Published online Jul 21, 2019. doi: 10.3748/wjg.v25.i27.3572
Peer-review started: April 10, 2019
First decision: May 9, 2019
Revised: May 27, 2019
Accepted: June 8, 2019
Article in press: June 8, 2019
Published online: July 21, 2019
Processing time: 101 Days and 21.1 Hours
Mucosal healing has become a therapeutic goal to achieve stable remission in patients with inflammatory bowel diseases. To achieve this objective, overlapping actions of complex cellular processes, such as migration, proliferation, and differentiation, are required. These events are longitudinally and tightly controlled by numerous factors including a wide range of distinct regulatory proteins. However, the sequence of events associated with colon mucosal repair after colitis and the evolution of the luminal content characteristics during this process have been little studied.
To document the evolution of colon mucosal characteristics during mucosal healing using a mouse model with chemically-induced colitis.
C57BL/6 male mice were given 3.5% dextran sodium sulfate (DSS) in drinking water for 5 d. They were euthanized 2 (day 7), 5 (day 10), 8 (day 13), and 23 (day 28) d after DSS removal. The colonic luminal environment and epithelial repair processes during the inflammatory flare and colitis resolution were analyzed with reference to a non-DSS treated control group, euthanized at day 0. Epithelial repair events were assessed histo-morphologically in combination with functional permeability tests, expression of key inflammatory and repairing factors, and evaluation of colon mucosa-adherent microbiota composition by 16S rRNA sequencing.
The maximal intensity of colitis was concomitant with maximal alterations of intestinal barrier function and histological damage associated with goblet cell depletion in colon mucosa. It was recorded 2 d after termination of the DSS-treatment, followed by a progressive return to values similar to those of control mice. Although signs of colitis were severe (inflammatory cell infiltrate, crypt disarray, increased permeability) and associated with colonic luminal alterations (hyperosmolarity, dysbiosis, decrease in short-chain fatty acid content), epithelial healing processes were launched early during the inflammatory flare with increased gene expression of certain key epithelial repair modulators, including transforming growth factor-β, interleukin (Il)-15, Il-22, Il-33, and serum amyloid A. Whereas signs of inflammation progressively diminished, luminal colonic environment alterations and microscopic abnormalities of colon mucosa persisted long after colitis induction.
This study shows that colon repair can be initiated in the context of inflamed mucosa associated with alterations of the luminal environment and highlights the longitudinal involvement of key modulators.
Core tip: When colitis was chemically induced with dextran sodium sulfate, 2 d after the end of the treatment, mice showed unequivocal sign of colitis, changes in the luminal environment of the large intestine, epithelial permeability loss, and dysbiosis. These inflammation-induced alterations progressively and partly resolved in the period of time following colitis induction. Early and long-term evaluation of the epithelial repairing process showed overlapping action of inflammatory and repairing markers, rather than successive actions.
- Citation: Vidal-Lletjós S, Andriamihaja M, Blais A, Grauso M, Lepage P, Davila AM, Gaudichon C, Leclerc M, Blachier F, Lan A. Mucosal healing progression after acute colitis in mice. World J Gastroenterol 2019; 25(27): 3572-3589
- URL: https://www.wjgnet.com/1007-9327/full/v25/i27/3572.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i27.3572
Inflammatory bowel diseases (IBD) are chronic inflammatory disorders of the gastrointestinal tract with unclear etiology. These pathologies display relapsing-remitting courses characterized by periods of inflammation and remission over long periods of time[1]. Maintaining stable remission without clinical symptoms and decreasing the incidence and duration of relapses are ultimate goals of IBD treatment[2]. A key step to achieving these objectives is the healing of inflamed mucosa, also known as mucosal healing (MH), described as the complete absence of blood, friability, erosion, and ulcerative lesions in all segments of the gut[3]. Indeed, clinical studies have shown the importance of MH on IBD outcomes, since MH is associated with lasting clinical remission, a decrease in intestinal complication risk, and less recourse to hospitalization and surgery[4].
The healing sequence starts with epithelial restitution, where epithelial cells surrounding the wounded area migrate to reseal the denuded basal membrane[5,6]. Within hours or days after injury, epithelial restitution is followed by additional steps in wound healing. These include increased epithelial cell proliferation to replenish the decreased cell pool, and later, maturation and differentiation of undifferentiated epithelial cells to recover colon physiological functions such as barrier function and water absorption[5,7]. These processes require tight control mechanisms that rely on modulation of numerous factors. They include a wide range of structurally distinct regulatory proteins, such as growth factors, cytokines, other peptide molecules like extracellular matrix (ECM) factors, and non-peptide molecules, including microbiota-derived metabolites[8]. Although their relevance as signal-transducers and modulators of cell function, migration, proliferation and differentiation has been well established individually, mostly by in vitro studies[8,9], the concomitant interaction of these factors with epithelial repair in vivo has been less ascertained. Furthermore, although alterations of microbiota composition during acute colitis have been repeatedly observed in IBD patients[10-12], the impact of epithelial changes during MH on mucosa-adherent microbiota has barely been evaluated. This is despite the observation that it may have a major impact on the colonic epithelium due to its proximity to epithelial cells. Lastly, changes in the bacterial metabolite composition in the luminal fluid facing the colonic and rectal epithelium are also likely to influence the course of inflammatory processes[13].
Given the clinical relevance of MH for IBD patients, it is important to understand the sequence of events and the modulators that intervene in the healing progression as well as how these modifications may influence mucosa-adherent microbiota and, ultimately, the MH outcome. Our aim was to further the current understanding of MH progression after an acute inflammatory episode. We used a kinetics approach with complementary methods to study events involved in epithelial repair and mucosa-adherent microbiota restitution that contribute to the restoration of colon barrier function after an acute inflammatory challenge, which was chemically induced by dextran sodium sulfate (DSS) in mice.
Male C57BL/6 OlaHsd mice [n = 64, 7-week-old, body weight (BW) mean: 22.4 ± 0.2 g, Envigo, France] were acclimated for 1 wk with free access to standard mouse chow and tap water under controlled temperature (23 °C), humidity (55% ± 10%), and light (12:12-h light-dark cycle) conditions. Each mouse was maintained in an individual cage with a grid and acclimated to the diet after 3 d of a standard mouse chow/fresh P14 diet (Table 1). The study was performed according to the European directive for the use and care of laboratory animals (2010/63/UE) and received the approval of the local animal ethics committee and the ministerial committee for animal experimentation (registration number: APAFIS#3987-2016012214388658). Animals were randomly distributed in each group.
| Ingredient, g/kg | P14 |
| Acid casein (Armor Protéines®, ref. 139860) | 112 |
| Whey protein (Armor Protéines®, Protarmor 80, ref. 139805) | 28 |
| Corn starch | 622.4 |
| Sucrose | 100.3 |
| Cellulose | 50 |
| Soybean oil | 40 |
| Mineral mixture (AIN 93-M) | 35 |
| Vitamin mixture (AIN 93-V) | 10 |
| Choline | 2.3 |
| Metabolizable energy, kJ/g | 14.5 |
A normoproteic diet (P14, 140 g/kg milk protein) was used throughout the study (Table 1). Healthy controls (untreated mice at day 0, n = 12) received fresh tap water. DSS-treated mice (n = 52) received 3.5% (w/v) DSS (36000-50000 MW, MP Biomedicals Illkirch-Graffenstaden, France) in drinking water for 5 d, from day 1 to day 5 (fresh DSS solution being prepared every 2 d), to induce an acute episode of colitis[14]. Drink and food were provided ad libitum. DSS, food consumption, and BW were measured daily. Mice were evaluated using an inflammatory score based on stool consistency and rectal bleeding. Each of these elements was rated on a 0-3 scale, 0 representing no disease symptoms and 3 representing severe disease symptoms. Presented data are the sum of both scores. Long-term assessment of mice in the day 28 group was performed by dual-energy x-ray absorptiometry to measure body fat and lean body mass every 9 d using a PIXImus imager (GE Lunar PIXImus, GE Healthcare, Waukesha, WI, United States). In parallel, an in vivo permeability assay was performed on the day 28 group by gavage of 4 kDa paracellular marker fluorescein isothiocyanate-labeled dextran (FD4, Sigma-Aldrich, St. Louis, MO, United States). Food was withdrawn for 4 hours, and mice were gavaged with permeability tracer (0.6 mg/100 g BW of FD4). Plasma was collected from the tail 4 hours later. Plasmatic FD4-concentrations were determined by fluorescence measurement from a standard curve using serial dilutions of FD4 gavage solution. Mice were euthanized at day 10 (n = 12), 13 (n = 12), and 28 (n = 16) to evaluate the impact of diet on epithelial repair kinetics or if they lost more than 20% BW, as per approved animal protocol guidelines, to meet the end point criteria.
At euthanasia, animals were anesthetized by isoflurane inhalation and euthanized by intracardiac puncture. Blood was collected in EDTA-containing tubes and plasma was frozen and kept at -80 °C for measurement of cytokines. The entire colon was removed, measured, and weighed. The colon was divided into six segments. One segment of the proximal colon was mounted in an Ussing chamber. Proximal colon mucosa was scraped, frozen in liquid nitrogen, and stored at -80 °C for subsequent analysis of adherent microbiota. Whole luminal colonic content was then removed for osmolarity and water content measurements. Another segment was harvested for RNA analysis and immediately frozen in TRIzol® Reagent (ThermoFisher Scientific, Waltham, MA, United States) and stored at -80 °C until further analysis. Two other segments were immediately frozen in liquid nitrogen and stored at -80 °C for myeloperoxidase (MPO) and protein expression assays. Two 1 cm segments of the distal colon were fixed in 4% buffered formaldehyde for histological analysis (longitudinal and transversal sections). Osmolarity of colonic content was measured with a freezing point depression osmometer (Löser Type 15 Roebling Osmometer, ThermoFisher Scientific), and water content was calculated as previously described[14].
Colonic samples were homogenized in TRIzol® Reagent using an ultra-turrax and phase separated. RNA was purified using an RNeasy Mini kit (Qiagen SAS, Courtabœuf, France) and DNase I treatment. qRT-PCR was performed with Fast SYBR Green MasterMix (Applied Biosystems, Foster City, CA, United States), gene-specific primers (sequences available on demand), and the StepOne Real-Time PCR system (Applied Biosystems, Life Technologies). Gene expression was determined using the 2-ΔΔCT formula, where ΔΔCT = (CT target gene - CT reference gene) using Hprt as the house-keeping gene and normalized to the day 7 group.
Intestinal tissue was assayed for MPO activity as a neutrophil infiltration marker. Activity analysis was performed using an O-dianisidine dihydrochloride assay as previously described[15]. Colonic cytokine concentrations were measured in total colon protein lysate by Luminex technology using Bio-Plex kits[14] (Bio-Rad, Marnes-La-Coquette, France). Results were expressed as nanograms per milligram of total protein. Plasmatic concentration of lipopolysaccharide-binding protein was determined with commercial solid-phase sandwich enzyme-linked immunosorbent assay from Tebu-Bio (LBP, Pikoline Elisa Kit Mouse, Set220EK1274; Boechout, Belgium).
