Published online Apr 7, 2019. doi: 10.3748/wjg.v25.i13.1580
Peer-review started: January 9, 2019
First decision: February 13, 2019
Revised: March 1, 2019
Accepted: March 11, 2019
Article in press: March 12, 2019
Published online: April 7, 2019
Processing time: 85 Days and 11.3 Hours
Early gastric cancer (EGC), compared with advanced gastric cancer (AGC), has a higher 5-year survival rate. However, due to the lack of typical symptoms and the difficulty in diagnosing EGC, no effective biomarkers exist for the detection of EGC, and gastroscopy is the only detection method.
To provide new biomarkers with high specificity and sensitivity through analyzed the differentially expressed microRNAs (miRNAs) in EGC and AGC and compared them with those in benign gastritis (BG).
We examined the differentially expressed miRNAs in the plasma of 30 patients with EGC, AGC, and BG by miRNA chip analysis. Then, we analyzed and selected the significantly different miRNAs using bioinformatics. Reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR) confirmed the relative transcription level of these miRNAs in another 122 patients, including patients with EGC, AGC, Helicobacter pylori (H. pylori)-negative gastritis (Control-1), and H. pylori-positive atrophic gastritis (Control-2). To establish a diagnostic model for the detection of plasma miRNA in EGC, we chose miRNAs that can be used to determine EGC and AGC from Control-1 and Control-2 and miRNAs in EGC from all other groups.
Among the expression profiles of the miRNA chips in the three groups in the discovery set, of 117 aberrantly expressed miRNAs, 30 confirmed target prediction, whereas 14 were included as potential miRNAs. The RT-qPCR results showed that 14 potential miRNAs expression profiles in the two groups exhibited no differences in terms of H. pylori-negative gastritis (Control-1) and H. pylori-positive atrophic gastritis (Control-2). Hence, these two groups were incorporated into the Control group. A combination of four types of miRNAs, miR-7641, miR-425-5p, miR-1180-3p and miR-122-5p, were used to effectively distinguish the Cancer group (EGC + AGC) from the Control group [area under the curve (AUC) = 0.799, 95% confidence interval (CI): 0.691-0.908, P < 0.001]. Additionally, miR-425-5p, miR-24-3p, miR-1180-3p and miR-122-5p were utilized to distinguish EGC from the Control group (AUC = 0.829, 95%CI: 0.657-1.000, P = 0.001). Moreover, the miR-24-3p expression level in EGC was lower than that in the AGC (AUC = 0.782, 95%CI: 0.571-0.993, P = 0.029), and the miR-4632-5p expression level in EGC was significantly higher than that in AGC (AUC = 0.791, 95%CI: 0.574-1.000, P = 0.024).
The differentially expressed circulatory plasma miR-425-5p, miR-1180-3p, miR-122-5p, miR-24-3p and miR-4632-5p can be regarded as a new potential biomarker panel for the diagnosis of EGC. The prediction and early diagnosis of EGC can be considerably facilitated by combining gastroscopy with the use of these miRNA biomarkers, thereby optimizing the strategy for effective detection of EGC. Nevertheless, larger-scale human experiments are still required to confirm our findings.
Core tip: Early gastric cancer (EGC) has no typical symptoms and difficulty to diagnosis. We filtrated the differentially expressed microRNAs (miRNAs) in the plasma of EGC, advanced gastric cancer and benign gastritis by miRNA chip analysis. Then, reverse transcription quantitative real-time polymerase chain reaction confirmed the relative transcription level of target miRNAs. The 5 plasma miRNAs can be used as new potential biomarkers for the diagnosis of EGC.
- Citation: Zhu XL, Ren LF, Wang HP, Bai ZT, Zhang L, Meng WB, Zhu KX, Ding FH, Miao L, Yan J, Wang YP, Liu YQ, Zhou WC, Li X. Plasma microRNAs as potential new biomarkers for early detection of early gastric cancer. World J Gastroenterol 2019; 25(13): 1580-1591
- URL: https://www.wjgnet.com/1007-9327/full/v25/i13/1580.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i13.1580
Gastric cancer (GC) is one of the most common upper digestive tract cancers, the fourth most commonly diagnosed cancer, and the second leading cause of cancer-related deaths worldwide[1,2]. China is well known to have a high incidence of GC. This incidence and mortality are the second and third highest, respectively, among all cancers, and the incidence and mortality rates are more than two-fold higher than world averages. Moreover, recent reports showed that the incidence rate of GC in Gansu Province, China, is significantly higher than the national rate, which is 55.25/100000, and the mortality rate is 36.94/100000[3], This province ranks first in terms of mortality caused by malignant tumors, accounting for 28.74% of all malignant tumor mortalities, far higher than the national average (30.00/100000, 21.48/100000, 12.80%)[4]. The prognosis of GC is related to the clinical progress and the early diagnosis, and its treatment is of critical significance to the prognosis. In early GC (EGC), the gastric mucosal lesion and the development of invasion cancer do not reach the submucosa, but their spread is limited to the mucous layer. This type of cancer is called intramucosal carcinoma. Most EGC can be treated with radical surgery with gastroscopy. The gastroscopy treatment using endoscopic submucosal dissection (ESD) can completely remove the gastric mucosal lesion, eliminating the pain of laparotomy and organ removal. The application of this approach can increase the survival rate to 90% within five years, whereas the survival rate from advanced GC (AGC) is less than 20%. However, the diagnosis of EGC is difficult due to the absence of typical symptoms. Furthermore, no clinically effective biomarkers have been established for EGC detection. Using simple methods to detect high-risk groups of GC and improve the diagnostic rate of EGC are some of the crucial strategies for prevention of GC. Currently, no reliable method or effective biomarkers for EGC that have high sensitivity and specificity exist for detection of high-risk groups of GC. The early detection and diagnosis of EGC at the subclinical stage would substantially improve the prognosis of GC patients.
