INTRODUCTION
Hepatitis C virus (HCV) belongs to the family Flaviviridae, a member of the genus hepaciviruses. HCV was identified in 1989 as one of the viruses capable of causing viral hepatitis, in addition to the previously known hepatitis A and B viruses (HAV, HBV)[1]. There are seven major HCV genotypes that differ in virulence and their geographic distribution[2]. Currently, it is estimated that worldwide more than 185 million people are infected with HCV, which represents 2.8% of the world’s population[3]. Infection usually takes place in the absence of obvious clinical symptoms, and the resulting inflammation of the liver often progresses to become chronic, usually lasting for years. In time, the chronic inflammation can lead to cirrhosis of the liver and ultimately liver failure or to the development of primary liver cancer[4]. Although the virus replicates mainly in hepatocytes, it also occurs in peripheral blood mononuclear and central nervous system microglia[5,6]. In people with simultaneous infection with HIV (human immunodeficiency virus), HCV replication was also observed in other tissues[7]. Sometimes HCV enters cells of the immune system, leading to very long-lasting effects which are extremely difficult to treat effectively[8].
While progress has been made in the treatment of some other common viral infections, HCV infection remains an important health problem in the world. Current standard treatment methods have the desired therapeutic effect on 40%-60% of patients[4]. The new highly effective drug, grazoprevir, is able to cure patients with 93% effectivity[9], unfortunately, the treatment is still very expensive and out of reach for the majority of the infected individuals, many of whom live in Third World countries. So far, no HCV vaccine has been developed, which is due to the high genetic variability of the virus, comparable to the genetic variability of HIV. In the absence of widely accessible conventional drugs and vaccines, numerous attempts have been made to design inhibitors of viral proteins, inhibitory oligomers of the antisense and ribozyme type, and more recently also of RNA interference tools directed against viral RNA[4,10,11].
HCV is a small, enveloped virus with a diameter of 40-60 nm, whose genome is a single-stranded, positive-sense RNA of about 9.6 kb in length[6,10,12]. In the viral replication process, the positive-sense RNA strand is transcribed into negative-sense counterpart, the replication intermediate, which serves as the template for the RNA synthesis of progeny genomes. The HCV genome has one very long open reading frame (ORF). It encodes a precursor polyprotein, which is digested in a series of cleavage processes to finally produce proteins: C, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B[13]. The coding sequence of the HCV genome is flanked by two untranslated regions: the 5’UTR