Colonic sections stained with hematoxylin-and-eosin were coded for blind microscopic assessment by an external histological platform (Histalim, Montpellier, France), and microscopic changes were qualitatively described and scored using a severity scale (0 to 3). The histological score was calculated as the sum of the score of four criteria: goblet cell depletion, ulceration and/or erosion, percentage of crypt damage, and edema. Epithelial repair score was calculated as the sum of the score of gland hyperplasia and presence of mitotic cells, re-epithelialization, and crypt repair. The inflammatory infiltrate (increase in mononuclear cells and/or neutrophils) was also scored depending on the location: 1- mucosa (lamina propria), 2- submucosa, 3- muscularis or serosa. Images were digitalized using a slide scanner (Lamina, Perkin Elmer, Waltham, MA, United States) and the CaseViewer software v. 2.2 (3DHISTECH, Budapest, Hungary). Colonic length of well-oriented epithelial crypt, mucosae, sub-mucosae, and muscularis was determined by image analysis using Pannoramic Viewer software v. 1.15.4 (3DHISTECH, Budapest, Hungary). Periodic Acid Schiff staining was used to visualize mucus-producing cells on 4-µm transversal colon sections counterstained with hematoxylin.
Proximal colon samples, opened along the mesenteric line, were mounted in EasyMount Ussing chambers within 15 minutes of dissection (Physiologic Instrument Inc, San Diego, CA, United States) with an exposed area of 0.1 cm2. Electric measurements were performed as previously described[16]. At the end of each procedure, tissue viability was assessed by adding the cholinergic drug carbachol (10-4 M) on the serosal side.
Total DNA was extracted from 20 mg of scraped mucosa samples using the PowerFecal DNA Isolation kit (MoBio Laboratories, Carlsbad, CA, United States) according to the manufacturer’s protocol. DNA extracts were used for qRT-PCR analysis of the 16S ribosomal genes. Total bacteria were quantified by real-time qPCR using specific primers (HAD-1: 5'- TGGCTCAGGACGAACGCTGGCGGC -3' and HAD-2: 5'- CCTACTGCTGCCTCCCGTAGGAGT-3', annealing at 59 ºC), Fast SYBR Green MasterMix (Applied Biosystems) and the StepOne Real-Time PCR system (Applied Biosystems, Life Technologies). A standard curve was generated from serial dilutions of a known copy number of the target gene cloned into a plasmid vector as previously described[17]. DNA were subjected to PCR amplification of the V3-V4 region of the 16S rDNA gene, and sequencing was performed at the GenoToul INRA platform (Castanet-Tolosan, France) using Illumina technology with MiSeq kit V2 2 x 250 bp. A total of 747538 high-quality sequences were produced in this study, with an average of 8100 reads per sample. Data analysis was performed as previously described[18].
The Kristensen et al[19] method was used to assay short chain fatty acids (SCFA) in cecal content. After bacterial metabolite extraction in supernatant, SCFA were derivatized by esterification and analyzed with a gas chromatograph equipped with a capillary column (30 m, 0.32 mm ID, RestekRtx 502.2) and fitted with a flame ionization detector. SCFA concentrations were determined by external standards with reference to internal standards.
The animal number for each group (n = 12) was the minimal number necessary to obtain statistically exploitable results according to the variables studied and the variability of the model. Results are expressed as means ± standard error. The mean of differential values for each time period was assessed by analysis of variance with Bonferroni post-hoc test. For repeated measurements, time was added as a repeated factor. All analyses were performed with RStudio software version 1.0.143 with packages lme4, car, and lsmeans. For all statistical tests, the level of significance was set to P < 0.05. Principal component analysis was performed at phylum and family-level taxonomy to assess the evolution of adherent-microbiota composition at day 0, 7, 10, 13, and 28. The analysis of similarity, conducted with the ANOSIM test, was used to assess the correlation between ecological distance (based on family composition) and time groups. 16S rDNA analysis was performed using R software and in-house pipeline as previously published[18].
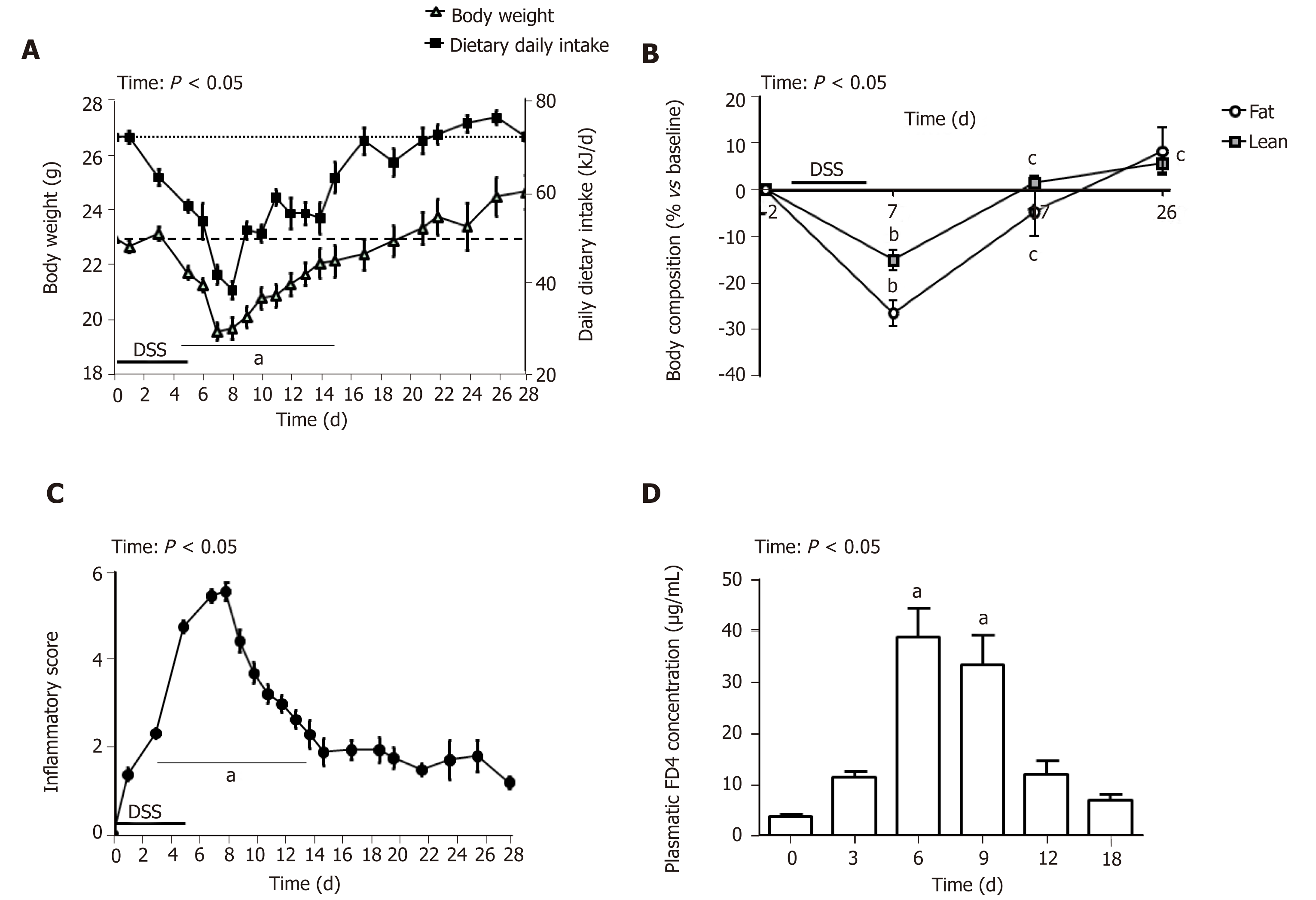
Administration of 3.5% DSS for 5 d induced severe BW loss (Figure 1A) and sharp diminution of the percentage of lean and fat body mass (Figure 1B) at day 7. Mice exposed to DSS experienced an average of 14% weight loss during the first week of DSS administration compared to their initial BW. Food intake follow-up showed a substantial decrease in dietary energy ingestion both during DSS-treatment and until day 7 (58%, P < 0.001) but then progressively increased to reach the baseline amount at day 17 (Figure 1A). The inflammatory score peaked at day 7 and diminished progressively to remain stable from day 15 until the end of the study. A total of 11 mice died or were euthanized because they reached endpoint criteria between day 2 and day 15. This inflammatory state was associated with intestinal barrier leakage that reached its maximum at days 6 and 9 and returned to its initial value at day 12 (Figure 1D). Altogether, follow-up and intestinal permeability data indicate that the maximal intensity of colitis appeared around day 7 (i.e. 2 d after DSS removal) and that inflammation resolution/epithelial repair occurred during the week following the inflammation peak. To understand the succession of events involved in colon healing, they will be described kinetically at critical stages: every 3 d from the inflammatory peak (day 7), therefore, at day 10 and day 13; and 21 d in (day 28) to assess evolution of the inflammatory process and restoration of the intestinal basal state.