It is estimated that approximately two-thirds of the genes in the human body are manipulated by specific microRNA (miRNA) or groups of miRNAs, and more than 60 percent of genes encoding human protein were manipulated by miRNAs. These small miRNAs always target one mRNA or multiple mRNAs and degrade or block the translation of mRNAs by base pairing RNA-induced silencing complex with the target gene mRNA. Additionally, they adjust the expression of genes at the translation level. Previous findings indicate that the results of the testing of the miRNA level in tissues, cells, and body fluids can be used as biomarkers for early diagnosis, treatment, and prognosis of tumors[5]. miRNAs can circulate in the blood in stable extracellular forms. The tests for circulating miRNAs can be employed to establish various disease statuses. Therefore, circulating miRNAs should be considered potential blood-based biomarkers, which are new tools for early diagnosis of cancer[6].
The purpose of this research was to identify the differentially expressed miRNAs in the plasma of patients with EGC, AGC, and benign gastritis (BG) through miRNAs chips and to determine their transcription levels at different stages of the disease through reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR). Finally, new specific biomarkers for detection of EGC were identified.
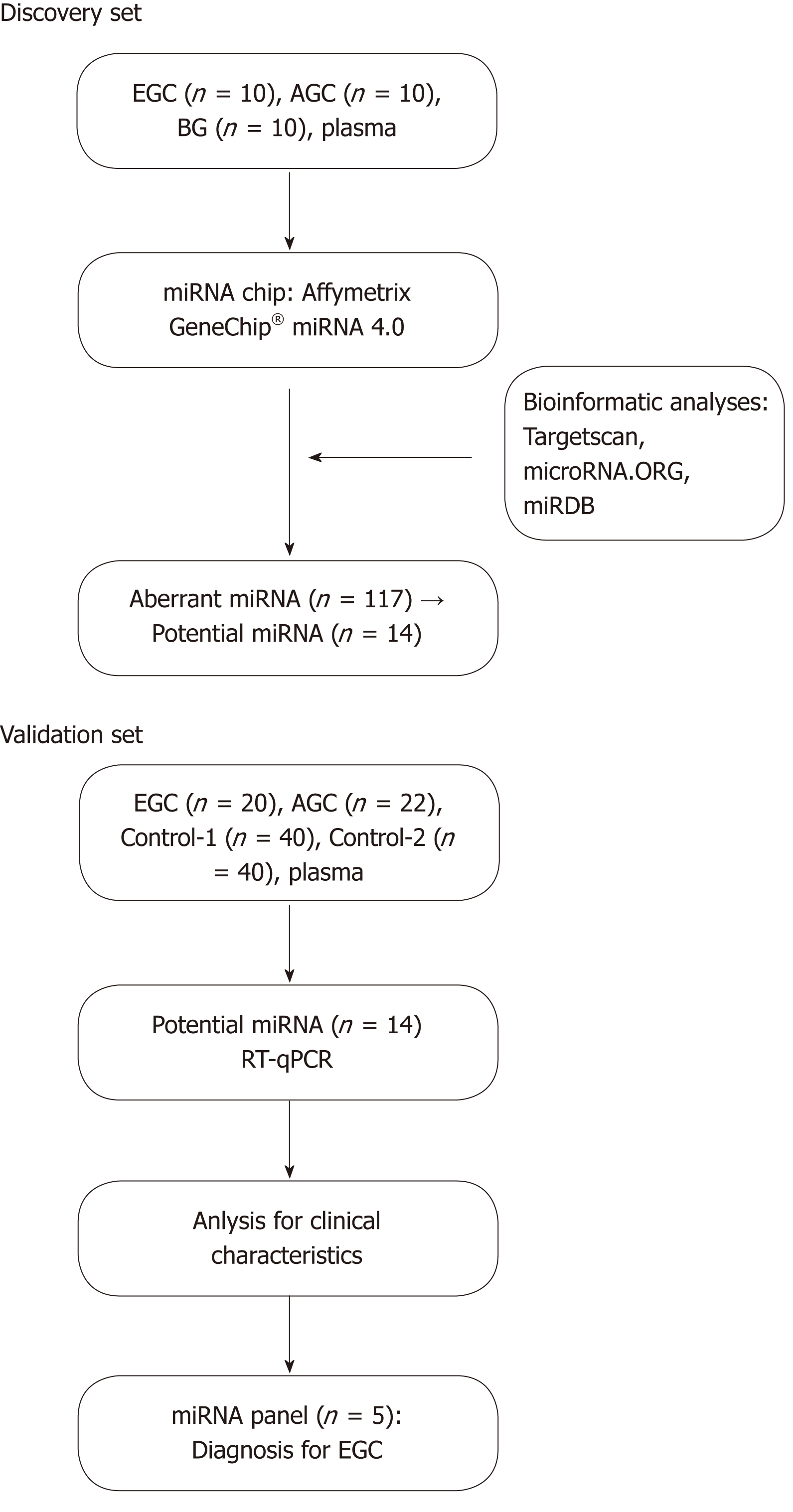
The protocol for this study was approved by the Ethics Committee of the First Hospital of Lanzhou University (Lanzhou, Gansu, China; ethic number: LDYYLL2018-60), and all patients signed informed consent forms. The present study is part of the project of “The Early Cancer Screening Program of the Upper Gastrointestinal Tract of Gansu Province” conducted from July 2016 to December 2017. The patients diagnosed with EGC, AGC, BG, Helicobacter pylori (H. pylori)-negative gastritis (Control-1), and H. pylori-positive atrophic gastritis (Control-2) received a gastroscopy examination and treatment using the OLYMPUS EVIS 290 electronic endoscope system. The biopsy specimen was analyzed and diagnosed by pathologists at the First Hospital of Lanzhou University using the Vienna classification. H. pylori infection was detected by 14C-urea breath test. Blood specimens were analyzed in the Key Laboratory of Biotherapy and Regenerative Medicine of Gansu Province. Patients who were diagnosed for the first time were included, whereas those with other types of previous malignant tumors were excluded (Figure 1).
