Published online Nov 21, 2019. doi: 10.3748/wjg.v25.i43.6386
Peer-review started: January 16, 2019
First decision: February 13, 2019
Revised: September 10, 2019
Accepted: September 13, 2019
Article in press: September 13, 2019
Published online: November 21, 2019
Processing time: 309 Days and 18.3 Hours
Hepatocellular carcinoma (HCC) is now the most common primary liver malignancy worldwide, and multiple risk factors attribute to the occurrence and development of HCC. Recently, increasing studies suggest that ubiquitin-conjugating enzyme E2T (UBE2T) serves as a promising prognostic factor in human cancers, although the molecular mechanism of UBE2T in HCC remains unclear.
To investigate the clinical relevance and role of UBE2T in HCC development.
UBE2T expression in HCC tissues from the TCGA database and its association with patient survival were analyzed. A lentivirus-mediated strategy was used to knock down UBE2T in HCC cells. qRT-PCR and Western blot assays were performed to check the effect of UBE2T silencing in HCC cells. Cell growth in vitro and in vivo was analyzed by multiparametric high-content screening and the xenograft tumorigenicity assay, respectively. Cell cycle distribution and apoptosis were determined by flow cytometry. The genes regulated by UBE2T were profiled by microarray assay.
UBE2T was overexpressed in HCC tissues compared with paired and non-paired normal tissues. High expression of UBE2T predicted a poor overall survival in HCC patients. In vitro, lentivirus-mediated UBE2T knockdown significantly reduced the viability of both SMMC-7721 and BEL-7404 cells. In vivo, the xenograft tumorigenesis of SMMC-7721 cells was largely attenuated by UBE2T silencing. The cell cycle was arrested at G1/S phase in SMMC-7721 and BEL-7404 cells with UBE2T knockdown. Furthermore, apoptosis was increased by UBE2T knockdown. At the molecular level, numerous genes were dysregulated after UBE2T silencing, including IL-1B, FOSL1, PTGS2, and BMP6.
UBE2T plays an important role in cell cycle progression, apoptosis, and HCC development.
Core tip: Ubiquitin-conjugating enzyme E2T (UBE2T) is a member of the ubiquitin-proteasome family. Although it has been reported that UBE2T promotes the growth of hepatocellular carcinoma (HCC) cells, the role of UBE2T in the HCC cell cycle, apoptosis, and tumorigenesis is unclear. UBE2T was highly expressed in HCC tissues, and its expression was correlated with the survival of HCC patients. Silencing of UBE2T reduced the viability and xenograft tumor growth of HCC cells. Additionally, cell cycle arrest and apoptosis were induced by UBE2T knockdown. Numerous genes were regulated by UBE2T silencing in HCC cells. Therefore, UBE2T is a novel diagnostic and therapeutic target for HCC.
- Citation: Guo J, Wang M, Wang JP, Wu CX. Ubiquitin-conjugating enzyme E2T knockdown suppresses hepatocellular tumorigenesis via inducing cell cycle arrest and apoptosis. World J Gastroenterol 2019; 25(43): 6386-6403
- URL: https://www.wjgnet.com/1007-9327/full/v25/i43/6386.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i43.6386
Hepatocellular carcinoma (HCC), which accounts for more than 90% of primary liver cancers, is the fifth most common malignancy and the third leading cause of cancer-related deaths worldwide[1]. Virus infections [including hepatitis B virus and hepatitis C virus (HCV)], obesity, inflammation, fibrosis, and cirrhosis are the major risk factors for HCC[2]. Genetic studies have identified that TP53, β-catenin, and TERT are the most frequently mutated genes in HCC[3-5]. However, effective drugs are lacking for this disease. Therefore, novel factors triggering HCC remain to be identified that may benefit the diagnosis and treatment of HCC patients.
Ubiquitin-conjugating enzyme E2T (UBE2T), also known as HSPC150, belongs to the E2 family of the ubiquitin-proteasome pathway. UBE2T activates FANCD2 mono-ubiquitination, and it was first identified in a patient with Fanconi anemia[6,7]. Recently, increasing evidence has reported that UBE2T is involved in cancer development. UBE2T is highly expressed in prostate cancer tissues and correlates with the metastatic stage and survival of patients. UBE2T promotes the proliferation, migration, and tumor development of prostate cancer cells[8]. Moreover, the upregulation of UBE2T is also found in other cancer types, such as breast and lung cancers[9]. In vitro, UBE2T accelerates the growth of HCC cells via destabilizing P53 protein abundance[10]. However, the role of UBE2T in HCC carcinogenesis requires more in-depth investigation.
In this study, we found that UBE2T was overexpressed in HCC specimens. We hypothesized that UBE2T functions as an oncogene in HCC. To this aim, we used a lentivirus to knock down UBE2T in two HCC cell lines and analyzed the cell proliferation and xenograft tumorigenesis. Furthermore, cell cycle distribution, apoptosis, and gene expression were analyzed to investigate the potential mechanisms. We found that UBE2T knockdown inhibited the viability and tumor development of HCC cells. Cell cycle arrest and apoptosis were induced by UBE2T silencing. Various genes were downregulated or upregulated by UBE2T knockdown. We suggest that UBE2T is a promising oncogene in HCC.
The transcript analysis of UBE2T in HCC patients was performed based on The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov). Fifty pairs of HCC and adjacent normal tissues and a total of 543 samples (373 tumor tissues and 169 normal tissues) were available for this study. The expression of UBE2T was analyzed in cancer and normal tissues. The patients were divided into UBE2T high expression and UBE2T low expression groups and subjected to overall survival analysis. The pathological characteristics of grade, T stage, and tumor stage were analyzed between these two groups.
The HCC cell lines BEL-7404 and SMMC-7721 were obtained from the American Type Culture Collection (United States) and the Cell Bank of the Chinese Academy of Sciences (China). The cells were cultured in Dulbecco modified Eagle’s medium (Invitrogen) containing 10% fetal bovine serum (Gibco) as well as 1% penicillin and streptomycin (Corning). The cells were cultured in a 37 °C incubator with 5% CO2.
UBE2T was knocked down using the lentivirus vector pGCSIL-GFP in BEL-7404 and SMMC-7721 cells. The targeted sequences (ShUBE2T, 5’-ACCTCCTCAGAT CCGATTT-3’ and ShCtrl, 5’-TTCTCCGAACGTGTCACGT-3’) were synthesized and inserted into the pGCSIL-GFP vector. pHelper1.0 and Helper2.0 served as the packaging vectors. Briefly, ShCtrl or ShUBE2T pGCSIL-GFP vectors were co-transfected with pHelper1.0 and Helper2.0 vectors into 293T cells using Lipofectamine 2000 (Invitrogen). After 48 or 72 h, the viral supernatants were harvested and filtered through 0.45 µm filters. The viral supernatants were used to infect the BEL-7404 and SMMC-7721 cells, and the infection efficiency was determined by qRT-PCR and Western blot assays.