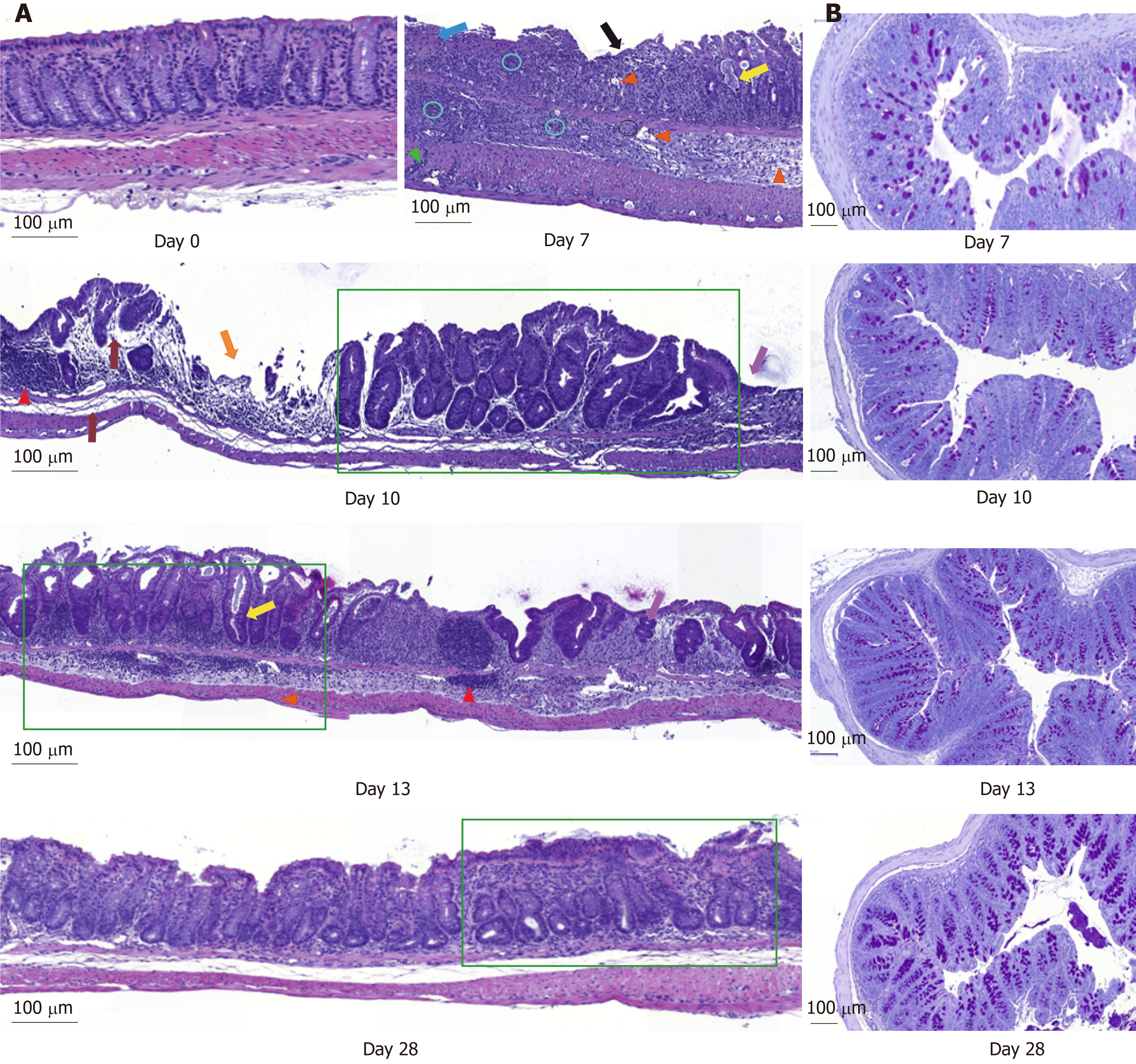
DSS impacts colon crypt architecture and permeability: Intestinal barrier permeability increase coincided with major changes of the colon structure, as evidenced by colon morphometry and histological analysis (Tables 2 and 3, Figure 2). DSS-treated colons were significantly shortened and thickened, as shown by the increase in the weight/length ratio (Table 2). The transmural potential of the proximal colon, measured in an Ussing chamber, was altered (-1.4 ± 0.4 mV in DSS-treated mice vs -4.7 ± 0.5 mV at day 0, P < 0.05). DSS treatment induced severe histological damage, as most mice presented with mucosal multifocal ulceration and/or erosion (Figure 2A) but with different degrees of severity according to the animals examined (severe for 5/11, moderate for 2/11, and minimal for 4/11 mice). All DSS-treated mice were subject to crypt disappearance associated with distension of the remaining crypts and cyst formation (large cystic spaces lined by flattened epithelial cells and filled with pale basophilic mucin) (Figure 2A). Unsurprisingly, DSS treatment induced goblet cell depletion (day 0: 13.36 ± 1.98 vs day 7: 4.95 ± 1.24 Periodic-acid Schiff-positive cells/well-oriented crypt, P < 0.05) and provoked modifications to their morphology. Goblet cell size was larger than usual, with abundant mucin-rich cytoplasm, peripheral nuclei, and open pole (Figure 2B). Accordingly, goblet cell markers were severely impacted by DSS (Table 4), such as the major intestinal secreted mucin Muc2 and the related transcription factor Klf4. These histo-morphological changes were associated with an increased colonic permeability, as evidenced by plasmatic FD4 concentrations (Figure 1D) and a decreased expression of several tight-junction proteins. Gene expression of Zonula Occludens 1 (Tjp1), Occludin (Ocln), and the permissive Claudin 2 (Cldn2) were indeed downregulated at day 7 compared to day 0 (Table 4).
| Parameter | d0 | d7 | d7 vs d0 | d10 | d10 vs d7 | d13 | d13 vs d10 | d28 | d28 vs d0 | Statistical effect (time) |
| Colon length (cm) | 6.5 ± 0.2 | 4.9 ± 0.3a | ↓ | 5.4 ± 0.2a | - | 5.5 ± 0.2a | - | 5.7 ± 0.2a | ↓ | < 0.001 |
| Colon weight/length (mg/cm) | 16 ± 0.3 | 24 ± 0.4a | ↑ | 31 ± 3.0ab | ↑ | 34 ± 3.0ab | - | 38 ± 8.0ab | ↑ | < 0.05 |
| Osmolarity of colonic content | 180 ± 8 | 311 ± 15a | ↑ | 278 ± 8.0a | - | 288 ± 13a | - | 281 ± 15a | ↑ | < 0.01 |
| Colonic water content (%) | 64.6 ± 2.5 | 87.5 ± 2.0a | ↑ | 75.5 ± 3.0ab | ↓ | 76.3 ± 1.5ab | - | 74.0 ± 1.9ab | ↑ | < 0.001 |
| IL-1β (ng/mg of total colon protein) | 9.4 ± 1.8 | 14.3 ± 2.4a | ↑ | 13.7 ± 1.8a | - | 16.2 ± 2.5a | - | 15.4 ± 2.2a | ↑ | < 0.01 |
| IL-6 (ng/mg of total colon protein) | 1.5 ± 0.5 | 3.6 ± 0.6a | ↑ | 4.2 ± 0.8a | - | 4.9 ± 1.2a | - | 2.3 ± 0.3bcd | - | < 0.001 |
| MPO activity (UA/mg total colon protein) | 1.2 ± 0.2 | 5.8 ± 1.2a | ↑ | 5.3 ± 0.8a | - | 3.1 ± 0.8abc | ↓ | 1.9 ± 0.6bcd | - | < 0.001 |
| Plasmatic LBP (µg/mL) | 2.0 ± 0.2 | 5.8 ± 0.4a | ↑ | 4.5 ± 0.5ab | ↓ | 4.9 ± 0.6ab | - | 4.6 ± 0.3c | ↑ | < 0.001 |
| Parameter | d7 | d10 | d10 vs d7 | d13 | d13 vs d10 | d28 | d28 vs d7 | Statistical effect (time) |
| Inflammatory score | 12.8 ± 0.8 | 9.3 ± 1.4b | ↓ | 8.4 ± 1.3b | - | 4.4 ± 1.7bcd | ↓ | < 0.001 |
| Goblet cell depletion | 2.27 ± 0.24 | 1.83 ± 0.21 | - | 1.08 ± 0.26bc | ↓ | 0.88 ± 0.30bc | ↓ | < 0.001 |
| Ulceration and erosion | 2.09 ± 0.29 | 1.34 ± 0.31 | - | 1.17 ± 0.27 | - | 0.63 ± 0.32bcd | ↓ | < 0.001 |
| Crypt damage | 2.72 ± 0.19 | 1.58 ± 0.34b | ↓ | 1.25 ± 0.30b | - | 1.25 ± 0.45b | ↓ | < 0.001 |
| Edema | 2.09 ± 0.09 | 1.25 ± 0.22b | ↓ | 1.42 ± 0.23b | - | 0.0 ± 0.0bcd | ↓ | < 0.001 |
| Inflammatory infiltrate (mononuclear cells) | 1.91 ± 0.27 | 1.83 ± 0.30 | - | 1.75 ± 0.18 | - | 1.40 ± 0.33bcd | - | < 0.001 |
| Inflammatory infiltrate (neutrophils) | 1.73 ± 0.13 | 1.50 ± 0.29 | - | 1.75 ± 0.19 | - | 0.63 ± 0.32bcd | ↓ | < 0.001 |
| Epithelial repair score | 0.82 ± 0.22 | 2.83 ± 0.42b | ↑ | 3.25 ± 0.43b | - | 3.25 ± 0.25b | ↑ | < 0.001 |
| Gland hyperplasia and presence of mitotic cells | 0.0 ± 0.0 | 0.83 ± 0.24b | ↑ | 0.92 ± 0.26b | - | 0.88 ± 0.35b | ↑ | < 0.001 |
| Re-epithelialization | 0.54 ± 0.16 | 0.58 ± 0.15 | - | 0.58 ± 0.15 | - | 0.63 ± 0.33 | - | NS |
| Crypt repair | 0.27 ± 0.19 | 1.42 ± 0.34b | ↑ | 1.75 ± 0.30b | - | 1.75 ± 0.45b | ↑ | < 0.001 |
| d0 | d7 | d7 vs d0 | d10 | d10 vs d7 | d13 | d13 vsd10 | d28 | d28 vsd0 | Statistical effect (time) | |
| Tight-junction protein | ||||||||||
| Cldn2 | 2.41 ± 0.29 | 1.24 ± 0.21a | ↓ | 2.90 ± 0.15b | ↑ | 1.94 ± 0.17b | - | 2.26 ± 0.31b | - | < 0.001 |
| Tjp1 | 1.45 ± 0.09 | 1.03 ± 0.08a | ↓ | 1.17 ± 0.10b | ↑ | 1.30 ± 0.06b | - | 1.29 ± 0.06b | - | < 0.01 |
| Ocln | 2.92 ± 0.37 | 1.18 ± 0.24a | ↓ | 1.36 ± 0.11a | - | 1.96 ± 0.18abc | ↑ | 2.21 ± 0.30bc | - | < 0.001 |
| ECM remodeling | ||||||||||
| Mmp7 | 0.58 ± 0.16 | 1.46 ± 0.40a | ↑ | 5.31 ± 1.37ab | ↑ | 5.21 ± 1.22ab | - | 3.69 ± 0.85ab | - | < 0.001 |
| Mmp9 | 0.13 ± 0.01 | 1.35 ± 0.24a | ↑ | 1.17 ± 0.27a | - | 0.89 ± 0.15a | - | 0.55 ± 0.20bcd | - | < 0.001 |
| Timp1 | 0.05 ± 0.00 | 1.75 ± 0.49a | ↑ | 0.84 ± 0.25ab | ↓ | 0.41 ± 0.05b | - | 0.23 ± 0.06bc | - | < 0.001 |
| Col3a1 | 0.77 ± 0.07 | 1.60 ± 0.33a | ↑ | 1.15 ± 0.15a | - | 1.20 ± 0.12a | - | 0.83 ± 0.16bcd | - | < 0.01 |
| Wisp | 0.31 ± 0.02 | 1.40 ± 0.35a | ↑ | 0.78 ± 0.17ab | ↓ | 0.52 ± 0.04b | - | 0.43 ± 0.06b | - | < 0.001 |
| Acta2 | 1.33 ± 0.17 | 1.25 ± 0.22 | - | 0.95 ± 0.16 | - | 0.79 ± 0.05a | - | 0.85 ± 0.11 | - | < 0.01 |
