Published online May 7, 2018. doi: 10.3748/wjg.v24.i17.1881
Peer-review started: March 12, 2018
First decision: March 29, 2018
Revised: April 6, 2018
Accepted: April 9, 2018
Article in press: April 9, 2018
Published online: May 7, 2018
Processing time: 56 Days and 4.7 Hours
Olfactomedin-4 (OLFM4, GW112, hGC-1) is a glycoprotein belonging to the olfactomedin family. The expression of OLFM4 is strong in the small intestine, colon and prostate, and moderate in the stomach and bone marrow. Previous studies have revealed that OLFM4 is closely associated with many digestive diseases. Up-regulation of OLFM4 has been detected in the Helicobacter pylori (H. pylori)-infected gastric mucosa, inflammatory bowel disease tissue and gastrointestinal malignancies, including gastric cancer, colorectal cancer, pancreatic cancer and gallbladder cancer. Down-regulation of OLFM4 has also been detected in some cases, such as in poorly differentiated, advanced-stage and metastatic tumors. Studies using OLFM4-deficient mouse models have revealed that OLFM4 acts as a negative regulator of H. pylori-specific immune responses and plays an important role in mucosal defense in inflammatory bowel disease. Patients with OLFM4-positive gastric cancer or colorectal cancer have a better survival rate than OLFM4-negative patients. However, the prognosis is worse in pancreatic cancer patients with high levels of expression of OLFM4. The NF-κB, Notch and Wnt signaling pathways are involved in the regulation of OLFM4 expression in digestive diseases, and its role in pathogenesis is associated with anti-inflammation, apoptosis, cell adhesion and proliferation. OLFM4 may serve as a potential specific diagnostic marker and a therapeutic target in digestive diseases. Further studies are required to explore the clinical value of OLFM4.
Core tip: This review is based on the currently available literature about olfactomedin-4 (OLFM4) and is intended to reveal the link between OLFM4 and digestive diseases, including Helicobacter pylori infection, inflammatory bowel disease and gastrointestinal malignancies. The data on the expression, function and regulatory pathways of OLFM4 in digestive diseases are summarized. The potential clinical value of OLFM4 in digestive diseases is also discussed.
- Citation: Wang XY, Chen SH, Zhang YN, Xu CF. Olfactomedin-4 in digestive diseases: A mini-review. World J Gastroenterol 2018; 24(17): 1881-1887
- URL: https://www.wjgnet.com/1007-9327/full/v24/i17/1881.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i17.1881
Olfactomedin-4 (OLFM4, also called GW112 or hGC-1) is a 72-kDa glycoprotein belonging to the olfactomedin family and is characterized by the presence of an olfactomedin domain with approximately 250 amino acids, which is located in the C-terminal region[1]. OLFM4 was initially cloned from human hematopoietic myeloid cells treated with granulocyte colony-stimulating factor[1]. The OLFM4 gene, located on chromosome 13q14.3, encodes a 510-amino acid N-linked glycoprotein with the olfactomedin domain[1,2]. OLMF4 can be expressed in the membrane, cytoplasm, nucleus, mitochondria and mature neutrophil granules[1,3-5]. OLFM4 is strongly expressed in the small intestine, colon and prostate, moderately expressed in the stomach and bone marrow, and weakly expressed or not expressed in other tissues[1].
