Published online Nov 7, 2018. doi: 10.3748/wjg.v24.i41.4643
Peer-review started: July 5, 2018
First decision: August 25, 2018
Revised: September 4, 2018
Accepted: October 5, 2018
Article in press: October 5, 2018
Published online: November 7, 2018
Processing time: 125 Days and 10.4 Hours
Autophagy is a “self-degradative” process and is involved in the maintenance of cellular homeostasis and the control of cellular components by facilitating the clearance or turnover of long-lived or misfolded proteins, protein aggregates, and damaged organelles. Autophagy plays a dual role in cancer, including in tumor progression and tumor promotion, suggesting that autophagy acts as a double-edged sword in cancer cells. Liver cancer is one of the greatest leading causes of cancer death worldwide due to its high recurrence rate and poor prognosis. Especially in China, liver cancer has become one of the most common cancers due to the high infection rate of hepatitis virus. In primary liver cancer, hepatocellular carcinoma (HCC) is the most common type. Considering the perniciousness and complexity of HCC, it is essential to elucidate the function of autophagy in HCC. In this review, we summarize the physiological function of autophagy in cancer, analyze the role of autophagy in tumorigenesis and metastasis, discuss the therapeutic strategies targeting autophagy and the mechanisms of drug-resistance in HCC, and provide potential methods to circumvent resistance and combined anticancer strategies for HCC patients.
Core tip: Liver cancer seriously threatens human health, due to its high recurrence rate and poor prognosis. Hepatocellular carcinoma is the most common type of primary liver cancer. Autophagy plays a dual role in cancer. Basic autophagy exerts a tumor suppression function by maintaining genomic stability in normal cells. When cancer occurs, activated autophagy benefits cancer cell survival and promotes cancer development. However, the mechanism and function of autophagy in human cancers, especially liver cancer, have not been clarified. We summarize the physiological function of autophagy and its role in tumorigenesis, metastasis, targeted therapy and drug-resistance in hepatocellular carcinoma.
- Citation: Huang F, Wang BR, Wang YG. Role of autophagy in tumorigenesis, metastasis, targeted therapy and drug resistance of hepatocellular carcinoma. World J Gastroenterol 2018; 24(41): 4643-4651
- URL: https://www.wjgnet.com/1007-9327/full/v24/i41/4643.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i41.4643
Since the Nobel Prize in Physiology or Medicine 2016 was awarded to Yoshinori Ohsumi “for his discoveries of mechanisms for autophagy”, there has been an increase in research worldwide to determine the mechanisms, functions and application of autophagy[1]. Autophagy, a “self-eating” phenomenon, maintains cellular homeostasis. In eukaryotic cells, there are two major cellular degradation organelles, the lysosome and the proteasome[2]. The proteasome generally recognizes and degrades only ubiquitinated substrates, with high selectivity. In contrast, lysosome degradation has a more complicated pattern. Extracellular material and plasma membrane proteins can be delivered to lysosomes for degradation via the endocytic pathway. However, cytosolic components and organelles can be delivered to the lysosome for degradation via autophagy. Thus, it is understood that autophagy is an intracellular catabolic degradative process targeting damaged and superfluous cellular proteins, organelles, and other cytoplasmic components, which plays a role in the renewal of cells and tissues.
Autophagy is an evolutionarily conserved multistep process involving a group of conserved gene family members known as autophagy-related genes (ATGs). The first step is the induction of nucleation of the autophagy-isolation membrane, which is initiated by assembly of the ULK1 complex, comprising ULK1, ATG13, ATG101, and FIP200[3]. During this assembly, an isolation membrane known as the phagophore is formed by the membrane derived from the endoplasmic reticulum, Golgi apparatus, and mitochondria. When the ULK1 complex translocates to the phagophore, the class III phosphoinositide 3-kinase (PI3K) complex is assembled with VPS34, TP150, BECLIN-1, ATG14L, and AMBRA1. The formed VPS34-BECLIN1-ATG14L complex promotes membrane nucleation[4]. After nucleation, elongation of the isolation membrane and autophagosome completion take place, which is involved in ATG5-ATG12 conjugation and LC3 processing. E1 and E2 ligases, ATG7 and ATG10, mediate the ATG12-ATG5 conjugation, which binds to ATG16L and aids phagophore elongation by recruitment of LC3-II to the membrane[5]. LC3 is widely used as an autophagosomal marker. Completion of the double-membrane autophagosome is dependent on elongation of the isolation membrane, sequestration of cargo, and closure of the membrane. The last event in the autophagic process is fusion of the autophagosome-lysosome and degradation of cargo. Lysosomal fusion is orchestrated by Rabs, SNAREs, and tethers, and the cargo is degraded in lysosomes[6,7]. Thus, the lysosome is often described as a ‘‘cellular garbage can’’, and plays a crucial role in autophagy for cellular renewal.
In the past two decades, the finding of a series of ATG genes in yeast and mammals has increased our understanding of the physiological and pathological roles of autophagy, particularly in many human diseases[8]. However, the mechanism and function of autophagy in human cancers, especially liver cancer, have not been clarified. In this review, we summarize the physiological function of autophagy and discuss the role of autophagy in tumorigenesis, metastasis, targeted therapy and drug resistance of hepatocellular carcinoma (HCC).
Under normal conditions, basal autophagy with a housekeeping function has physiological roles involved in the maintenance of cellular homeostasis and the control of cellular components by facilitating the clearance or turnover of long-lived or misfolded proteins, protein aggregates, and damaged organelles[9]. However, under cellular stress, such as nutrient starvation, oxidative stress, hypoxia, or infection, autophagy plays a cytoprotective or an adaptive role[10]. Thus, autophagy can degrade macromolecules including nucleic acid, proteins, carbohydrates, and triglycerides to nucleosides, amino acids, sugars, and free fatty acids, respectively, which are available for de novo synthesis of biomolecules or for the generation of ATP for energy cellular functions via the tricarboxylic acid cycle and other metabolic processes[11].