| Goblet cell markers and mucins | ||||||||||
| Tff3 | 1.80 ± 0.24 | 1.29 ± 0.32 | - | 1.05 ± 0.12ad | - | 1.62 ± 0.19c | ↑ | 1.87 ± 0.13c | - | < 0.05 |
| Muc2 | 3.77 ± 0.64 | 1.10 ± 0.16a | ↓ | 1.72 ± 0.22a | - | 3.03 ± 0.18bc | ↑ | 3.38 ± 0.21bc | - | < 0.001 |
| Klf4 | 2.31 ± 0.13 | 1.12 ± 0.22a | ↓ | 1.52 ± 0.15a | - | 1.73 ± 0.10a | - | 2.56 ± 0.24bcd | - | < 0.001 |
| Epithelial repair modulating factors | ||||||||||
| Igf-1 | 0.27 ± 0.02 | 1.23 ± 0.19a | ↑ | 1.14 ± 0.22ad | - | 0.77 ± 0.06ac | - | 0.55 ± 0.09abcd | - | < 0.001 |
| Il-15 | 4.59 ± 0.89 | 1.32 ± 0.51a | ↓ | 1.67 ± 0.18a | - | 3.61 ± 0.91bc | ↑ | 4.73 ± 0.86bc | - | < 0.01 |
| Il-22 | 0.33 ± 0.20 | 1.59 ± 0.44a | ↑ | 0.36 ± 0.08b | ↓ | 0.47 ± 0.14b | - | 0.18 ± 0.04b | - | < 0.001 |
| Il-33 | 0.08 ± 0.02 | 1.69 ± 0.43a | ↑ | 0.60 ± 0.11ab | ↓ | 0.43 ± 0.05ab | - | 0.26 ± 0.06ab | ↑ | < 0.001 |
| Saa | 1.51 ± 0.27 | 1.21 ± 0.23 | - | 1.05 ± 0.16 | - | 3.40 ± 0.40abc | ↑ | 3.18 ± 0.31abc | ↑ | < 0.001 |
| Tgf-ß1 | 0.50 ± 0.03 | 1.29 ± 0.28a | ↑ | 0.72 ± 0.07b | ↓ | 0.51 ± 0.07b | - | 0.43 ± 0.04b | - | < 0.001 |
| Tgf-ß3 | 1.08 ± 0.11 | 1.47 ± 0.37 | - | 0.72 ± 0.10b | ↓ | 0.93 ± 0.12 | - | 1.16 ± 0.16 | - | < 0.01 |
| Inflammatory markers | ||||||||||
| Il-1ß | 0.04 ± 0.01 | 1.71 ± 0.50a | ↑ | 0.39 ± 0.06ab | ↓ | 0.45 ± 0.07ab | - | 0.39 ± 0.09ab | ↑ | < 0.001 |
| Il-6 | 0.01 ± 0.00 | 1.73 ± 0.51a | ↑ | 0.84 ± 0.28ab | ↓ | 0.41 ± 0.08b | - | 0.15 ± 0.07b | - | < 0.001 |
| Il-10 | 0.23 ± 0.03 | 1.37 ± 0.27a | ↑ | 0.37 ± 0.04b | ↓ | 0.31 ± 0.06b | - | 0.25 ± 0.03b | - | < 0.001 |
| Il-13 | 3.81 ± 0.75 | 1.26 ± 0.23a | ↓ | 0.82 ± 0.08a | - | 0.78 ± 0.10a | - | 2.53 ± 0.39bcd | - | < 0.001 |
| Tnf-α | 0.13 ± 0.01 | 1.29 ± 0.22a | ↑ | 0.46 ± 0.05ab | ↓ | 0.50 ± 0.06ab | - | 0.47 ± 0.07ab | ↑ | < 0.001 |
| Ptgs | 0.14 ± 0.01a | 1.69 ± 0.39b | ↑ | 0.84 ± 0.30c | ↓ | 0.60 ± 0.10 c | - | 0.29 ± 0.05a | - | < 0.001 |
DSS causes inflammatory cell recruitment: DSS-treated animals suffered from subacute to chronic and segmental or diffuse lesions accompanied by a moderate to severe increase in edema (clear spaces). These lesions affected the entire intestinal wall, with particular effects on mucosa and submucosa at day 7 compared to untreated mice at day 0 (Figure 2A and Table 3). DSS-treatment provoked mucosal and submucosal infiltration of mononuclear cells (Figure 2A), mostly diffusely distributed and multifocally extended to the serosa, particularly around the blood vessels. Simultaneously, there were neutrophil clusters within lamina propria and submucosa. Colon MPO activity was indeed markedly increased by a 5-fold factor compared to untreated mice (Table 3), indicating a massive neutrophil infiltration. Analysis of relative mRNA levels showed that pro-inflammatory cell recruitment was associated with a marked upregulation of inflammatory genes such as Il-1β, Il-6, Tnf-α, and cyclooxygenase-2 (Ptgs gene) compared to untreated mice (Table 4). Colon pro-inflammatory cytokine (IL-1β and IL-6) concentrations were accordingly higher than in untreated animals (Table 2). Moreover, this coincided with a tremendous, increased expression of genes that encode key enzymes and structural constituents involved in the ECM remodeling process at day 7 (Table 4). There was also an increased expression of cytokines involved in epithelial repair. Gene expression of Il-22, Il-33, and Tgfβ-1 and -3 were correspondingly upregulated compared to untreated animals, whereas Il-13 and Il-15 gene expressions were downregulated at day 7 after DSS-treatment (Table 4).
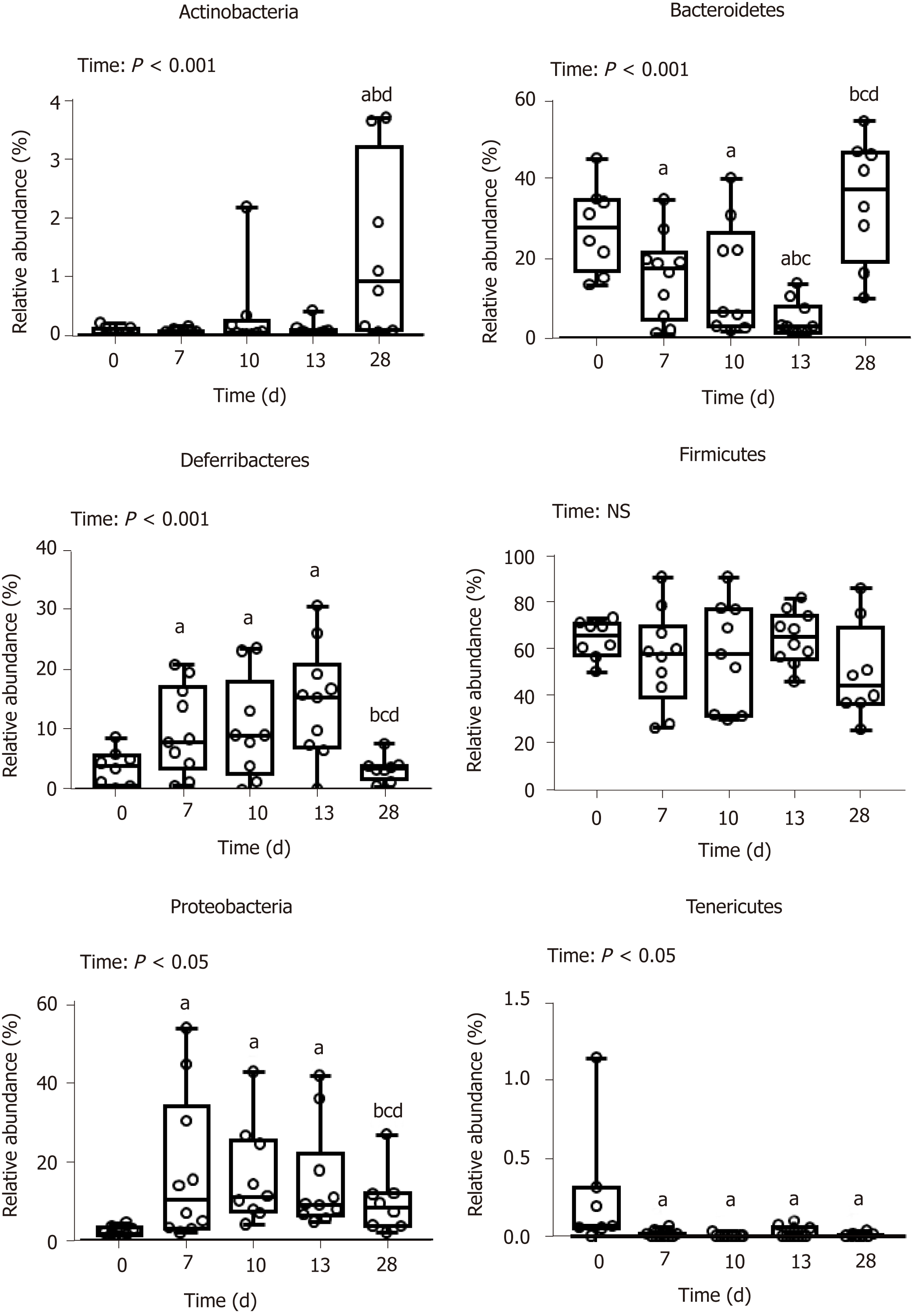
DSS-treatment alters luminal environment and induces microbiota dysbiosis: DSS intake induced luminal content hyperosmolarity and increased the percentage of water in colonic content (Table 2), consistent with the inflammatory score results (Figure 1). The cecal quantities of acetate, propionate, and butyrate (among the most important end products of bacterial fermentation) were significantly reduced at day 7 compared to day 0 (Table 5), while proportions between the three SCFA remained stable. These luminal environment modifications were associated with major changes in mucosa-adherent microbiota composition. While microbiota quantification showed an increase in total bacterial load (Table 5) after DSS-treatment, estimators of community richness (Chao) and diversity (Shannon) were markedly lower than those at day 0 (Table 5). In addition, the number of exclusive and shared species-level phylotypes were significantly reduced after inflammation (463.5 ± 31.4 at day 0 vs 318.1 ± 30.6 at day 7, P = 0.002). Furthermore, DSS-treatment heavily impacted the mucosa-adherent microbiota at a higher taxonomic level, as shown by a decreased relative abundance of over 30 genera, the most relevant presented in Table 5. Colitis also impacted the phyla proportions: Bacteroidetes and Tenericutes decreased by 2 and 20-fold, respectively (P < 0.05); while Proteobacteria and Deferribacteres increased by 8 and 3-fold, respectively (P < 0.05) (Figure 3). Within the Firmicutes phylum, the Eubacteriaceae, Lachnospiraceae, and Clostridiaceae families (the latter including Roseburia, Clostridium XIVa, and Butyrivibrio) were significantly decreased following DSS treatment (Table 5). Conversely, within the Proteobacteria, DSS treatment led to a massive increase in the Escherichia Shigella proportion, accounting for increased representation of the Enterobacteriaceae family (Table 5).