Samples of 5 mL of blood were collected into EDTA anticoagulant tubes after an overnight fast. After in vitro placement from 30 min to 2 h, each specimen was centrifuged at 3000 r/min for 10 min, followed by transfer of plasma into an EP tube in a liquid nitrogen canister and then transport into a -80 °C refrigerator. We extracted total RNA using TRIzol, and the total RNA was analyzed by NanoDrop 2000 and Agilent Bioanalyzer 2100.
The Affymetrix GeneChip® miRNA 4.0 system (Affymetrix, CA, United States) was used following the manufacturer’s protocol. FlashTagTM Biotin HSR Labeling Kit was utilized for Poly(A) biotin labeling and hybridization. Next, a GeneChip Hybridization Wash and Stain Kit was used to dye the array and pictures, and original data were obtained by scanning.
Differentially expressed miRNA were selected at logFC > 2. The microRNA target prediction was performed with three databases, TargetScan, microRNA.ORG, and miRDB, and the common target genes were obtained. We conducted pathway analysis, followed by enrichment analysis in accordance with the gene information from KEGG and BIOCARTA pathways and showing the results in order of P-value.
Total RNA was extracted from 200 μL plasma using TRIzol, and another 3 referential plasma samples were used for comparison. Synthetic short sequences of reference spike-in 1 (ID3EAL Spike-in control for Isolation, MiRXES, Singapore) RNA were put into lysis buffer. This process was considered quality control for the whole experiment, including extracting RNA, reverse transcription, pre-amplification, and the real-time fluorescent PCR. A target miRNAs sequence was obtained from miRBase 21 to design SYBR Green reverse transcription primers, while reverse transcription and miRNA pre-amplification were performed as described above. SYBR Green real-time fluorescent PCR provided the relative transcription and two technical duplications of every miRNAs in every specimen.
The Ct attenuation value of each type of miRNA in each of the samples was corrected by the internal reference spike-in 1 and three housekeeping genes, miR-454-3p, miR-423-3p, and miR-191-5p, which are stably expressed in humans. The normalized Ct values of the miRNAs are expressed as the mean ± SD. The Wilcoxon rank sum test was used for comparison between the two groups. Receiver-operating characteristics (ROC) curves were used to evaluate the diagnostic value of miRNAs, and logistic regression was used to calculate the weighting coefficients of miRNAs. P < 0.05 was considered to be statistically significant. The software uses SPSS 20.0.
Experiments were conducted with the miRNA chip expression profiles of the plasma of 30 patients with EGC, AGC, and BG. The validation set was based on the plasma RT-qPCR test results of 20 EGC patients, 22 AGC patients, 40 H. pylori-negative gastritis (Control-1) patients, and 40 H. pylori-positive atrophic gastritis (Control-2) patients (Table 1).
| Characteristics | Discovery set (n = 30) | Validation set (n = 122) | |||||
| EGC | AGC | BG | EGC | AGC | Control-1 | Control-2 | |
| Number | 10 | 10 | 10 | 20 | 22 | 40 | 40 |
| Age (yr) (mean ± SD) | 53.4 ± 2.6 | 57.0 ± 7.5 | 51.4 ± 10.0 | 63.3 ± 10.1 | 55.6 ± 13.9 | 50.2 ± 10.3 | 56.1 ± 11.3 |
| Gender | |||||||
| Male | 6 | 6 | 5 | 9 | 12 | 20 | 22 |
| Female | 4 | 4 | 5 | 11 | 10 | 20 | 18 |
| H. pylori | - | + | |||||
| Pathology | |||||||
| HGIN | 4 | 13 | |||||
| Intramucosal cancer | 6 | 7 | |||||
| TNM | |||||||
| HGIN | 4 | 13 | |||||
| T1a | 6 | 7 | |||||
| … | |||||||
| T4N2M0 | 2 | 2 | |||||
| T4N3M0 | 4 | 14 | |||||
| T4N3M1 | 4 | 6 | |||||
The logFC value of the chip expression profiles of the miRNAs from patients with EGC, AGC, and BG had to be larger than 2 (logFC > 2). Differential expression was found in 117 miRNAs of the three groups of miRNA chips. The miRNAs with the biggest differential multiple were screened to identify the target gene predictions in the three databases: TargetScan, microRNA.ORG, and miRDB. A total of 30 miRNAs passed the target gene predictions. Based on previous findings reported in the literature, 14 potential miRNAs were screened.
RT-qPCR tests were run for identification of 14 potential miRNAs, 3 housekeeping genes (miR-454-3p, miR-423-3p, and miR-191-5p), and a spike-in 1 of the internal references using the plasma of 20 EGC, 22 AGC, 40 H. pylori-negative gastritis (Control-1), and 40 H. pylori-positive atrophic gastritis (Control-2) patients. Reverse transcription and advanced amplification of 18 miRNAs mentioned above and the relative expression of miRNAs was read by real-time fluorescence PCR. The results indicated that no obvious differences were available between Control-1 and Control-2 in the relative expression of the 14 miRNAs with differential expression. The P-values were larger than 0.05 on the average. Thus, they were classified as the Control group.