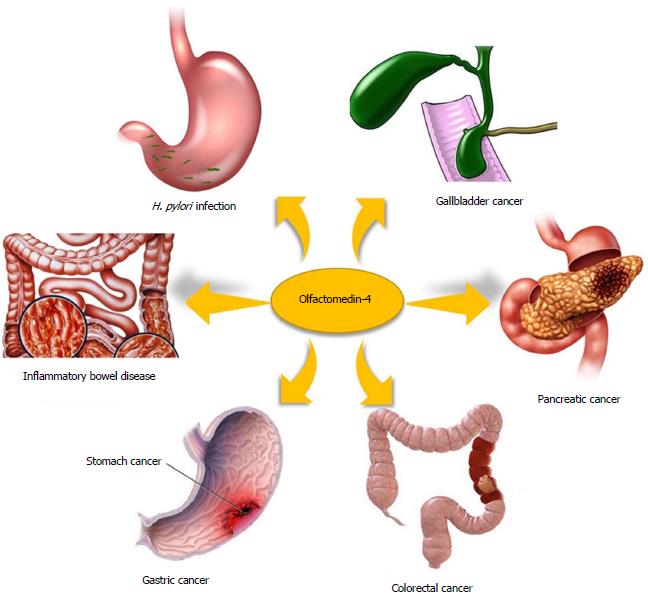
Compared with that in normal tissues, aberrant expression of OLFM4 has been detected in many pathological tissues, such as the gastric mucosa infected with Helicobacter pylori (H. pylori)[6,7], inflamed intestinal tissue in inflammatory bowel disease[8,9] and many types of gastrointestinal malignancies[10-14] (Figure 1). The primary function of OLFM4 in gastrointestinal malignancies is associated with its role as an antiapoptotic factor that promotes the tumor growth[4]. In addition, OLFM4 down-regulates innate immunity against H. pylori infection[7] and affects the anti-inflammatory function in inflammatory bowel disease[15]. In this review, we summarize the data on the expression, function and regulatory pathways of OLFM4 in digestive diseases.
H. pylori infection is a well-recognized risk factor for gastric diseases as well as extra-gastric diseases[16-18]. The host immune response plays a key role in the course and outcome of H. pylori infection[19,20]. The innate immune system serves as the first line of defense against H. pylori infection[21]. An adaptive immune response to H. pylori is also elicited in nearly all H. pylori-infected individuals[22]. OLFM4 is a novel glycoprotein that negatively regulates the host defense system against bacterial infection[23].
An early microarray study found that OLFM4 expression is significantly up-regulated in the gastric mucosa of H. pylori-infected patients compared with that in uninfected controls[6]. OLFM4 expression was also found to be significantly up-regulated in the gastric mucosa of H. pylori-infected mice. However, further study is warranted to determine whether eradication of H. pylori leads to the normalization of OLFM4 levels. The expression of OLFM4 is up-regulated in neutrophils, macrophages and epithelial cells after H. pylori infection, which suggests that overexpression of OLFM4 upon H. pylori infection is due to its direct action on epithelial cells as well as to activation of neutrophil and macrophage infiltration[7], thus suggesting a potential role for OLFM4 in the host immune response against H. pylori infection.
The exact function of OLFM4 in H. pylori infection has been demonstrated by generating an OLFM4-deficient mouse model. Colonization of H. pylori in the gastric mucosa is significantly reduced after knocking out the OLFM4 gene, as compared with that in wild-type mice[7]. In addition, in response to H. pylori infection, infiltration of inflammatory cells was significantly enhanced, the production of proinflammatory cytokines and chemokines was increased, and the bacterial load was reduced in OLFM4-deficient mice[7]. Therefore, OLFM4 acts as a negative regulator of the H. pylori-specific immune responses[7].
OLFM4 is a target gene of the NF-κB pathway and expression of the OLFM4 gene can be regulated by the transcription factor NF-κB[7,24]. The regulation is achieved by binding of NF-κB to the 5’-upstream region of the OLFM4 gene[24]. Moreover, OLFM4 exerts a negative feedback effect on the NF-κB pathway[7].
Mouse experiments have revealed that H. pylori infection up-regulates the OLFM4 expression in an NF-κB-dependent manner, and then, due to the negative feedback effect of OLFM4, the H. pylori-induced NF-κB activation is down-regulated[7]. Furthermore, OLFM4 inhibits the nucleotide oligomerization domain (NOD)-1/2-mediated NF-κB activation and subsequent cytokine and chemokine production through direct association with NOD1 and NOD2[7]. The reduced cytokine and chemokine production results in a weak inflammatory response and a high level of colonization of H. pylori in the gastric mucosa[7].