| Parameter | d0 | d7 | d7 vs d0 | d10 | d10 vs d7 | d13 | d13 vs d10 | d28 | d28 vs d0 | Statistical effect (time) | |
| Total bacteria (log/g mucosal content) | 9.36 ± 0.35 | 10.4 ± 0.14a | ↑ | 9.92 ± 0.16b | ↓ | 10.6 ± 0.22ac | ↑ | 10.2 ± 0.24ac | ↑ | < 0.001 | |
| Shannon index | 6.46 ± 0.07 | 4.75 ± 0.32a | ↓ | 4.56 ± 0.28a | - | 4.44 ± 0.26a | - | 5.03 ± 0.29a | ↓ | < 0.001 | |
| Chao index | 794 ± 55.2 | 633 ± 43.7a | ↓ | 593 ± 31.1a | - | 635 ± 34.4a | - | 606 ± 51.6a | ↓ | < 0.01 | |
| Actinobacteria (%) | |||||||||||
| Bifidobacteriaceae | 0.02 ± 0.01 | 0.01 ± 0.00 | - | 0.01 ± 0.01 | - | 0.00 ± 0.00 | - | 0.60 ± 0.28abcd | ↑ | < 0.001 | |
| Coriobacteriaceae | 0.01 ± 0.02 | 0.04 ± 0.02 | - | 0.07 ± 0.04 | - | 0.06 ± 0.04 | - | 0.83 ± 0.31abcd | ↑ | < 0.001 | |
| Bacteroidetes (%) | |||||||||||
| Bacteroidaceae | 4.35 ± 1.24 | 5.6 ± 1.34 | - | 11.55 ± 2.77b | ↑ | 3.08 ± 1.08c | ↓ | 20.1 ± 4.98cd | ↑ | < 0.05 | |
| Porphyromonadaceae | 12.1 ± 2.55 | 7.40 ± 2.08 | - | 2.53 ± 1.29 | - | 1.03 ± 0.35ab | - | 12.1 ± 5.18d | - | < 0.001 | |
| Prevotellaceae | 1.06 ± 0.33 | 0.01 ± 0.00a | ↓ | 0.01 ± 0.01a | - | 0.02 ± 0.01a | - | 0.01 ± 0.01a | ↓ | < 0.01 | |
| Alloprevotella | 1.04 ± 0.32 | 0.00 ± 0.00a | ↓ | 0.01 ± 0.01a | - | 0.01 ± 0.01a | - | 0.00 ± 0.00a | ↓ | < 0.05 | |
| Rikenelleceae | 9.64 ± 1.79 | 0.33 ± 0.24a | ↓ | 0.33 ± 0.24a | - | 0.31 ± 0.26a | - | 2.04 ± 1.20a | ↓ | < 0.001 | |
| Alistipes | 9.48 ± 0.72 | 1.04 ± 0.51a | ↓ | 0.24 ± 0.15a | - | 0.13 ± 0.09a | - | 1.98 ± 1.20a | ↓ | < 0.001 | |
| Deferribacterres (%) | ↓ | ||||||||||
| Deferribacteraceae | Mucispirillum | 3.53 ± 1.86 | 9.98 ± 2.33a | ↑ | 10.16 ± 3.04a | - | 14.82 ± 2.80a | - | 3.49 ± 0.76bcd | - | < 0.001 |
| Firmicutes (%) | |||||||||||
| Clostridiaceae | Roseburia | 0.80 ± 0.17 | 0.01 ± 0.00a | ↓ | 0.01 ± 0.01a | - | 0.01 ± 0.01a | - | 0.03 ± 0.01a | ↓ | < 0.001 |
| Clostridium XIVa | 6.07 ± 0.84 | 1.09 ± 0.35a | ↓ | 5.56 ± 2.73b | ↑ | 8.54 ± 2.80abc | ↑ | 4.01 ± 1.64b | - | < 0.05 | |
| Butyrivibrio | 0.28 ± 0.21 | 0.00 ± 0.00a | ↓ | 0.00 ± 0.00a | - | 0.00 ± 0.00a | - | 0.00 ± 0.00a | ↓ | < 0.01 | |
| Eubacteriaceae | 0.17 ± 0.05 | 0.04 ± 0.02a | ↓ | 0.00 ± 0.00a | - | 0.00 ± 0.00a | - | 0.00 ± 0.00a | ↓ | < 0.001 | |
| Lachnospiraceae | 35.0 ± 4.85 | 13.5 ± 2.91a | ↓ | 21.7 ± 4.25a | - | 23.2 ± 4.49a | - | 23.6 ± 4.69 | - | < 0.01 | |
| Acetitafactor | 5.46 ± 1.47 | 1.28 ± 0.55a | ↓ | 0.30 ± 0.13a | - | 1.11 ± 0.63a | - | 1.00 ± 0.63a | ↓ | < 0.001 | |
| Lactobacillaceae | 0.42 ± 0.19 | 0.24 ± 0.12 | - | 3.41 ± 2.82 | - | 0.63 ± 0.26 | - | 1.44 ± 0.88 | - | NS | |
| Ruminococcaceae | 24.7 ± 4.5 | 30.4 ± 6.4 | - | 18.2 ± 3.6b | ↓ | 26.7 ± 4.0 | - | 21.1 ± 4.0 | - | < 0.05 | |
| Proteobacteria (%) | |||||||||||
| Desulfovibrionaceae | 1.94 ± 0.39 | 1.68 ± 0.32 | - | 3.06 ± 0.64 | - | 3.64 ± 0.95ab | - | 1.30 ± 0.31d | - | < 0.01 | |
| Enterobacteriaceae | 0.24 ± 0.04 | 14.38 ± 5.93a | ↑ | 6.28 ± 2.59a | - | 5.99 ± 3.80a | - | 0.55 ± 0.16bcd | - | < 0.01 | |
| Escherichia Shigella | 0.05 ± 0.03 | 14.08 ± 5.83a | ↑ | 6.02 ± 2.53a | - | 5.47 ± 3.46bc | - | 0.30 ± 0.11bc | - | < 0.01 | |
| Pseudomonadaceae | 0.11 ± 0.04 | 0.06 ± 0.01 | - | 0.06 ± 0.02 | - | 0.18 ± 0.09 | - | 0.21 ± 0.12 | - | NS | |
| Sutterellaceae | 0.05 ± 0.40 | 1.02 ± 0.42 | - | 2.87 ± 0.87 | - | 4.97 ± 1.93 | - | 7.39 ± 2.97 | - | NS | |
| Rhodospirillaceae | 0.14 ± 0.05 | 0.51 ± 0.26a | ↑ | 0.02 ± 0.02b | ↓ | 0.00 ± 0.00b | - | 0.04 ± 0.02b | - | < 0.001 | |
| Tenericutes (%) | |||||||||||
| Anaeroplasmataceae | 0.23 ± 0.13 | 0.00 ± 0.00a | ↓ | 0.00 ± 0.00a | - | 0.01 ± 0.01a | - | 0.00 ± 0.00a | ↓ | < 0.01 | |
| Total SCFA (µmol/g) | 12.4 ± 1.63 | 1.13 ± 0.78a | ↓ | 0.06 ± 0.04a | - | 0.14 ± 0.07a | - | 4.88 ± 1.08abcd | ↓ | < 0.001 | |
| Acetate | 8.95 ± 1.16 | 0.94 ± 0.67a | ↓ | 0.04 ± 0.04ab | ↓ | 0.14 ± 0.07ab | - | 3.40 ± 0.71abcd | ↓ | < 0.001 | |
| Propionate | 1.93 ± 0.29 | 0.04 ± 0.04a | ↓ | 0.00 ± 0.00a | - | 0.00 ± 0.00a | - | 0.90 ± 0.26abcd | ↓ | < 0.001 | |
| Butyrate | 1.51 ± 0.22 | 0.15 ± 0.11a | ↓ | 0.01 ± 0.01a | - | 0.00 ± 0.00a | - | 0.59 ± 0.13abcd | ↓ | < 0.001 | |
Colon inflammation was still evident 5 d after DSS consumption arrest. At day 10, mice still suffered from severe ulceration and goblet cell depletion (Table 3). In one third of the animals, severe transmural inflammation continued to be observed with an increased gap between crypt bases and muscularis mucosae (Figure 2A), with a greater proportion of mononuclear cells. The neutrophils were mostly located at the edges of ulcerated areas showing epithelial exocytosis within the mucosa and/or crypt epithelium. In other animals, light to moderate inflammation was generally located within mucosa and mucosa/submucosa with an equilibrated proportion of mononuclear cells and neutrophils. Colonic MPO activity did indeed remain 4-fold greater than that in the untreated group, and IL-1β and IL-6 colonic concentrations were not different from day 7 (Table 2). However, gene expression of some pro-inflammatory markers (Il-1β, Il-6, Ptgs, Tnf-α) was decreased without reaching baseline values (Table 4). In vivo permeability increase continued to be observed at day 9 (Figure 1D) but was associated with a lower plasmatic concentration of the endotoxemic marker LBP when compared to day 7 (Table 2). This was concomitant and likely explained by increased gene expression of tight junction proteins that started to regain basal levels at day 10 (Table 4).
First signs of epithelial repair are measured in a context of active inflammation: Despite a high inflammatory score, the healing process became visible while the thickening of the colon wall increased (Table 2). Sharp crypt hypertrophy accompanied by mild hyperplasia with increased mitotic figures was indeed observed at day 10 compared to day 7 (Figure 2A and Table 3). The edges of ulcerated areas showed minimal healing by re-epithelization, and edema was graded from moderated to minimal (Table 3). However, qRT-PCR data indicated that most factors that contribute to colon remodeling and epithelial repair had already been induced at day 7, since the expression of corresponding genes was maintained or reduced at day 10. The Mmp7 gene expression was the notable exception with an almost 4–fold increase when compared to day 7 (Table 4).
First healing sequences are associated with major changes in microbiota-adherent mucosa composition: The osmolarity of luminal content remained high, while colon content was less watery at day 10 (Table 2). At that point, the three main SCFA concentrations were still severely diminished, notably the acetate concentration (Table 5). In addition, the relative abundance of the butyrate-producing Clostridium cluster XIVa species was significantly increased compared to day 7, reaching its initial proportion. Nonetheless, microbial dysbiosis remained present compared to day 7, as evidenced by similar proportions of Bacteroidetes, Deferribacteres, Tenericutes, and Proteobacteria phyla (Figures 3 and 4).
