Published online Mar 14, 2019. doi: 10.3748/wjg.v25.i10.1197
Peer-review started: December 14, 2018
First decision: January 18, 2019
Revised: February 13, 2019
Accepted: February 15, 2019
Article in press: February 16, 2019
Published online: March 14, 2019
Processing time: 90 Days and 7.7 Hours
It was shown in previous studies that high definition endoscopy, high magnification endoscopy and image enhancement technologies, such as chromoendoscopy and digital chromoendoscopy [narrow-band imaging (NBI), i-Scan] facilitate the detection and classification of colonic polyps during endoscopic sessions. However, there are no comprehensive studies so far that analyze which endoscopic imaging modalities facilitate the automated classification of colonic polyps. In this work, we investigate the impact of endoscopic imaging modalities on the results of computer-assisted diagnosis systems for colonic polyp staging.
To assess which endoscopic imaging modalities are best suited for the computer-assisted staging of colonic polyps.
In our experiments, we apply twelve state-of-the-art feature extraction methods for the classification of colonic polyps to five endoscopic image databases of colonic lesions. For this purpose, we employ a specifically designed experimental setup to avoid biases in the outcomes caused by differing numbers of images per image database. The image databases were obtained using different imaging modalities. Two databases were obtained by high-definition endoscopy in combination with i-Scan technology (one with chromoendoscopy and one without chromoendoscopy). Three databases were obtained by high-magnification endoscopy (two databases using narrow band imaging and one using chromoendoscopy). The lesions are categorized into non-neoplastic and neoplastic according to the histological diagnosis.
Generally, it is feature-dependent which imaging modalities achieve high results and which do not. For the high-definition image databases, we achieved overall classification rates of up to 79.2% with chromoendoscopy and 88.9% without chromoendoscopy. In the case of the database obtained by high-magnification chromoendoscopy, the classification rates were up to 81.4%. For the combination of high-magnification endoscopy with NBI, results of up to 97.4% for one database and up to 84% for the other were achieved. Non-neoplastic lesions were classified more accurately in general than non-neoplastic lesions. It was shown that the image recording conditions highly affect the performance of automated diagnosis systems and partly contribute to a stronger effect on the staging results than the used imaging modality.
Chromoendoscopy has a negative impact on the results of the methods. NBI is better suited than chromoendoscopy. High-definition and high-magnification endoscopy are equally suited.
Core tip: To determine which endoscopic imaging modalities are most suited for the automated diagnosis of colonic polyps, we apply a high number of state-of-the-art diagnosis systems to 5 endoscopic image databases obtained by different imaging modalities. We show that narrow-band imaging is well suited, whereas chromoendoscopy clearly decreases the results. High-definition and high-magnification endoscopy perform equally well. The image recording conditions have a great impact on the performance of the automated diagnosis systems.
- Citation: Wimmer G, Gadermayr M, Wolkersdörfer G, Kwitt R, Tamaki T, Tischendorf J, Häfner M, Yoshida S, Tanaka S, Merhof D, Uhl A. Quest for the best endoscopic imaging modality for computer-assisted colonic polyp staging. World J Gastroenterol 2019; 25(10): 1197-1209
- URL: https://www.wjgnet.com/1007-9327/full/v25/i10/1197.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i10.1197
The demand for cost saving strategies with respect to discovery, treatment and surveillance of precursor lesions of colorectal cancer is high.
To date, the discovery of areas or lesions of interest depends on the endoscopist’s awareness, skills, experience and knowledge. Subsequent evaluation relies mainly on a variety of established classification systems[1] that utilize different image enhancement technologies.
Detection, surveillance and treatment all require high diagnostic accuracy. Threshold criteria have been suggested by professional societies like the American Society for Gastrointestinal Endoscopy (ASGE)[2].
Although image enhancement technologies such as chromoendoscopy and digital chromoendoscopy [e.g., narrow-band imaging (NBI), Pentax’s i-Scan or Fujinon’s FICE] improve the detection, characterization and classification of colonic precursor lesions, it has been argued that this improvement applies for specialist centers only. Technical features of modern endoscopes and the body of information, which have to be applied during endoscopy, are rising steadily and, therefore, pose a challenge to the endoscopist. In fact, even after educational programs to train endoscopists, not all ASGE goals could be met by the majority of doctors, leading to misdiagnoses and inadequate surveillance strategies[3].
Chromoendoscopy can be subdivided into conventional chromoendoscopy (CC) and digital chromoendoscopy: (1) CC came into clinical use 40 years ago. Staining the mucosa using (indigo-carmine) dye spray enables an easier detection and staging of colonic polyps; and (2) digital chromoendoscopy is a technique to facilitate ’chromoendoscopy without dyes’[4] and can be subdivided into optical (NBI, blue laser imaging) and virtual chromoendoscopy (FICE, i-Scan).
In this work, we evaluate which of the image enhancement technologies (except for the FICE and the BLI system) and endoscopic modalities [high-definition (HD) or high-magnification (HM)] are most suited for computer-assisted colonic polyp staging. For this purpose, we utilize endoscopic imagery obtained from HD as well as HM endoscopes.
Clinical scenarios for the application of automated polyp staging systems are that the endoscopist either receives a staging suggestion after triggering the classification procedure to be applied to some captured area or that a continuous automated mucosa analysis is performed raising alarm in case of the identification of potentially malignant and precursor lesions.
In this work, we aim to answer the following two questions which were only partly or not at all answered in the previous literature: (1) which imaging modalities are best suited for automated colonic polyp staging systems? And (2) how strong is the influence of the image recording conditions on the results of automated diagnosis systems?
To answer these questions, we apply experiments with twelve different feature extraction methods on five colonic polyp image data sets from different imaging modalities.
To determine which imaging modality is best suited to stage colonic polyps, we use a specifically designed experimental setup to avoid biases in the outcome caused by differing numbers of images per modality. Only one prior work systematically analyzed the influence of imaging modalities on the results of automated diagnosis systems. However, only HD endoscopic imagery in combination with i-Scan technology, chromoendoscopy and traditional white light (WL) endoscopy was used. In this work, we additionally employ HM and NBI endoscopic imagery and a more systematical approach is taken.
To answer the second question, we analyze the influence of the image recording conditions, such as the viewpoint or the quality of the recorded images on the results of automated diagnosis systems. To the best of our knowledge, this has not been done before for automated polyp diagnosis systems.
In the following, we (1) briefly summarize findings from different clinical studies, comparing the effectiveness of endoscopic imaging modalities for the detection and/or staging of colonic polyps for human endoscopists; and (2) review related algorithms for the automated staging of colonic polyps.
Modality comparison - human endoscopist: Clinical studies have shown that i-Scan technology, NBI and chromoendoscopy can be considered equivalent and typically achieve better prediction results compared to traditional WL endoscopy in case of HD endoscopes[5,6].
It has been shown that HM NBI and HM chromoendoscopy achieve similar results for the diagnosis of colorectal polyps (non-neoplastic vs neoplastic)[7,8]. Additionally, HM NBI and HM chromoendoscopy achieve higher diagnosis accuracies than standard WL endoscopy[8].
