D1 and c-myc. In order to further verify the role of HULC in regulating the Wnt/β-catenin pathway, LiCl was added to pancreatic cancer cells, which showed that LiCl restored proliferation, invasion and apoptosis abilities in BxPC3 and PANC-1 cells with inhibited expression of HULC, and caused a significant increase in expression of β-catenin, cyclin D1 and c-myc proteins in these cells. Therefore, inhibiting HULC may inhibit proliferation and invasion of pancreatic cancer cells and promote apoptosis by inhibiting the Wnt/β-catenin signaling pathway.
However, our study has certain limitations. We only preliminarily studied the effects of inhibiting HULC on the biological function of pancreatic cancer cells, and it is unclear which target gene HULC regulated the biological function. In addition, we did not conduct tumor formation in nude mice; thus, the tumor killing effect of HULC administration is unclear. Further investigations are required to assess whether HULC may be a potential therapeutic target for pancreatic cancer.
In conclusion, HULC is an effective marker for the diagnosis and prognosis of pancreatic cancer, which may affect the biological function of pancreatic cancer cells through the Wnt/β-catenin signaling pathway.
Pancreatic cancer is a highly invasive malignant tumor in humans, which is relatively insidious in the early state and is usually confirmed in the late stage. Although significant progress has been made in the molecular diagnosis and treatment of tumors, pancreatic cancer still lacks effective biomarkers for prevention, diagnosis and prognosis. Thus, the identification of potential biomarkers related to the development, progression and prognosis of pancreatic cancer would be helpful for the diagnosis and treatment of pancreatic cancer.
Long-chain non-coding RNAs (lncRNAs) are involved in the development and progression of pancreatic cancer. In this study, we investigated the clinical value of lncRNA HULC in pancreatic cancer and determined its role in the progression of pancreatic cancer using in vitro experiments. However, the possible regulatory mechanism of HULC has not been well described.
This study aimed to determine the clinical value of HULC in pancreatic cancer and to identify its possible regulatory mechanism.
RT-qPCR was employed to determine the expression of HULC in pancreatic cancer tissues, serum and cells. ROC curves were used to evaluate the clinical diagnostic value of serum HULC for pancreatic cancer, and Cox regression analysis was used to analyze prognostic factors in pancreatic cancer patients. Additionally, the CCK-8, flow cytometry, and Transwell assay were conducted to evaluate the proliferation, apoptosis, and invasion of pancreatic cancer cells. We found that HULC affected the biological function of pancreatic cancer cells through the Wnt/β-catenin signaling pathway.
Our results revealed that HULC was up-regulated in pancreatic cancer tissues, serum, and cells, and was an effective marker for diagnosis and prognosis of the disease. In addition, down-regulation of HULC inhibited the proliferation and invasion of pancreatic cancer cells and induced apoptosis by inhibiting the Wnt/β-catenin signaling pathway.
Our study confirmed the clinical value of HULC in pancreatic cancer for the first time. HULC can affect the biological function of pancreatic cancer cells through the Wnt/β-catenin signaling pathway. Therefore, HULC may play an important role in the development and progression of pancreatic cancer. These results can provide a theoretical basis for the diagnosis, treatment and prognosis of pancreatic cancer.
Our research has proved the clinical value of HULC in pancreatic cancer and its possible regulatory mechanism. Future research may focus on the biological functions of potential target genes of HULC in vitro and in vivo, and the diagnostic and therapeutic effects of HULC in pancreatic cancer require verification in clinical practice.