Experiments in a MyD88 and OLFM4 double-knockout mouse model have demonstrated that the H. pylori colonization level in the model is similar to that in wild-type mice[25]. Even though the immune and inflammatory responses are enhanced compared with those in wild-type mice, infiltration of inflammatory cells in the gastric mucosa of double-knockout mice is lower than that in OLFM4 knockout mice[25]. Additionally, knocking out OLFM4 significantly up-regulates the MyD88 expression. It has been shown that deletion of OLFM4 indirectly increases the MyD88 expression by enhancing NOD2 expression, whereas the deficiency of MyD88 leads to a loss of the feedback inhibition of the NF-κB pathway and of the resulting response[25,26].
OLFM4 is a robust marker for murine intestinal stem cells as well as human intestinal stem cells[27]. Both OLFM4 mRNA and protein expression levels are significantly up-regulated in the intestinal epithelium in Crohn’s disease and ulcerative colitis[8,9]. Compared with that in inflamed tissue from Crohn’s disease patients, the OLFM4 expression is more obviously increased in inflamed tissue from patients with active ulcerative colitis[8,9]. Moreover, in active ulcerative colitis, the expression of OLFM4 expands to the surface of epithelial cells as well as to the crypt lumen, and OLFM4 seems to be secreted into the mucus[8,9]. In contrast, the OLFM4 gene expression is almost absent in luminal surface cells and mesenchymal cells and is confined to the lower third of the crypt in normal tissues[8,9].
OLFM4 plays an important role in the mucosal defense of the stomach and colon[9]. Experiments using OLFM4-deficient mice have revealed severe inflammation and proliferation in intestinal crypts in small intestines[15]. Serious inflammation and mucosa damage have also been found in the colon of OLFM4-deficient mice[15].
The anti-inflammatory function of OLFM4 in inflammatory bowel disease is consistent with that in the stomach. The function against inflammatory bowel disease may be related to the tissue-specific human beta-defensins (HBD)1, HBD2 and HBD3. As mucus components with different electric charges, OLFM4 and HBD1–3 can interact, and the binding ability of OLFM4 was ranked, from high to low, as HBD3 > HBD2 > HBD1[9]. Furthermore, OLFM4 binding leads to a decrease in the antimicrobial activities of HBD1-3[9].
OLFM4 is a target gene for the Notch signaling pathway, which regulates intestinal cell proliferation and differentiation[28]. The expression of OLFM4 increases after activation of Notch signaling[28]. Conversely, the expression of OLFM4 rapidly decreases after treatment with the Notch blocker dibenzazepine[9,28]. Researchers have found that after mesenchymal stem cell transplantation, the expression of OLFM4 is down-regulated, while that of Atoh1 is up-regulated[29]. This result suggests that the suppression of Notch signaling leads to decreased OLFM4 expression.
Although some studies have shown that cell incubation with TNF-α alone does not influence the OLFM4 expression, some other studies have found that TNF-α and components of the Notch pathways synergistically up-regulate the OLFM4 expression[9,30,31]. TNF-α is one of the most important proinflammatory cytokines promoting inflammatory bowel disease[30]. Microarray analysis has revealed that up to 21 genes are involved in the synergistic up-regulation of TNF-α and the Notch intracellular domain[30]. Further studies have suggested a markedly increased expression of OLFM4, reaching up to a 2500-fold increase in LS174T cells, when overexpression of Notch intracellular domain-1 (NICD1) or hairy and enhancer of split-1 (HES1) is combined with TNF-α stimulation[30,31]. Such a synergistic effect is mediated through transcriptional regulation, which is dependent on a proximal NF-κB binding site[31].
Increased OLFM4 expression has been reported in some gastrointestinal cancers, such as gastric cancer[10,11,32,33], pancreatic cancer[12] and early-stage colon cancer[13,14]. In addition, the expression of OLFM4 is correlated with the histological type of cancer, differentiation, lymphatic metastasis and prognosis[10,11,34]. Furthermore, OLFM4 is relevant to many cellular processes, including cell adhesion, apoptosis and proliferation[2,11,35]. Therefore, OLFM4 may serve as a candidate biomarker for these gastrointestinal cancers[36]. Here, we briefly summarize the recent advances in the expression, function and regulation of OLFM4 in gastrointestinal cancers.