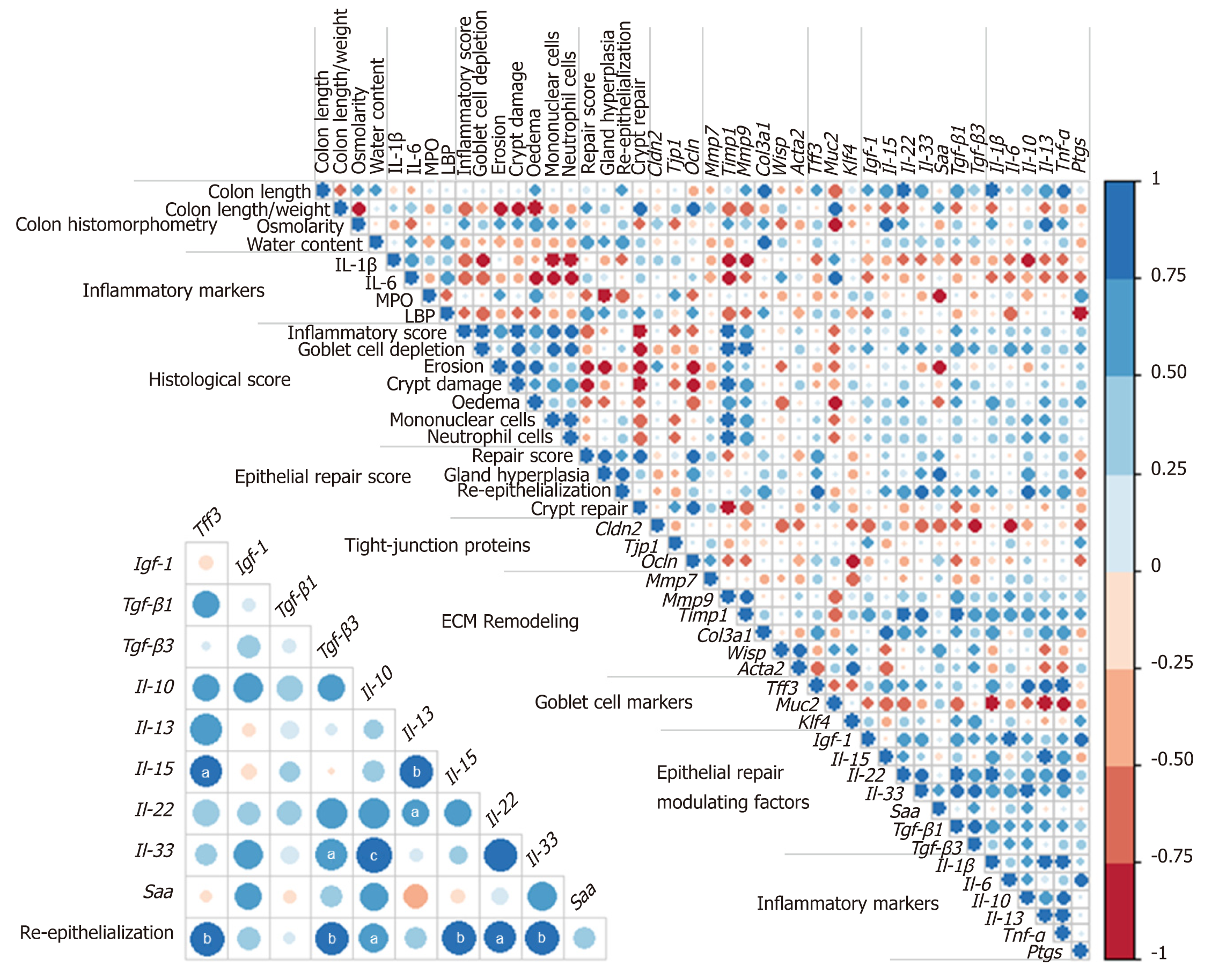
Colon DSS-induced inflammation diminishes in intensity: DSS-treated mice still presented with some crypt abscesses, but lesions were generally less severe than observed in the preceding days (although two animals still presented with a severe transmural leukocytic infiltrate) (Figure 2A). MPO activity was indeed significantly lower than at day 10, but remained higher than at day 0 (Table 2). Half of the mice showed healing by re-epithelization at the edges of ulcerated areas, and most animals showed marked architectural disarray and hyperplasia indicative of crypt regeneration (Figure 2A). These observations were confirmed by the epithelial repair score, which remained elevated at day 13 (Table 3). This was accompanied by improvement of both mechanical and chemical barrier functions, indicated by the increased mRNA expression of Ocln (Table 4) and goblet cell restoration (Figure 2B). Indeed, goblet cell depletion severity score was decreased (Table 3) in parallel with an increased expression of the goblet cell markers Tff3 and Muc2 (Table 4). The correlation analysis performed at day 13 also showed that Tgf-β3, Il-15, Il-22, and Il-33 gene expressions were statistically correlated with the re-epithelization score (Figure 5). The initial Il-15 colonic mRNA level was actually restored, and gene expression of colon Saa (serum amyloid A) was notably increased (Table 4). At day 12, the observed improvement of the intestinal barrier function (Figure 1D) may have been a consequence of these ongoing epithelial repair events.
Mucosa-adherent microbiota remains altered: The total biomass of mucosa-adherent microbiota at day 13 was higher than at day 10. Proteobacteria and Deferribacteres relative percentages remained high at day 13, while Bacteroidetes relative abundance was reduced 3-fold (Figure 3), with a clear reduction in the Bacteroidaceae family (4-fold less, P < 0.001, Table 5). In contrast, the Clostridium cluster XIVa proportion had increased compared to day 10. However, cecal SCFA concentrations remained very low at that time (Table 5).
DSS-treatment exerts long-term effects on colon crypt architecture and per-meability: While numerous parameters associated with colon inflammation did reach baseline values (Tables 2-4), several DSS-induced abnormalities such as colon environmental changes (hyperosmolarity and watery content) persisted. Colon histo-morphology also remained affected, as indicated by a high colon weight/length ratio (Table 2), crypt disarray (Figure 2A), and altered epithelium electrical parameters (transmural potential Vt: -1.2 ± 0.1 mV at day 28 vs -4.7 ± 0.5 mV at day 0, P < 0.05). Mice also still displayed inflammation traits, such as mild ulceration, mucosal erosion (Table 3), and increased gene expression of Tnf-α. Furthermore, higher concentrations of IL-1β in the colon, and of the endotoxemic marker in plasma, were recorded in DSS-treated mice when compared to untreated mice (Table 2). It is noteworthy that colon Il-33 and Saa gene expression remained elevated in DSS-treated mice (Table 4).
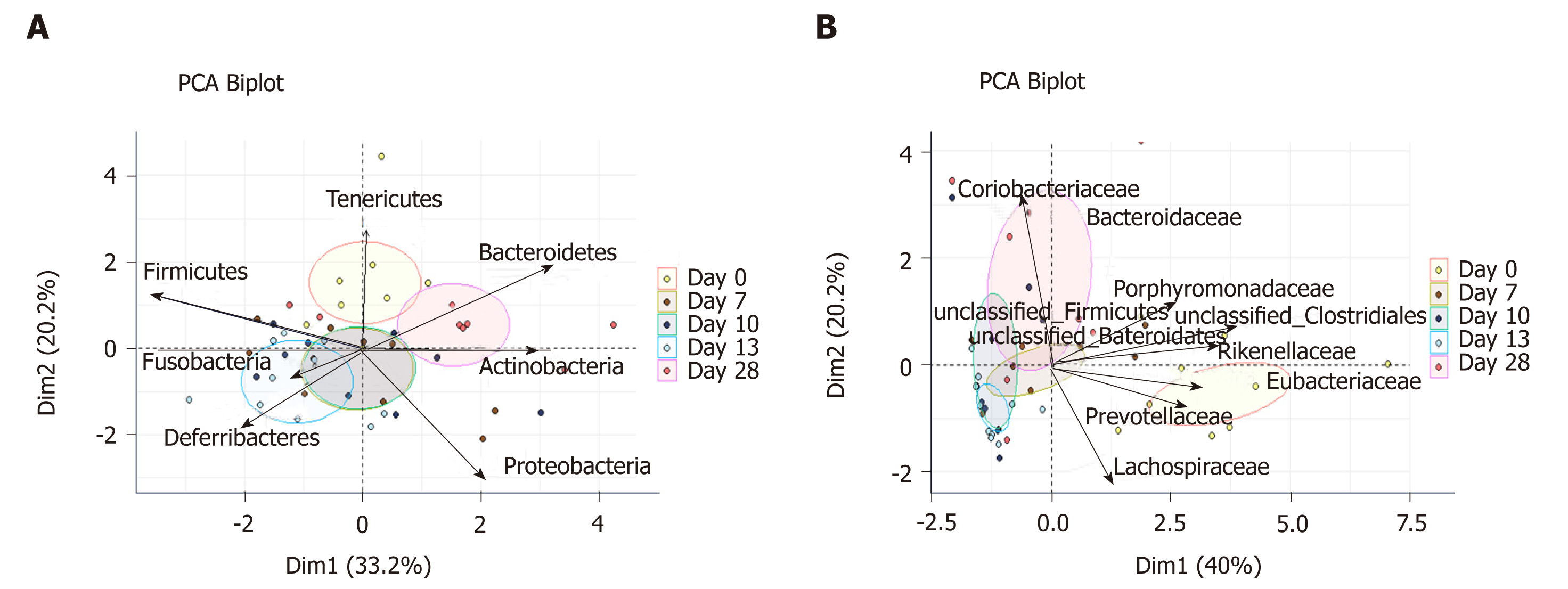
Inflammation alters mucosa-adherent microbiota in the long term: At day 28, dysbiosis was still noticeable, as evidenced by principal component analysis at both phylum and family levels. The different sample clustering between day 0 and day 28 highlighted a distinct microbial structure impairment (Figure 4). Indeed, the total bacterial load associated with the mucosa was slightly but significantly higher than at day 0, while both diversity and community richness indexes remained reduced (Table 5). In addition, while the cecal SCFA had returned to baseline proportions at day 28, the absolute concentrations of these metabolites were 3-fold lower than at day 0 (Table 5). Tenericutes and Actinobacteria were the only two phyla for which a significant difference was still observed at day 28. Proportions of Prevotellaceae and Rikenellaceae families (both from the Bacteroidetes phylum), Clostridiaceae, Eubacteriaceae and Lachnospiraceae (Firmicutes phylum), and Anaeroplasmataceae (Tenericutes phylum) remained reduced. Conversely, both Actinobacteria phylum (Figure 3) and Bacteroidaceae family proportions were strongly increased compared to day 0 (Table 5).
The present study examined the kinetics of molecular and cellular events in association with mucosa-adherent microbiota modifications involved in epithelial repair after acute colon inflammation induced by DSS administration. This study highlighted the heterogeneous responsiveness to DSS and evidenced long-term DSS-induced alterations on the colon luminal environment, notably on adherent microbiota composition, with associated changes in colon histo-morphology. This research also showed that epithelial healing processes were launched early during the inflammatory flare, supporting the concept of intricate involvement of certain key colon factors (Tgf-β, Il-15, Il-22, and Il-33) on epithelial repair modulation. Such factors could be potential therapeutic targets for MH enhancement. Few studies have investigated the mucosa healing signature after an acute intestinal inflammatory flare. Previous studies have rather focused on the evolution of the inflammation markers[20] than on the repairing factors. In addition, most studies were carried out over a shorter period of time after colitis induction[21-23].
DSS-induced colitis is one of the most commonly used animal models of IBD, reflecting many clinical features of ulcerative colitis, such as infiltration of inflammatory cells, crypt loss and erosion, local production of MPO, and inflammatory cytokines in the distal colon[24-27]. This model is generally described as reproducible once the mouse strain and DSS dose/duration have been defined[27-29]. However, this qualitative histological analysis demonstrated relatively high variability in DSS responsiveness among animals, in contrast to other studies[21,23,30]. The reasons for such heterogeneity between animals are not known, but it is tempting to speculate that it is related to animals having been individually housed in this study (in order to accurately follow the inflammatory score and food consumption). Such conditions may amplify differences in the microbiota composition between animals[31]. With this in mind, this study clearly shows that DSS treatment induced common modifications of luminal content physicochemical parameters, such as osmolarity, water, and short-chain fatty acid luminal contents.