RNA was harvested from indicated cells using TRIzol reagent (Invitrogen) and an RNA isolation kit (CWBIO, China) following the manufacturer’s instructions. Equal amounts of RNA were subjected to reverse transcription with M-MLV reverse transcriptase (Promega). The qRT-PCR experiment was performed using the SYBR master mixture (Takara) on a real-time PCR machine TP800 (Takara). The primer sequences are listed in Table 1. The expression of the targeted genes was normalized to GAPDH.
| Gene | Primer sequence | |
| UBE2T | Forward | TTGATTCTGCTGGAAGGATTTG |
| Reverse | CAGTTGCGATGTTGAGGGAT | |
| PTGS2 | Forward | CTCCTGTGCCTGATGATTGC |
| Reverse | CAGCCCGTTGGTGAAAGC | |
| IL1B | Forward | TCCTGTTGTCTACACCAATGCCCA |
| Reverse | GAACCAAATGTGGCCGTGGTTTCT | |
| RAC2 | Forward | GAAGCATCTACCCGTTCACTC |
| Reverse | AGTTGTGGCAGCAACCATCT | |
| ITGA2 | Forward | CCTACAATGTTGGTCTCCCAGA |
| Reverse | AGTAACCAGTTGCCTTTTGGATT | |
| IL1A | Forward | AGATGCCTGAGATACCCAAAACC |
| Reverse | CCAAGCACACCCAGTAGTCT | |
| FOSL1 | Forward | ACCCTCCCTAACTCCTTTCA |
| Reverse | CTGGAGTTGGATGTGGGATA | |
| SOD2 | Forward | TGGACAAACCTCAGCCCT |
| Reverse | TGAAACCAAGCCAACCC | |
| BMP6 | Forward | TCAACTTATCCCAGATTCCTGA |
| Reverse | CCATACTACACGGGTGTCCAA | |
| BCL2L1 | Forward | CTGAATCGGAGATGGAGACC |
| Reverse | GAGCTGGGATGTCAGGTCA | |
| GSTO1 | Forward | GCATACCCAGGGAAGAAGCT |
| Reverse | AGAATTGCCACCAAAGAAGG | |
| ITGA5 | Forward | GGCTTCAACTTAGACGCGGAG |
| Reverse | TGGCTGGTATTAGCCTTGGGT | |
| ENC1 | Forward | ACATGGTAGTGCAACTCTTGTC |
| Reverse | TTCAGGTCATAGCTGATCCAGT | |
| IL1RAP | Forward | CAAAGTGATGCCTCAGAACG |
| Reverse | CTGCCTAGTCCAATACCAGATC | |
| MET | Forward | AGTCATAGGAAGAGGGCATT |
| Reverse | CTTCACTTCGCAGGCAGA | |
| SOCS3 | Forward | GCCTCAAGACCTTCAGCTCCAA |
| Reverse | CTCCAGTAGAAGCCGCTCTCCT | |
| PRKAR1A | Forward | GTTTTCGGTCTCCTTTATCGC |
| Reverse | ACTGGTTGCCCATTCATTGTT | |
| RPL31 | Forward | CTCGGGCACTCAAAGAGATTC |
| Reverse | CGGATTCGGTATGGCACATTC | |
| PPARGC1A | Forward | TCTGAGTCTGTATGGAGTGACAT |
| Reverse | CCAAGTCGTTCACATCTAGTTCA | |
| RPL13A | Forward | GCCATCGTGGCTAAACAGGTA |
| Reverse | GTTGGTGTTCATCCGCTTGC | |
| NQO1 | Forward | AGGCAGTGCTTTCCATCA |
| Reverse | CAGGCGTTTCTTCCATCC | |
| ABCC1 | Forward | GTCGGGGCATATTCCTGGC |
| Reverse | CTGAAGACTGAACTCCCTTCCT | |
| RPL32 | Forward | ACCCAGAGGCATTGACAACA |
| Reverse | GAGCGATCTCGGCACAGTAA | |
| GCLM | Forward | ATCAGTGGGCACAGGTAAA |
| Reverse | CAGAAAGCAGTTCTTTTGGA | |
| FYN | Forward | GGATGCCAAGGCTTACCGAT |
| Reverse | GGGCTCCTCAGACACCACTG | |
| HMOX1 | Forward | AAGACTGCGTTCCTGCTCAAC |
| Reverse | AAAGCCCTACAGCAACTGTCG | |
| SCNN1A | Forward | AGTTCCACCGCTCCTACCGA |
| Reverse | GTCCGAGTTGAGGTTGATGTTG | |
| GSTA4 | Forward | ACTATCCCAACGGAAGAGGC |
| Reverse | CAGGTGGTTACCATCCTGCA | |
| MAP2K6 | Forward | GAAGCATTTGAACAACCTCAGAC |
| Reverse | CCTGGCTATTTACTGTGGCTC | |
| PIK3C2B | Forward | TTCCTCCACTGTAGACTTGCTT |
| Reverse | AGCCGAATGTCAATGTCAAACT | |
| GAPDH | Forward | TGACTTCAACAGCGACACCCA |
| Reverse | CACCCTGTTGCTGTAGCCAAA | |
The proteins were collected from the indicated cells with lysis buffer (Beyotime), and the concentration was analyzed with a BCA protein assay kit (Beyotime). The proteins were separated on a 10% or 12% SDS-PAGE gel and subsequently transferred to PVDF membranes (Millipore, United States). The membranes were then blocked with 5% nonfat milk for at least 60 min at room temperature. After incubating with primary antibodies overnight at 4 °C, the membranes were incubated with secondary antibodies and washed three times with PBS-T. The proteins on the membranes were detected using chemiluminescence (Thermo Fisher Scientific). The antibody against UBE2T (SAB1104968) was from Sigma. The antibodies against GAPDH (ab9485), ITGA2 (ab133557), FOSL1 (ab124722), PTGS2 (ab15191), IL1B (ab9722), and BMP6 (ab155963) were from Abcam. All the secondary antibodies were from Santa Cruz.
The HCS assay was performed to detect cell viability. ShCtrl and ShUBE2T SMMC-7721 or BEL-7404 cells were seeded in 96-well plates and cultured for 5 d. The viable cells were scanned by the detection of intensity and distribution of fluorescence. The images were collected using a 20 × objective fluorescence-imaging microscope and analyzed with ArrayScan HCS software (Cellomics Inc.).
A total of 5 × 106 ShCtrl and ShUBE2T SMMC-7721 cells were subcutaneously implanted into the immune-deficient nude mice (BALB/c, 4 wk old, n = 8 per group). The tumor volume was analyzed using the following formula: V = 0.5 × ab2 (a = long diameter of the tumor, b = short diameter of the tumor). The tumor weight was measured by day 42 after the implantation. This study was approved by the ethics committee of The Affiliated People’s Hospital of Shanxi Medical University.
Propidium iodide (PI) staining was used to detect the cell cycle distribution of ShCtrl and ShUBE2T SMMC-7721 or BEL-7404 cells. In brief, the SMMC-7721 or BEL-7404 cells expressing shRNA lentivirus against Ctrl or UBE2T were seeded in six-well plates. When reaching 80% density, the cell nuclei were stained with PI. PI absorbance was then measured by flow cytometry (FACSCalibur, Becton Dickinson).