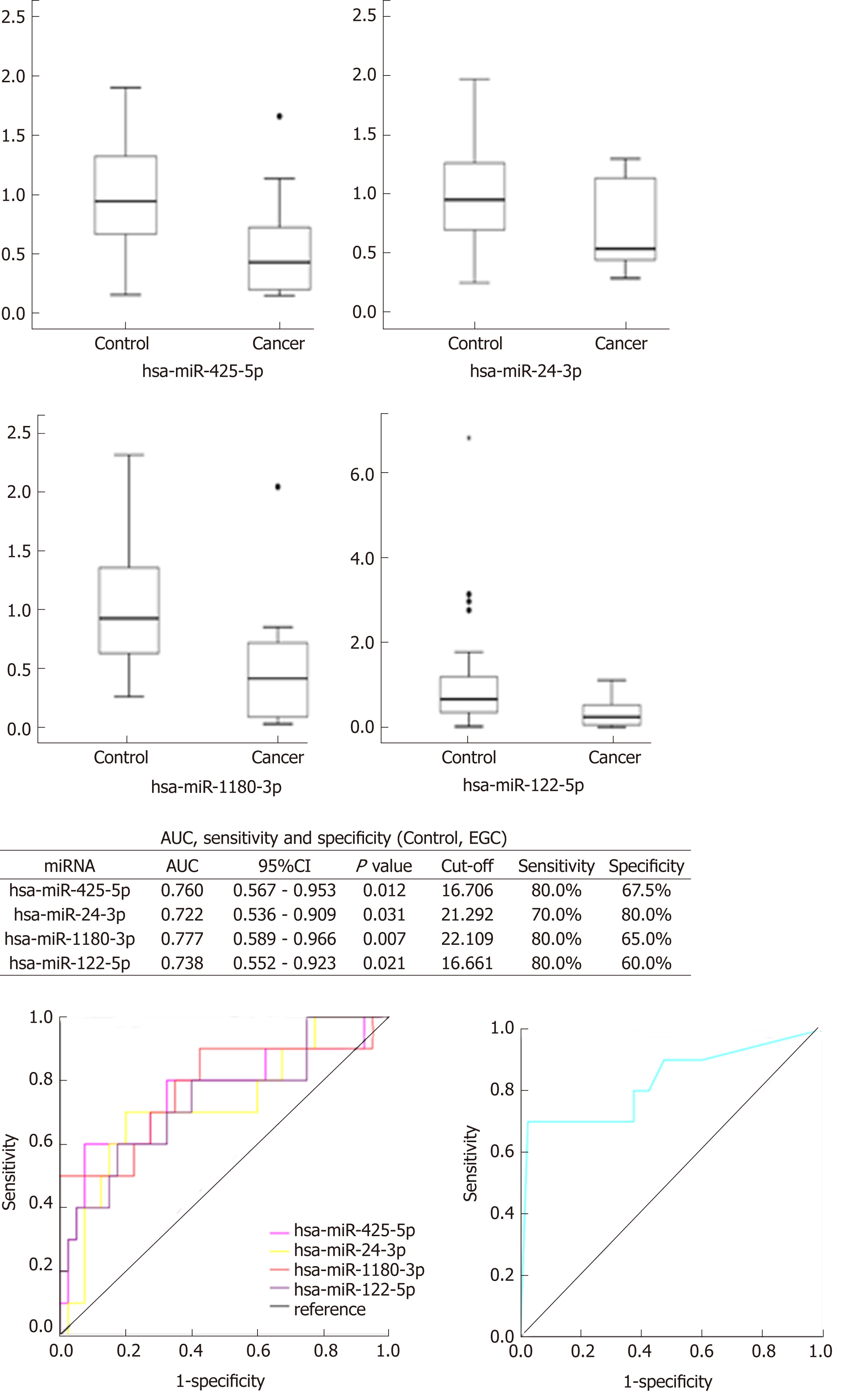
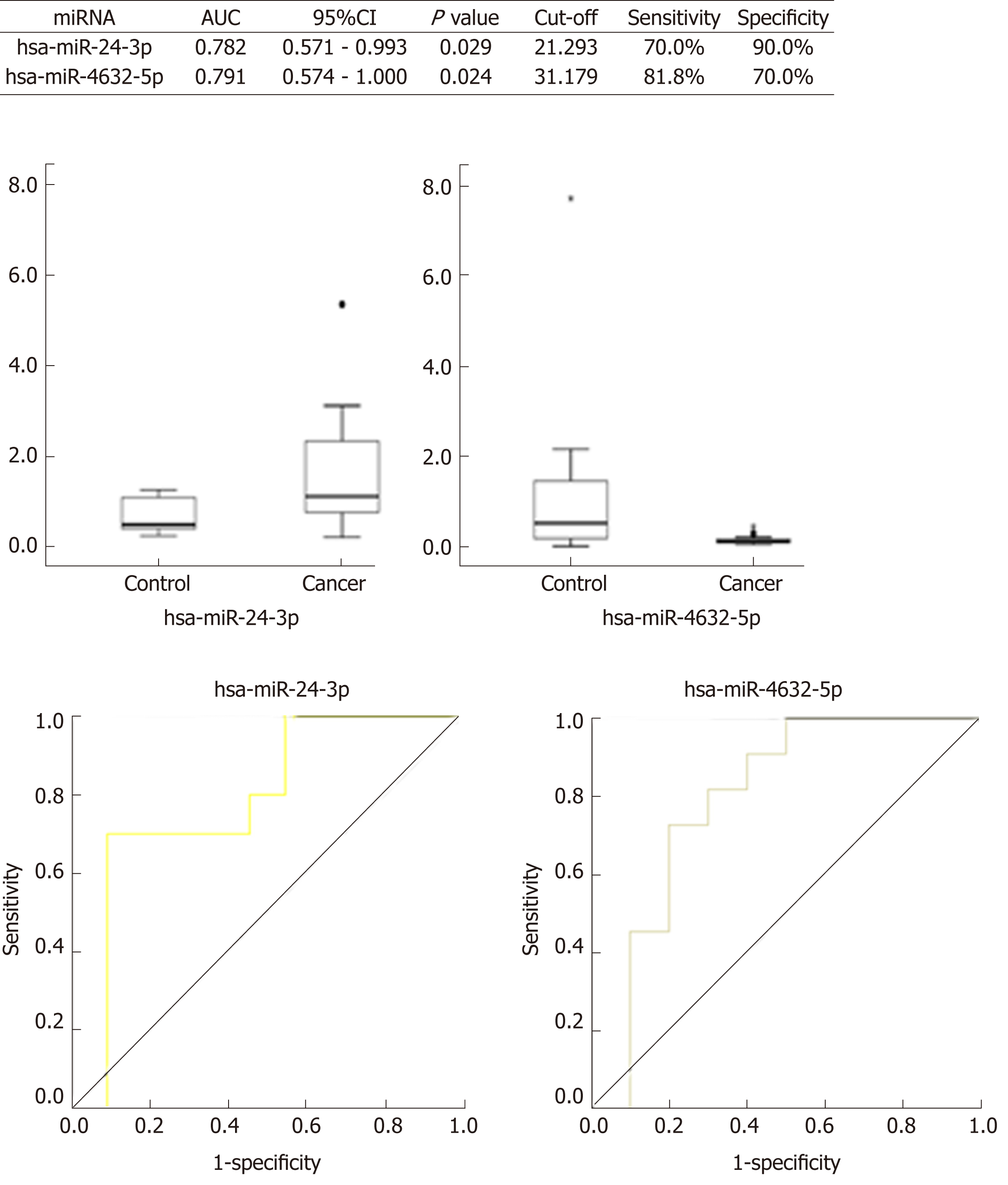
Four miRNAs distinguished the GC group (EGC + AGC) from the Control group, whose P-values, sensitivity, and specificity were as follows, respectively: miR-7641 (P = 0.006, 76.2%, 60.0%), miR-425-5p (P = 0.021, 66.7%, 65.0%), miR-1180-3p (P = 0.001, 81.0%, 60.0%) and miR-122-5p (P = 0.026, 71.4%, 60.0%). The equation was risk score factor (RSF) = 1.569 × miR-7641 + 1.312 × miR-425-5p + 1.852 × miR-1180-3p + 1.322 × miR-122-5p (Figure 2), and the area under the curve (AUC) was 0.799 [95% confidence interval (CI): 0.691-0.908, P < 0.001]. Additionally, four miRNAs distinguished EGC from the Control group, whose P-values, sensitivity, and specificity were as follows, respectively: miR-425-5p (P = 0.012, 80.0%, 67.5%), miR-24-3p (P = 0.031, 70.0%, 80.0%), miR-1180-3p (P = 0.007, 80.0%, 65.0%), miR-122-5p (P = 0.021, 80.0%, 60.0%). The equation was RSF = 2.117 × miR-425-5p + 2.234 × miR-24-3p + 2.005 × miR-1180-3p + 1.792 × miR-122-5p (Figure 3), and the value of AUC was 0.829 (95%CI: 0.657-1.000, P = 0.001). In addition, miR-24-3p and miR-4632-5p were used to distinguish EGC and AGC (P = 0.029, 70.0%, 90.0% and P = 0.024, 81.8%, 70.0%) (Figure 4).