The role of autophagy in cancer has been well studied in the past decade. Extensive interest has been focused on understanding the paradoxical roles of autophagy in tumor progression and tumor promotion, which suggests that autophagy acts as a double-edged sword in cancer cells[12,13]. On the one hand, basic autophagy has as tumor suppression function by maintaining genomic stability in normal cells. When cancer occurs, activated autophagy benefits cancer cell survival and promotes cancer development[14,15]. On the other hand, autophagy also appears to serve as a pro-survival or pro-death response in different cancers under different conditions[16]. For example, following radiation, chemotherapy and targeted therapy, autophagy in cancer cells is usually activated[17,18], and is considered to serve as an important mechanism of therapeutic resistance by supporting tumor cell survival[18]. Thus, inhibition of autophagy may be a candidate strategy for cancer therapy.
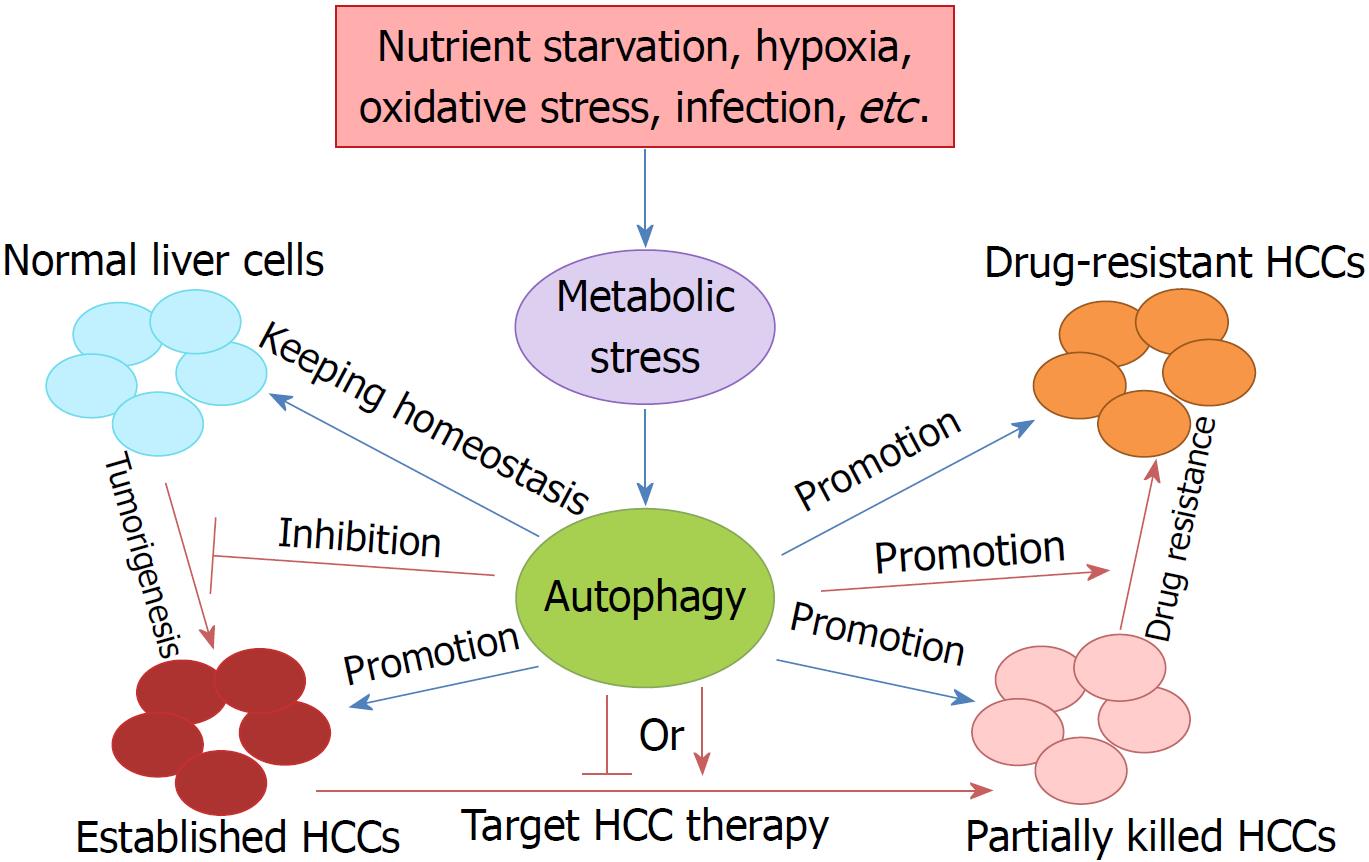
Liver cancer is the third leading cause of cancer death worldwide, due to its high recurrence rate and poor prognosis[9]. In China, liver cancer is the third most common cancer, having an incidence rate ranked fourth, a mortality rate ranked second, and 18.43 per 10000 patients diagnosed with it[19,20]. HCC is the most common type of primary liver cancer, and accounts for 85%-90% of all cases. Currently, surgical resection is recommended for very early stage and early stage HCC, following chemo/radiotherapy, but HCC is still prone to recurrence and metastasis after surgery, and there is still no effective treatment for patients with advanced, metastatic or drug-resistant HCC[21-23]. Therefore, it is essential to elucidate the mechanisms of tumorigenesis, metastasis, and drug resistance in HCC, and identify effective and safe therapeutic strategies and prognostic biomarkers (Figure 1). In view of the fact that activated autophagy plays an important role in the occurrence and progression of human neurodegenerative diseases, fatty liver disease, lupus disease, Crohn’s disease, and cancers, the regulation of autophagy may contribute to the treatment of tumors including HCC.
The function of autophagy in the liver is a topic of concern. Autophagy plays multiple roles in maintaining liver homeostasis, and contributes to the preservation of genome stability in liver cells and the prevention of malignant transformation by removing harmful mitochondria and transformed liver cells[9]. However, knock-out (KO) of ATG5 and ATG7, two key autophagy genes, results in the accumulation of nonfunctional proteins and organelles in liver cells[24]. The absence of ATG7 by conditional KO in mouse liver results in developing hepatomegaly and various metabolic liver disorders in these mice[25,26]. The role of autophagy in tumorigenesis is controversial and complex, as both up-regulation and down-regulation of autophagy have been found in cancers, suggesting that it has oncogenic and tumor suppressor properties during malignant transformation[9].