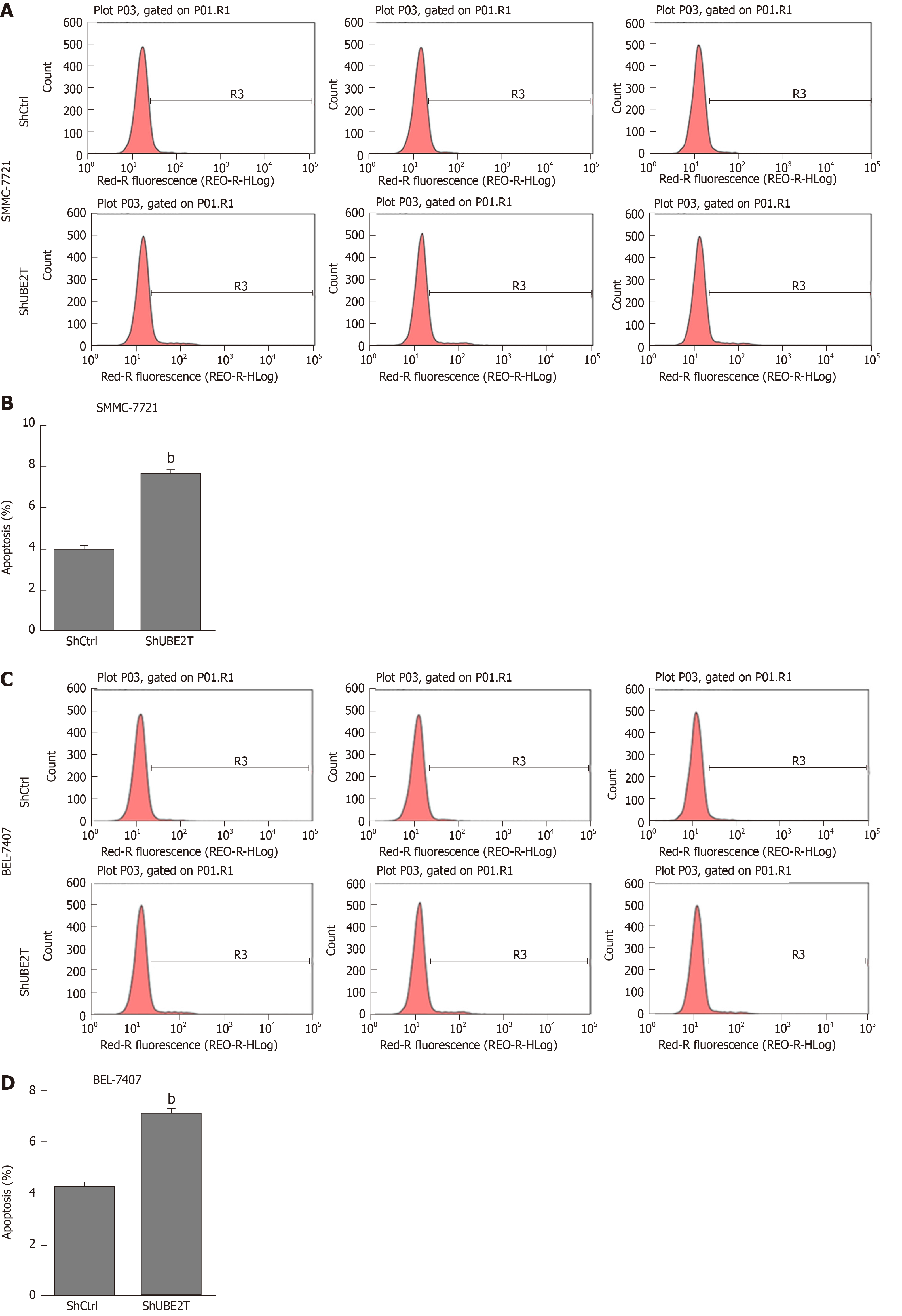
The apoptosis of ShCtrl and ShUBE2T SMMC-7721 or BEL-7404 cells was determined with an Annexin-V-APC kit (Ebioscience, United States) according to the manufacturer’s instructions. Briefly, the indicated cells were washed with PBS and resuspended with staining buffer. A total of 5 μL of annexin V-APC regent was added into a total of 100 μL cell suspension, and the mixture was maintained at room temperature for 15 min. Then, flow cytometry was used to detect cell apoptosis (FACSCalibur, Becton-Dickinson, United States).
Gene expression profiling was measured by the Affymetrix human GeneChip prime view. RNA was extracted from ShCtrl and ShUBE2T SMMC-7721 cells using TRIzol reagent. The difference in the gene expression was considered significant when the fold change > 2 and when P < 0.05. The pathway enrichment and gene networks were analyzed using the Ingenuity Pathway Analysis.
SPSS version 16.0 software was used to analyze the data as shown by the mean ± standard error of the mean (SEM) of three independent experiments. The difference between the two groups was analyzed by the unpaired Student’s t-test. One-way ANOVA was applied when more than two groups were analyzed. For survival analysis, the Kaplan-Meier method was applied to analyze the correction between UBE2T expression and the overall survival rate, which was determined using the log-rank test. The median value was used as the cutoff for classification of patients into high and low expression groups. The difference was considered significant at P < 0.05.
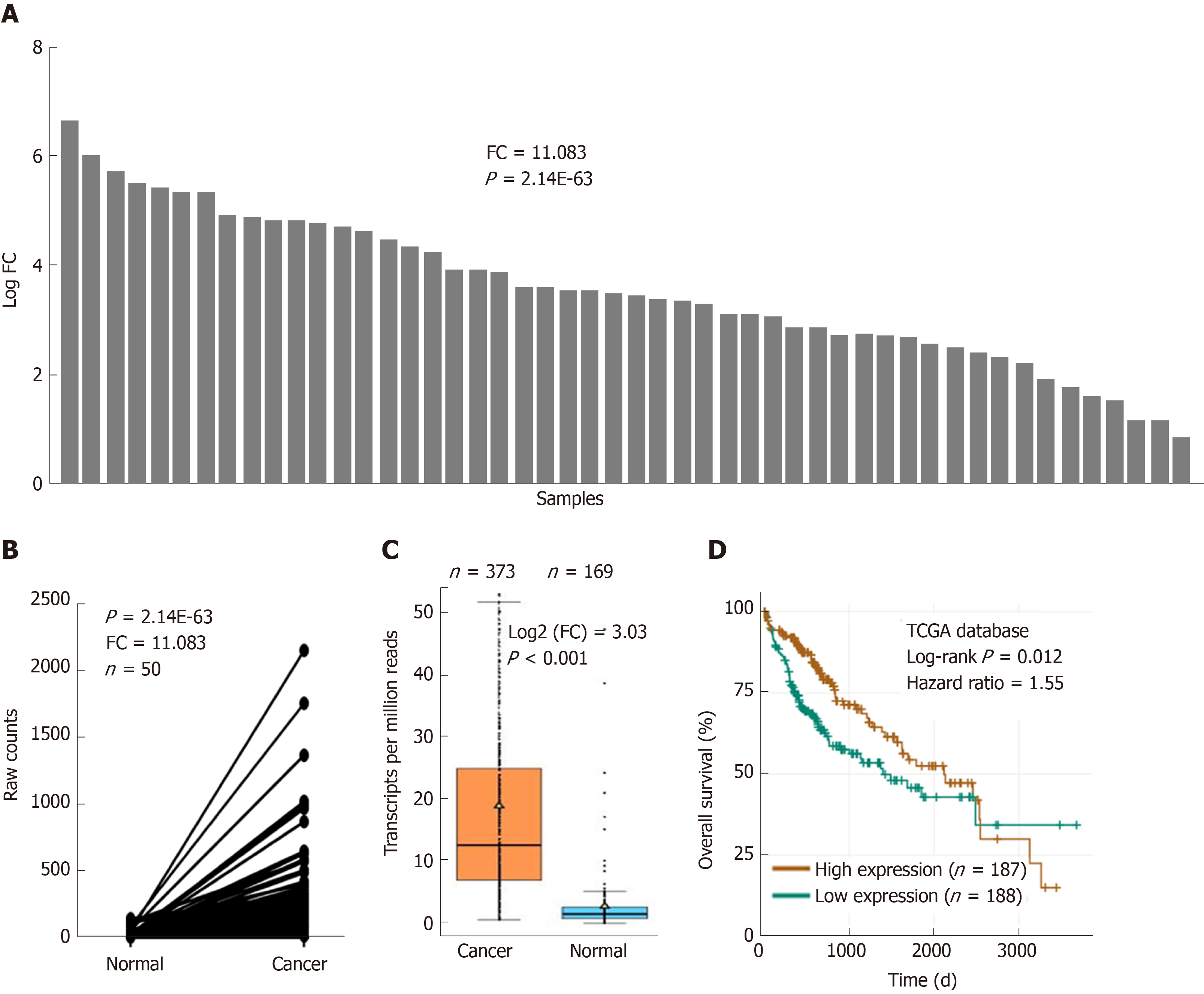
UBE2T is overexpressed in various cancer patients. We initially analyzed the abundance of UBE2T in HCC patients based on the TCGA database. UBE2T was highly expressed in 50 HCC specimens compared their adjacent normal tissues (Figure 1A and B). The expression of UBE2T was then analyzed in HCC and nonpaired normal tissues. The results showed that UBE2T was significantly overexpressed in 373 HCC tissues compared to 169 normal liver tissues (Figure 1C). In addition, high expression of UBE2T was positively correlated with grade, T stage, and the pathological stage of HCC patients (Table 2). The patients in the UBE2T high expression group exhibited a poorer overall survival than those in the low expression group (Figure 1D). Taken together, the results demonstrate that UBE2T is abundant in HCC tissues and correlates with HCC progression and survival.