Up-regulated OLFM4 expression is a frequent event in the gastric mucosa in gastric cancer[10,11,32,33]. Highly expressed OLFM4 is found in intestinal-type adenocarcinoma, while OLFM4 expression does not occur in diffuse-type adenocarcinoma[10]. Moreover, enhanced expression of OLFM4 occurs in well- or moderately differentiated and early-stage adenocarcinomas, and the expression is remarkably decreased or even lost in poorly differentiated and advanced-stage gastric cancer[10]. Furthermore, the OLFM4 expression is higher in patients without lymphatic metastasis than in those with lymphatic invasion[11,37]. OFLM4 expression is also related to the prognosis. OLFM4-positive gastric cancer patients have a better survival rate than do OLFM4-negative patients[34,37]. Using serum OLFM4 alone or in combination with human regenerating protein IV as biomarkers for gastric cancer patients is more sensitive than using CA199[32]. Down-regulation of OLFM4 suppresses the tumor proliferation, migration and invasion of gastric cancer cells in vitro[33,38].
The OLFM4 gene was found to be up-regulated via the NF-κB signaling pathway and to exert an antiapoptotic effect in gastric cancer[39]. The antiapoptotic effect caused by OLFM4 can be induced by reducing H2O2 or TNF-α[38]. Moreover, the antiapoptotic factor OLFM4 is a direct target of miR-486, which is a frequently lost microRNA (miRNA) in gastric cancer patients and may act as a tumor suppressor miRNA in gastric cancer[40]. miR-486 directly targets and inhibits OLFM4 and thereby induces antioncogenic effects against gastric cancer[40].
OLFM4 is enriched in human colon crypts, although it is not expressed in the murine colon[15,27,41,42]. It has been universally accepted that OLFM4 is a useful marker of intestinal stem cells (ISCs) in humans, similar to LGR5, which is a confirmed ISC marker[27,43,44]. Up-regulation of OLFM4 is detected more frequently in highly differentiated and early-stage colon cancers than in the normal colon mucosa, whereas it is often down-regulated or not expressed in poorly differentiated, late tumor-node-metastasis stage, and metastatic cancers[35]. OLFM4-positive colorectal cancer patients have a better survival rate than do OLFM4-negative patients[45]. In addition, precancerous colorectal lesions also show aberrant OLFM4 expression. For example, OLFM4 is expressed in a diffuse manner in traditional serrated adenomas, while other ISC markers such as LGR5 and ASCL2 are localized as in normal tissue[44]. OLFM4 silencing enhances the proliferation in intestinal crypts and inflammation initiated by azoxymethane/dextran sodium sulfate[15]. Moreover, systemic OLFM4 deletion promotes colon tumorigenesis, which may be associated with the loss of mucosal neutrophils[15].
There is an intimate connection between OLFM4, Wnt/β-catenin signaling, crypt biology[15,46-48] and colon cancer[27,49,50]. OLFM4 is a target gene that acts as a negative regulator of the Wnt/β-catenin signaling pathway and inhibits colon cancer progression by down-regulating the Wnt signaling pathway[15].
OLFM4 mRNA is expressed at higher levels in pancreatic cancer tissues than in noncancerous pancreatic tissue samples[12]. In addition, OLFM4 was found to be significantly over-expressed in peripheral blood mononuclear cells in pancreatic cancer patients compared with its expression in a control group[51]. Furthermore, OLFM4 has also been detected in pancreatic juice and ascites[52]. Pancreatic cancer may occur in a background of chronic pancreatitis. Whether OLFM4 is associated with chronic pancreatitis or acute pancreatitis flares is worth further investigation. In the PANC-1 cell line, OLFM4 is especially increased during the early S phase of the cell cycle and promotes proliferation by supporting the S to G2/M phase transition[12]. OLFM4 binds to the apoptosis-promoting factor GRIM-19 to induce antiapoptosis[4]. Pancreatic cancer patients with high levels of OLFM4 expression have a worse prognosis[53].