Autophagy also has dual roles during the progress of liver cancer. It has been confirmed that autophagy has a suppressive role in the very early or early stage of HCC tumorigenesis. Direct evidence of the tumor-suppressing role of autophagy in HCC was derived from the key autophagy gene BECLIN1 KO mice[27]. BECLIN1 is the only dual function molecule which acts as both a tumor suppressor and autophagy regulator[28,29]. Mice with heterozygous disruption of BECLIN1 showed reduced autophagy activity and tended to initiate the spontaneous formation of malignant lesions, including HCC[27]. Further studies showed that decreased expression of BECLIN1 was correlated with HCC grade, which demonstrated the possible use of BECLIN1 as an HCC prognostic biomarker[30].
Takamura et al[24] used mice with liver-specific KO of ATG5 to study the role of autophagy in carcinogenesis and found that abolishing the expression of ATG5 impaired autophagy in the liver and led to oxidative DNA damage and the development of benign hepatic tumors with no visible carcinoma. This inability to develop HCC was correlated with the induction of tumor suppressors, such as TP53, TP16, TP21, and TP27, which negatively modulated the progression of tumorigenesis when autophagy was impaired, with the features of mitochondrial swelling, TP62 accumulation, oxidative stress, and genomic damage responses[31]. Simultaneously, liver specific ATG7 KO mice also developed liver tumors that were smaller in size after TP62 deletion[24,31], which indicated that the accumulation of p62 caused by autophagy deficiency contributes to tumor progression.
More recently, Liu et al[32] reported that autophagy is required for benign hepatic tumors to progress into malignant HCC. Their studies indicated that autophagy, or more specifically mitophagy, was required to suppress TP53 and induce the expression of the transcription factor NANOG to maintain hepatic cancer stem cells and promote hepatocarcinogenesis[32]. Thus, although autophagy functions as a tumor suppressor in nontumor cells or the early stages of tumor cell development, autophagy becomes important for cancer cell survival once tumors, including HCC, are established. It has been shown that autophagy can enhance the survival of tumor cells in the hypoxic regions of solid tumors[33], and autophagy is required to promote tumorigenesis by maintaining oxidative metabolism or facilitating glycolysis in cells expressing oncogenic Ras[34,35].
Tumor metastasis causes more than 90% of cancer deaths. Tumor cells grow in close quarters and then break away from the primary tumor under stressful conditions, such as exhaustion of available oxygen and nutrients, and then the migrating cancer cells develop into metastatic nodules[16,36,37]. This process involves the assembly of cell membrane surface proteins, activation of cell adhesion signals, and changes in the extracellular environment[38,39]. There is increasing evidence to show that autophagy plays an important role in the process of tumor metastasis[40]. Firstly, tumor cells gain energy through autophagy, which improves their survival ability and promotes migration outward[41,42]. Furthermore, autophagy can induce cell adhesion signal changes, and promote tumor cell invasion and migration[43]. More specific inhibition of focal adhesion kinase (commonly known as FAK) activates SRC kinase and thus inhibits autophagy[44]. In turn, inhibition of autophagy prevents the migration of SRC-driven metastatic tumor cells[43]. Further evidence suggests that focal adhesion inhibits the level of autophagic flux due to the accumulation of paxillin (PXN), a core component of focal adhesions[45,46].
Autophagy may play a positive role in HCC metastasis due to its pro-survival effect. Peng et al[47] found that autophagy inhibition by lentivirus-mediated silencing of BECN1 and ATG5 genes suppressed HCC metastasis by facilitating anoikis resistance and lung colonization of HCC cells, and autophagy-based HCC tissue-specific targeted therapy efficiently inhibited autophagy of HCC cells and tissue in a tissue-specific manner in vitro and in vivo. Further results showed that the expression of LC3 in metastases was significantly higher than in patients with primary HCC, which suggested a higher level of autophagy in HCC metastases. Moreover, autophagy is activated in metastatic colonization but not in invasion, migration and detachment of HCC cells[48].
Epithelial-mesenchymal transition (EMT) is an important intermediate stage of cancer metastasis[49]. Invasion of HCC cells is a leading cause of intrahepatic dissemination and metastasis. Autophagy is considered to be an important mediator in the invasion of cancer cells. One study showed that starvation-induced autophagy promoted the expression of EMT markers and invasion in HCC cells in a transforming growth factor beta (commonly known as TGF-β)/Smad3 signaling-dependent manner[50]. This suggests that activation of TGF-β/Smad3-dependent signaling plays a key role in regulating autophagy-induced EMT. Another study also highlighted the importance of the Wnt/β-catenin signaling pathway in regulating the role of autophagy in HCC metastasis[51]. They found that activation of autophagy can promote metastasis and glycolysis accompanied by MCT1 up-regulation in HCC cells, and autophagy induced MCT1 expression by activating Wnt/β-catenin signaling[51]. Thus, the study described the relationship between autophagy and glucose metabolism in HCC cell metastasis and may provide a potential therapeutic target for HCC treatment.
Compared to traditional therapy, the role of autophagy in targeted therapy of liver cancer seems to show potential in improving the lethality of tumors while having a lesser effect on normal tissue cells[25]. For example, chemotherapy lacks cell type specificity and acts on both normal and cancer cells; thus, its efficacy is largely decreased. Autophagy is usually considered to play a suppressive role in tumorigenesis, including of HCC, and acts as an oncogenic factor in advanced HCC[52]. The best example (reported by Wu et al[53]) is autophagic LC3-II over-expression being positively correlated with malignant progression and predictive of poor prognosis in HCC. Autophagy may provide the energy and materials for growth and survival of cancer cells in the tumor microenvironment, which may include hypoxia, nutrient deficiency, and therapeutic stress[54]. This also provides the foundation for promising targeted therapy of established HCC through autophagy inhibition.
However, autophagy has a dual function in the pathophysiology of liver cancer, and it is necessary to clarify which function benefits cancer therapy. Then, a targeting therapeutic strategy for HCC can be designed according to the particular role of autophagy, either autophagy induction or inhibition. A recent study showed that over-expression of lncRNA PTENP1 (a pseudogene of the tumor suppressor gene PTEN) in HCC cells induces onco-miRs miR-17, miR-19b and miR-20a, elevating the levels of PTENP1 and PTEN, and suppressing the oncogenic PI3K/AKT pathway, cell proliferation, migration/invasion and inducing both autophagy and apoptosis[55]. These miRNAs increased the expression of autophagy genes including ULK1, ATG7, or TP62, triggered autophagy, and suppressed HCC tumor growth. These findings indicate that the lncRNA/miRNAs/autophagy axis is important in targeted HCC therapy.