In a survey on HD and HM endoscopes[9], it was shown that HM endoscopes as well as HD endoscopes, with or without mucosal enhancement techniques, improve the detection and diagnosis of mucosal lesions compared to low definition endoscopes. In a survey about digital chromoendoscopy techniques in colonoscopy[10], it was found that NBI, i-Scan and FICE show encouraging results in predicting the histopathology of colonic polyps.
In summary, HM and HD- endoscopy both improve the detection and diagnosis of colonic polyps compared to low-definition endoscopy. Furthermore, image enhancement technologies such as i-Scan, NBI and chromoendoscopy generally improve the diagnosis of colonic polyps compared to standard WL endoscopy.
Computer-assisted polyp staging: The topic of this work is the computer-assisted staging of colonic polyps using endoscopic image data gathered from flexible endoscopes with different imaging enhancement technologies. We do not deal with automated detection of colonic polyps based on geometric properties (which is more often conducted for capsule endoscopic data due to the high effort required for human annotation of these data).
Previous works on computer-assisted staging of colonic polyps in combination with different image enhancement technologies can be divided in three categories depending on the used imaging modality: (1) HD endoscopes combined with or without staining the mucosa and the i-Scan technology: Previous works employed shape and contrast features extracted from blobs[11], fractal analysis-based features[12] and convolutional neural networks[13]. (2) HM chromoendoscopy: Former studies estimated the pit density using Delaunay triangulation[14], applied local binary patterns (LBP)[15] and extracted features from wavelet transform subbands[16-18]. And (3) HM endoscopy with NBI: Dense SIFT features were applied[19,20], and features based on the vessel structure were extracted[21].
In this work, we use the two-class classification scheme differentiating between non-neoplastic (normal mucosa and hyperplastic polyps) and neoplastic lesions (adenomatous and malignant polyps). A sub-classification between adenomatous and malignant lesions cannot be performed because of missing label information for the two NBI databases and the very limited amount of malignant polyps for the remaining three databases (Table 1).
| Non-neoplastic | Neoplastic | Total | ||
| HDNC | Images | 144 | 334 (301 A, 33 M) | 478 |
| Patients | 39 | 77 | 81 | |
| HDC | Images | 213 | 307 (276 A, 31 M) | 520 |
| Patients | 50 | 78 | 84 | |
| CCHM | Images | 198 | 518 (420 A, 98 M) | 716 |
| Patients | 14 | 32 | 40 | |
| NBI-A | Images | 173 | 214 | 387 |
| Patients | 98 | 135 | 211 | |
| NBI-H | Images | 112 | 186 | 298 |
| Patients | 84 | 120 | 182 |
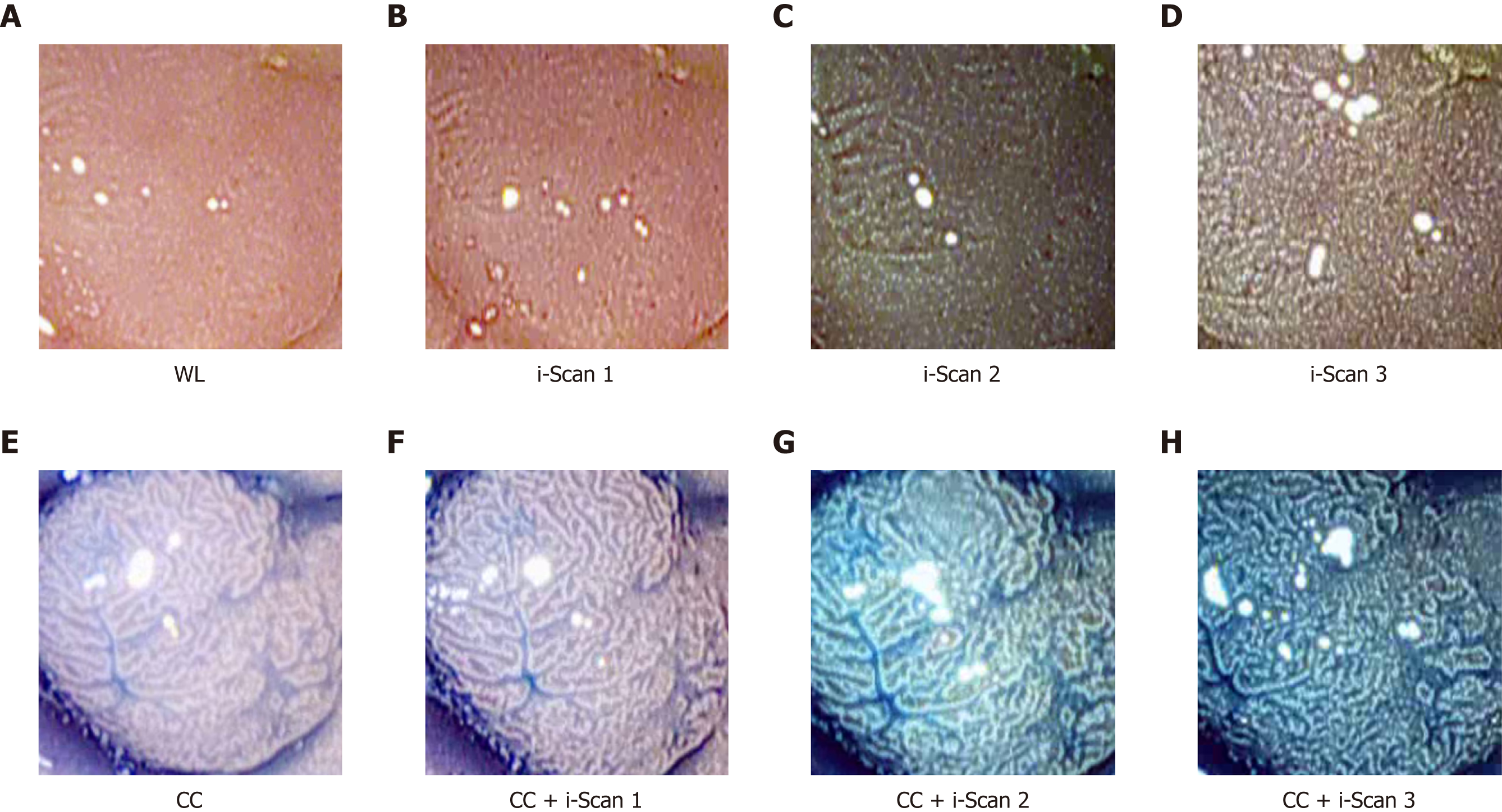
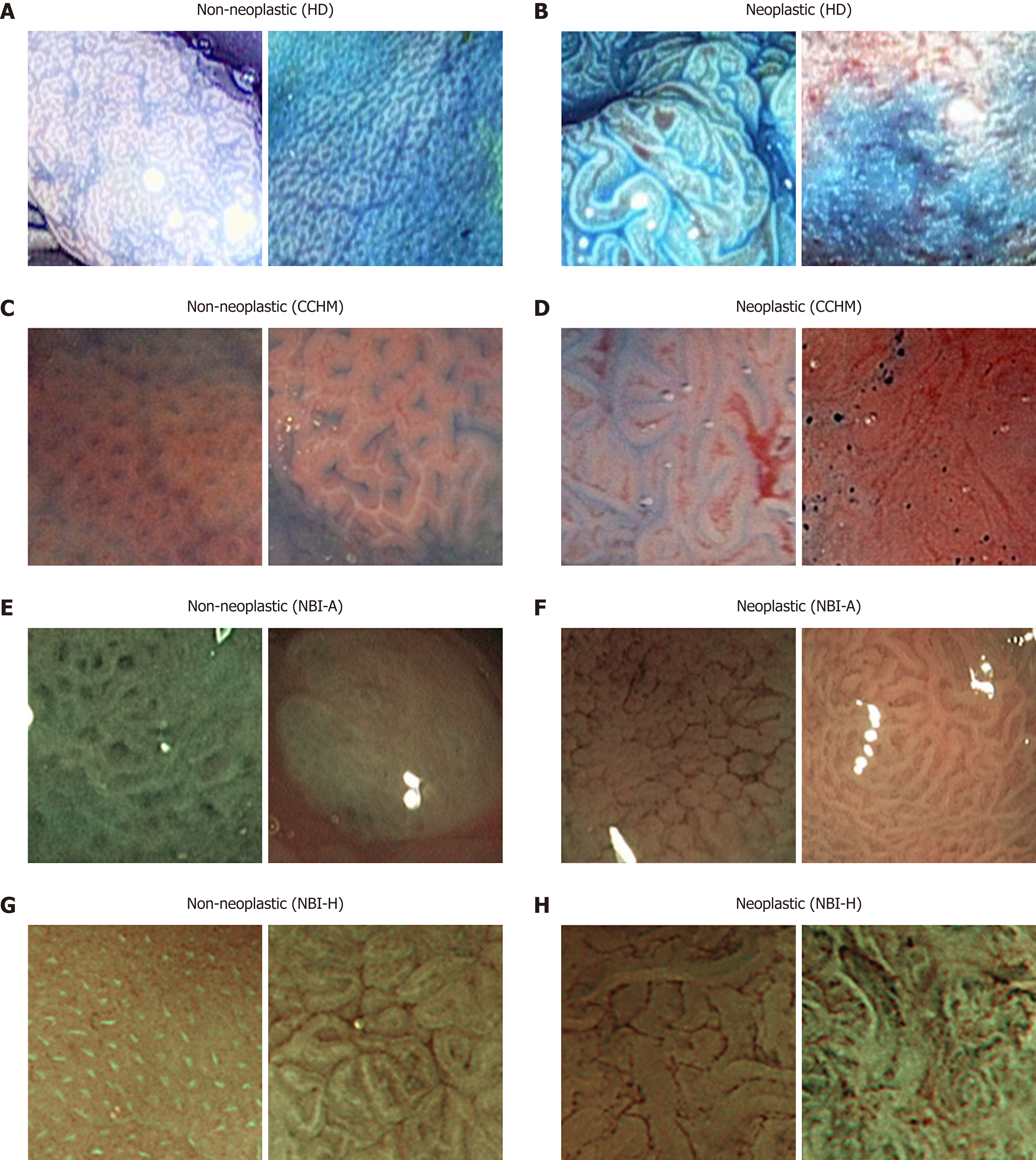
HD-endoscopy has the advantage of a higher resolution compared to standard definition endoscopes. Our HD-endoscopic images are gathered by traditional WL and three i-Scan modes, both with and without CC. The i-Scan (Pentax) image processing technology[21,22] is a digital contrast enhancement method that consists of combinations of surface enhancement, contrast enhancement and tone enhancement. Figure 1 shows images of an adenomatous polyp using WL endoscopy (Figure 1A), i-Scan (Figure 1B-D), CC (Figure 1E) and combinations of CC and i-Scan (Figure 1F-H). Figure 2A and B shows exemplar images of the two classes using CC and i-Scan mode 2.