| 1. | Zhou M, Diao Z, Yue X, Chen Y, Zhao H, Cheng L, Sun J. Construction and analysis of dysregulated lncRNA-associated ceRNA network identified novel lncRNA biomarkers for early diagnosis of human pancreatic cancer. Oncotarget. 2016;7:56383-56394. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 121] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 2. | Li DD, Fu ZQ, Lin Q, Zhou Y, Zhou QB, Li ZH, Tan LP, Chen RF, Liu YM. Linc00675 is a novel marker of short survival and recurrence in patients with pancreatic ductal adenocarcinoma. World J Gastroenterol. 2015;21:9348-9357. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 28] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 3. | Wu B, Wang K, Fei J, Bao Y, Wang X, Song Z, Chen F, Gao J, Zhong Z. Novel three‑lncRNA signature predicts survival in patients with pancreatic cancer. Oncol Rep. 2018;40:3427-3437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 4. | Kishikawa T, Otsuka M, Ohno M, Yoshikawa T, Takata A, Koike K. Circulating RNAs as new biomarkers for detecting pancreatic cancer. World J Gastroenterol. 2015;21:8527-8540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 122] [Cited by in RCA: 120] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 5. | Liu WT, Lu X, Tang GH, Ren JJ, Liao WJ, Ge PL, Huang JF. LncRNAs expression signatures of hepatocellular carcinoma revealed by microarray. World J Gastroenterol. 2014;20:6314-6321. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 21] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 6. | Ou ZL, Zhang M, Ji LD, Luo Z, Han T, LuYB, Li YX. Long noncoding RNA FEZF1-AS1 predicts poor prognosis and modulatespancreatic cancer cell proliferation and invasion through miR-142/HIF-1alphaand miR-133a/EGFR upon hypoxia/normoxia. J Cell Physiol. 2019;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 7. | Huang X, Zhi X, Gao Y, Ta N, Jiang H, Zheng J. LncRNAs in pancreatic cancer. Oncotarget. 2016;7:57379-57390. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 83] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 8. | Wei W, Liu Y, Lu Y, Yang B, Tang L. LncRNA XIST Promotes Pancreatic CancerProliferation Through miR-133a/EGFR. J Cell Biochem. 2017;118:3349-3358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 168] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 9. | Qu S, Yang X, Song W, Sun W, Li X, Wang J, Zhong Y, Shang R, Ruan B, Zhang Z, Zhang X, Li H. Downregulation of lncRNA-ATB correlates with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 2016;37:3933-3938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 10. | Giulietti M, Righetti A, Principato G, Piva F. LncRNA co-expression network analysis reveals novel biomarkers for pancreatic cancer. Carcinogenesis. 2018;39:1016-1025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 123] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 11. | Li SP, Xu HX, Yu Y, He JD, Wang Z, Xu YJ, Wang CY, Zhang HM, Zhang RX, Zhang JJ, Yao Z, Shen ZY. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget. 2016;7:42431-42446. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 174] [Cited by in RCA: 219] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 12. | Zhang Y, Song X, Wang X, Hu J, Jiang L. Silencing of LncRNA HULC Enhances Chemotherapy Induced Apoptosis in Human Gastric Cancer. J Med Biochem. 2016;35:137-143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 64] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 13. | Yu X, Zheng H, Chan MT, Wu WK. HULC: an oncogenic long non-coding RNA in human cancer. J Cell Mol Med. 2017;21:410-417. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 102] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 14. | Freelove R, Walling AD. Pancreatic cancer: diagnosis and management. Am Fam Physician. 2006;73:485-492. [PubMed] |