Similar to the above findings, expression of the OLFM4 gene has been found to be increased in gallbladder cancer tissues[54]. In addition, the expression level of OLFM4 is significantly related to the age of gallbladder cancer patients[54]. However, further studies are needed to clarify the precise role of OLFM4 in gallbladder cancer.
Since the initial discovery of OLFM4, researchers have explored many aspects of OLFM4, including its aberrant expression, biological functions and related mechanisms (Table 1). The expression of OLFM4 has been relatively well studied in normal tissues as well as in numerous diseases. The anti-inflammatory and antiapoptotic roles of OLFM4 are generally accepted. However, the exact mechanism for its effects in gastrointestinal diseases remains to be determined. Moreover, the clinical applications of OLFM4 as a specific detection marker or a therapeutic target need to be defined in the future.
| Disease | Expression | Function | Regulation |
| H. pylori infection | Up-regulated in the H. pylori–infected gastric mucosa | Negative regulator of H. pylori-specific immune responses | NF-κB, NOD-1/2, MyD88 |
| Inflammatory bowel disease | Up-regulated in the intestinal epithelium in Crohn’s disease and ulcerative colitis | Mucosal defense, anti-inflammatory effects | Notch, TNF-α |
| Gastrointestinal malignancies | Up-regulated in well/moderately differentiated, early-stage gastrointestinal malignancies without lymphatic metastasis | Biomarker, candidate therapeutic target | NF-κB, TNF-α, miR-486, Wnt/β-catenin |
| 1. | Zhang J, Liu WL, Tang DC, Chen L, Wang M, Pack SD, Zhuang Z, Rodgers GP. Identification and characterization of a novel member of olfactomedin-related protein family, hGC-1, expressed during myeloid lineage development. Gene. 2002;283:83-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 116] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 2. | Liu W, Chen L, Zhu J, Rodgers GP. The glycoprotein hGC-1 binds to cadherin and lectins. Exp Cell Res. 2006;312:1785-1797. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 78] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 3. | Liu W, Lee HW, Liu Y, Wang R, Rodgers GP. Olfactomedin 4 is a novel target gene of retinoic acids and 5-aza-2’-deoxycytidine involved in human myeloid leukemia cell growth, differentiation, and apoptosis. Blood. 2010;116:4938-4947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 48] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 4. | Zhang X, Huang Q, Yang Z, Li Y, Li CY. GW112, a novel antiapoptotic protein that promotes tumor growth. Cancer Res. 2004;64:2474-2481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 115] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 5. | Liu W, Yan M, Liu Y, McLeish KR, Coleman WG Jr, Rodgers GP. Olfactomedin 4 inhibits cathepsin C-mediated protease activities, thereby modulating neutrophil killing of Staphylococcus aureus and Escherichia coli in mice. J Immunol. 2012;189:2460-2467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 77] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 6. | Mannick EE, Schurr JR, Zapata A, Lentz JJ, Gastanaduy M, Cote RL, Delgado A, Correa P, Correa H. Gene expression in gastric biopsies from patients infected with Helicobacter pylori. Scand J Gastroenterol. 2004;39:1192-1200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 49] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 7. | Liu W, Yan M, Liu Y, Wang R, Li C, Deng C, Singh A, Coleman WG Jr, Rodgers GP. Olfactomedin 4 down-regulates innate immunity against Helicobacter pylori infection. Proc Natl Acad Sci U S A. 2010;107:11056-11061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 121] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 8. | Shinozaki S, Nakamura T, Iimura M, Kato Y, Iizuka B, Kobayashi M, Hayashi N. Upregulation of Reg 1alpha and GW112 in the epithelium of inflamed colonic mucosa. Gut. 2001;48:623-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 81] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 9. | Gersemann M, Becker S, Nuding S, Antoni L, Ott G, Fritz P, Oue N, Yasui W, Wehkamp J, Stange EF. Olfactomedin-4 is a glycoprotein secreted into mucus in active IBD. J Crohns Colitis. 