The images were acquired at St. Elisabeth Hospital in Vienna by extracting patches 256 × 256 in size from frames of HD-endoscopic (Pentax HiLINE HD+90i Colonoscope) videos. Most of the videos contain sequences from eight imaging modalities [with or without i-Scan (modes 1,2,3) and with or without staining], but in some of the videos, only a subset of the eight imaging modalities occurs.
A previous work[23] showed that it is generally favorable to combine images of different i-Scan modes and WL endoscopy for classifier training, but the chromoendoscopy mode (chromoendoscopy or no chromoendoscopy) should be identical for training and evaluation images. This outcome motivated us to group the HD images into two separate image databases, one consisting of images without using chromoendoscopy (no matter which i-Scan mode or WL) and the other one consisting of images using chromoendoscopy. The two HD databases are further denoted as ’HD-no-chromo’ (HDNC) and ’HD-chromo’ (HDC). Table 1 lists the number of images and patients per class. The two HD databases consist of between 102 and 154 images per i-Scan mode and WL endoscopy, respectively.
HM endoscopes are defined by the capacity to perform optical zoom/magnification by using a moveable lens in the tip of the endoscope. In that way, magnified images are obtained without losing display quality. HM endoscopy enables the visualization of mucosal details that cannot be seen with standard endoscopy. The CCHM database was acquired using zoom-endoscopy with 150-fold magnification in combination with chromoendoscopy. The CCHM image data set was acquired at the Medical University of Vienna using a zoom-colonoscope (Olympus Evis Exera CF-Q160ZI/L) with a magnification factor of 150 and indigocarmine dye-spraying. The database is acquired by extracting patches 256 × 256 in size from 327 endoscopic color images (either 624 × 533 pixels or 586 × 502 pixels in size) of 40 patients (Table 1). Example images of the two classes can be seen in Figure 2C and D.
NBI is a videoendoscopic system using RGB rotary filters placed in front of a WL source to narrow the bandwidth of the spectral transmittance. NBI enhances the visibility of microvessels and their fine structure on the colorectal surface. The pits are also indirectly observable since the microvessels between the pits are enhanced in black, while the pits remain white.
In this work, we employ two different data sets gathered by HM endoscopy using NBI. The images of the NBI-HM database Aachen (NBI-A) were acquired at the University Hospital Aachen using an NBI zoom endoscope, which can magnify the image to a maximum of 150-fold (CF-Q160ZI, Olympus Medical Systems Europe). The database was acquired by extracting patches 256 × 256 in size from 387 endoscopic color images 502 × 586 in size from 211 patients (Table 1). Exemplar images of the two classes are shown in Figure 2E and F.
The NBI-HM database Hiroshima (NBI-H) was acquired at the Hiroshima University Hospital using an NBI zoom endoscope, which can magnify the image to a maximum of 75-fold (CF-H260AZ/I, Olympus Optical Co.). Care was taken that the lighting conditions, zooming and optical magnification were kept as similar as possible across different images. The database (Table 1) is acquired by extracting patches 256 × 256 in size from captured images 1440 × 1080 pixels in size. Exemplar images of the two can be seen in Figure 2G and H.