Reduced autophagy can induce growth inhibition in liver cancer, and autophagy activation may be beneficial in HCC treatment[25]. The antitumor agent rapamycin and its derivatives also induce autophagy by inhibiting the mTOR pathway[56]. In clinical trials, rapamycin treatment resulted in an obvious antitumor effect along with improved overall survival rates in post-liver transplantation patients with HCC[57]. Similar results were observed following sirolimus (a derivative of rapamycin) treatment in patients with advanced HCC but without liver transplantation[58]. Another derivative of rapamycin, everolimus (RAD001), also had a significant inhibitory effect on tumor growth in human HCC xenograft models[59]. However, in the clinical phase III trial EVOLVE-1, administration of everolimus for advanced HCC after failure of sorafenib did not show a significant difference in the overall survival rate[60]. Egan et al[61] screened a potent ULK1 small molecule inhibitor, SBI-0206965, and the PI3K/mTOR inhibitor BEZ235 which down-regulated autophagy and inhibited HCC cell survival, and further found that SBI-0206965 markedly synergized with the mTOR inhibitor everolimus or BEZ235 to kill HCC cells. Thus, the suitable combined use of different autophagic pathway targets may provide a promising therapeutic strategy for HCC in the clinic.
As a multi-tyrosine kinase inhibitor of the mTOR pathway, sorafenib is also an autophagy inducer and is the first United States Food and Drug Administration-approved drug for targeted HCC treatment[62]. Sorafenib has been widely used to treat advanced HCC patients and improved the survival rate in preclinical studies and several clinical trials[63,64]. A possible issue with sorafenib is drug resistance in HCC patients when sorafenib induces autophagy[65], as autophagy is required for HCC cell survival, especially in the early stages. In addition, sorafenib has a synergic antitumor effect on HCC growth when combined with Beclin1 or metformin and vorinostat, and selumetinib via autophagy induction[66-69]. To date, 225 studies on sorafenib and liver cancer were found on the clinicaltrials.gov website. Of these, an ongoing clinical trial is investigating whether sorafenib/hydroxychloroquine (HCQ)-regulated autophagy will have improved efficacy in HCC treatment when compared with sorafenib alone, and in patients progressing on sorafenib the addition of HCQ could lead to disease stability in those with advanced HCC[70].
Interestingly, in our previous studies, we found that autophagy modulation participated in oncolytic virotherapy in a liver cancer stem cell model[71]. The inhibition of autophagy by HCQ in cancer stem-like cells affected the invasion and migration of HCC cells, and the EMT process[72]. Further studies by our group showed that a novel oncolytic adenovirus targeting Wnt signaling effectively inhibited cancer-stem like cell growth via metastasis, apoptosis and autophagy in HCC models[71,72]. Thus, we also suggest that the inhibition of tumor metastasis by regulating autophagy may be one of the most effective therapies for HCC.
Although autophagy induction or inhibition can efficiently treat advanced HCC patients, either alone or combined with other anticancer agents, drug resistance is a challenge. It was shown that autophagy acted as a tumor promoter in an established metastatic tumor, as it facilitated cancer cell survival against anticancer therapy by providing nutrients and energy to cancer cells under metabolic and oxidative stress[73]. Studies have found that various chemotherapeutic drugs for HCC treatment can increase autophagic flux of HCC cells, and it may be related with enhancing drug resistance and promoting cell survival[23]. Thus, autophagy induction at the tumor development stage promotes resistance to cancer therapy, while inhibition of autophagy promotes cancer cell death during cancer therapy[15,74,75]. However, enhancement of autophagic flux may also induce tumor cell death in some cases such as oncolytic virotherapy[76], which contributes to targeting HCC therapy.
It has been proved that multiple signaling pathways and some key regulators are involved in cancer drug resistance. Notably, the role of autophagy in cancer is also controlled by regulatory elements, including proto-oncogenes (PI3K, AKT, MTOR, RAS, RAF and BCR–ABL), tumor suppressors (BECLIN1, TP53, FOXO1 and BCL-2), and noncoding RNAs (miRNAs and lncRNAs)[77-82]. These autophagy regulatory elements are divided into autophagy inducers and inhibitors, according to their functions. The existence or over-expression of autophagy inducers often results in therapeutic resistance to cancer therapy, and autophagy inhibitors can overcome resistance to anticancer drug therapies. The noncoding RNA and autophagy inhibitors are also used to resensitize resistant cancer cells to cancer therapy[74]. A study reported by Gao et al[83] showed that miR-520b increased doxorubicin sensitivity in HCC and inhibited ATG7-dependent autophagy, suggesting that the miR-520b/ATG7 pathway is a potential target for chemosensitive therapy in HCC.
Sorafenib is currently the only systemic agent with an autophagy induction role for the treatment of advanced-stage HCC, and exerts a significant prolonged overall survival rate in HCC patients[84]. However, the appearance of intrinsic and acquired resistance to sorafenib remains a huge challenge for the prognosis of stage III/IV HCC patients, with only approximately 30% responsive to sorafenib. The potential underlying mechanism may derive from autophagic induction by sorafenib with prosurvival functions[65]. However, a recent randomized, multicenter, multinational phase II trial found no evidence that sorafenib combined with everolimus (a potent inhibitor of mTOR) has higher efficacy compared with sorafenib alone[85]. A recent study indicated that the receptor for advanced glycation end products (RAGE) induced HCC proliferation and sorafenib resistance by modulating autophagy, and RAGE deficiency contributed to sorafenib response by activating the AMPK/mTOR signaling pathway, indicating that Rage may be a potential target for therapeutic interventions in HCC and a biomarker of sorafenib resistance[86]. RAGE ligands, such as the high mobility group box 1 (HMGB1), play a key role in the proliferation, apoptosis, metastasis and angiogenesis of HCC cells, which is similar to the role of RAGE in HCC[87,88]. Furthermore, HMGB1 also contributed to chemotherapy resistance in multiple tumors, such as lung cancer, osteosarcoma and neuroblastoma. Especially, in a recent study, a novel potential role of HMGB1 in the regulation of sorafenib therapy resistance in HCC was suggested, implying the positive association between HMGB1 and sorafenib resistance in HCC[89]. Moreover, the cell surface molecule CD24 regulates sorafenib resistance by activating autophagy in HCC, and depletion of CD24 or inhibition of autophagy caused a notable increase in sorafenib sensitivity[90]. Thus, the combination of autophagy modulation and sorafenib or other targeted therapy is a promising therapeutic strategy for the treatment of HCC.