Major changes in mucosal-associated microbiota at the peak of colitis and during the days after may have resulted from a luminal increase in oxygen concentration caused by blood presence, in turn induced by inflammatory lesions. For instance, the increased bacterial biomass at mucosal sites at day 7 may be the consequence of an increase in redox potential that promoted development of facultative anaerobic bacteria such as Proteobacteria[32]. At day 10, relative abundance of Lachnospiraceae and Ruminococcaceae was reduced, similar to the microbial dysbiosis observed in IBD patients[33-35]. Eubacteriaceae and Clostridiaceae showed no improvement compared to proportions found in mice euthanized at day 7. This was particularly true for the Roseburia and Butyrivibrio genera, which are generally considered beneficial due to their role in butyrate production. Meanwhile, the proportion of Escherichia Shigella (Proteobacteria), strongly linked to intestinal inflammation[36], increased until day 13. These taxonomic shifts replicated what is generally observed in IBD patient microbiota[11,34,37,38], characterized by chronic dysbiosis even during remission[39]. These results did indeed show a severe colitis-associated dysbiosis of mucosa-associated microbiota, with an α-diversity loss that persisted throughout the resolution phase (from day 10 to day 28). As in ulcerative colitis, where longitudinal variations in mucosal bacterial populations are associated with disease severity[40], depletion of these bacterial families may be linked to observed functional disturbances during the resolution phase, such as altered epithelial barrier and inflammatory flare[41]. The drastic and persistent reduction of SCFA concentrations in the cecum was also likely due to disturbance of the cecal microbiota composition and/or metabolic activity, thus altering SCFA production. Acute lesions in the cecum induced by DSS have been described[42,43], though it is not one of the most common features[27].
This work indicates two important findings: Epithelial repair induction started rapidly even while inflammation was still severe; and although permeability was largely restored 6 d after the inflammatory peak, colon alterations persisted long after, likely because of consequential tissue remodeling. This is supported by observation of the over-expression of inflammatory markers and ECM remodeling factors as well as factors that influence epithelial repair, namely Tgfβ[44], Il-22[45], Il-33[46], and Igf-1[47,48]. This study showed that Tgf-β3, Il-22, Il-33, and Il-15, but not Igf-1, positively correlated with the re-epithelization score at day 13. The expression of Il-33 remained higher than in control animals throughout the experiment; it may, in particular, be a promising therapeutic target in the MH process. While conflicting results were obtained regarding its role in colitis, IL-33 has recently been shown to attenuate colitis and to favor colon repair in mice by promotion of M2 macrophage development and stimulation of goblet cell differentiation[49]. Moreover, the acute phase protein SAA, usually analyzed in plasma, displayed an increased gene expression in the colon 6 d after the maximal intensity of colitis. Importantly, SAA was recently identified as a protective factor against colon epithelium acute injury[50].
Several results obtained in this study regarding Il-15 expression deserve additional attention. This interleukin is considered a deleterious factor in cases of colitis[51]. IL-15 actually promotes intestinal dysbiosis associated with butyrate deficiency and increased susceptibility to colitis[52]. Although severely decreased in this study in the course of colitis induction, IL-15 followed the same expression kinetics as those of epithelial repair, this process being characterized by the restoration of goblet cells and expression of genes coding for tight-junction proteins. Notably, in other pathological contexts, this cytokine has been shown to promote wound repair via TGFβ[53] and IGF-1[54] production, suggesting that IL-15 may be a therapeutic agent to manage wound healing. Additional, specific immunohistochemical assays, outside the scope of the present study, are necessary to identify which cells produce these modulating epithelial repair factors, since they may be synthesized by many cell types in the colon mucosa (colonocytes, innate cells, macrophages, myofibroblasts, or fibroblasts). Unsurprisingly, epithelial repair coincides with goblet cell restoration, as evidenced in this study by Muc2 expression increase and a decreased goblet cell depletion score. This was associated with increased expression of gene coding for the major epithelial repair factor Tff3 6 d after the peak colitis. However, this also indicates that in this model, epithelial repair was triggered by pathways independent of Tff3 since histological improvements had already been observed 3 d before changes in Tff3 expression.
Overall, this histo-morphological analysis, the measurement of several inflammatory markers, the luminal parameters related to the microbiota composition, and SCFA concentrations indicate effects of DSS on colon mucosa that lasted more than 3 wk after DSS removal from drinking water. Although MH is usually considered attained when lesions are no longer visible in the colon after an inflammatory flare, microscopic colon mucosa abnormalities were still present 3 wk after the colitis peak in this study. These alterations might be related to residual inflammatory infiltrate with high expression of pro-inflammatory cytokines (Il-1β and Tnf-α) and Mmp7, whereas other factors involved in ECM remodeling returned to their basal levels. Future studies using an Mmp-7 inhibitor would be useful to evaluate the involvement of this potentially therapeutic modulator in colitis progression[55]. Finally, the resulting mild colon mucosa disarray associated with microscopic inflammation raises questions about clinical consequences for patients without endoscopic lesions, specifically in terms of relapse risk and/or associated complications.
In conclusion, the present study showed the longitudinal evolution of several key parameters associated with inflammation and healing during and after inflammatory flare in a DSS model of colitis. As previously proposed[56], our study offers additional information regarding the imbrication of mucosal inflammation and repairing processes. Such a concept is worth considering for further research aiming at improving intestinal MH by means of therapeutic and nutritional interventions.
Rapid mucosal healing after an intestinal inflammatory episode is considered beneficial to diminishing the relapse risk in inflammatory bowel disease patients. However, the event progression of colon mucosal repair after a colitic flare has barely been studied.
A better understanding of the events associated with inflammation resolution and mucosal healing are necessary to identify potential targets for colon mucosal healing enhancement.
To document longitudinal modifications of the colon mucosa and luminal ecosystem following an episode of chemically-induced colitis.
Evolution of colon mucosa inflammation and healing indicators, as well the changes in colonic luminal environment, were assessed in dextran sodium sulfate-treated mice during the 3 wk after the maximal intensity of colitis. Complementary approaches such as measurement of physicochemical parameters in colonic luminal content, mucosa-adherent microbiota composition and activity, colon mucosa histo-morphological analysis, permeability tests, and expression of numerous factors involved in epithelial inflammation and/or repair were used.
Indications of epithelial repair were observed early, while inflammation was still active. However, colitis-induced luminal colonic environment alterations and microscopic abnormalities of colon mucosa persisted even though inflammation had been resolved.
The longitudinal evolution study of the overlapping events that participated in epithelial repair revealed modulation factors (Il-15, Il-33, and Saa) that may prove to be potential therapeutic targets for mucosal healing enhancement.
Since repairing processes were launched by mucosal inflammation, the interventional time window is an important parameter to take into account in clinical trials aiming to accelerate intestinal mucosal healing.
The authors gratefully acknowledge R. Onifarasoaniaina and M. Favier from the Cochin HistIM Facility, Morgane Dufay who took care of the animals, and Luc Gérard and Gayatri Kessavane for their involvement in the experiment. They also thank Armor Protéines (Saint-Brice-en-Cogles, France) for providing the dietary proteins used to prepare diets.
| 1. | Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2693] [Cited by in RCA: 2770] [Article Influence: 115.4] [Reference Citation Analysis (3)] |
| 2. | Kozuch PL, Hanauer SB. Treatment of inflammatory bowel disease: A review of medical therapy. World J Gastroenterol. 2008;14:354-377. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 165] [Cited by in RCA: 171] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 3. | Pineton de Chambrun G, Peyrin-Biroulet L, Lémann M, Colombel JF. Clinical implications of mucosal healing for the management of IBD. Nat Rev Gastroenterol Hepatol. 2010;7:15-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 359] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 4. | Neurath MF, Travis SP. Mucosal healing in inflammatory bowel diseases: A systematic review. Gut. 2012;61:1619-1635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 676] [Cited by in RCA: 676] [Article Influence: 48.3] [Reference Citation Analysis (1)] |
| 5. | Iizuka M, Konno S. Wound healing of intestinal epithelial cells. World J Gastroenterol. 2011;17:2161-2171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 205] [Cited by in RCA: 207] [Article Influence: 13.8] [Reference Citation Analysis (2)] |
| 6. | Dignass AU, Stow JL, Babyatsky MW. Acute epithelial injury in the rat small intestine in vivo is associated with expanded expression of transforming growth factor alpha and beta. Gut. 1996;38:687-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 51] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 7. | Neurath MF. New targets for mucosal healing and therapy in inflammatory bowel diseases. Mucosal Immunol. 2014;7:6-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 261] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 8. | Sturm A, Dignass AU. Epithelial restitution and wound healing in inflammatory bowel disease. World J Gastroenterol. 2008;14:348-353. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 237] [Cited by in RCA: 267] [Article Influence: 14.8] [Reference Citation Analysis (1)] |
| 9. | Dignass AU. Mechanisms and modulation of intestinal epithelial repair. Inflamm Bowel Dis. 2001;7:68-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 183] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 10. | Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Fölsch UR, Timmis KN, Schreiber S. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 875] [Cited by in RCA: 956] [Article Influence: 43.5] [Reference Citation Analysis (0)] |
| 11. | Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780-13785. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3731] [Cited by in RCA: 3507] [Article Influence: 184.6] [Reference Citation Analysis (1)] |
| 12. | Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: Causation or correlation? Nat Rev Gastroenterol Hepatol. 2017;14:573-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1021] [Cited by in RCA: 1295] [Article Influence: 143.9] [Reference Citation Analysis (0)] |