In contrast to the other databases, whose image labels were provided according to their histological diagnosis, the image labels of the NBI-H database were provided according to the optical appearance of the polyps (by at least two experts).
In this section, we briefly describe the twelve feature extraction methods. All methods have proven to be well suited for the diagnosis of colonic polyps in the literature for certain imaging settings.
Non-adapted CNN and adapted CNN: Two convolutional neural networks. One is learned on the ImageNet database [non-adapted CNN (NA-CNN)] and the other one is adapted to colonic polyp classification (A-CNN) by training it on our polyp databases[24].
Delaunay[14]: This approach analyzes the pit-pattern structure and was developed for HM-endoscopic imagery using chromoendoscopy.
BlobShape[11], BALFD[25] and BSAGLFD[12]: Three methods that are analyzing the pit-pattern structure and were designed for HD-endoscopic image data.
Dual-tree complex wavelet transform, Gabor wavelet transform-Weibull and Shearlet-Weibull: Three wavelet based transforms [dual-tree complex wavelet transform (DT-CWT), Gabor wavelet transform (GWT) and shearlet transform] whose subbands are fitted by the two-parameter Weibull distribution[18].
Local color vector patterns and multiscale block binary patterns: Two methods based on LBP. Multiscale local color vector patterns (LCVP) [15] were developed for HM-endoscopic imagery using chromoendoscopy, and the multiscale block binary patterns (MB-LBP) operator[26] is a general purpose image descriptor.
VesselFeature: This approach[21] analyzes the blood vessel structure and is designed for NBI images.
For our experiments classifying the five endoscopic image databases, we rely on a protocol based on the selection of repeated random splits, as is common practice in machine learning. In one iteration, the classification model is trained and evaluated with randomly selected samples from the overall data set. This selection is performed in a way to ensure that the images of one patient are either all in the training or in the evaluation data set (similar to the leave-one-patient-out protocol[15]). To facilitate a fair comparison between image data sets, the amount of training samples is fixed and balanced over the two classes. Specifically, we randomly select 80 samples per class for training and 20 samples per class for evaluation.
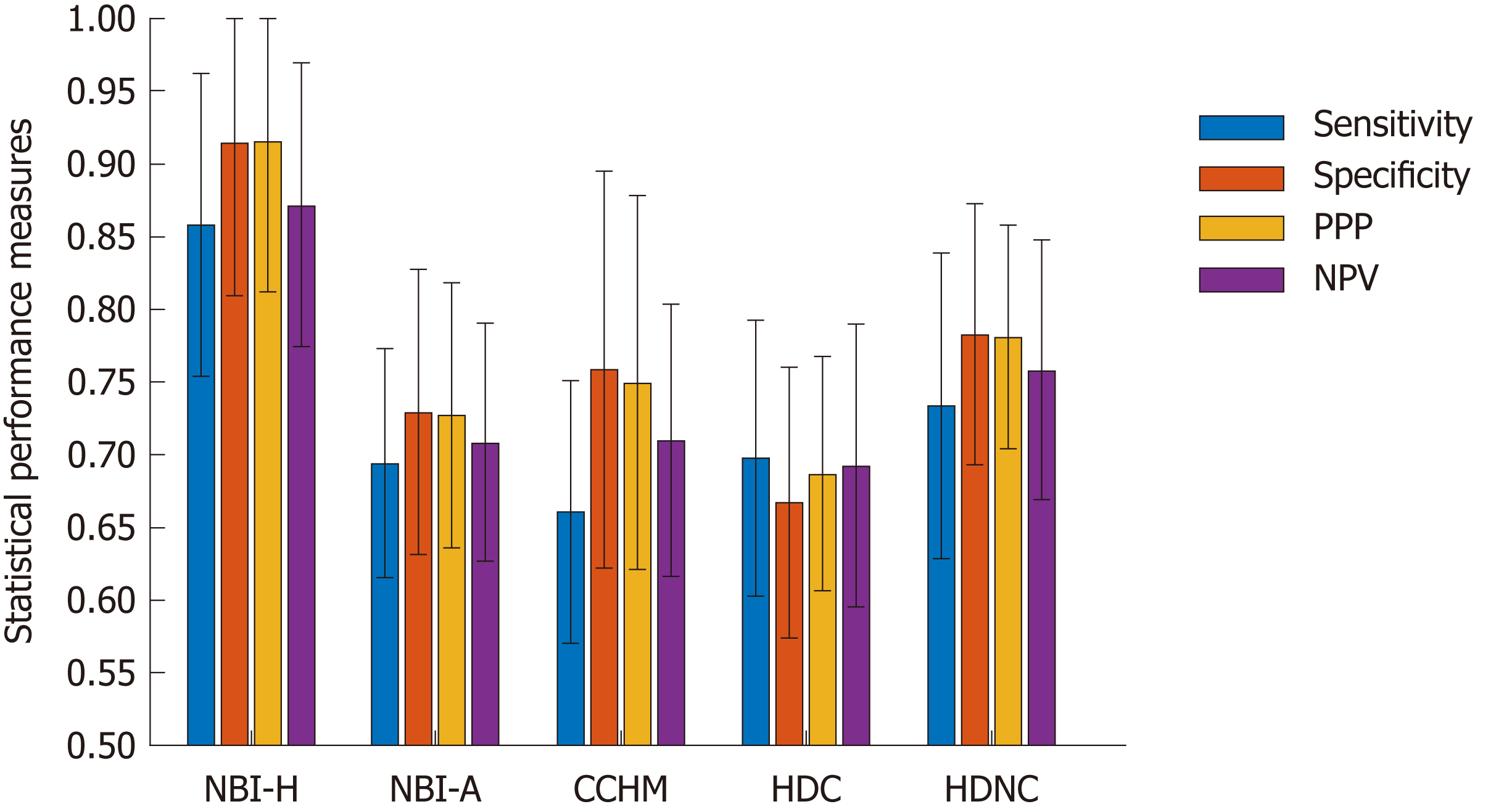
To obtain stable outcomes, we apply 32 random splits and report the mean accuracies as well as the standard deviations. Additionally, we report the sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV).
Classification is performed using linear support vector machines (SVMs)[27]. The optimal SVM’s c-value is assessed during inner cross-validation using the training data only.