| Expression of UBE2T | Total | P value | |||
| Low | High | ||||
| Grade | 0.000 | ||||
| G1/2 | 137 | 95 | 232 | ||
| G3/4 | 47 | 87 | 134 | ||
| Total | 184 | 182 | 366 | ||
| T stage | 0.001 | ||||
| T1 | 108 | 73 | 181 | ||
| T2 | 38 | 56 | 94 | ||
| T3/4 | 38 | 55 | 93 | ||
| Total | 184 | 184 | 368 | ||
| Pathological stage | 0.002 | ||||
| I | 101 | 70 | 171 | ||
| II | 36 | 50 | 86 | ||
| III/IV | 37 | 53 | 90 | ||
| Total | 174 | 173 | 347 | ||
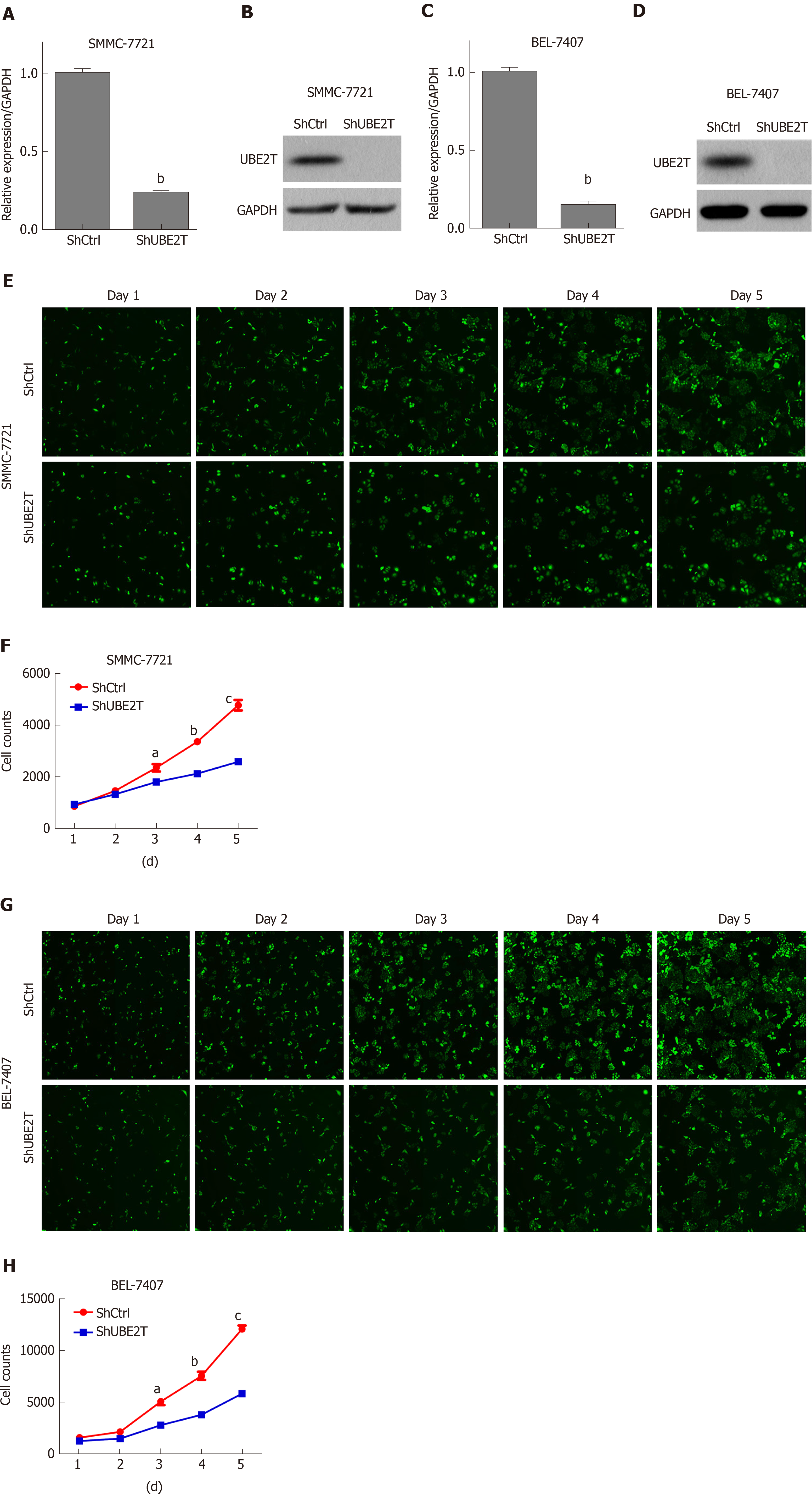
We next determined the effect of UBE2T on HCC cell viability using the multi-parametric HCS assay. UBE2T was knocked down using the lentivirus-mediated strategy. The qRT-PCR and Western blot assays showed that UBE2T was efficiently silenced in SMMC-7721 and BEL-7404 cells (Figure 2A-D). The HCS results indicated that UBE2T knockdown clearly decreased the viability of SMMC-7721 and BEL-7404 cells (Figure 2E-H).