Autophagy plays a pivotal role in tumorigenesis, metastasis, targeted therapy and drug resistance in HCC. Autophagy functions as a tumor suppressor by removing oxidative stress, maintaining genomic stability and preventing uncontrolled inflammation in the early stage of HCC. However, when a malignant tumor is established, autophagy induction elicits tumor promotion and drug tolerance. Thus, targeted drugs to regulate autophagy could be a novel therapeutic strategy to suppress HCC metastasis and resistance.
Recently, the role of autophagy in immunity has been extensively investigated. Autophagy is involved in the differentiation, development and activation of multiple immune cells, and is also critical in antigen presentation[91]. Defective autophagy severely affects T cell proliferation, and tumor-associated macrophage differentiation, and disturbs the function of terminally differentiated lymphocytes, senescent CD8+ T cells and dendritic cells[91,92]. Considering that immune cells are crucial in HCC development and cancer immunotherapies such as chimeric antigen receptor expressing T cells and immune checkpoint inhibitors (PD-1, PD-L1), which represent a breakthrough in hematological malignancy, autophagy may be an excellent candidate target for tumor immunotherapy. With future investigations on autophagic mechanisms in HCC, the combination of an autophagy regulator with multiple kinase inhibitors, signal pathway inhibitors, immunotherapy and oncolytic virotherapy may exert synergic therapeutic efficacy in advanced-stage HCC patients.
| 1. | Levine B, Klionsky DJ. Autophagy wins the 2016 Nobel Prize in Physiology or Medicine: Breakthroughs in baker’s yeast fuel advances in biomedical research. Proc Natl Acad Sci USA. 2017;114:201-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 125] [Article Influence: 12.5] [Reference Citation Analysis (1)] |
| 2. | Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728-741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3678] [Cited by in RCA: 5117] [Article Influence: 341.1] [Reference Citation Analysis (0)] |
| 3. | Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2780] [Cited by in RCA: 2999] [Article Influence: 136.3] [Reference Citation Analysis (0)] |
| 4. | Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 549] [Cited by in RCA: 663] [Article Influence: 41.4] [Reference Citation Analysis (0)] |
| 5. | Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:1845-1846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 260] [Cited by in RCA: 472] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 6. | Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823-830. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1245] [Cited by in RCA: 1262] [Article Influence: 78.9] [Reference Citation Analysis (0)] |
| 7. | Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256-1269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 805] [Cited by in RCA: 1027] [Article Influence: 79.0] [Reference Citation Analysis (0)] |
| 8. | Deretic V, Levine B. Autophagy balances inflammation in innate immunity. Autophagy. 2018;14:243-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 445] [Article Influence: 55.6] [Reference Citation Analysis (6)] |
| 9. | Liu L, Liao JZ, He XX, Li PY. The role of autophagy in hepatocellular carcinoma: friend or foe. Oncotarget. 2017;8:57707-57722. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 129] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 10. | Gatica D, Lahiri V, Klionsky DJ. Cargo recognition and degradation by selective autophagy. Nat Cell Biol. 2018;20:233-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 554] [Cited by in RCA: 863] [Article Influence: 107.9] [Reference Citation Analysis (0)] |
| 11. | Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344-1348. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1632] [Cited by in RCA: 1613] [Article Influence: 100.8] [Reference Citation Analysis (0)] |
| 12. | Goldsmith J, Levine B, Debnath J. Autophagy and cancer metabolism. Methods Enzymol. 2014;542:25-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 115] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 13. | Onorati AV, Dyczynski M, Ojha R, Amaravadi RK. Targeting autophagy in cancer. Cancer. 2018;124:3307-3318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 509] [Cited by in RCA: 571] [Article Influence: 71.4] [Reference Citation Analysis (0)] |
| 14. | Liu B, Wen X, Cheng Y. Survival or death: disequilibrating the oncogenic and tumor suppressive autophagy in cancer. Cell Death Dis. 2013;4:e892. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 101] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 15. | Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther. 2011;10:1533-1541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 830] [Cited by in RCA: 1010] [Article Influence: 67.3] [Reference Citation Analysis (0)] |
| 16. | Singh SS, Vats S, Chia AY, Tan TZ, Deng S, Ong MS, Arfuso F, Yap CT, Goh BC, Sethi G. Dual role of autophagy in hallmarks of cancer. Oncogene. 2018;37:1142-1158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 283] [Cited by in RCA: 438] [Article Influence: 48.7] [Reference Citation Analysis (0)] |
| 17. | Thorburn A, Thamm DH, Gustafson DL. Autophagy and cancer therapy. Mol Pharmacol. 2014;85:830-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 244] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 18. | Cao Z, Zhang H, Cai X, Fang W, Chai D, Wen Y, Chen H, Chu F, Zhang Y. Luteolin Promotes Cell Apoptosis by Inducing Autophagy in Hepatocellular Carcinoma. Cell Physiol Biochem. 2017;43:1803-1812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 103] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 19. | Wang YG, Huang PP, Zhang R, Ma BY, Zhou XM, Sun YF. Targeting adeno-associated virus and adenoviral gene therapy for hepatocellular carcinoma. World J Gastroenterol. 2016;22:326-337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 32] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 20. | Zhang X, Meng S, Zhang R, Ma B, Liu T, Yang Y, Xie W, Liu X, Huang F, Liu T. GP73-regulated oncolytic adenoviruses possess potent killing effect on human liver cancer stem-like cells. Oncotarget. 2016;7:29346-29358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 21. | He G, Lei W, Wang S, Xiao R, Guo K, Xia Y, Zhou X, Zhang K, Liu X, Wang Y. Overexpression of tumor suppressor TSLC1 by a survivin-regulated oncolytic adenovirus significantly inhibits hepatocellular carcinoma growth. J Cancer Res Clin Oncol. 2012;138:657-670. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 22. | Ma B, Wang Y, Zhou X, Huang P, Zhang R, Liu T, Cui C, Liu X, Wang Y. Synergistic suppression effect on tumor growth of hepatocellular carcinoma by combining oncolytic adenovirus carrying XAF1 with cisplatin. J Cancer Res Clin Oncol. 2015;141:419-429. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 23. | Sheng J, Qin H, Zhang K, Li B, Zhang X. Targeting autophagy in chemotherapy-resistant of hepatocellular carcinoma. Am J Cancer Res. 2018;8:354-365. [PubMed] |