The relative expressions of miR-7641, miR-425-5p, miR-1180-3p and miR-122-5p in the GC group (EGC + AGC) were lower than those in the Control (P < 0.05), with miR-7641 (AUC = 0.714, 95%CI: 0.563-0.865, P = 0.006, sensitivity 76.2%, specificity 60.0%), miR-425-5p (AUC = 0.681, 95%CI: 0.534-0.828, P = 0.021, sensitivity 66.7%, specificity 65.0%), miR-1180-3p (AUC = 0.767, 95%CI: 0.635-0.899, P = 0.001, sensitivity 81.0%, specificity 60.0%) and miR-122-5p (AUC = 0.675, 95%CI: 0.515-0.835, P = 0.026, sensitivity 71.4%, specificity 60.0%), The AUC was significantly increased after combining the four miRNAs (AUC = 0.799, 95%CI: 0.691-0.908, P < 0.001), and the equation was RSF = 1.569 × miR-7641 + 1.312 × miR-425-5p + 1.852 × miR-1180-3p + 1.322 × miR-122-5p. The relative expression levels of miR-425-5p, miR-24-3p, miR-1180-3p and miR-122-5p in the EGC were lower than those in the Control Group, and combining the miRNAs significantly improved the diagnostic value (AUC = 0.829, 95%CI: 0.657-1.000, P = 0.001). The equation was RSF = 2.117 × miR-425-5p + 2.234 × miR-24-3p + 2.005 × miR-1180-3p + 1.792 × miR-122-5p. The expression of miR-24-3p in the EGC group was significantly lower than that in the AGC group (AUC = 0.782, 95%CI: 0.571-0.993, P = 0.029). The expression of miR-4632-5p in the EGC group was significantly higher than that in the AGC (AUC = 0.791, 95%CI: 0.574-1.000, P = 0.024).