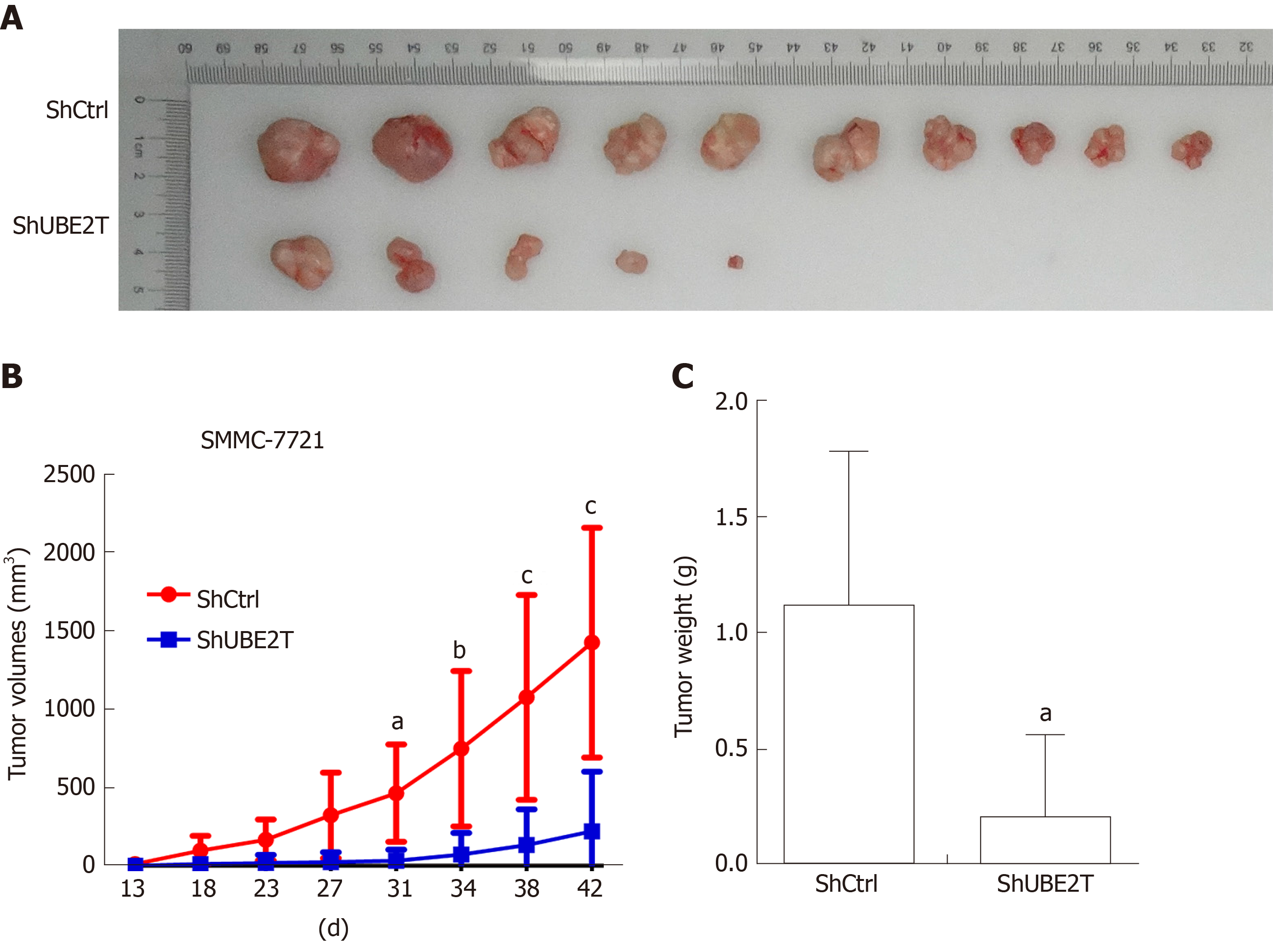
To explore the role of UBE2T in HCC tumorigenesis, we implanted SMMC-7721 cells carrying the ShCtrl or ShUBE2T lentivirus. The tumors were harvested and photographed 42 d after implantation. The images showed that the tumor size of the ShUBE2T group was clearly smaller than that of the ShCtrl group (Figure 3A). The tumor growth curve suggested that UBE2T knockdown suppressed the tumor initiation and progression of SMMC-7721 cells (Figure 3B). Likewise, the tumor weight was decreased in UBE2T-silenced tumors compared with ShCtrl tumors (Figure 3C). Therefore, UBE2T is essential for the proliferation and carcinogenesis of both SMMC-7721 and BEL7404 cells.
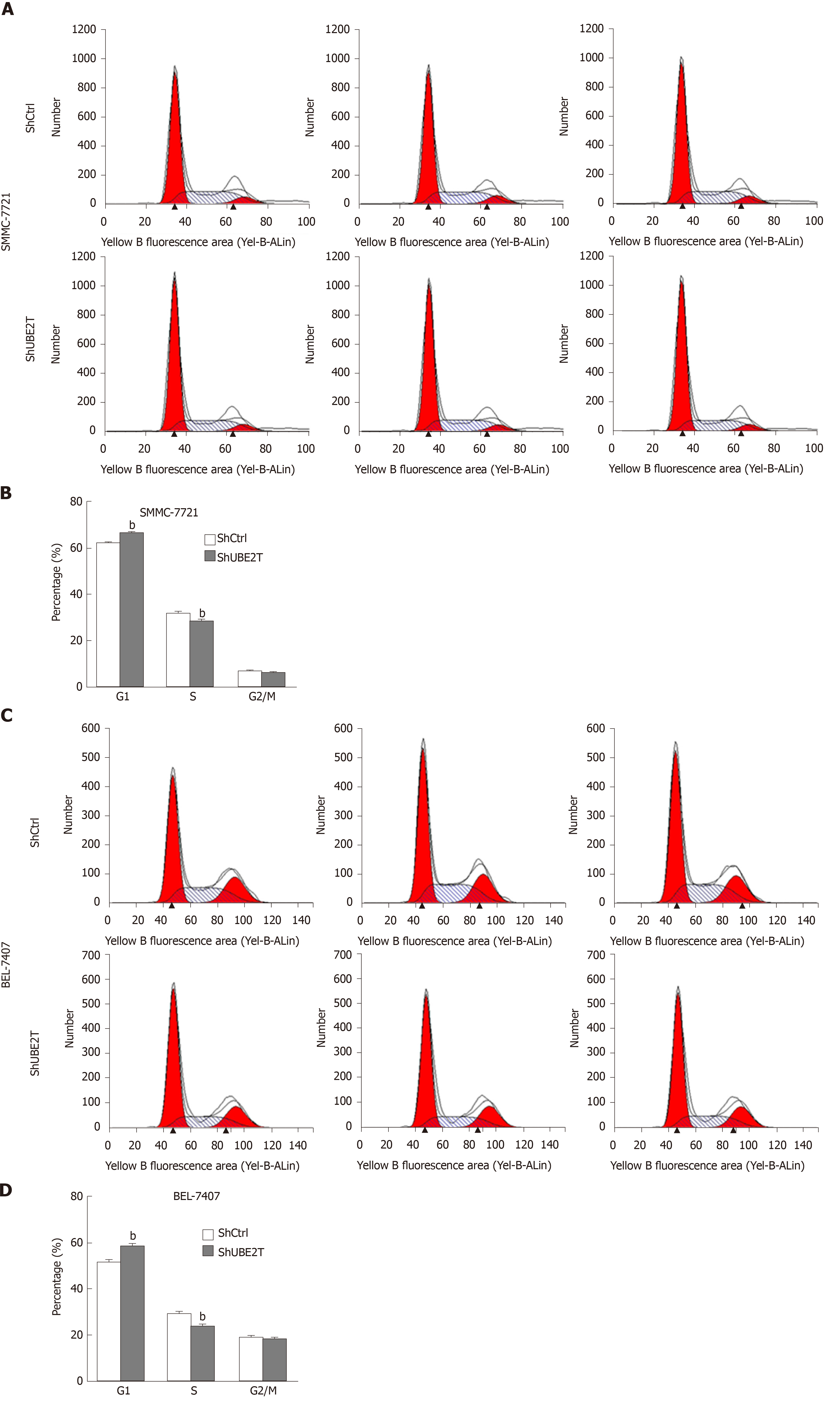
Aberrant cell cycle progression and suppressed apoptosis are hallmarks of cancer. We then analyzed whether UBE2T regulated the cell cycle and apoptosis of HCC cells. Using PI staining and analysis by flow cytometry, we found that UBE2T knockdown in SMMC-7721 cells resulted in increased G1 phase and decreased S phase distribution. The G2/M phase remained unchanged (Figure 4A). Consistent results were observed in ShCtrl and ShUBE2T BEL-7404 cells (Figure 4B).