| 24. | Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795-800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 923] [Cited by in RCA: 1089] [Article Influence: 72.6] [Reference Citation Analysis (0)] |
| 25. | Marinković M, Šprung M, Buljubašić M, Novak I. Autophagy Modulation in Cancer: Current Knowledge on Action and Therapy. Oxid Med Cell Longev. 2018;2018:8023821. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 155] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 26. | Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425-434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1755] [Cited by in RCA: 1961] [Article Influence: 93.4] [Reference Citation Analysis (0)] |
| 27. | Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809-1820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1582] [Cited by in RCA: 1856] [Article Influence: 80.7] [Reference Citation Analysis (0)] |
| 28. | Liang XH, Yu J, Brown K, Levine B. Beclin 1 contains a leucine-rich nuclear export signal that is required for its autophagy and tumor suppressor function. Cancer Res. 2001;61:3443-3449. [PubMed] |
| 29. | Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2446] [Cited by in RCA: 2726] [Article Influence: 101.0] [Reference Citation Analysis (0)] |
| 30. | Qiu DM, Wang GL, Chen L, Xu YY, He S, Cao XL, Qin J, Zhou JM, Zhang YX, E Q. The expression of beclin-1, an autophagic gene, in hepatocellular carcinoma associated with clinical pathological and prognostic significance. BMC Cancer. 2014;14:327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 102] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 31. | Tian Y, Kuo CF, Sir D, Wang L, Govindarajan S, Petrovic LM, Ou JH. Autophagy inhibits oxidative stress and tumor suppressors to exert its dual effect on hepatocarcinogenesis. Cell Death Differ. 2015;22:1025-1034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 98] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 32. | Liu K, Lee J, Ou JJ. Autophagy and mitophagy in hepatocarcinogenesis. Mol Cell Oncol. 2018;5:e1405142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 33. | Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621-1635. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 672] [Cited by in RCA: 692] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 34. | Mathew R, Karantza-Wadsworth V, White E. Assessing metabolic stress and autophagy status in epithelial tumors. Methods Enzymol. 2009;453:53-81. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 35. | Mathew R, White E. Autophagy in tumorigenesis and energy metabolism: friend by day, foe by night. Curr Opin Genet Dev. 2011;21:113-119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 203] [Cited by in RCA: 202] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 36. | Langley RR, Fidler IJ. The seed and soil hypothesis revisited--the role of tumor-stroma interactions in metastasis to different organs. Int J Cancer. 2011;128:2527-2535. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 728] [Cited by in RCA: 696] [Article Influence: 46.4] [Reference Citation Analysis (0)] |
| 37. | May CD, Sphyris N, Evans KW, Werden SJ, Guo W, Mani SA. Epithelial-mesenchymal transition and cancer stem cells: a dangerously dynamic duo in breast cancer progression. Breast Cancer Res. 2011;13:202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 230] [Cited by in RCA: 267] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 38. | Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol. 2010;11:633-643. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1665] [Cited by in RCA: 1529] [Article Influence: 95.6] [Reference Citation Analysis (9)] |
| 39. | Ridley AJ. Life at the leading edge. Cell. 2011;145:1012-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 714] [Cited by in RCA: 744] [Article Influence: 49.6] [Reference Citation Analysis (0)] |
| 40. | Koustas E, Karamouzis MV, Mihailidou C, Schizas D, Papavassiliou AG. Co-targeting of EGFR and autophagy signaling is an emerging treatment strategy in metastatic colorectal cancer. Cancer Lett. 2017;396:94-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 62] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 41. | Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gélinas C, Fan Y. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51-64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1605] [Cited by in RCA: 1611] [Article Influence: 80.6] [Reference Citation Analysis (0)] |
| 42. | Sandilands E, Serrels B, Mcewan DG, Morton JP, Macagno JP, Mcleod K, Stevens C, Brunton VG, Langdon WY, Vidal M. Autophagic targeting of Src promotes cancer cell survival following reduced FAK signalling. Nat Cell Biol. 2012;14:51-60. [RCA] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 149] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 43. | Sharifi M, Mowers E, Drake L, Collier C, Hong C, Zamora M, Mui S, Macleod K. Autophagy Promotes Focal Adhesion Disassembly and Cell Motility of Metastatic Tumor Cells through the Direct Interaction of Paxillin with LC3. Cell Rep. 2016;15:1660-1672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 263] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 44. | Schoenherr C, Byron A, Sandilands E, Paliashvili K, Baillie GS, Garcia-Munoz A, Valacca C, Cecconi F, Serrels B, Frame MC. Ambra1 spatially regulates Src activity and Src/FAK-mediated cancer cell invasion via trafficking networks. Elife. 2017;6:pii: e23172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 45. | Deakin NO, Turner CE. Paxillin comes of age. J Cell Sci. 2008;121:2435-2444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 417] [Cited by in RCA: 412] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 46. | Chen GC, Lee JY, Tang HW, Debnath J, Thomas SM, Settleman J. Genetic interactions between Drosophila melanogaster Atg1 and paxillin reveal a role for paxillin in autophagosome formation. Autophagy. 2008;4:37-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 51] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 47. | Peng YF, Shi YH, Ding ZB, Ke AW, Gu CY, Hui B, Zhou J, Qiu SJ, Dai Z, Fan J. Autophagy inhibition suppresses pulmonary metastasis of HCC in mice via impairing anoikis resistance and colonization of HCC cells. Autophagy. 2013;9:2056-2068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 233] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 48. | Peng YF, Shi YH, Shen YH, Ding ZB, Ke AW, Zhou J, Qiu SJ, Fan J. Promoting colonization in metastatic HCC cells by modulation of autophagy. PLoS One. 2013;8:e74407. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 56] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 49. | Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2533] [Cited by in RCA: 3565] [Article Influence: 396.1] [Reference Citation Analysis (0)] |