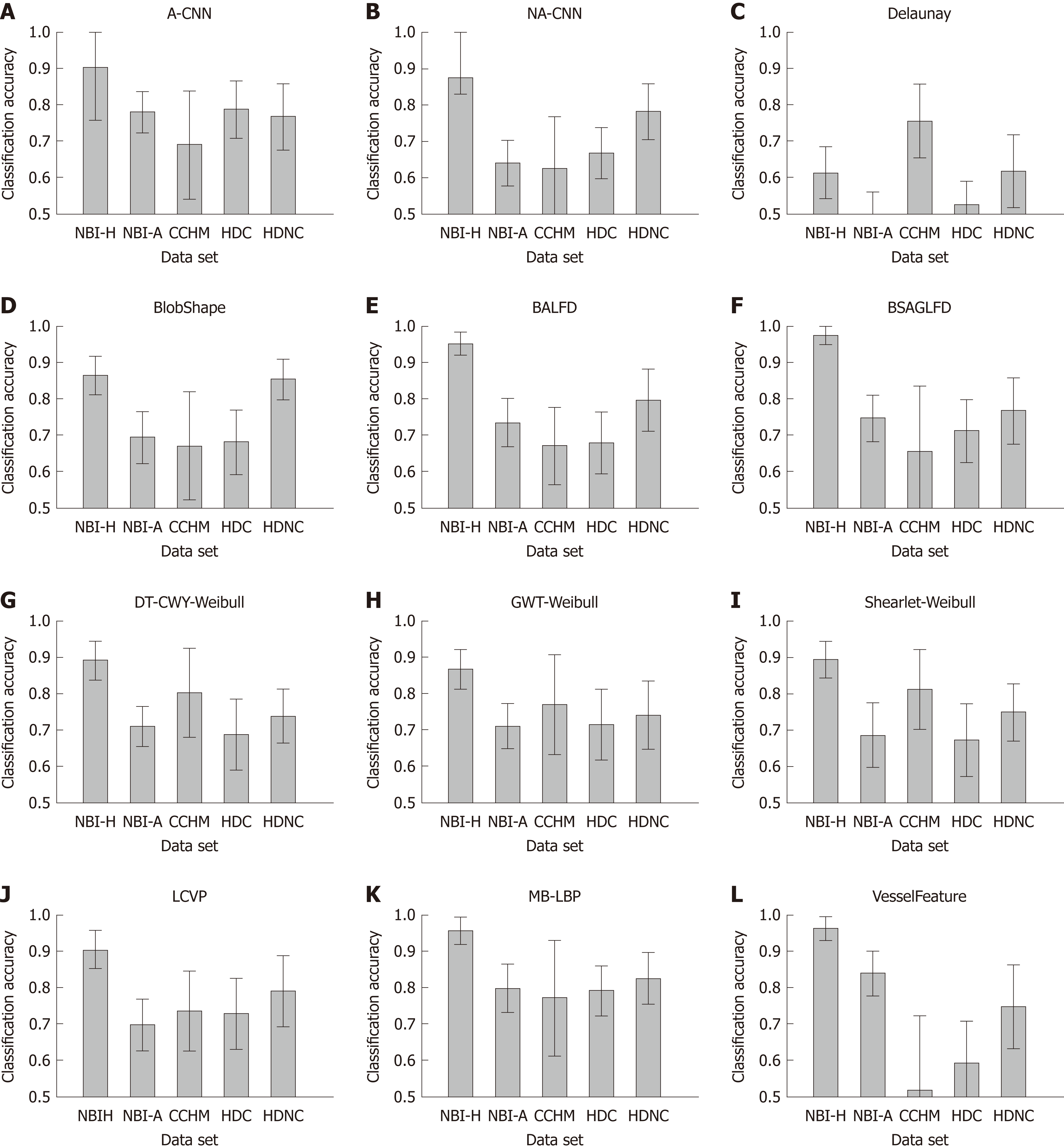
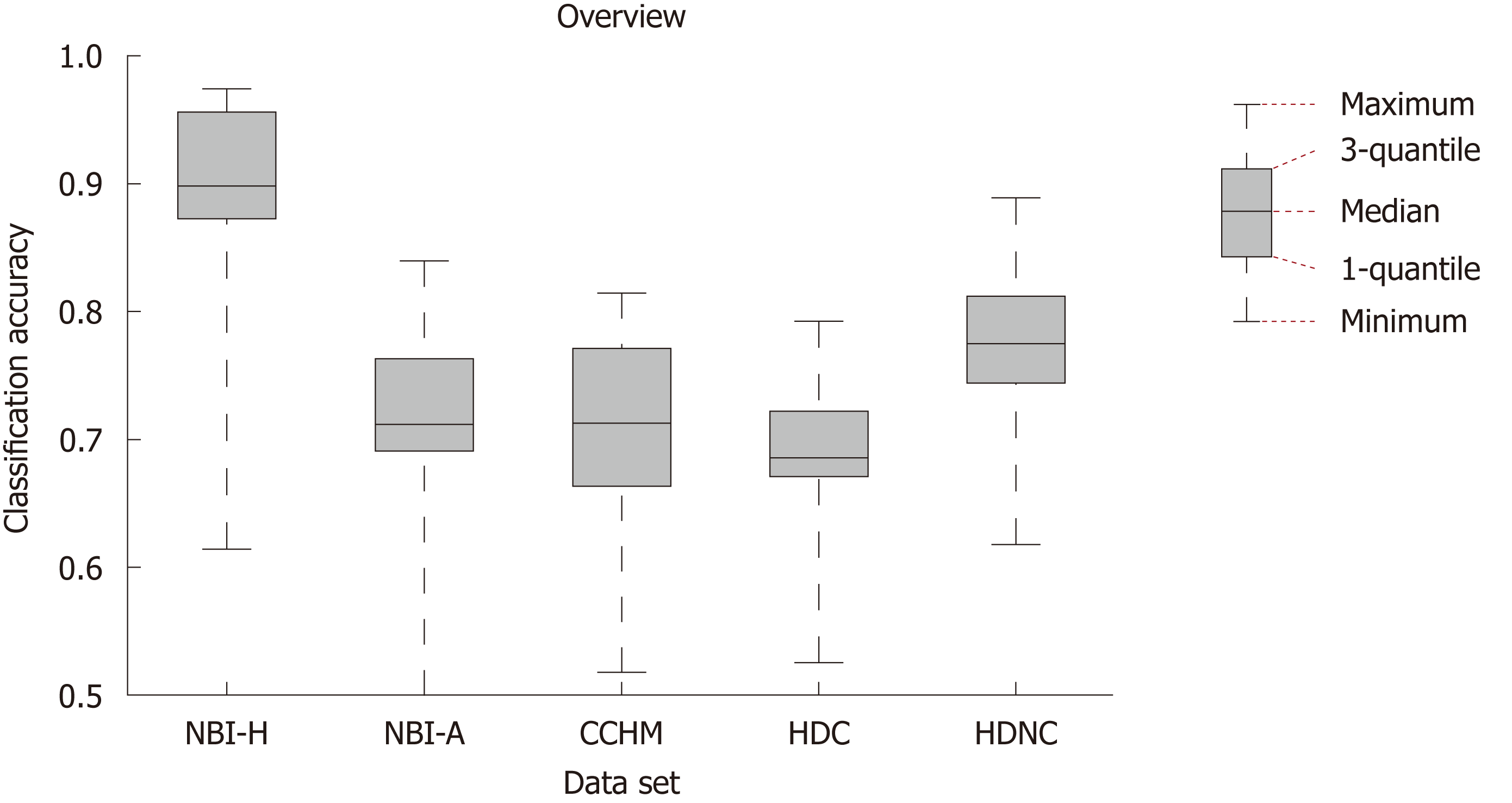
In Figure 3, we show the SVM classification results of our 12 feature extraction methods on each of our 5 colonic polyp image data sets. In each subplot, we see the classification accuracies on the 5 image data sets using one certain image representation. Figure 4 shows a boxplot that summarizes the results in Figure 3. This figure gives an overview of the results of the different feature extraction techniques per data set.