and the 3’UTR (5’ and 3’ untranslated regions). Both these regions play key functions in regulating HCV life cycle and determine its expression level. The 5’UTR contains a highly structured regulatory element, an IRES (internal ribosome entry site), that enables cap-independent translation. The 3’UTR is engaged in the replication process and in the regulation of translation. In addition, control of other processes as, for example, assembly of virions and switching between different developmental phases takes place with the participation of the structural RNA elements present at the very 3’ end of the HCV genome.
This article presents the current state of knowledge about the structure and functions of the most terminal section of the 3’UTR of hepatitis C virus, the 3’X-tail.
STRUCTURE OF THE 3’UTR REGION
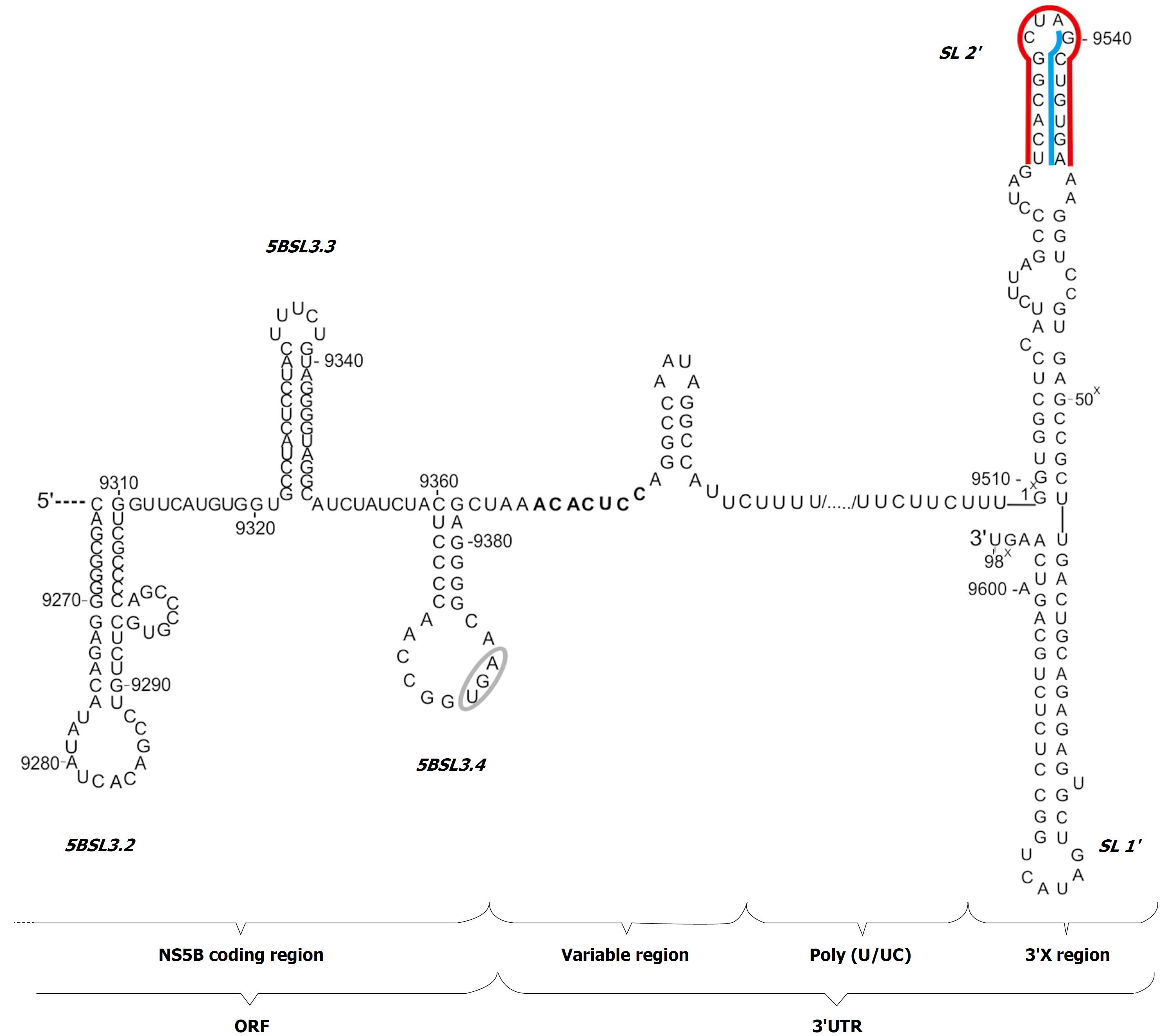
HCV 3’UTR has a variable length of 170 to 250 nucleotides (Figure 1). Three characteristic sections have been recognized in this region: immediately after the stop codon there is a variable region about 25-130 nucleotides in length, characterized by high sequence heterogeneity between various genotypes, but conserved within the same genotype of the virus. Next there is a poly-pyrimidine segment of varying length, independent of the type, or even a subtype of virus, ranging from about 30 to 130 nucleotides. At the very end of the genome, there is an 3’X region, discovered 6 years after cloning of the virus, which is 98 nucleotides long and is an almost absolutely conserved sequence[6].
Figure 1 Secondary structure model of the 3'UTR of hepatitis C virus genome with adjacent 3' terminal sequence of the coding region.
The stop codon is indicated by a gray ellipse. The regions involved in the kissing-interactions are marked with blue lines; the region involved in dimerization is indicated by a red line; the seed region for miR122 is highlighted in bold.
Within the first, variable region of 3’UTR, two sequence motifs are present that are found in all genotypes of HCV. These are: the ACACUCC section, which represents a seed-region for miR-122[14], and the UG dinucleotide located at the very end of the region, directly upstream the poly (U/UC) section[6]. The stop codon is located in the apical loop of the stem-loop motif, named 5BSL3.4 or SL9360, which is created partly from the terminal nucleotides of the ORF encoding the NS5B protein and partly from non-coding nucleotides. The poly-pyrimidine section can be divided into a two poly (UC) parts uneven in length, which are separated by the one poly (U) region. The poly-pyrimidine segment is heterogeneous, not only in terms of length, but also in nucleotide sequence. In genotypes 2a, 3a, 3b there are several conserved adenosine residues in this region that are missing in genotypes 1b and 2b[6]. Individual guanosine residues are also observed on rare occasions.