| 50. | Li J, Yang B, Zhou Q, Wu Y, Shang D, Guo Y, Song Z, Zheng Q, Xiong J. Autophagy promotes hepatocellular carcinoma cell invasion through activation of epithelial-mesenchymal transition. Carcinogenesis. 2013;34:1343-1351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 242] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 51. | Fan Q, Yang L, Zhang X, Ma Y, Li Y, Dong L, Zong Z, Hua X, Su D, Li H. Autophagy promotes metastasis and glycolysis by upregulating MCT1 expression and Wnt/β-catenin signaling pathway activation in hepatocellular carcinoma cells. J Exp Clin Cancer Res. 2018;37:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 175] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 52. | Fu XT, Shi YH, Zhou J, Peng YF, Liu WR, Shi GM, Gao Q, Wang XY, Song K, Fan J. MicroRNA-30a suppresses autophagy-mediated anoikis resistance and metastasis in hepatocellular carcinoma. Cancer Lett. 2018;412:108-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 89] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 53. | Wu DH, Jia CC, Chen J, Lin ZX, Ruan DY, Li X, Lin Q, Min-Dong , Ma XK, Wan XB. Autophagic LC3B overexpression correlates with malignant progression and predicts a poor prognosis in hepatocellular carcinoma. Tumour Biol. 2014;35:12225-12233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 54. | Janji B, Berchem G, Chouaib S. Targeting Autophagy in the Tumor Microenvironment: New Challenges and Opportunities for Regulating Tumor Immunity. Front Immunol. 2018;9:887. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 69] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 55. | Chen CL, Tseng YW, Wu JC, Chen GY, Lin KC, Hwang SM, Hu YC. Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA PTENP1 and MicroRNA regulation. Biomaterials. 2015;44:71-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 183] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 56. | Huang S, Houghton PJ. Inhibitors of mammalian target of rapamycin as novel antitumor agents: from bench to clinic. Curr Opin Investig Drugs. 2002;3:295-304. [PubMed] |
| 57. | Ashworth RE, Wu J. Mammalian target of rapamycin inhibition in hepatocellular carcinoma. World J Hepatol. 2014;6:776-782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 58. | Decaens T, Luciani A, Itti E, Hulin A, Roudot-Thoraval F, Laurent A, Zafrani ES, Mallat A, Duvoux C. Phase II study of sirolimus in treatment-naive patients with advanced hepatocellular carcinoma. Dig Liver Dis. 2012;44:610-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 59. | Huynh H, Chow KH, Soo KC, Toh HC, Choo SP, Foo KF, Poon D, Ngo VC, Tran E. RAD001 (everolimus) inhibits tumour growth in xenograft models of human hepatocellular carcinoma. J Cell Mol Med. 2009;13:1371-1380. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 110] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 60. | Zhu AX, Kudo M, Assenat E, Cattan S, Kang YK, Lim HY, Poon RT, Blanc JF, Vogel A, Chen CL. Effect of everolimus on survival in advanced hepatocellular carcinoma after failure of sorafenib: the EVOLVE-1 randomized clinical trial. JAMA. 2014;312:57-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 436] [Cited by in RCA: 488] [Article Influence: 40.7] [Reference Citation Analysis (1)] |
| 61. | Egan DF, Chun MG, Vamos M, Zou H, Rong J, Miller CJ, Lou HJ, Raveendra-Panickar D, Yang CC, Sheffler DJ. Small Molecule Inhibition of the Autophagy Kinase ULK1 and Identification of ULK1 Substrates. Mol Cell. 2015;59:285-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 450] [Cited by in RCA: 588] [Article Influence: 53.5] [Reference Citation Analysis (0)] |
| 62. | Chen J, Lu S, Zhang Y, Xu L, Chen J, Wang J, Chen M, Zhang R, Zhou Z. Sorafenib Monotherapy Versus Sorafenib Combined with Regional Therapies for Hepatocellular Carcinoma Patients with Pulmonary Oligometastases: A Propensity Score-matched Analysis. J Cancer. 2018;9:1745-1753. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 63. | Meyer T. Treatment of advanced hepatocellular carcinoma: beyond sorafenib. Lancet Gastroenterol Hepatol. 2018;3:218-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 64. | Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, Luo R, Feng J, Ye S, Yang TS. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3854] [Cited by in RCA: 4739] [Article Influence: 263.3] [Reference Citation Analysis (0)] |
| 65. | Sun T, Liu H, Ming L. Multiple Roles of Autophagy in the Sorafenib Resistance of Hepatocellular Carcinoma. Cell Physiol Biochem. 2017;44:716-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 97] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 66. | Tai WM, Yong WP, Lim C, Low LS, Tham CK, Koh TS, Ng QS, Wang WW, Wang LZ, Hartono S. A phase Ib study of selumetinib (AZD6244, ARRY-142886) in combination with sorafenib in advanced hepatocellular carcinoma (HCC). Ann Oncol. 2018;29:526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 67. | Tai WT, Shiau CW, Chen HL, Liu CY, Lin CS, Cheng AL, Chen PJ, Chen KF. Mcl-1-dependent activation of Beclin 1 mediates autophagic cell death induced by sorafenib and SC-59 in hepatocellular carcinoma cells. Cell Death Dis. 2013;4:e485. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 135] [Cited by in RCA: 177] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 68. | Bareford MD, Hamed HA, Tang Y, Cruickshanks N, Burow ME, Fisher PB, Moran RG, Nephew KP, Grant S, Dent P. Sorafenib enhances pemetrexed cytotoxicity through an autophagy-dependent mechanism in cancer cells. Autophagy. 2011;7:1261-1262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 69. | Ling S, Song L, Fan N, Feng T, Liu L, Yang X, Wang M, Li Y, Tian Y, Zhao F. Combination of metformin and sorafenib suppresses proliferation and induces autophagy of hepatocellular carcinoma via targeting the mTOR pathway. Int J Oncol. 2017;50:297-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 76] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 70. | Epp Goodwin, Sukeshi Patel Arora. Sorafenib Induced Autophagy Using Hydroxychloroquine in Hepatocellular Cancer. US National Library of Medicine. Available from: https://www.clinicaltrials.gov/ct2/show/NCT03037437 ClinicalTrials.gov Identifier: NCT03037437. |