2012;6:425-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 10. | Liu W, Zhu J, Cao L, Rodgers GP. Expression of hGC-1 is correlated with differentiation of gastric carcinoma. Histopathology. 2007;51:157-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 58] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 11. | Grover PK, Hardingham JE, Cummins AG. Stem cell marker olfactomedin 4: critical appraisal of its characteristics and role in tumorigenesis. Cancer Metastasis Rev. 2010;29:761-775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 59] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 12. | Kobayashi D, Koshida S, Moriai R, Tsuji N, Watanabe N. Olfactomedin 4 promotes S-phase transition in proliferation of pancreatic cancer cells. Cancer Sci. 2007;98:334-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 74] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 13. | Koshida S, Kobayashi D, Moriai R, Tsuji N, Watanabe N. Specific overexpression of OLFM4(GW112/HGC-1) mRNA in colon, breast and lung cancer tissues detected using quantitative analysis. Cancer Sci. 2007;98:315-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 60] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 14. | Duan C, Liu X, Liang S, Yang Z, Xia M, Wang L, Chen S, Yu L. Oestrogen receptor-mediated expression of Olfactomedin 4 regulates the progression of endometrial adenocarcinoma. J Cell Mol Med. 2014;18:863-874. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 15. | Liu W, Li H, Hong SH, Piszczek GP, Chen W, Rodgers GP. Olfactomedin 4 deletion induces colon adenocarcinoma in ApcMin/+ mice. Oncogene. 2016;35:5237-5247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 68] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 16. | Malfertheiner P, Megraud F, O’Morain CA, Atherton J, Axon AT, Bazzoli F, Gensini GF, Gisbert JP, Graham DY, Rokkas T. Management of Helicobacter pylori infection--the Maastricht IV/ Florence Consensus Report. Gut. 2012;61:646-664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1719] [Cited by in RCA: 1614] [Article Influence: 115.3] [Reference Citation Analysis (7)] |
| 17. | McColl KE. Clinical practice. Helicobacter pylori infection. N Engl J Med. 2010;362:1597-1604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 529] [Cited by in RCA: 560] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 18. | Sung KC, Rhee EJ, Ryu SH, Beck SH. Prevalence of Helicobacter pylori infection and its association with cardiovascular risk factors in Korean adults. Int J Cardiol. 2005;102:411-417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 41] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 19. | Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19:449-490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1446] [Cited by in RCA: 1580] [Article Influence: 79.0] [Reference Citation Analysis (1)] |
| 20. | Borody T, Ren Z, Pang G, Clancy R. Impaired host immunity contributes to Helicobacter pylori eradication failure. Am J Gastroenterol. 2002;97:3032-3037. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 41] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 21. | Algood HM, Gallo-Romero J, Wilson KT, Peek RM Jr, Cover TL. Host response to Helicobacter pylori infection before initiation of the adaptive immune response. FEMS Immunol Med Microbiol. 2007;51:577-586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 96] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 22. | Portal-Celhay C, Perez-Perez GI. Immune responses to Helicobacter pylori colonization: mechanisms and clinical outcomes. Clin Sci (Lond). 2006;110:305-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 62] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 23. | Liu W, Yan M, Sugui JA, Li H, Xu C, Joo J, Kwon-Chung KJ, Coleman WG, Rodgers GP. Olfm4 deletion enhances defense against Staphylococcus aureus in chronic granulomatous disease. J Clin Invest. 2013;123:3751-3755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 54] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 24. | Chin KL, Aerbajinai W, Zhu J, Drew L, Chen L, Liu W, Rodgers GP. The regulation of OLFM4 expression in myeloid precursor cells relies on NF-kappaB transcription factor. Br J Haematol. 2008;143:421-432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 25. | Yan M, Liu WL, Joo J, Sun JF, Datta S, Yang A, Rodgers G, Coleman WG. MyD88 Has a Key Role for OLFM4, a Novel Anti-Inflammatory Mediator in H. pylori Infection. Gastroenterology. 2012;142:S-686. [DOI] [Full Text] |