3'X-tail
Initially, it was suspected that at the 3' terminus of the HCV genome a poly (U) or poly (A) sequence was present. The existence of the 98-nt long 3'X region was discovered by Tanaka and colleagues in 1995[15]. Almost simultanously the presence of the 3'X at the end of the HCV genome identified Kolykhalov et al[16]. Comparison of the sequence of this RNA segment in different viral isolates indicated 96%-100% sequence conservation of the 3'X region, with only single substitutions in the 3' terminal 46-nt sequence[6]. This is an unusual feature for this dynamically changing virus, that suggests its extremely important function.
Several different secondary structure models of the 3'X region were proposed, based on computer predictions and experimental structure probing by chemical modification, enzyme digestion, RNA cleavage induced by Pb2+ ions, NMR (nuclear magnetic resonance) and SAXS (small angle X-ray scattering). A stable structure of the SL1 hairpin was proposed for the 3' part of the 3'X RNA, which is common to different structural models (Figure 2)[17-21]. The 52-nucleotide segment making up the 5' part of the 3'X region could not be assigned an unambiguous structure based on the experimental results obtained. The proposed models were only partially confirmed by the results of experimental studies[6,17,18]. Poor ordering of this fragment or formation of more than one structural form have been suggested[17,18,20].
Figure 2 Diverse secondary structure models proposed for the 3'X region of hepatitis C virus genome.
A: The 3xSL model[17,18]; B: The 4xSL model[20]; C: The 2xSL model[21]. The region involved in the kissing-interactions is indicated with blue line, the region involved in dimerization is marked with red line; possible alternative folding of separate fragments are displayed as gray rectangles.
One of the first structural models of the 3'X region suggested the presence of three hairpin motifs: SL1, SL2, and SL3 (Figure 2A)[17,18]. Another structural model suggested a set of four hairpins: SL1, SL2a, SL2b and SL3, where SL1 and SL3 did not differ from the first model, but two shorter hairpins replaced the SL2 motif[20]. The main reason for the proposed change was the observation of strong DMS modifications, as well as Pb2+ ion-induced cleavages, in the middle of the SL2 double-stranded stem (C44-C45), which indicated a single-stranded or highly flexible region there. In this four stem-loop model (4xSL), the reactive cytidine residues are located in the apical loop of the short SL2b motif (Figure 2B). In addition, it was noted that the SL2a and SL2b hairpins have the potential to create a pseudoknot after rearrangement of their base pairing[20]. However, discovery of functionally important long-range kissing interactions between a sequence located in the apical loop of the SL2 hairpin and the upstream sequence in the region encoding NS5B[22] (see next section), seemed to support the three- (3xSL), but not the four- (4xSL) stem-loop model for the 3'X region. Thus, the 3xSL structure model of the 3'X-tail became the favored idea during the following decade.
Recently, based on NMR and SAXS studies, a two-hairpin model (2xSL) has been proposed for the 3'X region, consisting of the SL1 and SL2' (named elsewhere also SL2/3) (Figure 2C)[21,23,24]. In addition, in this model the SL1 hairpin may fold in two different ways: the closed structure - SL1 or the open one - SL1', differing by the three terminal base-pairs of the hairpin being paired or unpaired, respectively (Figure 2C)[21,23,25]. In the 2xSL model the previously identified flexible cytosine residues (C44-C45) are located within the internal loop of the SL2' motif. The closed conformation of SL1 is associated with long-range kissing interactions with another part of the genome, while the open conformation, SL1' with the 3' overhang is associated with dimerization (described in the next section).
The role of selected metal ions was investigated regarding its influence on the structure of the 3'X-tail but it seems that neither magnesium, nor sodium ion concentration determine its folding, within the range of normal physiological conditions[26]. However, at higher ionic strength, extended homodimers are preferentially formed over 2xSL monomers[24]. A chaperone role of the viral C protein (core protein), has also been suggested[23,27]. Long-range RNA-RNA interactions with 5’ sequences in the genome or with a second genomic RNA molecule seems to influence the structure of the 3'X region more than the presence of specific metal ions.
It is very likely that the 3'X region can adopt more than one structural form in infected cells and that a specific equilibrium between these forms regulates several processes of the viral life cycle. These different structural forms may be favored by distinct viral genotypes what can help to explain their differential virulence and drug resistance[28,29].