Next, Annexin-V-APC staining was used to determine the apoptosis of ShCtrl and ShUBE2T HCC cells. The results showed that UBE2T knockdown led to increased apoptosis of both SMMC-7721 and BEL-7404 cells (Figure 5). Taken together, the results show that UBE2T silencing induces G1/S cell cycle arrest and enhances apoptosis in HCC cells.
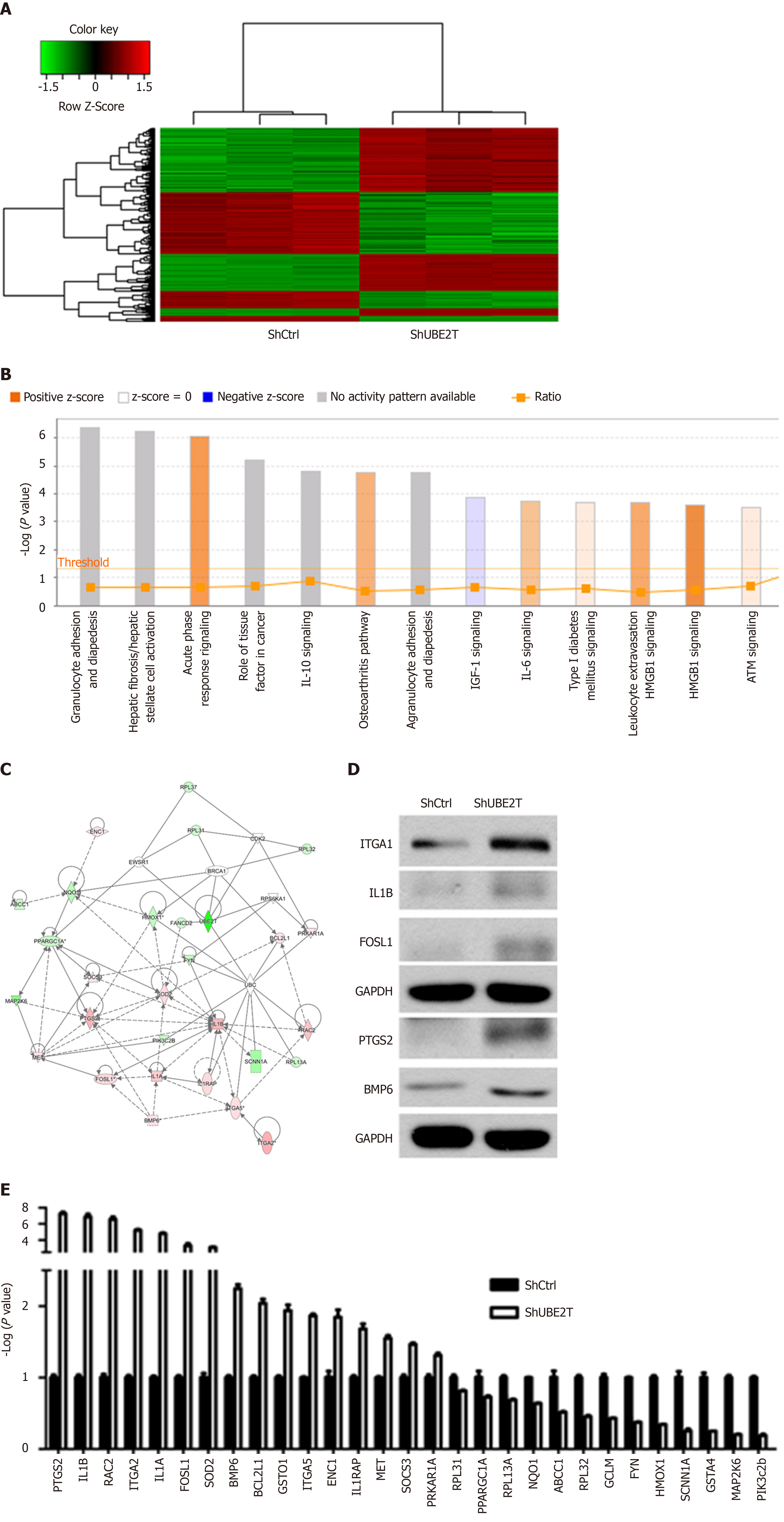
To explore the downstream factors regulated by UBE2T in HCC, ShCtrl and ShUBE2T SMMC-7721 cells were subjected to gene chip analysis. A total of 354 genes were upregulated while 276 downregulated in the UBE2T knockdown cells compared with ShCtrl cells (Figure 6A). The pathway enrichment analysis showed that acute phase response signaling, osteoarthritis pathway, interleukin (IL)-6 signaling, type I diabetes mellitus signaling, and HMGB1 signaling were significantly activated by UBE2T knockdown, while IGF-1 signaling was suppressed (Figure 6B). Based on the previous studies and our data, a possible regulation network for UBE2T is presented in Figure 6C. We then performed Western blot and qRT-PCR assays to verify the GeneChip results. We found that PTGS2, IL-1B, RAC2, ITGA2, IL-1A, FOSL1, SOD2, bone morphogenetic protein (BMP) 6, BCL2L1, GSTO1, IGTA5, ENC1, IL-1RAP, MET, SOCS3, and PRKAR1A were upregulated, while RPL31, PPARGC1A, RPL13A, NQO1, ABCC1, RPL32, GCLM, FYN, HMOX1, SCNN1A, GSTA4, MAP2K6, and PIK3c2b were downregulated by UBE2T knockdown (Figure 6D and E ).
HCC is one of the most malignant tumors, with a poor 5-year survival rate. It has been shown that HCC is a chronic disease, where non-alcoholic fatty liver disease and non-alcoholic steatohepatitis are risk factors for HCC[11,12]. Even though a large number of studies have identified that some critical genes, such as TP53 and β-catenin, and several important signaling pathways, including Wnt/β-catenin and PI3K/ AKT/mTOR, play essential roles in HCC[5,13,14], effective drugs targeting these genes or signaling pathways are still scarce. Therefore, identifying novel genes contributing to this disease may help us develop effective targeted treatments.