Generally, it is feature dependent which imaging modalities achieve high results and which do not. This outcome was to be expected since many of the employed feature extraction methods were specifically developed for certain imaging modalities.
For the two HD image databases, overall classification rates were achieved of up to 79.2% (HDC) and up to 88.9% (HDNC). The classification rates for the database obtained by CCHM reached up to 81.4%. For the two databases combining HM endoscopy with NBI, results of up to 97.4% (NBI-H) and 84% (NBI-A) were achieved.
In Figure 3, we can observe that the NBI-H set achieves the highest overall classification rates for 11 out of 12 feature extraction methods (all except the Delaunay feature).
Comparing the results of the two NBI-based data sets, we notice that the NBI-A image data shows distinctly lower classification rates compared to the NBI-H data set. Considering the results of the two HD-endoscopic data sets HDC and HDNC, it can be observed that the obtained rates using chromoendoscopy are inferior in 11 out of 12 cases (all except the A-CNN approach).
Figure 5 gives an overview of the results of the methods for the statistical performance measures specificity, sensitivity, PPV and NPV per data set. The specificity rates are in general clearly higher than the sensitivity rates (in four of five databases) which means that non-neoplastic lesions are classified more correctly than neoplastic lesions.
The classification results of the considered computer-assisted staging systems depend on not only the used feature extraction methods and image modalities, but also to a considerable extent on the quality of the gathered images (blur, lighting conditions, in focus vs out of focus, visibility of the mucosal structures, visibility of markers for the respective classes, and other factors) and the uniformity of the image recording conditions such as the viewpoint (distance and angle of view of the endoscope towards the mucosa wall) and the lighting conditions across the images of a database.
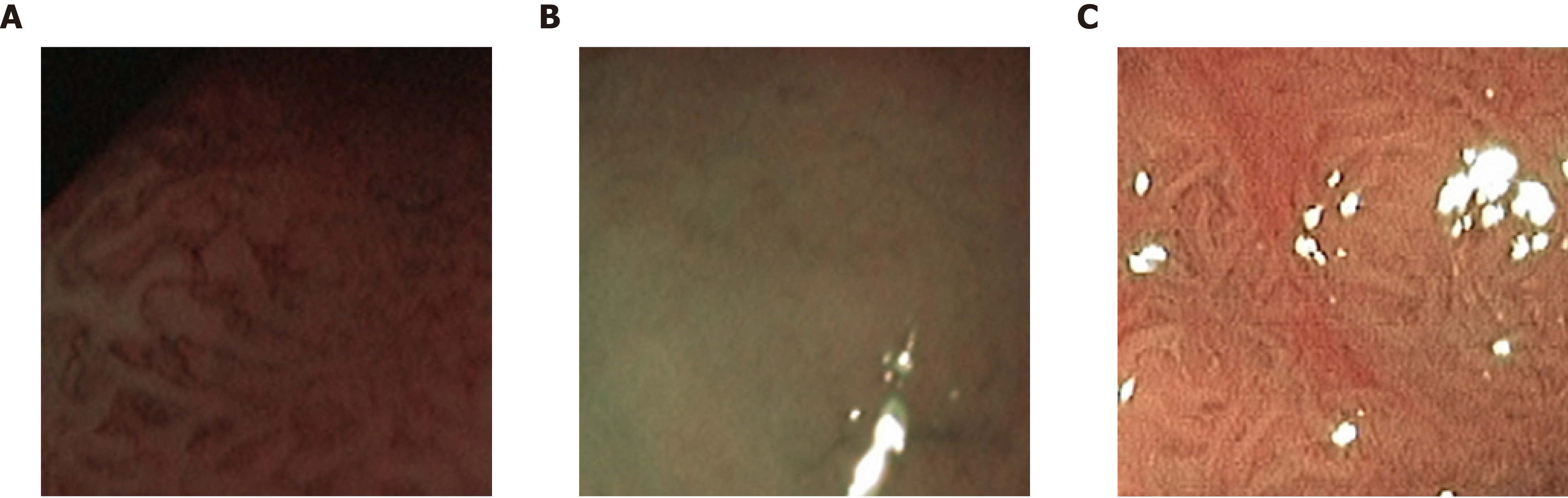
When we compare our two NBI databases, there are clear advantages with respect to the visibility of markers for the respective classes and the consistency of the image recording conditions for the NBI-H database. For the NBI-H database, special care was taken to keep lighting conditions, zoom and optical magnification as similar as possible across different images. The NBI-H database is the only database whose labeling is based on the optical appearance of the polyps[19] (especially the microvessels); thus, special care was taken to ensure that class-specific characteristics were well visible on each image. In Figure 6, we can see some example images from the NBI-A database that were acquired with poor illumination, poor visibility of mucosal structures or reflections.
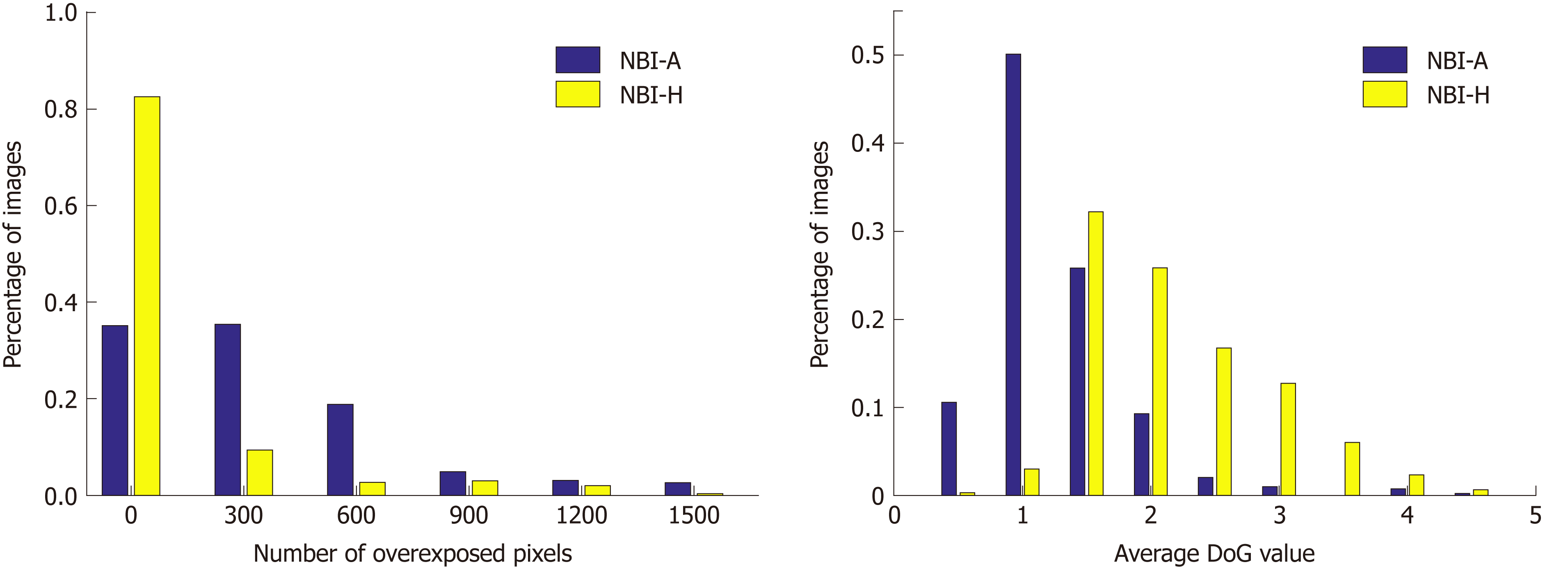
To quantify the differences in the image quality between the two NBI databases we conduct two experiments that will show us differences in the visibility of mucosal structures and the amount of reflections in the images. To show the amount of reflections in an image we simply add up the number of overexposed pixels (gray value bigger than 240) per image. Our approach to show the visibility of mucosal structures per image is based on difference of Gaussians (DoG). We apply DoG by subtracting a Gaussian blurred image from the original image. By using a Gaussian filter kernel with σ = 3 and size 8 × 8, the resulting DoG image of a grayscale endoscopic image highlights mucosal structures, such as the pit-pattern structure. The mean over the absolute values of the DoG image gives an indication about the visibility of mucosal structures in an image. The higher the mean value, the better the visibility of mucosal structures. In Figure 7, we present two histograms showing differences in image quality between the two NBI databases.