Long range RNA-RNA interactions
Previous investigation of genomic HCV RNA and a construct containing only 5'UTR and 3'UTR did not show any interaction of the X region with other regions of the molecules studied. This suggested the structural independence of these two regions of viral RNA from each other[17]. Later tertiary[30-32] interactions at the 3' end of the HCV genomic strand were proposed by Friebe et al[22]. The kissing interactions between the absolutely conserved “k” segment of the X region: 32X-GCUGUGA-38X and the “k′” segment of the NS5B coding sequence: 9281-UCACAGC-9287 do not require a protein chaperone. These sequence stretches are located within apical parts of SL elements called SL2 or SL2' and 5BSL3.2, which is one of the domains in the CRE (cis-acting replication element) (Figure 1)[22,28,31,33]. It is easy to imagine that kissing interactions could be initiated by any complementary stretch of nucleotides located within two apical loops of RNA hairpins. This scenario was previously suggested for the 3xSL model of the 3'X-tail[22,28,31,33]. However, recent studies with the use of mutagenesis, NMR and SAXS methods, indicate that before kissing-interactions are formed, the “k”- sequence in the X region is involved in base pairing within the SL2' element[21,24]. How the 5BSL3.2 element is able to induce the conformational transition from SL2' to SL2, or more globally: from 2xSL to 3xSL form of the 3'X, remains unclear. The next great challenge is to elucidate step by step how this transition occurs.
Two replicon systems, Con1b and JF-H1, have been investigated with a SHAPE method, which is based on a chemical modification of single-stranded RNA residues. The replicons are constructed on the basis of two different viral genotypes 1b and 2a, respectively. The experimental results showed that the proposed kissing interactions were detectable only for replicon JF-H1 (genotype 2a). The results obtained for replicon Con1b were in agreement with data obtained for the genotype 1a (strain H) of the virus and favors the open conformation of SL1'. In this open conformation the very 3' end of the SL' remains single-stranded, and that is associated with an increase in the efficiency of RNA synthesis initiation. In fact, subtype 1b is more virulent and resistant to interferon-based therapy than other genotypes, including subtype 2a[29,34]. This suggests how virus virulence and drug response is significantly influenced by the long-range kissing interactions, which likely cause changes in the base-pairing character of the very 3’ terminal nucleotides.
Dimerization
Another intriguing set of tertiary interactions were proposed for the 3’X region of HCV genome by Ivanyi-Nagy et al[23]. Primarily in vitro investigations showed that the apical part of the SL2 hairpin in the 3xSL model, 29X-CUAG-32X, is able to interact with the respective palindromic sequence in the second 3’X RNA molecule thus inducing the formation of a homodimer[35]. The 16-nt palindromic sequence, called also DLS (dimerization leading stretch) is absolutely conserved among all HCV genotypes[36]. The homodimer, consisting of two isolated 3’X-RNA molecules, was characterized in vitro by NMR and SAXS. It was proposed that the resulting homoduplex could involve shorter (SL2) or extended (SL2’) sequence fragments (Figure 3)[21,23-25,35]. The core protein supports the formation and stabilizes the extended homodimer[24].
Figure 3 Long-range RNA-RNA interactions proposed for the 3'X region of hepatitis C virus genome.
A: The homodimeric interactions between two 3'X regions embedded into two RNA molecules, model according to Cantero-Camacho et al[24]; B: The kissing-interactions with SL5B3.2[22,28,31,33]. Nucleotide sequence involved in dimerization is additionally indicated with red line, while those involved in the kissing-interactions are marked with blue lines.
The dimerization of the HCV genome still remains unproved in vivo and its function remains to be elucidated. It was suggested that the dimerization could be helpful in ensuring that only full-length progeny RNA molecules are encapsidated[24]. Moreover, the unwinding of the 3’-end of the genome, could greatly facilitate the minus RNA synthesis[37]. Masante et al[38] suggested that the homodimeric genome operates as a preferred template for the HCV polymerase (NS5B). Additionally, dimerization might enhance the rate of RNA recombination between two homologue RNA strands (resumed in[39]). The equilibrium between 2xSL monomers and dimers would likely also be tuned by the local concentration of RNA and the presence of core protein[24].