A recent profiling study based on the database of TCGA showed that most of the cancer types harbor at least one driver signaling pathway. In HCC, the cell cycle and Wnt signaling pathways are the most frequently altered signaling[15]. Based on the TCGA database, we initially found that UBE2T was clearly overexpressed in HCC patients. And a previous study suggested that UBE2T expression was elevated in HCC tissues with higher pathological grade and vascular invasion according to TCGA cohort[10]. UBE2T acts as a member of ubiquitin-conjugating enzymes[16]. Notably, UBE2T locates at 1q32.1 and the gain of 1q in most human cancers may contribute to the aberrant upregulation of UBE2T[8]. Previously, several studies have illustrated the function of UBE2T in cancer development. For example, UBE2T expression is linked with the poor outcome of breast and lung cancers[9]. UBE2T is also overexpressed in prostate cancer and contributes to the growth of prostate cancer[8]. Furthermore, the knockdown of upregulated UBE2T suppresses cell and/or tumor growth in bladder cancer and gastric cancer[17,18]. The involvement of UBE2T was also reported in HCC cell growth in vitro. UBE2T potentiated HCC cell growth by increasing the ubiquitination level of p53[10]. However, the in vivo function of UBE2T in HCC is still unknown. Based on the TCGA database, UBE2T was highly expressed in HCC tissues; hence, we knocked down UBE2T in HCC SMMC-7721 and BEL-7404 cells. We showed that UBE2T silencing suppressed the growth of both cells in vitro and the xenograft tumor development of SMMC-7721 cells in vivo. These results indicate that UBE2T may be a functional oncogene in HCC. Moreover, UBE2T is critical for HCC cell growth and proliferation.
Cancer cells have accelerated cell cycle progression and reduced apoptosis[19]. Targeting cell cycle proteins has been used in cancer patients[20]. In bladder cancer, UBE2T has been found to negatively regulate the cell cycle process of G2/M transition and apoptosis[17]. Therefore, we investigated the role of UBE2T in the HCC cell cycle. We found that, although changes in the G2/M phase of the cell cycle in HCC cells remained minimal after UBE2T knockdown, the percentage of cells in the G1 phase was increased and that in the S phase was decreased in both ShUBE2T SMMC-7721 and BEL-7404 cells. These results suggest that UBE2T knockdown suppresses the cell cycle progression in both bladder cancer and HCC cells, while it exhibits a distinct function in regulating cell cycle phases in different cancers. Consistent with the results in bladder cancer, UBE2T silencing enhanced apoptosis in HCC cells.
Some studies have illustrated the downstream substrates of UBE2T in various cancers. UBE2T activates the AKT/GSK3β/β-catenin pathway and triggers nasopharyngeal carcinoma progression[21]. UBE2T knockdown suppresses the phosphorylation of both PI3K and AKT in osteosarcoma[22]. In addition, UBE2T interacts with the BRCA1/BARD1 complex and promotes the development and progression of breast cancer[16]. To explore the downstream targets regulated by UBE2T, we performed GeneChip analysis in ShCtrl and ShUBE2T SMMC-7721 cells. We identified hundreds of genes that are regulated by UBE2T. The pathway enrichment and regulation network analyses showed the potential signaling pathways and the central genes downstream of UBE2T. As the GeneChip results indicated, multiple genes were dys-regulated upon UBE2T silencing, including ITGA2, FOSL1, PTGS2, IL-1β, and BMP6. ITGA2 subunit forms a heterodimer with the beta 1 subunit to mediate the adhesion of extracellular matrix, involved in tumor cell proliferation, apoptosis, and migration. Blockade of ITGA2 induces apoptosis and inhibits cell migration in gastric cancer, which might partially explain the ability of UBE2T silencing to inhibit HCC growth[23]. FOSL1 is a member of the FOS family, involved in cancer cell progression and maintenance of the transformed state in several cell types. Further studies suggest that FOSL1 is mainly involved in late tumor stages, including epithelial-to-mesenchymal transition and tumor invasion[24]. Prostaglandin endoperoxide synthases (also known as COX) are the rate-limiting enzymes in the conversion of arachidonic acid into prostaglandins. As one of the isoforms, PTGS2 could activate PEG2, which functions as a regulator in cell proliferation, apoptosis, and invasion[25]. The IL-1 family are proinflammatory cytokines involved in tumor growth, invasion, and distant metastasis in various cancers. A study revealed that the genotypes of IL-1 beta is associated with the prognosis of HCV-related HCC[26]. BMPs belong to the superfamily of the transforming growth factor-β. BMP6 is required to mediate the self-renewal and differentiation of several types of stem cells, and the deficiency of BMP6 may lead to cancer[27]. Taken together, all of these factors may contribute to the UBE2T silencing-induced deceleration of progression in HCC. These results provide clues for further mechanistic studies of UBE2T in HCC and other cancers.
In summary, UBE2T is overexpressed in HCC specimens. Downregulation of UBE2T suppresses HCC cell and tumor growth, at least partly through arrested cell cycle progression and enhanced apoptosis. Numerous genes are regulated by UBE2T. Therefore, UBE2T acts as a potential oncogene and is significantly involved in HCC.
Hepatocellular carcinoma (HCC) is one of most common malignant tumors with a poor prognosis. Increasing studies indicated that dysregulation of ubiquitin-conjugating enzyme E2T (UBE2T) contributes to the development of human cancers. However, whether UBE2T is involved in the progression of HCC is unclear.
Previous studies have illustrated the critical role of UBE2T in the progression of various tumors, and our study will suggest the role of UBE2T in regulating the occurrence and development of HCC.
To investigate the biological function of UBE2T in HCC cell proliferation and progression in vitro by gain-of-function and loss-of-function strategies, and provides significant insights into the underlying molecular mechanisms of UBE2T involved in the development of HCC, which may contribute to the future research of more effective diagnosis and treatment.
The expression of UBE2T in HCC tissues was obtained from The Cancer Genome Atlas database. In vitro experiments using lentivirus-medicated approach were performed to examine cell growth, cell cycle distribution, and apoptosis by CCK8 assay and flow cytometry, respectively. The xenograft tumorigenicity assay was performed to determine the capacity of cell proliferation in vivo. The whole genome expression profile was analyzed by microarray assay.
In this study, we identified remarkable overexpression of UBE2T in HCC tissues, which closely corrected with a poor overall survival in HCC patients. The results of in vitro experiments suggested a critical role of UBE2T in cell viability, cell cycle, and apoptosis of HCC. In both SMMC-7721 and BEL-7404 cells, the HCC cell proliferation was obviously inhibited, cell cycle was arrested at G1/S phase, and apoptosis was significantly promoted by lentivirus-medicated UBE2T knockdown. Moreover, the growth of SMMC-7721 cells with UBE2T knockdown in xenografts was significantly inhibited in vivo. The microarray assay confirmed that UBE2T silencing contributed to the dysregulation of numerous genes, including IL-1B, FOSL1, PTGS2, and BMP6.
In conclusion, UBE2T is significantly involved in HCC cell proliferation, cell cycle, and apoptosis.
Our study may provide a novel potential diagnostic and therapeutic target for HCC intervention.