We can clearly see in Figure 7 that the NBI-A database clearly contains more reflections in the images as the NBI-H database. The average DoG values are clearly higher in the NBI-H database, which indicates a better visibility of mucosal structures in the NBI-H database.
A third experiment comparing the number of underexposed pixels per image showed that the NBI-A database has a higher percentage of images where large areas of the image are underexposed, but the differences between the two NBI databases are rather small, which is why we only show figures for reflections and the visibility of mucosal structures, where clear differences occur.
Thus, it is not surprising that the NBI-H database achieved distinctly better results than the NBI-A database in our experiments. Therefore, the image recording conditions do have a high impact on automated diagnosis systems.
The fact that the image recording conditions have significant impact on the classification results and that it is difficult to quantify the quality of the images and the uniformity of the image recording conditions in the different databases makes it difficult to assess which imaging modality is best suitable for the staging of colonic polyps.
However, for example the two HD databases are gathered from the same endoscopic videos (taken by the same endoscopist); thus, their quality and the uniformity of the image recording conditions are quite similar. The images from the CCHM database are taken from the same endoscopist as the videos of the HD database; therefore, their quality could also be assumed to be at least similar. The images of the CCHM database as well as the two HD databases are acquired under varying viewpoint and lighting conditions, but the mucosal structure (the pit-pattern structure) is always visible for these three databases. Only the NBI-A database contains images without visible mucosal structures.
When we compare the results on the two NBI-HM databases (NBI-H and NBI-A) with the results on the CCHM, we see that the results for the NBI-H database are clearly better than those for the CCHM database and that the results for the NBI-A database are quite similar to those for the CCHM database. Therefore, we cannot clearly answer the question which of the two imaging modalities, chromoendoscopy or NBI, is better suited for the automated staging of colonic polyps. However, because of the similar results of the NBI-A and CCHM databases, despite the higher image quality of the CCHM database, and the distinctly higher results on the NBI-H database it seems safe to assume that NBI is better suited than chromoendoscopy.
When we compare the results of the two HD-endoscopic data sets, then we can observe that chromoendoscopy decreases the classification rates for all feature extraction methods except for one. Therefore, chromoendoscopy should not be used for automated diagnosis systems.
Based on the overall similar results of the two chromoendoscopic databases (CCHM and HDC) shown in Figures 3 and 4, HD endoscopy and HM endoscopy are approximately equally well suited for automated diagnosis systems. However, the results are not strictly conclusive since most of the HD-endoscopic images are gathered using different i-Scan modes, in contrast to the HM images which are gathered without digital chromoendoscopy.
Colorectal cancer is the second most common cause of cancer death worldwide. Computer-aided decision support systems (CADSSs) aim at helping physicians to detect and classify colonic polyps more accurately. Since about two decades, there is a rising interest in CADSSs and a rising number on publications on CADSSs for colonic polyp staging.
Clinical studies showed that high definition endoscopy, high magnification endoscopy and image enhancement technologies, such as chromoendoscopy and digital chromoendoscopy [narrow-band imaging (NBI), i-Scan] facilitate the detection and classification of colonic polyps during endoscopic sessions. However, there are no comprehensive studies so far that analyze which endoscopic imaging modalities facilitate CADSSs for colonic polyp staging.
In this work, we assess which endoscopic imaging modalities are best suited for the computer-assisted classification of colonic polyps.
In our experiments, we apply twelve automated polyp staging systems to five endoscopic image databases of colonic lesions. The image databases were obtained using different endoscopic imaging modalities. By comparing the classification results of the different image databases, we aim to find out which imaging modalities are most suited for the automated classification of colonic polyps.
The high-definition (HD) image databases obtained with chromoendoscopy achieved overall classification rates of up to 79.2% whereas the HD image databases without chromoendoscopy achieved results up to 88.9%. The classification rates of the image database obtained by high-magnification (HM) chromoendoscopy were up to 81.4%. For the two image databases obtained by HM endoscopy in combination with NBI, one database achieved classification rates up to 97.4%, whereas the other one only achieved classification rates up to 84%.
The results strongly indicate that chromoendoscopy has a negative impact on the automated diagnosis. The results also indicate that HD and HM endoscopy perform equally well, although the results are not strictly conclusive. Given the higher costs of HM systems and the difficulty in acquiring high quality imagery due to the HM (which definitely requires a well-trained endoscopist), HD systems should be the better option in clinical computer-assisted staging practice. In case of the comparison of chromoendoscopy vs NBI, there are strong indications that NBI is favorable. However, it turned out that the image recording conditions partly contribute to a stronger effect on the staging results than the used imaging modality.