FUNCTIONS OF THE 3’UTR REGION
Hepatitis C virus can only infect humans and chimpanzees; there is no experimental model of its infectivity among small animals. For this reason, very few studies on the spread of the virus have been carried out in vivo. An experiment was carried out by Yanagi et al[40], in which a number of viral constructs containing deletions within the 3’UTR were injected into the liver of the chimpanzee at time intervals, and then the animal was examined for the presence of HCV RNA, anti-HCV antibodies and liver enzymes in its serum. Viral mutants lacking the entire 3’X region or parts thereof (nt: 1-50 and 57-98) were not able to replicate. Also viral infection was not observed when a mutant containing no poly (U/UC) segment was used. Only the construct devoid of 24 nucleotides within the variable region turned out to be infectious, indicating that this region is not essential for the viral life cycle under these conditions[40]. A similar experiment was carried out by Kolykhalov et al[41], and the obtained results were in line with previous observations of Yanagi et al[40].
One of the possible explanations of 3’X region function was its influence on genomic RNA stability. This has been shown in vitro but is not equally important for all genotypes of the virus[42].
Replication process
The information on the involvement of individual parts of the 3’UTR in the life cycle of the virus presented in the previous section was confirmed in research with the replicating Huh7 and HeLa cell systems[19,22,42,43]. Namely, constructs devoid of the entire poly (U/UC) section, were not able to replicate[42,43], and the minimum length of the polypyrimidine segment was 50 nt in one study[43], and only 26 nt in the other[42]. In contrast, the deletion of the 3’X region or any one of its parts, SL1, SL2 or SL3, led to a complete failure to replicate[42,43] and only few point mutations in this region were tolerated[19,43]. In turn, the removal of the variable region from the 3’UTR of the viral genome only reduced the efficiency of the process leading to decrease in the rate of replication[22,42,43]. In addition, it has been shown that the region directly upstream the stop codon plays a key role in viral replication[22].
The site of the NS5B polymerase attachment within region 3’X has been mapped to be within the SL2 sequence and within the SL1[44]. They are protected against digestion with RNase T1 at guanosine residues at positions 41, 42, 50 and 53. The direct interaction of the 3’X RNA with the NS5B protein has been found in studies conducted in vitro[6,12,44-46]. The specificity of the viral polymerase to the model RNA template is relatively low and the presence of the 3’X region is not always necessary for RNA synthesis[6,12,37,45]. However, in the case of matrices containing genomic or subgenomic RNA, the presence of the 3’X region is necessary for the efficiency and the specificity of the process. It has been shown that the lack of this sequence, or a part thereof, almost completely inhibits replication, and in the case of deletion of nucleotides in positions 31-40, the product is too long[12,44].
The NS5B polymerase catalytic center probably only interacts with a single-stranded RNA fragment. The initiation of replication seems to take place in the SL1 loop, 21 nucleotides from the 3’ end of the 3’X RNA[12]. However, another research group, Kim et al[46] indicates that the process begins near the 3’ terminus, in a region rich in purines. However, in studies carried out in a cellular system, it has been shown that the presence of the 3’ terminal GU dinucleotide is preferred in the reaction[19]. Similar preferences of the replicase for the U at the 3’-end of the template RNA were observed by Shimm et al[47] in the in vitro system. Also, Kao et al[37] postulate that the enzyme requires a stable secondary template structure and at least one unpaired cytidine residue at its 3’ end to initiate RNA synthesis. Butcher et al[48] proposed a replication initiation scenario, one common to various polymerases, in which the synthesis of a new strand of RNA begins with a nucleotide complementary to the penultimate of the 3’ end of the nucleotide of the template molecule. Secondly, a complementary nucleotide is added to the remainder of the template at the 3' end, and only after the synthesis of this dinucleotide-primer is the complementary strand of viral RNA synthesized. Many observations suggest that NS5B is a non-specific enzyme, i.e. it can recognize more than one nucleotide sequence. The presence of specific I-, II- and tertiary structures at the 3’-end of the template RNA may allow modulation of the specificity of the enzyme for better yield and/or greater precision in the selection of the origin. In addition, both viral and host proteins can be involved in the replication process[6,49].
In summary, it should be noted that this replication phase of the life cycle of the virus has not yet been precisely understood. In addition to interacting with the viral polymerase, the 3’X region also interacts with other proteins one of them being the NS3 viral protein[49]. Acting as a helicase, it is probably a very important component of the replication complex, because the NS5B polymerase tends to detach from the RNA template when it encounters very stable RNA secondary structures[50]. The specific interaction between NS3 and the HCV 3’UTR probably involves a large part of this region containing the sequence 3’X and the poly (U/UC) section, since none of these elements alone is sufficient to form a stable complex with this protein[49].