According to clinical data for Singapore, just one patient is diagnosed with GC out of every 170 patients who are subjected to gastroscopy. Due to its large population and unbalanced development of the medical career, China provides no universal screening programs or early cancer screening activities in medical institutions at the primary level. Liquid biopsy is used to identify related markers from blood and various body fluid samples instead of invasive tests or biopsy[7]. The risk of gastric mucosa lesion can be found through a test of small quantities of a few bodily fluids, such as 2 mL of venous blood. No classical symptoms are evident in most EGC patients. Thus, high-risk patients can be detected through liquid biopsy as early as possible and then subjected to meticulous gastroscopy. Meticulous examinations such as magnifying endoscopy and dye gastroscopy should be used. In addition to comprehensive pathological examination, the diagnosis rate of EGC will be improved. Therefore, the use of liquid biopsy in EGC screening is minimally invasive, safe, economical, and convenient. In addition, it is suitable for the screening of a wide range of people, which makes it valuable for the improvement of EGC diagnosis and treatment.
In a previous study, Li et al[8] used a miRNA microarray chip analysis with GC patients and discovered the simultaneous presence of upregulated and downregulated miRNA expression profiles. In addition, Tsai et al[9] reported that miR-196a/b was upregulated in both the plasma and tissue of GC patients. Moreover, Fang et al[10] found that some carcinogenesis-related miRNAs (miR-10b, miR-21, miR-223, and miR-338) and tumor suppressor miRNAs (miR-30a-5p, miR-126, and let-7a) can be used as prognosis markers in GC patients. The findings from Zhou et al[11] show that the differential expression of plasma miRNAs can be utilized as diagnostic markers of GC. Furthermore, plasma miR-106b, miR-20a, and miR-221 can be employed as new types of noninvasive markers for the early diagnosis of GC, whereas miR-21 can be used as a maker of early (stage I) and advanced (stage IV) GC. Jiang detected high expression of miR-421 in EGC and suggested that it could serve as a diagnostic marker[12]. Additionally, Fehmida et al[13] discovered that miR-200c-3p was considerably downregulated in EGC tissue, which can be used for early diagnosis of the disease. Racial differences exist in the expression of GC-related miRNAs. It is noteworthy that Li discovered that the level of miRNA-199a-3p in the plasma of EGC patients changed with the treatment, but its expression level was not related to depth of tumor infiltration[14]. In many studies, the use of combinations of circulating miRNAs can lead to high diagnostic accuracy, which is indicated by an area under the ROC curve larger than 0.8. Although, limited research has been conducted, similar diagnostic accuracy was reported using long noncoding RNAs or proteins of extracellular vesicles[15].
At present, a number of studies have been carried out on the use of miRNAs in GC patients related to the occurrence, development, diagnosis, treatment, and prognosis of the disease. Therefore, miRNAs can be used as early diagnostic markers of GC. However, substantial discrepancies exist among the findings of those studies, and there is no consistent conclusion. Relatively few studies have been conducted on the application of miRNAs in the diagnosis and screening of EGC. In the present investigation, we used “The Early Cancer Screening Program of the Upper Gastrointestinal Tract of Gansu Province” biobank. The differences in the expression profiles of the plasma miRNAs in EGC, AGC, and BG patients were established through miRNA chip analysis, and 14 potential miRNAs were screened through bioinformatics analysis. Since H. pylori infection is a well-known GC carcinogen, and atrophic gastritis is an exceedingly common precancerous disease, EGC screening should eliminate the negative influence of test sensitivity and specificity to which precancerous diseases such as H. pylori infection, atrophic gastritis, and precancerous lesion might contribute to. Thus, H. pylori-negative gastritis (Control-1) and H. pylori-positive atrophic gastritis (Control-2) groups should be available in the validation set to replace the BG group in the discovery set to eliminate the influence that precancerous diseases such as H. pylori infection and atrophic gastritis might have on miRNA metabolism.
Initially, this study found no statistically significant difference among the relative expression levels of 14 potential miRNAs from the groups of H. pylori-negative gastritis and H. pylori-positive atrophic gastritis patients. Hence, these patients were combined and classified as a Control group. The combination of miR-7641, miR-425-5p, miR-1180-3p and miR-122-5p distinguished the Cancer group, which included EGC and AGC, from the Control group, whose AUC was 0.799 (95%CI: 0.691-0.908, P < 0.001). On the other hand, the combination of miR-425-5p, miR-24-3p, miR-1180-3p and miR-122-5p distinguished the EGC group from the Control group. The AUC was 0.829 (95%CI: 0.657-1.000, P = 0.001). Additionally, miR-24-3p and miR-4632-5p can be used to distinguish EGC from AGC. Of note, screening strategies can be employed during the statistical analysis of the relative expression and pathological results of RT-qPCR testing of target miRNAs. First, miR-7641, miR-425-5p, miR-1180-3p and miR-122-5p should be used to distinguish EGC and AGC from the Control group. In addition, EGC can be compared to and distinguished from the Control group using miR-425-5p, miR-24-3p, miR-1180-3p and miR-122-5p. Finally, miR-425-5p, miR-1180-3p and miR-122-5p are shared potential miRNAs, whereas miR-24-3p and miR-4632-5p can distinguish EGC from AGC. Thus, a joint testing mode that contains miR-425-5p, miR-1180-3p, miR-122-5p, miR-24-3p and miR-4632-5p can be established and used for predictive EGC diagnosis.
No relevant research is present in the existing literature on the value of miRNA as a biomarker for screening and diagnosis of EGC. At present, the correlation between hsa-miR-1180-3p and hsa-miR-4632-5p and GC, especially EGC, has not been reported. miR-425-5p is upregulated in human GC and promotes its invasion and metastasis in vitro and in vivo, which may serve as a predictor of poor prognosis[16,17]. Earlier studies found that miR-425-5p passed the mechanism of ubiquitinating enzyme (CYLD) as an oncogene and promoted the progression of GC, whereas competitive endogenous RNA (ceRNA) targeting miR-425-5p inhibited the development of GC by p53[18,19]. Moreover, Xu et al[20] discovered that the expression of miR-122-5p was downregulated in GC tissues and cells, whereas the overexpression of miR-122-5p inhibited the migration and invasion of GC cells and the occurrence of lung metastases by downregulating DUSP4. These findings indicate that miR-122-5p inhibits GC cell proliferation and induces apoptosis by targeting MYC[21]. miR-24-3p targets and negatively regulates Prdx-6, which significantly inhibits the growth, migration, and invasion of GC cell lines and promotes apoptosis. Of note, H. pylori infection may decrease the expression of miR-24-3p[22].