| 13. | Blachier F, Beaumont M, Andriamihaja M, Davila AM, Lan A, Grauso M, Armand L, Benamouzig R, Tomé D. Changes in the Luminal Environment of the Colonic Epithelial Cells and Physiopathological Consequences. Am J Pathol. 2017;187:476-486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 96] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 14. | Lan A, Blais A, Coelho D, Capron J, Maarouf M, Benamouzig R, Lancha AH, Walker F, Tomé D, Blachier F. Dual effects of a high-protein diet on DSS-treated mice during colitis resolution phase. Am J Physiol Gastrointest Liver Physiol. 2016;311:G624-G633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 15. | Lan A, Blachier F, Benamouzig R, Beaumont M, Barrat C, Coelho D, Lancha A, Kong X, Yin Y, Marie JC, Tomé D. Mucosal healing in inflammatory bowel diseases: Is there a place for nutritional supplementation? Inflamm Bowel Dis. 2015;21:198-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 16. | Blachier F, Beaumont M, Kim E. Cysteine-derived hydrogen sulfide and gut health: A matter of endogenous or bacterial origin. Curr Opin Clin Nutr Metab Care. 2019;22:68-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 135] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 17. | Lee C, Kim J, Shin SG, Hwang S. Absolute and relative QPCR quantification of plasmid copy number in Escherichia coli. J Biotechnol. 2006;123:273-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 451] [Cited by in RCA: 507] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 18. | Chemouny JM, Gleeson PJ, Abbad L, Lauriero G, Boedec E, Le Roux K, Monot C, Bredel M, Bex-Coudrat J, Sannier A, Daugas E, Vrtovsnik F, Gesualdo L, Leclerc M, Berthelot L, Ben Mkaddem S, Lepage P, Monteiro RC. Modulation of the microbiota by oral antibiotics treats immunoglobulin A nephropathy in humanized mice. Nephrol Dial Transplant. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 74] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 19. | Kristensen NB, Gäbel G, Pierzynowski SG, Danfaer A. Portal recovery of short-chain fatty acids infused into the temporarily-isolated and washed reticulo-rumen of sheep. Br J Nutr. 2000;84:477-482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 20. | De Fazio L, Cavazza E, Spisni E, Strillacci A, Centanni M, Candela M, Praticò C, Campieri M, Ricci C, Valerii MC. Longitudinal analysis of inflammation and microbiota dynamics in a model of mild chronic dextran sulfate sodium-induced colitis in mice. World J Gastroenterol. 2014;20:2051-2061. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 59] [Cited by in RCA: 66] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 21. | Yan Y, Kolachala V, Dalmasso G, Nguyen H, Laroui H, Sitaraman SV, Merlin D. Temporal and spatial analysis of clinical and molecular parameters in dextran sodium sulfate induced colitis. PLoS One. 2009;4:e6073. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 264] [Cited by in RCA: 315] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 22. | Bento AF, Leite DF, Marcon R, Claudino RF, Dutra RC, Cola M, Martini AC, Calixto JB. Evaluation of chemical mediators and cellular response during acute and chronic gut inflammatory response induced by dextran sodium sulfate in mice. Biochem Pharmacol. 2012;84:1459-1469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 23. | Laroui H, Ingersoll SA, Liu HC, Baker MT, Ayyadurai S, Charania MA, Laroui F, Yan Y, Sitaraman SV, Merlin D. Dextran sodium sulfate (DSS) induces colitis in mice by forming nano-lipocomplexes with medium-chain-length fatty acids in the colon. PLoS One. 2012;7:e32084. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 205] [Cited by in RCA: 263] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 24. | Wirtz S, Neurath MF. Mouse models of inflammatory bowel disease. Adv Drug Deliv Rev. 2007;59:1073-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 368] [Article Influence: 19.4] [Reference Citation Analysis (1)] |
| 25. | Rose WA 2nd. Sakamoto K, Leifer CA. Multifunctional role of dextran sulfate sodium for in vivo modeling of intestinal diseases. BMC Immunol. 2012;13:41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 26. | Sha T, Igaki K, Yamasaki M, Watanabe T, Tsuchimori N. Establishment and validation of a new semi-chronic dextran sulfate sodium-induced model of colitis in mice. Int Immunopharmacol. 2013;15:23-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 27. | Chassaing B, Aitken JD, Malleshappa M, Vijay-Kumar M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr Protoc Immunol. 2014;104:Unit 15.25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 755] [Cited by in RCA: 1428] [Article Influence: 119.0] [Reference Citation Analysis (1)] |
| 28. | Okayasu I, Hatakeyama S, Yamada M, Ohkusa T, Inagaki Y, Nakaya R. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology. 1990;98:694-702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1733] [Cited by in RCA: 1857] [Article Influence: 51.6] [Reference Citation Analysis (8)] |
| 29. | Eichele DD, Kharbanda KK. Dextran sodium sulfate colitis murine model: An indispensable tool for advancing our understanding of inflammatory bowel diseases pathogenesis. World J Gastroenterol. 2017;23:6016-6029. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 621] [Cited by in RCA: 630] [Article Influence: 70.0] [Reference Citation Analysis (20)] |
| 30. | Mine Y, Zhang H. Anti-inflammatory Effects of Poly-L-lysine in Intestinal Mucosal System Mediated by Calcium-Sensing Receptor Activation. J Agric Food Chem. 2015;63:10437-10447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 31. | Thoene-Reineke C, Fischer A, Friese C, Briesemeister D, Göbel UB, Kammertoens T, Bereswill S, Heimesaat MM. Composition of intestinal microbiota in immune-deficient mice kept in three different housing conditions. PLoS One. 2014;9:e113406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 32. | Litvak Y, Byndloss MX, Tsolis RM, Bäumler AJ. Dysbiotic Proteobacteria expansion: A microbial signature of epithelial dysfunction. Curr Opin Microbiol. 2017;39:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 471] [Article Influence: 52.3] [Reference Citation Analysis (0)] |
| 33. | Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: Current status and the future ahead. Gastroenterology. 2014;146:1489-1499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1145] [Cited by in RCA: 1353] [Article Influence: 112.8] [Reference Citation Analysis (0)] |
| 34. | Matsuoka K, Kanai T. The gut microbiota and inflammatory bowel disease. Semin Immunopathol. 2015;37:47-55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 407] [Cited by in RCA: 601] [Article Influence: 50.1] [Reference Citation Analysis (1)] |
| 35. | Halfvarson J, Brislawn CJ, Lamendella R, Vázquez-Baeza Y, Walters WA, Bramer LM, D'Amato M, Bonfiglio F, McDonald D, Gonzalez A, McClure EE, Dunklebarger MF, Knight R, Jansson JK. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol. 2017;2:17004. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 605] [Cited by in RCA: 862] [Article Influence: 95.8] [Reference Citation Analysis (0)] |
| 36. | Kleessen B, Kroesen AJ, Buhr HJ, Blaut M. Mucosal and invading bacteria in patients with inflammatory bowel disease compared with controls. Scand J Gastroenterol. 2002;37:1034-1041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 249] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 37. | Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 668] [Cited by in RCA: 775] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 38. | Rajilić-Stojanović M. Function of the microbiota. Best Pract Res Clin Gastroenterol. 2013;27:5-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 39. | Walujkar SA, Kumbhare SV, Marathe NP, Patangia DV, Lawate PS, Bharadwaj RS, Shouche YS. Molecular profiling of mucosal tissue associated microbiota in patients manifesting acute exacerbations and remission stage of ulcerative colitis. World J Microbiol Biotechnol. 2018;34:76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 40. | Fite A, Macfarlane S, Furrie E, Bahrami B, Cummings JH, Steinke DT, Macfarlane GT. Longitudinal analyses of gut mucosal microbiotas in ulcerative colitis in relation to patient age and disease severity and duration. J Clin Microbiol. 2013;51:849-856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 95] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 41. | Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: The role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1657] [Cited by in RCA: 1918] [Article Influence: 106.6] [Reference Citation Analysis (0)] |
| 42. | Ohkawara T, Nishihira J, Takeda H, Hige S, Kato M, Sugiyama T, Iwanaga T, Nakamura H, Mizue Y, Asaka M. Amelioration of dextran sulfate sodium-induced colitis by anti-macrophage migration inhibitory factor antibody in mice. Gastroenterology. 2002;123:256-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 127] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 43. | Nakanishi M, Tazawa H, Tsuchiya N, Sugimura T, Tanaka T, Nakagama H. Mouse strain differences in inflammatory responses of colonic mucosa induced by dextran sulfate sodium cause differential susceptibility to PhIP-induced large bowel carcinogenesis. Cancer Sci. 2007;98:1157-1163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 44. | Beck PL, Rosenberg IM, Xavier RJ, Koh T, Wong JF, Podolsky DK. Transforming growth factor-beta mediates intestinal healing and susceptibility to injury in vitro and in vivo through epithelial cells. Am J Pathol. 2003;162:597-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 156] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 45. | Pickert G, Neufert C, Leppkes M, Zheng Y, Wittkopf N, Warntjen M, Lehr HA, Hirth S, Weigmann B, Wirtz S, Ouyang W, Neurath MF, Becker C. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206:1465-1472. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 791] [Cited by in RCA: 885] [Article Influence: 52.1] [Reference Citation Analysis (0)] |
| 46. | Hodzic Z, Schill EM, Bolock AM, Good M. IL-33 and the intestine: The good, the bad, and the inflammatory. Cytokine. 2017;100:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 82] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 47. | Chen T, Zheng F, Tao J, Tan S, Zeng L, Peng X, Wu B. Insulin-Like Growth Factor-1 Contributes to Mucosal Repair by β-Arrestin2-Mediated Extracellular Signal-Related Kinase Signaling in Experimental Colitis. Am J Pathol. 2015;185:2441-2453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 48. | Howarth GS, Xian CJ, Read LC. Insulin-like growth factor-I partially attenuates colonic damage in rats with experimental colitis induced by oral dextran sulphate sodium. Scand J Gastroenterol. 1998;33:180-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 61] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 49. | Seo DH, Che X, Kwak MS, Kim S, Kim JH, Ma HW, Kim DH, Kim TI, Kim WH, Kim SW, Cheon JH. Interleukin-33 regulates intestinal inflammation by modulating macrophages in inflammatory bowel disease. Sci Rep. 2017;7:851. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 89] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 50. | Zhang G, Liu J, Wu L, Fan Y, Sun L, Qian F, Chen D, Ye RD. Elevated Expression of Serum Amyloid A 3 Protects Colon Epithelium Against Acute Injury Through TLR2-Dependent Induction of Neutrophil IL-22 Expression in a Mouse Model of Colitis. Front Immunol. 2018;9:1503. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 51. | Yoshihara K, Yajima T, Kubo C, Yoshikai Y. Role of interleukin 15 in colitis induced by dextran sulphate sodium in mice. Gut. 2006;55:334-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 87] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 52. | Meisel M, Mayassi T, Fehlner-Peach H, Koval JC, O'Brien SL, Hinterleitner R, Lesko K, Kim S, Bouziat R, Chen L, Weber CR, Mazmanian SK, Jabri B, Antonopoulos DA. Interleukin-15 promotes intestinal dysbiosis with butyrate deficiency associated with increased susceptibility to colitis. ISME J. 2017;11:15-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 53. | Kagimoto Y, Yamada H, Ishikawa T, Maeda N, Goshima F, Nishiyama Y, Furue M, Yoshikai Y. A regulatory role of interleukin 15 in wound healing and mucosal infection in mice. J Leukoc Biol. 2008;83:165-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 54. | Wang Y, Bai Y, Li Y, Liang G, Jiang Y, Liu Z, Liu M, Hao J, Zhang X, Hu X, Chen J, Wang R, Yin Z, Wu J, Luo G, He W. IL-15 Enhances Activation and IGF-1 Production of Dendritic Epidermal T Cells to Promote Wound Healing in Diabetic Mice. Front Immunol. 2017;8:1557. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 55. | Hayden DM, Forsyth C, Keshavarzian A. The role of matrix metalloproteinases in intestinal epithelial wound healing during normal and inflammatory states. J Surg Res. 2011;168:315-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 40] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 56. | Liu X, Beaumont M, Walker F, Chaumontet C, Andriamihaja M, Matsumoto H, Khodorova N, Lan A, Gaudichon C, Benamouzig R, Tomé D, Davila AM, Marie JC, Blachier F. Beneficial effects of an amino acid mixture on colonic mucosal healing in rats. Inflamm Bowel Dis. 2013;19:2895-2905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 1.8] [Reference Citation Analysis (1)] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: France
Peer-review report classification
Grade A (Excellent): A, A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Howarth GS, Saraiva Camara NO, Tang ZP, Yang MS S-Editor: Yan JP L-Editor: Filipodia E-Editor: Wang J