Translation process
One of the earliest known cellular proteins interacting with HCV 3’UTR was the PTB protein. The interaction of PTB with the 3’X region may be related to the regulation of the translation process. The proposed site of PTB binding is 21 nucleotides from the 5’ end of the 3’X region (SL3), containing a part of the consensus sequence recognized by this protein[18]. Similar results are presented by Tsuchihara et al[51] suggesting that the region involved in the interaction with PTB is 19 nucleotides from the 5’ end of the 3’X region and extends 7 nucleotides upstream this sequence. The mutagenesis data showed that both secondary structure and the nucleotide sequence in the SL2 and SL3 regions are important for this interaction[18,51].
The 3’UTR has been ascribed as a translation enhancer[52-56]. One of mechanisms proposed by which enhancing the process is achieved, is the interaction between the 5’ and 3’ ends of the HCV genome, mediated by the PTB protein and another, hypothetical Y protein[57]. The effect of enhancing translation through genome cyclization is observed in many viruses[58]. However, not all results indicate PTB enhances HCV translation, sometimes the protein seems to be even inhibitory[59]. There is also no consensus on the impact of the 3’UTR on the efficiency of translation. For example, Murakami et al[59] observed that in an in vitro system, deletion of SL3 in the 3’X region and/or the poly (U/UC) section resulted in an increase in the amount of protein produced. A possible explanation for this observation would be that these sequence fragments, by interacting with other protein factors, inhibit the translation process. In other studies, however, it was observed that the presence of 3’UTR had no effect on the increase of the product amount and the efficiency of the polyprotein cleavage process[43,60]. It appears the choice of the experimental model plays a key role in this kind of research. For example, while in the lysate from rabbit reticulocytes no effect of 3’UTR on translation efficiency is observed[42,53], in HeLa cells, hepatocytes and in an in vivo mouse model the presence of the 3’UTR stimulates this process[53]. Recently, the role of 3’UTR in enhancing translational efficiency has been reported both in the rabbit reticulocyte lysate and in Huh7 cells[61].
Finally, it has been shown that the region X interacts with the ribosomal proteins L22, L3, S3 and mL3[62]. Similar viral RNA - L22 protein interactions have been observed for EBV (Epstein-Barr virus) and HPV-1 (human type 1 papilloma virus), showing a positive effect of L22 on translational efficiency[62]. Meanwhile, in the case of Qβ virus, ribosomal host proteins are involved in the formation of the viral replication complex[62]. It is unclear whether the interaction of ribosomal proteins with the HCV X region is related to the replication or translation process, or to both of these processes. The demonstrated ability of HCV protein NS5B to bind to the ribosome[63] is reminiscent of the strategy used by the Qβ virus, which uses ribosomal proteins to build its replication complex.
CONCLUSION
The most intriguing feature of the 3’X region is its extremely high sequence conservation. The maintenance of a 98-nt long stretch of RNA at over 95% conservation places severe constraints on such a fast-mutating RNA virus. Apparently almost any change to this stretch result in progeny incapable of reproducing which rapidly disappear from the population.
The high sequence conservation might be explained by existence of different structural forms of the 3’X region. Within one structure there is usually a way to neutralize point mutations by adaptive mutations in another structure, thus restoring important base-pairing. Simultaneous compensation of mutations in two (or more) different structures/forms is much less probable. However, instead of several different structural forms of the X-region, the long-range RNA-RNA interactions with the involvement of that region could explain its high sequence conservation.
Evidence indicates the 3’X region is involved in a number of interactions including kissing 5BSL3.2 (required for replication), an interaction with the second RNA molecule carrying the X-region sequence (homodimerization), binding of the NS5B protein (for initiation of RNA synthesis), an interaction with NS3 helicase (for elongation of RNA synthesis), binding of PTB (for proposed translation regulation). Is it possible that one structural form of the 3’X region supports such different interactions? The answer is: yes, it is possible, but unlikely. On the other hand, it is easy to imagine that different structural forms of this region are responsible for different processes and interactions. For instance: 2xSL - structure for dimerization and enhanced RNA synthesis, 3xSL - structure for kissing interactions that supports translation.
Hopefully in-depth in vivo structure mapping of the 3’X-region at the various replication stages, in different cellular compartments will answer these questions to give us a better understanding of the virus and lead to additional means to control it.