We showed that CADSSs for colonic polyp staging should not be applied to endoscopic image data obtained by chromoendoscopy, whereas image data obtained by NBI is suited for automated diagnosis systems. Important factors for the success of CADSSs are the image quality of the endoscopic image data and the uniformity of the image recording conditions.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Austria
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: De Silva AP, Kwon KA S- Editor: Ma RY L- Editor: A E- Editor: Yin SY
| 1. | Berr F, Oyama T, Ponchon T, Yahagi N. Early Neoplasias of the Gastrointestinal Tract: Endoscopic Diagnosis and Therapeutic Decisions. Boston: Springer 2014; . [DOI] [Full Text] |
| 2. | Rex DK, Kahi C, O'Brien M, Levin TR, Pohl H, Rastogi A, Burgart L, Imperiale T, Ladabaum U, Cohen J, Lieberman DA. The American Society for Gastrointestinal Endoscopy PIVI (Preservation and Incorporation of Valuable Endoscopic Innovations) on real-time endoscopic assessment of the histology of diminutive colorectal polyps. Gastrointest Endosc. 2011;73:419-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 406] [Cited by in RCA: 480] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 3. | Patel SG, Schoenfeld P, Kim HM, Ward EK, Bansal A, Kim Y, Hosford L, Myers A, Foster S, Craft J, Shopinski S, Wilson RH, Ahnen DJ, Rastogi A, Wani S. Real-Time Characterization of Diminutive Colorectal Polyp Histology Using Narrow-Band Imaging: Implications for the Resect and Discard Strategy. Gastroenterology. 2016;150:406-418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 70] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 4. | ASGE Technology Committee; Manfredi MA, Abu Dayyeh BK, Bhat YM, Chauhan SS, Gottlieb KT, Hwang JH, Komanduri S, Konda V, Lo SK, Maple JT, Murad FM, Siddiqui UD, Wallace MB, Banerjee S. Electronic chromoendoscopy. Gastrointest Endosc. 2015;81:249-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 5. | Hoffman A, Kagel C, Goetz M, Tresch A, Mudter J, Biesterfeld S, Galle PR, Neurath MF, Kiesslich R. Recognition and characterization of small colonic neoplasia with high-definition colonoscopy using i-Scan is as precise as chromoendoscopy. Dig Liver Dis. 2010;42:45-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 76] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 6. | Bouwens MW, de Ridder R, Masclee AA, Driessen A, Riedl RG, Winkens B, Sanduleanu S. Optical diagnosis of colorectal polyps using high-definition i-scan: an educational experience. World J Gastroenterol. 2013;19:4334-4343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 19] [Cited by in RCA: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 7. | Tischendorf JJ, Wasmuth HE, Koch A, Hecker H, Trautwein C, Winograd R. Value of magnifying chromoendoscopy and narrow band imaging (NBI) in classifying colorectal polyps: a prospective controlled study. Endoscopy. 2007;39:1092-1096. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 155] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 8. | Chang CC, Hsieh CR, Lou HY, Fang CL, Tiong C, Wang JJ, Wei IV, Wu SC, Chen JN, Wang YH. Comparative study of conventional colonoscopy, magnifying chromoendoscopy, and magnifying narrow-band imaging systems in the differential diagnosis of small colonic polyps between trainee and experienced endoscopist. Int J Colorectal Dis. 2009;24:1413-1419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 9. | ASGE Technology Committee. High-definition and high-magnification endoscopes. Gastrointest Endosc. 2014;80:919-927. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 72] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 10. | Rameshshanker R, Wilson A. Electronic Imaging in Colonoscopy: Clinical Applications and Future Prospects. Curr Treat Options Gastroenterol. 2016;14:140-151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 11. | Häfner M, Uhl A, Wimmer G, Medical Computer Vision. A novel shape feature descriptor for the classification of polyps in HD colonoscopy. Medical Computer Vision. Large Data in Medical Imaging. Cham: Springer 2014; 205-213. [RCA] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 12. | Häfner M, Tamaki T, Tanaka S, Uhl A, Wimmer G, Yoshida S. Local fractal dimension based approaches for colonic polyp classification. Med Image Anal. 2015;26:92-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (1)] |
| 13. | Ribeiro E, Uhl A, Wimmer G, Häfner M. Exploring Deep Learning and Transfer Learning for Colonic Polyp Classification. Comput Math Methods Med. 2016;2016:6584725. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 81] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 14. | Häfner M, Liedlgruber M, Uhl A, Vécsei A, Wrba F. Delaunay triangulation-based pit density estimation for the classification of polyps in high-magnification chromo-colonoscopy. Comput Methods Programs Biomed. 2012;107:565-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 15. | Häfner M, Liedlgruber M, Uhl A, Vécsei A, Wrba F. Color treatment in endoscopic image classification using multi-scale local color vector patterns. Med Image Anal. 2012;16:75-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 54] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 16. | Häfner M, Kwitt R, Uhl A, Gangl A, Wrba F, Ve´csei A. Computer-assisted pit-pattern classification in different wavelet domains for supporting dignity assessment of colonic polyps. Pattern Recognition. 2008;42:1180-1191. [RCA] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 20] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 17. | Häfner M, Liedlgruber M, Uhl A. Colonic polyp classification in high-definition video using complex wavelet-packets. Bildverarbeitung für die Medizin. 2015;365-370. [RCA] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 18. | Wimmer G, Tamaki T, Tischendorf JJ, Häfner M, Yoshida S, Tanaka S, Uhl A. Directional wavelet based features for colonic polyp classification. Med Image Anal. 2016;31:16-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 19. | Tamaki T, Yoshimuta J, Kawakami M, Raytchev B, Kaneda K, Yoshida S, Takemura Y, Onji K, Miyaki R, Tanaka S. Computer-aided colorectal tumor classification in NBI endoscopy using local features. Med Image Anal. 2013;17:78-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 20. | Kominami Y, Yoshida S, Tanaka S, Sanomura Y, Hirakawa T, Raytchev B, Tamaki T, Koide T, Kaneda K, Chayama K. Computer-aided diagnosis of colorectal polyp histology by using a real-time image recognition system and narrow-band imaging magnifying colonoscopy. Gastrointest Endosc. 2016;83:643-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 162] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 21. | Gross S, Palm S, Tischendorf JJW, Behrens A, Trautwein C, Aach T. Automated classification of colon polyps in endoscopic image data. SPIE Proceedings Medical Imaging 2012: Computer-Aided Diagnosis, 83150W. . [DOI] [Full Text] |
| 22. | Kodashima S, Fujishiro M. Novel image-enhanced endoscopy with i-scan technology. World J Gastroenterol. 2010;16:1043-1049. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 130] [Cited by in RCA: 151] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 23. | Wimmer G, Gadermayr M, Kwitt R, Häfner M, Merhof D, Uhl A. Evaluation of i-scan virtual chromoendoscopy and traditional chromoendoscopy for the automated diagnosis of colonic polyp. Proceedings of the 3rd International Workshop on Computer-Assisted and Robotic Endoscopy (CARE). Cham: Springer 2016; 59-71. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 24. | Wimmer G, Ve´csei A, Uhl A. Cnn transfer learning for the automated diagnosis of celiac disease. Proceedings of the Sixth International Conference on Image Processing Theory, Tools and Applications (IPTA). 2016;1-6. [DOI] [Full Text] |
| 25. | Häfner M, Uhl A, Wimmer G. Shape and size adapted local fractal dimension for the classification of polyps in HD colonoscopy. 2014;Oct 27-30; Paris, France. IEEE, 2014. [DOI] [Full Text] |
| 26. | Liao S, Zhu X, Lei Z, Zhang L, Li SZ. Learning multi-scale block local binary patterns for face recognition. In: Proceedings of the 2007; Springer, 2007: 828-837. [RCA] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 19] [Reference Citation Analysis (1)] |
| 27. | Fan RE, Chang KW, Hsieh CJ, Wang XR, Lin CJ. LIBLINEAR: A library for large linear classification. J Mach Learn Res. 2008;9:1871-1874. |