In the present investigation, we found that the abnormal expression levels of plasma miR-425-5p, miR-1180-3p, miR-122-5p, miR-24-3p, and miR-4632-5p can be used as a new combination of specific biomarkers for predictive diagnosis of EGC. In addition, we established a predictive diagnosis mode for EGC circulating plasma miRNAs markers, which can be used as a new specific combination of biomarkers for predictive diagnosis of EGC. In such a way, more EGC patients will be diagnosed after combining the application of this mode with gastroscopy, and the strategies for EGC screening will be considerably improved. It has a working mode, in which liquid biopsy can be used to identify high-risk groups and shrink the screening scope, while using gastroscopy can be implemented for diagnosis of EGC patients. Moreover, the screening efficiency should be improved, which requires verification of our findings in a larger sample size of people. This replication will be our objective in the next screening project of “The Early Cancer Screening Program of the Upper Gastrointestinal Tract of Gansu Province”.
In summary, there are increasing demand on early diagnosis of GC, especially the EGC for the more higher survival rate than AGC after radical treatment. The stability of miRNAs makes it suitable biomarker in liquid biopsy, and previous and this study showed the miRNAs pannel maybe have better sensitivity and specificity. We first identified that five miRNAs can be used in early detection of EGC, the pannel will useful to screening high risk population, nevertheless, larger-scale human experiments are still required to confirm our findings.
The early gastric cancer means the gastric mucosal lesion and the development of invasion cancer do not reach the submucosa, but their spread is limited to the mucous layer. Compared with advanced gastric cancer, it performs better in prognosis. Yet due to the lack of typical symptoms and biomarkers, the early gastric cancer is hard to be diagnosed and the golden standard is gastroscope. However, due to the patient's acceptance with certain risks and the difficulty in gastroscopic diagnosis, the detection rate of early gastric cancer in China is still low. Some existed research suggests that microRNA (miRNA) in the peripheral blood can be used as biomarkers in the diagnosis and prognosis of gastric cancer especially for advanced gastric cancer, but few shows whether miRNA can be used as the biomarker for predictive diagnosis of early gastric cancer.
MiRNA is relatively stable in the circulation system, and the detection technique is simple and easy to popularize. The several combinations of miRNAs can improve the accuracy of diagnosis which found in kinds of cancers. Through the predictive diagnosis model of the combination of several miRNAs, screening out high-risk or suspect patients and then being confirmed by gastroscopy combined with biopsy, will improve the accuracy and efficiency of gastroscopy, and promote the detection of early gastric cancer and early diagnosis of advanced gastric cancer, thereby improving the overall treatment effect and prognosis of gastric cancer.
The focus of the study is whether miRNA in peripheral blood can be used as sensitive and specific biomarkers for predictive diagnosis of early gastric cancer. First of all, it is to be studied that whether there is one or there are several miRNAs used to diagnose gastric cancer and non-cancer. Furthermore, it is to be known that whether such miRNAs can be used to suggest the occurrence of early gastric cancer. If there are indeed such miRNAs, it needs to be known that whether it can be applied in the screening of early gastric cancer and the sensitivity and specificity as well as influence factors. All these are possibly applied to clinical practice.
First, in the discovery set, miRNA array was applied to detect the differential expressions of plasma miRNA in the early and advanced gastric cancer patients as well as the control group. Then through the bioinformatics, miRNAs possibly related to disease staging were screened out. In the validation set, in order to rule out the effects of Helicobacter pylori (H. pylori) infection, atrophic gastritis and other diseases on miRNA, the control group was divided into H. pylori infection with atrophic gastritis and H. pylori-negative superficial gastritis. Then RT-qPCR was used to verify target miRNAs selected in the last stage and miRNAs or combinations that may be used for predictive diagnosis of early gastric cancer are selected.
Fourteen target miRNAs were screened from the miRNA array by bioinformatics and they show differential expressions in early and advanced gastric cancer and control group. Subsequent reverse transcription quantitative real-time polymerase chain reaction verification suggested that five miRNAs combinations might be used for predictive diagnosis of early and advanced gastric cancer while not being affected by diseases such as H. pylori infection and atrophic gastritis.
In this article, we found that miRNAs in early and advanced gastric cancer as well as control group show differential expressions. Through further confirmatory experiment, it is found that combinations of several miRNAs may suggest the occurrence of early and advanced gastric cancer. Gastroscopy combined with biopsy can be used to further confirm the diagnosis and then this combination of miRNAs may be regarded as the biomarker of predictive diagnosis of early gastric cancer.