| 15. | Peng JF, Zhuang YY, Huang FT, Zhang SN. Noncoding RNAs and pancreatic cancer. World J Gastroenterol. 2016;22:801-814. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 48] [Cited by in RCA: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 16. | Xie Z, Chen X, Li J, Guo Y, Li H, Pan X, Jiang J, Liu H, Wu B. Salivary HOTAIR and PVT1 as novel biomarkers for early pancreatic cancer. Oncotarget. 2016;7:25408-25419. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 128] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 17. | Chen X, Lun L, Hou H, Tian R, Zhang H, Zhang Y. The Value of lncRNA HULC as a Prognostic Factor for Survival of Cancer Outcome: A Meta-Analysis. Cell Physiol Biochem. 2017;41:1424-1434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 18. | Lu Y, Li Y, Chai X, Kang Q, Zhao P, Xiong J, Wang J. Long noncoding RNA HULC promotes cell proliferation by regulating PI3K/AKT signaling pathway in chronic myeloid leukemia. Gene. 2017;607:41-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 64] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 19. | Peng W, Wu J, Feng J. Long noncoding RNA HULC predicts poor clinical outcome and represents pro-oncogenic activity in diffuse large B-cell lymphoma. Biomed Pharmacother. 2016;79:188-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 20. | Peng W, Gao W, Feng J. Long noncoding RNA HULC is a novel biomarker of poor prognosis in patients with pancreatic cancer. Med Oncol. 2014;31:346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 127] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 21. | Court CM, Hines OJ. The New American Joint Committee on Cancer TNM Staging System for Pancreatic Cancer-Balancing Usefulness With Prognostication. JAMA Surg. 2018;153:e183629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 22. | Adsay NV, Bagci P, Tajiri T, Oliva I, Ohike N, Balci S, Gonzalez RS, Basturk O, Jang KT, Roa JC. Pathologic staging of pancreatic, ampullary, biliary, and gallbladder cancers: pitfalls and practical limitations of the current AJCC/UICC TNM staging system and opportunities for improvement. Semin Diagn Pathol. 2012;29:127-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 98] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 23. | Shi W, Lu Y, Gong R, Sun JJ, Liu G. Serum miR-629 is a novel molecular marker for diagnosis and the prognosis of pancreatic cancer. Eur Rev Med Pharmacol Sci. 2018;22:5187-5193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 24. | Ma YB, Li GX, Hu JX, Liu X, Shi BM. Correlation of miR-494 expression with tumor progression and patient survival in pancreatic cancer. Genet Mol Res. 2015;14:18153-18159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 25. | Basturk O, Saka B, Balci S, Postlewait LM, Knight J, Goodman M, Kooby D, Sarmiento JM, El-Rayes B, Choi H, Bagci P, Krasinskas A, Quigley B, Reid MD, Akkas G, Maithel SK, Adsay V. Substaging of Lymph Node Status in Resected Pancreatic Ductal Adenocarcinoma Has Strong Prognostic Correlations: Proposal for a Revised N Classification for TNM Staging. Ann Surg Oncol. 2015;22 Suppl 3:S1187-S1195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 66] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 26. | Wang ZY, Hu M, Dai MH, Xiong J, Zhang S, Wu HJ, Zhang SS, Gong ZJ. Upregulation of the long non-coding RNA AFAP1-AS1 affects the proliferation, invasion and survival of tongue squamous cell carcinoma via the Wnt/β-catenin signaling pathway. Mol Cancer. 2018;17:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 27. | Pei Z, Du X, Song Y, Fan L, Li F, Gao Y, Wu R, Chen Y, Li W, Zhou H, Yang Y, Zeng J. Down-regulation of lncRNA CASC2 promotes cell proliferation and metastasis of bladder cancer by activation of the Wnt/β-catenin signaling pathway. Oncotarget. 2017;8:18145-18153. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 92] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 28. | Gui X, Li H, Li T, Pu H, Lu D. Long Noncoding RNA CUDR Regulates HULC and β-Catenin to Govern Human Liver Stem Cell Malignant Differentiation. Mol Ther. 2015;23:1843-1853. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 68] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 29. | Zhao H, Hou W, Tao J, Zhao Y, Wan G, Ma C, Xu H. Upregulation of lncRNA HNF1A-AS1 promotes cell proliferation and metastasis in osteosarcoma through activation of the Wnt/β-catenin signaling pathway. Am J Transl Res. 2016;8:3503-3512. [PubMed] |
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:
P-Reviewer: Chow WK, Dragoteanu MN S-Editor: Wang J L-Editor: Webster JR E-Editor: Zhang YL