| 26. | Yan M, Liu W, Xu C, Joo J, Yu C, Zhu Y, Yan S, Rodgers G, Coleman WG. Olfactomedin 4 Deletion Enhances Host Pro-Inflammatory Immune Responses Against Helicobacter pylori Infection Through a MyD88 Dependent Mechanism. Gastroenterology. 2014;146:S287-S288. [DOI] [Full Text] |
| 27. | van der Flier LG, Haegebarth A, Stange DE, van de Wetering M, Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology. 2009;137:15-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 384] [Cited by in RCA: 440] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 28. | VanDussen KL, Carulli AJ, Keeley TM, Patel SR, Puthoff BJ, Magness ST, Tran IT, Maillard I, Siebel C, Kolterud Å, Grosse AS, Gumucio DL, Ernst SA, Tsai YH, Dempsey PJ, Samuelson LC. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488-497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 377] [Cited by in RCA: 458] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 29. | Xing Y, Chen X, Cao Y, Huang J, Xie X, Wei Y. Expression of Wnt and Notch signaling pathways in inflammatory bowel disease treated with mesenchymal stem cell transplantation: evaluation in a rat model. Stem Cell Res Ther. 2015;6:101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 30. | Kawamoto A, Nakata T, Fujii S, Suzuki K, Ishibashi F, Tsuchiya K, Nakamura T, Okamoto R, Watanabe M. Notch Signaling and TNF-alpha Synergistically Up-regulate Stem Cell-Specific Genes, OLFM4 and UBD, in the Inflamed Intestinal Epithelia of IBD Patients. Inflammat Bowel Dis. 2017;23:S92-S92. [DOI] [Full Text] |
| 31. | Okamoto R, Akiyama J, Murano T, Shimizu H, Tsuchiya K, Nakamura T, Watanabe M. Notch-Hes1 Pathway and TNF-α Synergistically up-Regulates OLFM4 Expression in the Inflamed Mucosa of the Human Intestine. Gastroenterology. 2011;140:S-173. [DOI] [Full Text] |
| 32. | Oue N, Sentani K, Noguchi T, Ohara S, Sakamoto N, Hayashi T, Anami K, Motoshita J, Ito M, Tanaka S. Serum olfactomedin 4 (GW112, hGC-1) in combination with Reg IV is a highly sensitive biomarker for gastric cancer patients. Int J Cancer. 2009;125:2383-2392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 83] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 33. | Ran X, Xu X, Yang Y, She S, Yang M, Li S, Peng H, Ding X, Hu H, Hu P. A quantitative proteomics study on olfactomedin 4 in the development of gastric cancer. Int J Oncol. 2015;47:1932-1944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 34. | Oue N, Sentani K, Sakamoto N, Yasui W. Clinicopathologic and molecular characteristics of gastric cancer showing gastric and intestinal mucin phenotype. Cancer Sci. 2015;106:951-958. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 68] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 35. | Liu W, Liu Y, Zhu J, Wright E, Ding I, Rodgers GP. Reduced hGC-1 protein expression is associated with malignant progression of colon carcinoma. Clin Cancer Res. 2008;14:1041-1049. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 64] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 36. | Guette C, Valo I, Vétillard A, Coqueret O. Olfactomedin-4 is a candidate biomarker of solid gastric, colorectal, pancreatic, head and neck, and prostate cancers. Proteomics Clin Appl. 2015;9:58-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 37. | Luo Z, Zhang Q, Zhao Z, Li B, Chen J, Wang Y. OLFM4 is associated with lymph node metastasis and poor prognosis in patients with gastric cancer. J Cancer Res Clin Oncol. 2011;137:1713-1720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 39] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 38. | Liu RH, Yang MH, Xiang H, Bao LM, Yang HA, Yue LW, Jiang X, Ang N, Wu LY, Huang Y. Depletion of OLFM4 gene inhibits cell growth and increases sensitization to hydrogen peroxide and tumor necrosis factor-alpha induced-apoptosis in gastric cancer cells. J Biomed Sci. 2012;19:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 39. | Kim KK, Park KS, Song SB, Kim KE. Up regulation of GW112 Gene by NF kappaB promotes an antiapoptotic property in gastric cancer cells. Mol Carcinog. 