Currently, there is a lack of effective tumor biomarker in the diagnosis of gastric cancer, which is related to the heterogeneity of tumors. Although gastroscopy and biopsy are the golden standards for the diagnosis of gastric cancer, they are currently difficult to spread in China due to the large population. This study hopes to find high-risk patients with early gastric cancer in the population through simple and economical liquid biopsy of new miRNA biomarkers. Then gastroscope and pathological examination can be used to confirm the diagnosis and treatment of early gastric cancer, and early diagnosis and early treatment to advanced gastric cancer, so as to improve the overall prognosis and curative effect of gastric cancer. Certainly, the new combinations of these miRNAs biomarkers need to be further validated in a larger sample population.
| 1. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9172] [Cited by in RCA: 9980] [Article Influence: 907.3] [Reference Citation Analysis (15)] |
| 2. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18694] [Cited by in RCA: 21466] [Article Influence: 1951.5] [Reference Citation Analysis (6)] |
| 3. | Liu Y, Zhang X, Chen L, Zhao Q, Xia X. Cancer incidence and mortality in Gansu province, 2012. Chin J Cancer Res. 2016;28:301-310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 4. | Yang Z, Zeng H, Xia R, Liu Q, Sun K, Zheng R, Zhang S, Xia C, Li H, Liu S, Zhang Z, Liu Y, Guo G, Song G, Zhu Y, Wu X, Song B, Liao X, Chen Y, Wei W, Zhuang G, Chen W. Annual cost of illness of stomach and esophageal cancer patients in urban and rural areas in China: A multi-center study. Chin J Cancer Res. 2018;30:439-448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 5. | Cheng G. Circulating miRNAs: roles in cancer diagnosis, prognosis and therapy. Adv Drug Deliv Rev. 2015;81:75-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 258] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 6. | Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11:145-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 704] [Cited by in RCA: 857] [Article Influence: 71.4] [Reference Citation Analysis (0)] |
| 7. | Tsujiura M, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Otsuji E. Liquid biopsy of gastric cancer patients: circulating tumor cells and cell-free nucleic acids. World J Gastroenterol. 2014;20:3265-3286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 45] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 8. | Li X, Luo F, Li Q, Xu M, Feng D, Zhang G, Wu W. Identification of new aberrantly expressed miRNAs in intestinal-type gastric cancer and its clinical significance. Oncol Rep. 2011;26:1431-1439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 45] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 9. | Tsai MM, Wang CS, Tsai CY, Huang CG, Lee KF, Huang HW, Lin YH, Chi HC, Kuo LM, Lu PH, Lin KH. Circulating microRNA-196a/b are novel biomarkers associated with metastatic gastric cancer. Eur J Cancer. 2016;64:137-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (1)] |
| 10. | Fang Y, Shen H, Li H, Cao Y, Qin R, Long L, Zhu X, Xie C, Xu W. miR-106a confers cisplatin resistance by regulating PTEN/Akt pathway in gastric cancer cells. Acta Biochim Biophys Sin (Shanghai). 2013;45:963-972. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 11. | Zhou X, Zhu W, Li H, Wen W, Cheng W, Wang F, Wu Y, Qi L, Fan Y, Chen Y, Ding Y, Xu J, Qian J, Huang Z, Wang T, Zhu D, Shu Y, Liu P. Diagnostic value of a plasma microRNA signature in gastric cancer: a microRNA expression analysis. Sci Rep. 2015;5:11251. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 110] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 12. | Jiang Z, Guo J, Xiao B, Miao Y, Huang R, Li D, Zhang Y. Increased expression of miR-421 in human gastric carcinoma and its clinical association. J Gastroenterol. 2010;45:17-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 115] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 13. | Bibi F, Naseer MI, Alvi SA, Yasir M, Jiman-Fatani AA, Sawan A, Abuzenadah AM, Al-Qahtani MH, Azhar EI. microRNA analysis of gastric cancer patients from Saudi Arabian population. BMC Genomics. 2016;17:751. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 14. | Li C, Li JF, Cai Q, Qiu QQ, Yan M, Liu BY, Zhu ZG. MiRNA-199a-3p: A potential circulating diagnostic biomarker for early gastric cancer. J Surg Oncol. 2013;108:89-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 70] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 15. | Matsuzaki J, Ochiya T. Circulating microRNAs and extracellular vesicles as potential cancer biomarkers: a systematic review. Int J Clin Oncol. 2017;22:413-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 90] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 16. | Zhang Z, Li Y, Fan L, Zhao Q, Tan B, Li Z, Zang A. microRNA-425-5p is upregulated in human gastric cancer and contributes to invasion and metastasis in vitro and in vivo. Exp Ther Med. 2015;9:1617-1622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 17. | Zhang Z, Wen M, Guo J, Shi J, Wang Z, Tan B, Zhang G, Zheng X, Zhang A. Clinical value of miR-425-5p detection and its association with cell proliferation and apoptosis of gastric cancer. Pathol Res Pract. 2017;213:929-937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 18. | Yan YF, Gong FM, Wang BS, Zheng W. MiR-425-5p promotes tumor progression via modulation of CYLD in gastric cancer. Eur Rev Med Pharmacol Sci. 2017;21:2130-2136. [PubMed] |
| 19. | Chen Z, Ju H, Yu S, Zhao T, Jing X, Li P, Jia J, Li N, Tan B, Li Y. Prader-Willi region non-protein coding RNA 1 suppressed gastric cancer growth as a competing endogenous RNA of miR-425-5p. Clin Sci (Lond). 2018;132:1003-1019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 20. | Xu X, Gao F, Wang J, Tao L, Ye J, Ding L, Ji W, Chen X. MiR-122-5p inhibits cell migration and invasion in gastric cancer by down-regulating DUSP4. Cancer Biol Ther. 2018;19:427-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 76] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 21. | Pei ZJ, Zhang ZG, Hu AX, Yang F, Gai Y. miR-122-5p inhibits tumor cell proliferation and induces apoptosis by targeting MYC in gastric cancer cells. Pharmazie. 2017;72:344-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 22. | Li Q, Wang N, Wei H, Li C, Wu J, Yang G. miR-24-3p Regulates Progression of Gastric Mucosal Lesions and Suppresses Proliferation and Invasiveness of N87 Via Peroxiredoxin 6. Dig Dis Sci. 2016;61:3486-3497. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Burada F S-Editor: Ma RY L-Editor: A E-Editor: Ma YJ