| 1. | Sia D, Villanueva A, Friedman SL, Llovet JM. Liver Cancer Cell of Origin, Molecular Class, and Effects on Patient Prognosis. Gastroenterology. 2017;152:745-761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 619] [Cited by in RCA: 867] [Article Influence: 96.3] [Reference Citation Analysis (2)] |
| 2. | Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer. 2013;13:123-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 581] [Cited by in RCA: 641] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 3. | Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F, Calatayud AL, Pinyol R, Pelletier L, Balabaud C, Laurent A, Blanc JF, Mazzaferro V, Calvo F, Villanueva A, Nault JC, Bioulac-Sage P, Stratton MR, Llovet JM, Zucman-Rossi J. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505-511. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1006] [Cited by in RCA: 1404] [Article Influence: 127.6] [Reference Citation Analysis (0)] |
| 4. | Tornesello ML, Buonaguro L, Tatangelo F, Botti G, Izzo F, Buonaguro FM. Mutations in TP53, CTNNB1 and PIK3CA genes in hepatocellular carcinoma associated with hepatitis B and hepatitis C virus infections. Genomics. 2013;102:74-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 138] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 5. | Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1461] [Cited by in RCA: 1575] [Article Influence: 78.8] [Reference Citation Analysis (0)] |
| 6. | Machida YJ, Machida Y, Chen Y, Gurtan AM, Kupfer GM, D'Andrea AD, Dutta A. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589-596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 218] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 7. | Alpi A, Langevin F, Mosedale G, Machida YJ, Dutta A, Patel KJ. UBE2T, the Fanconi anemia core complex, and FANCD2 are recruited independently to chromatin: a basis for the regulation of FANCD2 monoubiquitination. Mol Cell Biol. 2007;27:8421-8430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 73] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 8. | Wen M, Kwon Y, Wang Y, Mao JH, Wei G. Elevated expression of UBE2T exhibits oncogenic properties in human prostate cancer. Oncotarget. 2015;6:25226-25239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 68] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 9. | Perez-Peña J, Corrales-Sánchez V, Amir E, Pandiella A, Ocana A. Ubiquitin-conjugating enzyme E2T (UBE2T) and denticleless protein homolog (DTL) are linked to poor outcome in breast and lung cancers. Sci Rep. 2017;7:17530. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 55] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 10. | Liu LP, Yang M, Peng QZ, Li MY, Zhang YS, Guo YH, Chen Y, Bao SY. UBE2T promotes hepatocellular carcinoma cell growth via ubiquitination of p53. Biochem Biophys Res Commun. 2017;493:20-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 73] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 11. | Font-Burgada J, Sun B, Karin M. Obesity and Cancer: The Oil that Feeds the Flame. Cell Metab. 2016;23:48-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 280] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 12. | Alexander J, Torbenson M, Wu TT, Yeh MM. Non-alcoholic fatty liver disease contributes to hepatocarcinogenesis in non-cirrhotic liver: a clinical and pathological study. J Gastroenterol Hepatol. 2013;28:848-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 96] [Article Influence: 7.4] [Reference Citation Analysis (2)] |
| 13. | Villanueva A, Chiang DY, Newell P, Peix J, Thung S, Alsinet C, Tovar V, Roayaie S, Minguez B, Sole M, Battiston C, Van Laarhoven S, Fiel MI, Di Feo A, Hoshida Y, Yea S, Toffanin S, Ramos A, Martignetti JA, Mazzaferro V, Bruix J, Waxman S, Schwartz M, Meyerson M, Friedman SL, Llovet JM. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology. 2008;135:1972-1983, 1983.e1-1983.11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 564] [Cited by in RCA: 599] [Article Influence: 33.3] [Reference Citation Analysis (1)] |
| 14. | Kenerson HL, Yeh MM, Kazami M, Jiang X, Riehle KJ, McIntyre RL, Park JO, Kwon S, Campbell JS, Yeung RS. Akt and mTORC1 have different roles during liver tumorigenesis in mice. Gastroenterology. 2013;144:1055-1065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 15. | Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, Dimitriadoy S, Liu DL, Kantheti HS, Saghafinia S, Chakravarty D, Daian F, Gao Q, Bailey MH, Liang WW, Foltz SM, Shmulevich I, Ding L, Heins Z, Ochoa A, Gross B, Gao J, Zhang H, Kundra R, Kandoth C, Bahceci I, Dervishi L, Dogrusoz U, Zhou W, Shen H, Laird PW, Way GP, Greene CS, Liang H, Xiao Y, Wang C, Iavarone A, Berger AH, Bivona TG, Lazar AJ, Hammer GD, Giordano T, Kwong LN, McArthur G, Huang C, Tward AD, Frederick MJ, McCormick F, Meyerson M; Cancer Genome Atlas Research Network, Van Allen EM, Cherniack AD, Ciriello G, Sander C, Schultz N. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell. 2018;173:321-337.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2333] [Cited by in RCA: 2288] [Article Influence: 286.0] [Reference Citation Analysis (0)] |
| 16. | Ueki T, Park JH, Nishidate T, Kijima K, Hirata K, Nakamura Y, Katagiri T. Ubiquitination and downregulation of BRCA1 by ubiquitin-conjugating enzyme E2T overexpression in human breast cancer cells. Cancer Res. 2009;69:8752-8760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 104] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 17. | Gong YQ, Peng D, Ning XH, Yang XY, Li XS, Zhou LQ, Guo YL. UBE2T silencing suppresses proliferation and induces cell cycle arrest and apoptosis in bladder cancer cells. Oncol Lett. 2016;12:4485-4492. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 18. | Luo C, Yao Y, Yu Z, Zhou H, Guo L, Zhang J, Cao H, Zhang G, Li Y, Jiao Z. UBE2T knockdown inhibits gastric cancer progression. Oncotarget. 2017;8:32639-32654. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 19. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 48654] [Article Influence: 3243.6] [Reference Citation Analysis (11)] |
| 20. | Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17:93-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1413] [Cited by in RCA: 1424] [Article Influence: 158.2] [Reference Citation Analysis (0)] |
| 21. | Hu W, Xiao L, Cao C, Hua S, Wu D. UBE2T promotes nasopharyngeal carcinoma cell proliferation, invasion, and metastasis by activating the AKT/GSK3β/β-catenin pathway. Oncotarget. 2016;7:15161-15172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 61] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 22. | Wang Y, Leng H, Chen H, Wang L, Jiang N, Huo X, Yu B. Knockdown of UBE2T Inhibits Osteosarcoma Cell Proliferation, Migration, and Invasion by Suppressing the PI3K/Akt Signaling Pathway. Oncol Res. 2016;24:361-369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 52] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 23. | Chuang YC, Wu HY, Lin YL, Tzou SC, Chuang CH, Jian TY, Chen PR, Chang YC, Lin CH, Huang TH, Wang CC, Chan YL, Liao KW. Blockade of ITGA2 Induces Apoptosis and Inhibits Cell Migration in Gastric Cancer. Biol Proced Online. 2018;20:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 24. | Maurus K, Hufnagel A, Geiger F, Graf S, Berking C, Heinemann A, Paschen A, Kneitz S, Stigloher C, Geissinger E, Otto C, Bosserhoff A, Schartl M, Meierjohann S. The AP-1 transcription factor FOSL1 causes melanocyte reprogramming and transformation. Oncogene. 2017;36:5110-5121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 73] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 25. | Kunzmann AT, Murray LJ, Cardwell CR, McShane CM, McMenamin UC, Cantwell MM. PTGS2 (Cyclooxygenase-2) expression and survival among colorectal cancer patients: a systematic review. Cancer Epidemiol Biomarkers Prev. 2013;22:1490-1497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 26. | Okamoto K, Ishida C, Ikebuchi Y, Mandai M, Mimura K, Murawaki Y, Yuasa I. The genotypes of IL-1 beta and MMP-3 are associated with the prognosis of HCV-related hepatocellular carcinoma. Intern Med. 2010;49:887-895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 47] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 27. | Wang H, Yuan Q, Sun M, Niu M, Wen L, Fu H, Zhou F, Chen Z, Yao C, Hou J, Shen R, Lin Q, Liu W, Jia R, Li Z, He Z. BMP6 Regulates Proliferation and Apoptosis of Human Sertoli Cells Via Smad2/3 and Cyclin D1 Pathway and DACH1 and TFAP2A Activation. Sci Rep. 2017;7:45298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (1)] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Gougelet A, Lim SJ S-Editor: Yan JP L-Editor: Wang TQ E-Editor: Ma YJ