2010;49:259-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 40. | Oh HK, Tan AL, Das K, Ooi CH, Deng NT, Tan IB, Beillard E, Lee J, Ramnarayanan K, Rha SY. Genomic loss of miR-486 regulates tumor progression and the OLFM4 antiapoptotic factor in gastric cancer. Clin Cancer Res. 2011;17:2657-2667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 168] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 41. | Kosinski C, Li VS, Chan AS, Zhang J, Ho C, Tsui WY, Chan TL, Mifflin RC, Powell DW, Yuen ST. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc Natl Acad Sci USA. 2007;104:15418-15423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 411] [Cited by in RCA: 470] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 42. | van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, Begthel H, van den Born M, Guryev V, Oving I. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136:903-912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 536] [Cited by in RCA: 574] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 43. | Jang BG, Lee BL, Kim WH. Intestinal Stem Cell Markers in the Intestinal Metaplasia of Stomach and Barrett’s Esophagus. PLoS One. 2015;10:e0127300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 44. | Jang BG, Kim HS, Kim KJ, Rhee YY, Kim WH, Kang GH. Distribution of intestinal stem cell markers in colorectal precancerous lesions. Histopathology. 2016;68:567-577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 45. | Seko N, Oue N, Noguchi T, Sentani K, Sakamoto N, Hinoi T, Okajima M, Yasui W. Olfactomedin 4 (GW112, hGC-1) is an independent prognostic marker for survival in patients with colorectal cancer. Exp Ther Med. 2010;1:73-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 46. | Kuhnert F, Davis CR, Wang HT, Chu P, Lee M, Yuan J, Nusse R, Kuo CJ. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc Natl Acad Sci U S A. 2004;101:266-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 483] [Cited by in RCA: 512] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 47. | Pinto D, Gregorieff A, Begthel H, Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709-1713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 758] [Cited by in RCA: 809] [Article Influence: 35.2] [Reference Citation Analysis (0)] |
| 48. | Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379-383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1207] [Cited by in RCA: 1218] [Article Influence: 43.5] [Reference Citation Analysis (0)] |
| 49. | Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784-1787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2584] [Cited by in RCA: 2687] [Article Influence: 92.7] [Reference Citation Analysis (0)] |
| 50. | Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787-1790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2986] [Cited by in RCA: 3126] [Article Influence: 107.8] [Reference Citation Analysis (0)] |
| 51. | Yan H, Lu D, Xu L, Xie Q, Dong X, Wu Y. Increased expression level of Olfactomedin4 in peripheral blood mononuclear cells of pancreatic adenocarcinoma patients. Hepatogastroenterology. 2011;58:1354-1359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 52. | Makawita S, Smith C, Batruch I, Zheng Y, Rückert F, Grützmann R, Pilarsky C, Gallinger S, Diamandis EP. Integrated proteomic profiling of cell line conditioned media and pancreatic juice for the identification of pancreatic cancer biomarkers. Mol Cell Proteomics. 2011;10:M111.008599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 93] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 53. | Takadate T, Onogawa T, Fukuda T, Motoi F, Suzuki T, Fujii K, Kihara M, Mikami S, Bando Y, Maeda S. Novel prognostic protein markers of resectable pancreatic cancer identified by coupled shotgun and targeted proteomics using formalin-fixed paraffin-embedded tissues. Int J Cancer. 2013;132:1368-1382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 54. | Gu X, Li B, Jiang M, Fang M, Ji J, Wang A, Wang M, Jiang X, Gao C. RNA sequencing reveals differentially expressed genes as potential diagnostic and prognostic indicators of gallbladder carcinoma. Oncotarget. 2015;6:20661-20671. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Ierardi E, Vorobjova T S- Editor: Gong ZM L- Editor: Filipodia E- Editor: Huang Y