| 71. | Zhang J, Lai W, Li Q, Yu Y, Jin J, Guo W, Zhou X, Liu X, Wang Y. A novel oncolytic adenovirus targeting Wnt signaling effectively inhibits cancer-stem like cell growth via metastasis, apoptosis and autophagy in HCC models. Biochem Biophys Res Commun. 2017;491:469-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 57] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 72. | Jian Z, Lai WJ, Qiang LI, Jin J, Xiao BD, Wan G, Wang YG. Inhibition of Hepatocellular Stem Cells by Oncolytic Virus Targeting Wnt Signaling Pathway. Shengwu Huaxue Yu Shengwu Wuli Jinzhan. 2017;44:326-337. [DOI] [Full Text] |
| 73. | Das CK, Mandal M, Kögel D. Pro-survival autophagy and cancer cell resistance to therapy. Cancer Metastasis Rev. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 128] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 74. | Kumar A, Singh UK, Chaudhary A. Targeting autophagy to overcome drug resistance in cancer therapy. Future Med Chem. 2015;7:1535-1542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 117] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 75. | Aredia F, Scovassi AI. Manipulation of autophagy in cancer cells: an innovative strategy to fight drug resistance. Future Med Chem. 2013;5:1009-1021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 76. | Tong Y, You L, Liu H, Li L, Meng H, Qian Q, Qian W. Potent antitumor activity of oncolytic adenovirus expressing Beclin-1 via induction of autophagic cell death in leukemia. Oncotarget. 2013;4:860-874. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 77. | Sooro MA, Zhang N, Zhang P. Targeting EGFR-mediated autophagy as a potential strategy for cancer therapy. Int J Cancer. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 78. | Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600-606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 662] [Cited by in RCA: 647] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 79. | Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, Codogno P, Debnath J, Gewirtz DA, Karantza V. Autophagy in malignant transformation and cancer progression. EMBO J. 2015;34:856-880. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 905] [Cited by in RCA: 974] [Article Influence: 88.5] [Reference Citation Analysis (0)] |
| 80. | Yang J, He Y, Zhai N, Ding S, Li J, Peng Z. MicroRNA-181a inhibits autophagy by targeting Atg5 in hepatocellular carcinoma. Front Biosci (Landmark Ed). 2018;23:388-396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 81. | Paquette M, El-Houjeiri L, Pause A. mTOR Pathways in Cancer and Autophagy. Cancers (Basel). 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 175] [Cited by in RCA: 229] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 82. | Denisenko TV, Pivnyuk AD, Zhivotovsky B. p53-Autophagy-Metastasis Link. Cancers (Basel). 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 83. | Gao AM, Zhang XY, Hu JN, Ke ZP. Apigenin sensitizes hepatocellular carcinoma cells to doxorubic through regulating miR-520b/ATG7 axis. Chem Biol Interact. 2018;280:45-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 84. | Finn RS, Zhu AX, Farah W, Almasri J, Zaiem F, Prokop LJ, Murad MH, Mohammed K. Therapies for advanced stage hepatocellular carcinoma with macrovascular invasion or metastatic disease: A systematic review and meta-analysis. Hepatology. 2018;67:422-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 167] [Article Influence: 20.9] [Reference Citation Analysis (1)] |
| 85. | Koeberle D, Dufour JF, Demeter G, Li Q, Ribi K, Samaras P, Saletti P, Roth AD, Horber D, Buehlmann M, Wagner AD, Montemurro M, Lakatos G, Feilchenfeldt J, Peck-Radosavljevic M, Rauch D, Tschanz B, Bodoky G; Swiss Group for Clinical Cancer Research (SAKK). Sorafenib with or without everolimus in patients with advanced hepatocellular carcinoma (HCC): a randomized multicenter, multinational phase II trial (SAKK 77/08 and SASL 29). Ann Oncol. 2016;27:856-861. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 98] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 86. | Li J, Wu PW, Zhou Y, Dai B, Zhang PF, Zhang YH, Liu Y, Shi XL. Rage induces hepatocellular carcinoma proliferation and sorafenib resistance by modulating autophagy. Cell Death Dis. 2018;9:225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 87. | Chen R, Hou W, Zhang Q, Kang R, Fan XG, Tang D. Emerging role of high-mobility group box 1 (HMGB1) in liver diseases. Mol Med. 2013;19:357-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 93] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 88. | Yaser AM, Huang Y, Zhou RR, Hu GS, Xiao MF, Huang ZB, Duan CJ, Tian W, Tang DL, Fan XG. The Role of receptor for Advanced Glycation End Products (RAGE) in the proliferation of hepatocellular carcinoma. Int J Mol Sci. 2012;13:5982-5997. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 53] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 89. | Xiao Y, Sun L, Fu Y, Huang Y, Zhou R, Hu X, Zhou P, Quan J, Li N, Fan XG. High mobility group box 1 promotes sorafenib resistance in HepG2 cells and in vivo. BMC Cancer. 2017;17:857. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 90. | Lu S, Yao Y, Xu G, Zhou C, Zhang Y, Sun J, Jiang R, Shao Q, Chen Y. CD24 regulates sorafenib resistance via activating autophagy in hepatocellular carcinoma. Cell Death Dis. 2018;9:646. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 112] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 91. | Yu S, Wang Y, Jing L, Claret FX, Li Q, Tian T, Liang X, Ruan Z, Jiang L, Yao Y. Autophagy in the “inflammation-carcinogenesis” pathway of liver and HCC immunotherapy. Cancer Lett. 2017;411:82-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 56] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 92. | Shibutani ST, Saitoh T, Nowag H, Münz C, Yoshimori T. Autophagy and autophagy-related proteins in the immune system. Nat Immunol. 2015;16:1014-1024. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 443] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Corrales FJ, Hashimoto N, Lim SC, Tomizawa M S- Editor: Ma RY L- Editor: Filipodia E- Editor: Yin SY